Abstract
Reovirus attaches to cellular receptors with the σ1 protein, a fiber-like molecule protruding from the 12 vertices of the icosahedral virion. The crystal structure of a receptor-binding fragment of σ1 reveals an elongated trimer with two domains: a compact head with a new β-barrel fold and a fibrous tail containing a triple β-spiral. Numerous structural and functional similarities between reovirus σ1 and the adenovirus fiber suggest an evolutionary link in the receptor-binding strategies of these two viruses. A prominent loop in the σ1 head contains a cluster of residues that are conserved among reovirus serotypes and are likely to form a binding site for junction adhesion molecule, an integral tight junction protein that serves as a reovirus receptor. The fibrous tail is mainly responsible for σ1 trimer formation, and it contains a highly flexible region that allows for significant movement between the base of the tail and the head. The architecture of the trimer interface and the observed flexibility indicate that σ1 is a metastable structure poised to undergo conformational changes upon viral attachment and cell entry.
Keywords: adenovirus/evolution/reovirus/σ1/virus–receptor interactions
Introduction
Reoviruses form non-enveloped, icosahedral particles (Dryden et al., 1993) that contain a segmented double-stranded (ds) RNA genome. The virions measure ∼850 Å in diameter and are composed of eight structural proteins. Five of these (λ1, λ2, λ3, µ2 and σ2) form the ‘core’ or inner capsid particle, the crystal structure of which has been determined (Reinisch et al., 2000). A second layer of proteins (µ1, σ1 and σ3) forms the reovirus outer capsid, with µ1 and σ3 comprising the bulk of this capsid and σ1 protruding from the 12 vertices of the icosahedron. The σ3 protein, whose crystal structure is known (Olland et al., 2001), is thought to serve as a protective cap for µ1, and cleavage of σ3 by endosomal proteases during viral infection results in the loss of σ3 and generation of infectious subvirion particles. The reovirus σ1 protein serves as the viral attachment protein (Weiner et al., 1980; Lee et al., 1981). Rotary shadowing studies show that σ1 is a long, fiber-like molecule with head-and-tail morphology and several defined regions of flexibility within its tail (Fraser et al., 1990). The σ1 tail partially inserts into the virion via ‘turrets’ formed by the pentameric λ2 protein, whereas the σ1 head projects away from the virion surface (Furlong et al., 1988; Dryden et al., 1993).
Reoviruses have been isolated from many mammalian species, including humans (Tyler and Fields, 1996). Three major reovirus serotypes have been described, which are represented by the prototype strains type 1 Lang (T1L), type 2 Jones (T2J) and type 3 Dearing (T3D). Reoviruses infect most children and can cause mild respiratory or gastrointestinal illnesses (Tyler and Fields, 1996). They also serve as important models for studies of viral replication and pathogenesis and, in particular, for analysis of viral determinants of central nervous system (CNS) injury. After infection of newborn mice, reoviruses disseminate to the CNS and produce serotype-specific patterns of disease (Tyler and Fields, 1996). Type 1 reovirus strains spread by hematogenous routes to the CNS where they infect ependymal cells, leading to non-lethal hydrocephalus (Weiner et al., 1977, 1980; Tyler et al., 1986). In contrast, type 3 reoviruses spread primarily by neural routes to the CNS and infect neurons, causing fatal encephalitis (Weiner et al., 1977, 1980; Tyler et al., 1986; Morrison et al., 1991). The σ1 protein plays a pivotal role in these disease patterns (Weiner et al., 1980; Tyler et al., 1986), most likely through the selective recognition of cell-surface receptors.
The σ1 protein is also responsible for the efficiency of virus-induced apoptosis (Tyler et al., 1995). Reovirus induces the biochemical and morphological hallmarks of apoptosis in cultured cells (Tyler et al., 1995) and in vivo (Oberhaus et al., 1997; DeBiasi et al., 2001). Reovirus strain T3D induces apoptosis to a substantially greater extent than T1L, and differences in the capacity of these strains to induce apoptosis are determined primarily by the σ1-encoding S1 gene (Tyler et al., 1995; Rodgers et al., 1997; Connolly et al., 2000).
T3D σ1 contains two receptor-binding domains: one in the tail that binds α-linked sialic acid (Chappell et al., 1997, 2000) and another in the head that binds junction adhesion molecule (JAM; Barton et al., 2001b). T1L and T2J strains also bind JAM (Barton et al., 2001b and J.A.Campbell and T.S.Dermody, unpublished observations), but neither binds sialic acid. JAM is a transmembrane protein with two immunoglobulin-type domains in the extracellular region (Martin-Padura et al., 1998). It is located at tight junctions and mediates homophilic interactions between cells. Binding of reovirus to JAM is required for activation of NF-κB and apoptotic cell death (Barton et al., 2001b).
To understand better the structure–function relationships in σ1 that facilitate receptor binding and to gain further insight into mechanisms that underlie reovirus pathogenesis at the molecular level, we undertook structural studies of the T3D σ1 protein. The structure described here has been refined to 2.6 Å resolution and includes the JAM-binding σ1 head domain and a portion of the fibrous tail. This structure provides insights into σ1–receptor interactions and intramolecular conformational dynamics that characterize σ1 (Furlong et al., 1988; Fraser et al., 1990; Dryden et al., 1993). Moreover, the σ1 structure reveals a striking similarity to the adenovirus attachment protein, fiber. Although both adenoviruses and reoviruses are non-enveloped and icosahedrally shaped viruses, they differ in design and capsid composition. Most importantly, adenoviruses have a dsDNA genome, whereas reoviruses contain dsRNA. However, both viruses interact with cell-surface receptors using a long, fiber-like molecule located at the 12 vertices of the icosahedron. The structural similarity between these two attachment proteins suggests a distant evolutionary relationship.
Results and discussion
Determination of σ1 structure and model accuracy
The crystallized σ1 fragment was obtained through tryptic cleavage of σ1 deletion mutant 3-Δ-3-3-3 (Chappell et al., 2000). N-terminal sequencing of the proteolyzed protein confirmed that, as expected, trypsin cleaved after Arg245, a site in σ1 that is also cleaved during trypsin treatment of T3D virions (Chappell et al., 1998). The cleavage product contains amino acids 246–455 and is functional, as judged by its capacity to form trimers in solution and bind reovirus receptor JAM (Barton et al., 2001b). The crystallized fragment does not include residues involved in binding the type 3 σ1 co-receptor sialic acid (Chappell et al., 2000). The structure was determined by X-ray crystallography with the use of multiple isomorphous replacement and non-crystallographic symmetry averaging (Table I). The model was built into 3-fold averaged maps using O (Jones et al., 1991) and refined with XPLOR (Brünger et al., 1987). The crystallographic R-factor for the final model and all available data between 20 and 2.6 Å is 17.2%; the corresponding free R-factor (Brünger, 1992) for 5% of the data (817 reflections) is 23.5%. The model has good geometry, with small root mean square deviations (r.m.s.d.) from ideal values for bond lengths and bond angles (Table I). PROCHECK (CCP4, 1994) analysis shows two residues (Ser256 of chain B and His388 of chain C) in disallowed regions in the Ramachandran plot. His388 is in a β-turn and has good electron density. Its main chain conformation is similar to that of the other two His388 residues in the trimer, which are just inside the allowed region of the Ramachandran plot. Ser256 is in a less well-defined, flexible region at the N-terminus of molecule B. The refined model contains residues 250–455 of chains A and C, residues 251–455 of chain B, and 209 water molecules. In each chain, three prolines (Pro260, Pro299 and Pro377) are in the cis-configuration.
Table I. Data collection and refinement statistics.
| Data set | Native | CH3HgNO3 | K2HgI4 |
|---|---|---|---|
| Diffraction dataa | |||
| resolution range (Å) | 20–2.6 | 20–3.0 | 20–2.9 |
| completeness (%) | 91.0 (64.0) | 84.0 (67.4) | 98.0 (87.3) |
| total reflections | 56 260 | 24 196 | 53 855 |
| unique reflections | 16 338 | 9920 | 12 769 |
| Rmerge(%)b | 8.7 (28.4) | 12.5 (24.0) | 13.8 (30.0) |
| I/σ | 9.0 (2.2) | 6.4 (2.5) | 8.3 (2.1) |
| Phasing | |||
| Rnat (%)c | – | 21.4 | 20.2 |
| heavy atom binding sites | – | 8 | 7 |
| phasing power | – | 1.69 | 1.67 |
| Refinement statistics | |||
| Rcryst (%); working setd | 17.2 (no I/σI cutoff) | ||
| Rcryst (%); free setd | 23.5 (no I/σI cutoff) | ||
| r.m.s.d. bond lengths (Å) | 0.006 | ||
| r.m.s.d. bond angles (degrees) | 1.4 |
aData sets were collected at 293K using CuKα radiation. Values in parentheses refer to the outermost 0.1 Å resolution shell.
bRmerge = Σhkl|I – <I>|/ΣhklI, where I is the intensity of a reflection hkl and <I> is the average over symmetry-related observations of hkl.
cRnat = Σhkl|Fder – Fnat|/Σhkl(Fder + Fnat), where Fder and Fnat are derivative and native structure factors, respectively.
dRcryst = Σhkl|Fobs – Fcalc|/ΣhklFobs, where Fobs and Fcalc are observed and calculated structure factors, respectively. Free set (Brünger, 1992) contains 5% of the data.
Overall structure of σ1
The crystallized σ1 trimer has an elongated shape, with maximum dimensions of 120 Å in length and 50 Å in width. Each monomer contains two domains: a slender tail and a compact head (Figure 1A). Both domains are involved in trimer formation; contacts between monomers extend from the base of the tail to almost the top of the head. Significant bending of the trimer occurs at a defined region within the tail, which results in a kink between the 3-fold axes of the head and the tail (Figure 1A).
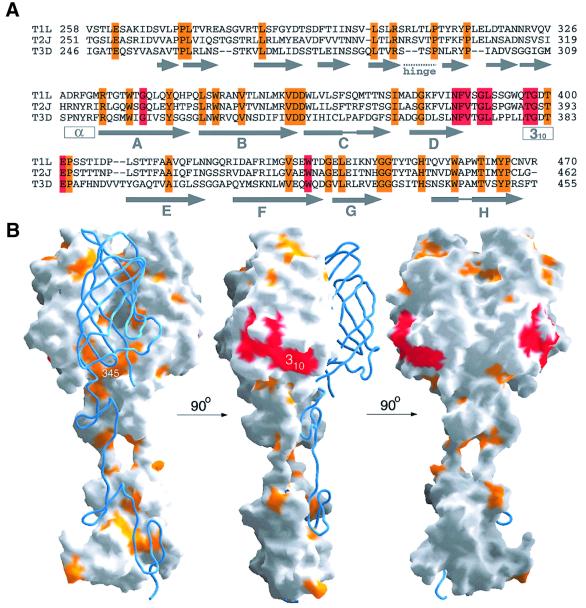
Fig. 1. Structure of reovirus σ1. (A) Ribbon drawing of the σ1 trimer. The three σ1 monomers are shown in red, orange and blue. Each monomer consists of a head domain formed by a compact β-barrel and a fibrous tail with three β-spiral repeats. Note the kink between the 3-fold axes of the head and tail domains. (B) Enlarged view of the σ1 head domain. The two Greek key motifs, shown in red and orange, form a compact, cylindrical β-sheet that contains eight β-strands (A–H). With the exception of the DE loop, the connections between the β-strands are very tight. The head domain also contains two short helices (blue): one 310 and one α-helix. (C) Schematic view of the β-strand arrangement in the σ1 head domain. Colors are as in (B); an additional D strand is shown in gray to depict the circular nature of the barrel. Strand D forms β-sheet-type hydrogen bonds with strands A and G.
Residues 246–309 form the tail, which contains repeating units of two anti-parallel β-strands connected by short loops. The β-strands run roughly parallel to the trimer axis and are arranged in a manner that resembles a spiral staircase. In the σ1 trimer, three tails assemble into a fibrous structure, which has been described as a triple β-spiral (Figure 1A; van Raaij et al., 1999). Triple β-spirals contain sequence repeats characterized by conserved hydrophobic and glycine or proline residues. Each repeat consists of two β-strands connected by a four-residue β-turn that has either a proline or glycine residue at its third position. A surface-exposed variable loop links one repeat to the next, and trimerization then generates the highly regular and rigid β-spiral structure. The adenovirus fiber (van Raaij et al., 1999) is thus far the only other example of a protein formed using a triple β-spiral fold. The crystallized fragment of fiber revealed four β-spiral repeats, all of the glycine type. The β-spiral of the σ1 fragment presented here contains three repeats with several unique features. (i) It is the first example with proline-type β-turns; the first and third σ1 repeats contain prolines (Pro260 and Pro299), and both prolines adopt the cis-configuration, which allows them to form type 2 β-turns similar to those in the glycine-type β-turns of the adenovirus fiber. (ii) The σ1 β-spiral shows that a residue other than glycine or proline can be accommodated in a β-turn; the second repeat contains a threonine (Thr278) at the position usually occupied by a glycine or a proline, and here the β-turn is of the type 1 class. (iii) Most importantly, the σ1 tail contains an insertion (Ser291–Pro294) that interrupts progression of the spiral between the second and third repeats. Amino acids in this insertion assume an extended structure with slightly different main chain conformations in each monomer. The insertion introduces a kink into the σ1 trimer, causing the head to bend with respect to the base of the tail (Figure 1A). Thus, the Ser291–Pro294 insertion is a site of substantial flexibility in σ1.
The remaining σ1 residues (310–455) fold into two Greek key motifs (β-sheets DABC and HEFG), which form the σ1 head (Figure 1B and C). The two Greek keys assemble into a compact β-barrel that features an uninterrupted cylindrical β-sheet. The β-barrel is 50 Å long, 35 Å wide and 25 Å thick; thus, its cross-section is slightly oval. A short α-helix prior to strand A links the β-spiral of the tail to the head. The loops connecting the β-strands are extremely short, with the notable exception of the loop between strands D and E; this loop also includes a short 310 helix. The adjacent strands C and H are interrupted by prolines (Pro353 and Pro444, respectively), which introduce β-bulges at the side of the barrel that faces towards the trimer axis. Single Greek keys frequently participate in the formation of five- and six-stranded β-barrels (Zhang and Kim, 2000). However, a circular barrel formed by two Greek keys is a new fold that is unique to the σ1 head. The two Greek key motifs may have arisen from gene duplication, although such a relationship is no longer evident at the sequence level.
Structural basis of σ1 flexibility
Inspection of the σ1 trimer reveals a distinct kink between the 3-fold axes of the head and tail domains (Figure 1A). Superposition of the three independent copies of σ1 present in our crystals shows that this kink occurs within the four-residue insertion (residues 291–294) between the second and third β-spiral repeat in the σ1 tail (Figure 2A). The insertion allows the head to move with respect to the base of the tail by as much as 23°. There is no single hinge residue to which this flexibility can be attributed; rather, the differences are the cumulative effect of small changes in main chain conformation of all four residues within the insertion. Three of these residues, Ser291, Thr292 and Ser293, do not form any contacts with each other or with other amino acids in an isolated σ1 trimer. However, all four insertion residues participate extensively in crystal lattice formation, which probably accounts for their assuming an ordered structure in the crystals. Thus, the observed conformation of each σ1 monomer appears to be primarily determined by crystal packing forces. We therefore expect the σ1 trimer to exhibit a significantly higher degree of flexibility in solution; electron microscope images of full-length σ1 do in fact show high flexibility in a region of the molecule that corresponds to the insertion within the σ1 tail (Fraser et al., 1990). The flexibility of the σ1 trimer is shown in Figure 2B. We envisage that σ1 in virions is firmly anchored into the λ2 turrets of the capsid by the tail. The protruding σ1 head would rest solely on the three extended polypeptide segments of the insertion, which would promote substantial conformational mobility of the head with respect to the virion surface.
Fig. 2. Flexibility of σ1. (A) Superposition of the three σ1 monomers present in the crystals. The superposition is based on the N-terminal two β-spiral repeats and reveals flexibility in the linker region (residues 291–294) between repeats 2 and 3. (B) Superposition, as in (A), using σ1 trimers. This representation highlights the degree of flexibility within the σ1 trimer as it protrudes from the virion. Three σ1 trimers are shown in red, orange and blue.
Flexibility in the tail of the σ1 trimer probably plays an important role in viral attachment, most likely by facilitating interactions between the head and its receptor JAM. The crystal structure of the murine homolog of JAM has led to the development of a model of the role for JAM in the formation of intercellular tight junctions (Kostrewa et al., 2001). In such tight junctions, the JAM ectodomains from two opposing cells are hypothesized to engage in homophilic interactions, leaving a distance of only ∼100 Å between the two cell membranes. Given the dimensions of each σ1 head domain of ∼25 × 35 × 50 Å, it seems that flexibility in a region just below the σ1 head would have to be required for it to bind the JAM ectodomain.
Interaction of σ1 with JAM
The three prototype reovirus strains T1L, T2J and T3D differ primarily in σ1 sequence (Duncan et al., 1990; Nibert et al., 1990). However, all three strains bind to JAM (Barton et al., 2001b and J.A.Campbell and T.S.Dermody, unpublished observations), suggesting that the JAM-binding site of σ1 is conserved among the three serotypes. Alignment of the T1L, T2J and T3D σ1 sequences shows that only 42 of the 210 residues of the crystallized T3D σ1 fragment are conserved in all three reovirus strains (Figure 3A). Many of these residues are buried, especially those located in the tail and at the base of the head trimer interface. A surprisingly large fraction of the remaining residues cluster in a single, solvent-exposed region at the lower edge of the β-barrel (Figure 3B). We propose that this region interacts with JAM. The predicted JAM-binding surface is formed primarily by a stretch of conserved residues (Asn369–Glu384) within the 310 helix and the long loop between β-strands D and E (Figure 3B). It also includes Trp421 at the end of strand F. The conserved region is fairly hydrophobic, with the side chains of Val371, Leu379 and Trp421 accounting for a large portion of the surface area predicted to be involved in JAM binding. A second, smaller cluster of conserved residues (Leu331, Trp333, Ile360 and His438) lies above this putative JAM-binding surface, near the top of the trimer (Figure 3B). The side chains of these residues are mostly buried and therefore unlikely to participate in receptor binding. The remaining surface area of the σ1 trimer, especially near the top of the head and the head-to-head contacts, is almost entirely devoid of conserved residues.
Fig. 3. Conservation of σ1 residues and predicted interaction with JAM. (A) Sequence alignment of T1L, T2J and T3D σ1. Conserved residues predicted to interact with JAM are shown in red; other conserved residues are shown in orange. (B) Surface representation of two σ1 monomers, with the third monomer shown as a blue ribbon. The conserved residues from (A) were mapped onto the σ1 surface using the same color code. The three views differ by rotations of 90° and 180°, respectively, along a vertical axis.
The conserved region at the lower edge of the β-barrel (Figure 3B) is easily accessible to a ligand. This region is somewhat recessed into the protein surface and surrounded by protruding, non-conserved residues on three sides. Its borders are formed by Gln422, Asp423 and Val425 in the FG loop above it, the highly hydrophobic Pro376-Pro377-Leu378 sequence on the left, and His388 and the C-terminal residue Thr455 on the right (Figure 3B). The recessed nature of the proposed JAM-binding site suggests that the region of JAM engaged by σ1 protrudes from the JAM surface. Only residues from a single monomer contribute to each region and its borders, and these regions are not involved in σ1 intersubunit contacts. Thus, the location of the conserved regions within the trimer suggests that each σ1 monomer can independently bind a JAM molecule.
Reovirus variants
Neutralization-resistant variants of reovirus T3D, selected using monoclonal antibodies to the σ1 head, exhibit diminished neurovirulence and altered CNS tropism in mice (Spriggs et al., 1983). These phenotypes have been mapped genetically to a point mutation at position 419 in the head (Bassel-Duby et al., 1986; Kaye et al., 1986). Glu419 of T3D σ1 is not conserved in the other two serotypes and it is not part of the proposed JAM-binding site. However, Glu419 lies in close proximity to the trimer interface (not shown) and a mutation at this position could distort the conformation of σ1 or alter its trimeric arrangement, perhaps affecting its receptor-binding properties. Additional neutralization-resistant variants have point mutations at Asp340 (Bassel-Duby et al., 1986), a residue also near the trimer interface (not shown).
The σ1 trimer interface
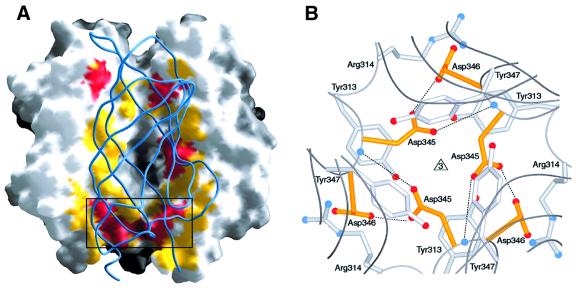
The intersubunit contacts within the σ1 trimer extend from the base of the tail to almost the very top of the head. The total buried surface area is ∼1100 Å2 for each monomer, of which almost 60% is accounted for by the tail (∼650 Å2). As expected, the intersubunit contacts within the triple β-spiral involve many conserved and hydrophobic residues (Figure 3B). The head trimer interface, on the other hand, buries a much smaller surface (∼450 Å2). Surprisingly, this interface almost completely lacks conserved residues (Figure 3B) and is relatively hydrophilic. Residues involved in contacts between the head domains are shown in Figure 4A. These contacts are discontinuous and involve few residues; moreover, the contact area is interrupted by a large water-filled cavity at the center of the trimer (Figure 4A). The largest contact area at the head trimer interface is at the base of the β-barrel and involves the conserved residues Val344, Asp345, Asp346 and Pro451 (Figures 3B and 4B). This contact is unusual since it is centered around six buried aspartic acid side chains (two from each monomer). The Asp346 side chains form salt bridges with Arg314 from neighboring monomers. However, the Asp345 side chains face each other and do not engage in interactions that would neutralize their charges. We also do not observe any electron density that would suggest the presence of a neutralizing metal ion in their vicinity. Although the σ1 crystals were grown at pH 7.5, the Asp345 side chains are probably protonated to avoid an unfavorable accumulation of negative charge in a solvent-excluded environment. A protonated Asp345 side chain would be able to form two hydrogen bonds with Asp346 (Figure 4B): one of these involves the protonated hydroxyl group of Asp345 and the carboxylate group of Asp346, and the other involves the carbonyl oxygen of Asp345 and the main chain amide of Asp346. The Asp345 side chains are sandwiched between two tyrosine side chains (Tyr313 and Tyr347) and, in addition to the hydrogen bonds, this arrangement presumably helps to lock them in this position.
Fig. 4. The σ1 head trimer interface. (A) View into the head trimer interface. Two monomers are shown as surface representations, and the third monomer is shown as a blue ribbon. Surface residues that are within 4 Å of residues in the third monomer are shown in red (residues conserved in T1L, T2J and T3D σ1) and yellow (residues unique to T3D σ1). The contact area involving conserved residues Val344, Asp345 and Asp346 is boxed, and this region is shown in more detail in (B). (B) View along the trimer axis, centered at conserved residues Asp345 and Asp346 (yellow) located at the base of the head. Residues Tyr313, Arg314 and Tyr347 engage in contacts with the two aspartic acids. The side chains of Asp345 are likely to be protonated to avoid an accumulation of negative charge at the interface. Hydrogen bonds involving protonated Asp345 are indicated. Oxygen and nitrogen atoms of side chains are shown as red and blue spheres, respectively, and the Asp346 main chain amides are shown as blue spheres as well.
The nature of the interactions at the head trimer interface suggests that the head contributes minimally to the overall oligomeric stability of σ1 and perhaps even destabilizes the trimer. The few intersubunit contacts between the σ1 head domains, the unusual cluster of aspartic acid residues at the center of the head trimer interface, and the observed flexibility in the σ1 tail (Figure 2A) suggest that σ1 is a high-energy structure poised for strategic movements. During viral disassembly, proteolytic cleavage of reovirus outer-capsid protein σ3, which lies in close proximity to σ1 on the virion surface, coincides with a change in σ1 from a retracted to an extended conformation (Dryden et al., 1993; Nibert et al., 1995). Given the conspicuously small and unusual interface between the σ1 heads, we speculate that such a change might involve the transition from monomer to trimer in the C-terminal region of σ1. In such a scenario, the triple β-spiral would act as a zipper, similar in a structural sense to the α-helical coiled-coil sequences that mediate dramatic conformational changes in influenza virus (Bullough et al., 1994). Trimerization of the head domain might alter σ1–JAM interactions in a way essential for viral disassembly or activation of intracellular responses required for subsequent steps in the viral life cycle.
Structural relationship between reovirus σ1 and adenovirus fiber
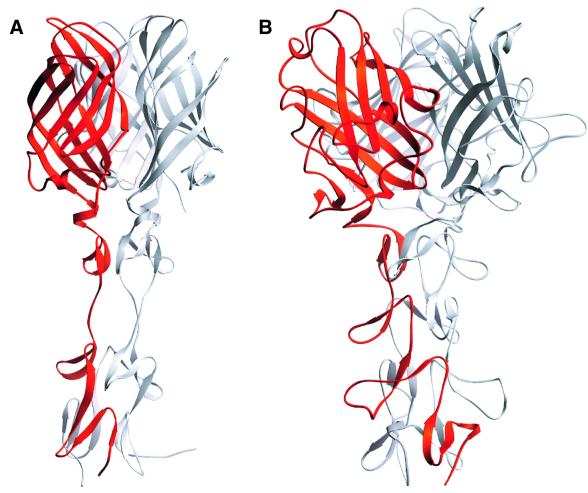
The σ1 structure exhibits a striking similarity to that of the adenovirus fiber (Figure 5; van Raaij et al., 1999). Both proteins are trimers that contain fibrous tails and globular heads; these are called ‘shaft’ and ‘knob’, respectively, in the adenovirus fiber. To date, the triple β-spiral motif has been observed only in the adenovirus fiber shaft and the reovirus σ1 tail. Although the σ1 spiral has an insertion that is absent in the adenovirus fiber, the two spiral structures are otherwise similar and emerge from the head domains in similar orientations and directions (Figure 5).
Fig. 5. Comparison of reovirus σ1 and adenovirus fiber structures. Trimeric structures of reovirus σ1 (A) and adenovirus fiber (B; van Raaij et al., 1999). In each case, one of the monomers is shown in red. Both attachment proteins have head-and-tail morphology, with a triple β-spiral forming the tail. The spirals of the crystallized reovirus σ1 fragment and adenovirus fiber (van Raaij et al., 1999) contain three and four repeats, respectively. (C) Superposition, in stereo, of σ1 head (red) and adenovirus fiber knob (blue; van Raaij et al., 1999). A portion of the β-spiral is shown in each case. The spirals emerge from the head domains in similar orientations and directions. In addition to the conserved β-sheet topology, the head and knob structures share a short helix (indicated in orange for σ1) in a long loop at the domain base.
The closest structural homolog for the σ1 head is again found in the corresponding region of the adenovirus fiber; a DALI (Holm and Sander, 1993) search using the σ1 head coordinates yielded the adenovirus fiber knob (Xia et al., 1994) as the only significant match (Z-score = 6.8). Both σ1 head and fiber knob contain eight anti-parallel β-strands with identical connectivity. The β-strands of σ1 circularize to form a β-barrel, whereas those of the adenovirus fiber knob form two separate four-stranded β-sheets that face each other in a β-sandwich-type arrangement (Figure 5C). The core regions of the two domains can be superimposed with an r.m.s.d. of 2.0 Å (76 Cα atoms). Both domains also contain a long loop (the DE loop in σ1) with a short helix at almost identical locations (Figure 5C). While the core structures of the two domains are similar, their surface structures are different. The σ1 head is a highly compact domain in which the β-strands are connected by extremely short loops. The fiber head has much more elaborate loops, which contain ∼50 additional residues.
The structural similarities between reovirus σ1 and adenovirus fiber correspond to common functional properties. Both σ1 head and fiber knob contain sequences that are involved in binding to cell-surface receptors (Bewley et al., 1999; Barton et al., 2001b) and, in both cases, are members of the immunoglobulin superfamily with two extracellular immunoglobulin-like domains (Bergelson et al., 1997; Barton et al., 2001b). Both attachment proteins also contain a slender fibrous tail that inserts into a pentameric slot in the virion capsid (the λ2 pentamer of reovirus and the penton base of adenovirus). In both cases, a large portion of the fibrous tail is likely to form a triple β-spiral; the adenovirus fiber shaft is thought to contain β-spiral repeats throughout (van Raaij et al., 1999), and the reovirus σ1 sequence indicates the presence of eight β-spiral repeats in the head-proximal portion of the tail (see below). Deviations from β-spiral sequence repeats correlate with enhanced flexibility in both proteins (Figure 2; Ruigrok et al., 1990, 1994; van Raaij et al., 1999).
The numerous structural and functional parallels between σ1 and fiber provide evidence for an evolutionary relationship. Recognizing common ancestry among viruses is complicated by their rapid divergence in sequence. Conservation at the structural level is often the only way to reveal such relationships. However, it is not always clear whether observed structural similarities result from convergent or divergent evolutionary processes because apparently unrelated viruses have a tendency to make use of similar designs. A well-known example is the ‘jelly-roll’ motif, a β-sandwich structure that contains two four-stranded anti-parallel β-sheets with unique topology. The jelly-roll motif is present in the structures of a large number of viruses studied to date, without obvious functional similarities or evolutionary relationships (Harrison, 2001).
However, in some cases, structural relationships between viruses or viral components do indicate common ancestry. One such example is the recently noted structural similarity between glycoprotein E1 of alphaviruses (Lescar et al., 2001; Pletnev et al., 2001) and glycoprotein E of flaviviruses (Rey et al., 1995). The conservation of key structural and functional features between these proteins supports the hypothesis that alphaviruses and flaviviruses have diverged from a common progenitor (Strauss and Strauss, 2001). Another example is the triple-helical coiled-coil structure found in the glycoproteins of many enveloped viruses and thought to be involved in initiation of the fusion process (Weissenhorn et al., 1999). A third example is the structural similarity between the adenovirus hexon protein and the P3 capsid protein of bacteriophage PRD1 (Benson et al., 1999). In all three cases, the structural similarities extend beyond the conservation of a single domain and translate into common functional properties. The relationship between reovirus σ1 and adenovirus fiber clearly belongs to this class.
Structure of full-length σ1
The short triple β-spiral structure in the crystallized portion of the σ1 tail (residues 246–309) is characterized by the presence of hydrophobic amino acids at specific locations and by conserved β-turn residues (Figure 6A). Examination of the sequence prior to residue 246 strongly suggests that the β-spiral extends for another five repeats towards the N-terminus in all three σ1 serotypes (Figure 6B). The sequence alignment also indicates that, with the exception of the insertion between the C-terminal 2 repeats (Figure 2), the spiral lacks any other insertions that would introduce additional flexibility into the molecule. Thus, we predict that residues 167–309 of T3D σ1 assemble into a continuous and mostly rigid triple β-spiral similar to the model shown in Figure 6C. Several observations support this model. First, the T3D σ1 protein can be cleaved with trypsin after Arg245 (Chappell et al., 1998). According to our prediction, Arg245 lies in a relatively long, surface-exposed loop between repeats 5 and 6 and would be easily accessible to trypsin. Secondly, almost all the insertions and deletions in the alignment shown in Figure 6B lie within surface-exposed loops linking two consecutive spiral repeats, thus allowing for variation of surface structure while maintaining the spiral core. Thirdly, the model offers a plausible explanation for the observed flexibility of the σ1 trimer at its midpoint (Fraser et al., 1990). The β-spiral probably does not extend beyond eight repeats, as the N-terminal third of σ1 is thought to form an extended α-helical coiled-coil structure (Bassel-Duby et al., 1985; Duncan et al., 1990; Fraser et al., 1990; Nibert et al., 1990). The transition between the coiled-coil and β-spiral segments probably introduces flexibility into the σ1 trimer. Fourthly, the dimensions predicted by our model agree with those observed by rotary shadowing studies. The overall length of a soluble, fully extended σ1 trimer is ∼480 Å (Fraser et al., 1990). The β-spiral/head portion of σ1 (Figure 6C) measures ∼200 Å in length, and a 165 amino acid α-helical coiled-coil structure would span ∼240 Å.
Fig. 6. Structure of the β-spiral repeats in the reovirus σ1 tail. (A) Detailed view of a section of the triple β-spiral of σ1. One σ1 chain is shown in orange; the other two are in gray. Residues Leu270, Met272, Ile274, Leu279 and Ile281 (blue) are located in β-strands and participate in the formation of a hydrophobic core that stabilizes the spiral. Residues Ser277 (blue) and Thr278 (magenta) are located in a β-turn that connects the two β-strands. The position of Thr278 is usually occupied by a proline or a glycine residue in β-spirals (van Raaij et al., 1999). (B) Sequence alignment of a region of the σ1 tail, indicating that the tail contains at least eight β-spiral repeats. The hydrophobic residues characteristic of β-spirals are indicated in blue, and the residues (usually proline or glycine) in the β-turns are shown in magenta. The consensus sequence for β-spiral repeats is shown at the bottom (h, hydrophobic residue; G, glycine; P, proline). Residues Asn198, Arg202 and Pro204 (shown in red) have been implicated in the interaction of T3D σ1 with sialic acid (Chappell et al., 1997). Arg245 (shown in blue) is the cleavage site for trypsin (Chappell et al., 1998). (C) Model of the complete spiral of σ1. Based on the sequence analysis shown in (B), the β-spiral probably begins at residue 167 of T3D σ1 and comprises eight repeats. The N-terminal five repeats are shown in gray. These repeats are not included in the crystal structure; the spiral has been extended using translated and rotated σ1 repeats to generate a model that depicts the approximate dimensions of the molecule. Residues prior to 167 are not shown; these residues are predicted to form an α-helical coiled-coil structure (Bassel-Duby et al., 1985; Duncan et al., 1990; Fraser et al., 1990; Nibert et al., 1990).
In addition to a binding site for JAM (Figure 2), T3D σ1 also contains a site for interaction with sialic acid (Chappell et al., 1997, 2000). The two binding sites are distinct (Nibert et al., 1995), and sialic acid is thought to serve as a co-receptor that enhances reovirus attachment (Barton et al., 2001a). Analysis of reovirus strains that vary in sialic acid-binding capacity has implicated σ1 residues Asn198, Arg202 and Pro204 in the interaction with sialic acid (Chappell et al., 1997). Although these residues are not included in the crystallized σ1 fragment, they are located within the segment of the protein that is expected to form a triple β-spiral (Figure 6B and C). Because the β-spiral is a highly regular structure, the location of specific residues can be predicted with reasonable accuracy. In the model shown in Figure 6C, the three residues important for sialic acid binding are all surface exposed and lie near each other on one side of the tail, supporting the hypothesis that they form part of a sialic acid-binding site (Chappell et al., 2000). The σ1 protein of reovirus T1L also binds cell-surface carbohydrate. Although the identity of the carbohydrate bound by T1L σ1 is not known, this interaction has been mapped to a region close to the C-terminal end of the β-spiral (Chappell et al., 2000). Hence, the triple β-spiral provides a novel framework for protein–carbohydrate interactions.
Conclusions
Our structural analysis of a receptor-binding fragment of reovirus attachment protein σ1 offers the first view of how this molecule interacts with its receptor JAM and provides a platform for studies to probe this interaction. In addition, the σ1 structure reveals several unexpected features. First, the short β-spiral appears to be the principal means of trimerization, whereas the contacts between the head domains are minimal and involve an unusual cluster of potentially destabilizing acidic residues. Secondly, the trimer has a flexible hinge within its tail that allows for substantial mobility between head and tail. Both of these features suggest that σ1 is a high-energy molecule that undergoes conformational rearrangements during viral attachment and cell entry. Thirdly, the structure offers a plausible structural model, a triple β-spiral, for much of the tail region. This model explains the observed flexibility of the trimer at defined positions and also provides a first view of how T3D σ1 might interact with its carbohydrate co-receptor. Fourthly, we show that reovirus σ1 and adenovirus fiber have remarkably similar structures. The similarities extend well beyond the conservation of a single domain, and they correlate with common functional properties. These observations suggest common ancestry, although the sequences of the two proteins have now diverged past a recognizable relationship. The alternative would be an amazing convergence on an optimal design for a fiber-like viral attachment protein.
Materials and methods
Protein production, purification and crystallization
The σ1 fragment containing amino acids 246–455 was obtained by tryptic cleavage of σ1 deletion mutant (3-Δ-3-3-3) expressed in insect cells and purified as described (Chappell et al., 2000). For cleavage, 3-Δ-3-3-3 σ1 protein was incubated with 1:1 trypsin at 17°C for 75 min. The cleavage products were separated using anion-exchange chromatography followed by gel filtration on Superdex 200 (Pharmacia). The C-terminal fragment comprising residues 246–455 eluted as a homogeneous peak corresponding to a trimer from the gel filtration column. N-terminal sequencing of the fragment confirmed that cleavage had occurred after Arg245. The protein was concentrated to 6 mg/ml for crystallization trials. Crystals were grown by vapor diffusion at 20°C by mixing equal volumes of protein solution and precipitant (10% isopropanol, 20% polyethylene glycol 4000, 100 mM HEPES pH 7.5). Heavy atom derivatives were obtained by soaking crystals overnight in either 0.1 mM CH3HgNO3 or 1 mM K2HgI4.
Data collection
Crystals belong to the space group P212121 (a = 52.7 Å, b = 89.0 Å, c = 119.8 Å) and contain one σ1 trimer in their asymmetric unit. Diffraction images were collected at 298K using a rotating anode generator equipped with an RAXIS IV image plate detector (Molecular Structure Corporation). The native data set was assembled from diffraction images collected from two crystals. A single crystal was used to collect each of the two derivative data sets. Data were integrated and scaled using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997), respectively.
Structure determination
Heavy atom binding sites were determined manually using difference Patterson and difference Fourier techniques. Difference Patterson maps were calculated with normalized structure factors. An initial electron density map based on two derivative data sets was calculated to 3.0 Å using MLPHARE (CCP4, 1994). This map was then averaged 3-fold (Kleywegt and Jones, 1994) using non-crystallographic symmetry operators derived from the refined heavy atom positions. The two main heavy atom binding sites (at residues Cys351 and His388) are located in the σ1 head; therefore, the averaging procedure improved the electron density for this domain substantially, allowing the entire domain to be traced. However, tail-forming residues 246–309 were not visible in this electron density map, and inspection of the unaveraged map revealed a kink between the 3-fold symmetry axes of head and tail. Thus, portions of the tail were initially traced using unaveraged maps, and these coordinates were then used to calculate a preliminary set of non-crystallographic symmetry operators. These symmetry operators were refined with the program IMP (Kleywegt and Jones, 1994) and then used to average the σ1 tail density. The three tails could then be traced. The model was built using O (Jones et al., 1991) and refinement was performed with XPLOR (Brünger et al., 1987) using bulk solvent correction and non-crystallographic symmetry restraints. Figures were prepared using RIBBONS (Carson, 1987) and GRASP (Nicholls et al., 1991). Buried surface areas were calculated using ACCESS (CCP4, 1994) with a probe radius of 1.4 Å.
Coordinates
The refined coordinates of the σ1 trimer and structure factor amplitudes have been deposited in the RCSB Protein Data Bank (http://www.rcsb.org) with the accession number code 1KKE. The coordinates are also available from the authors.
Acknowledgments
Acknowledgements
We thank members of our laboratories for review of the manuscript. This research was supported by Public Health Service awards AI38296 (T.S.D.) and AI45716 (T.S.) and the Elizabeth B.Lamb Center for Pediatric Research. Additional support was provided by Public Health Service awards CA68485 to the Vanderbilt Cancer Center and DK20593 to the Vanderbilt Diabetes Research and Training Center, by the Milton Foundation at Harvard Medical School and by the Structural Biology Core at Massachusetts General Hospital.
References
- Barton E.S., Connolly,L., Forrest,J.C., Chappell,J.D. and Dermody,T.S. (2001a) Utilization of sialic acid as a co-receptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem., 276, 2200–2211. [DOI] [PubMed] [Google Scholar]
- Barton E.S., Forrest,J.C., Connolly,J.L., Chappell,J.D., Liu,Y., Schnell,F.J., Nusrat,A., Parkos,C.A. and Dermody,T.S. (2001b) Junction adhesion molecule is a receptor for reovirus. Cell, 104, 441–451. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R., Jayasuriya,A., Chatterjee,D., Sonenberg,N., Maizel,J.V.,Jr and Fields,B.N. (1985) Sequence of reovirus haemagglutinin predicts a coiled-coil structure. Nature, 315, 421–423. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R., Spriggs,D.R., Tyler,K.L. and Fields,B.N. (1986) Identification of attenuating mutations on the reovirus type 3 S1 double-stranded RNA segment with a rapid sequencing technique. J. Virol., 60, 64–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S.D., Bamford,J.K., Bamford,D.H. and Burnett,R.M. (1999) Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell, 98, 825–833. [DOI] [PubMed] [Google Scholar]
- Bergelson J.M., Cunningham,J.A., Droguett,G., Kurt-Jones,E.A., Krithivas,A., Hong,J.S., Horwitz,M.S., Crowell,R.L. and Finberg,R.W. (1997) Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science, 275, 1320–1323. [DOI] [PubMed] [Google Scholar]
- Bewley M.C., Springer,K., Zhang,Y.B., Freimuth,P. and Flanagan,J.M. (1999) Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science, 286, 1579–1583. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Kuriyan,J. and Karplus,M. (1987) Crystallographic R-factor refinement by molecular dynamics. Science, 235, 458–460. [DOI] [PubMed] [Google Scholar]
- Bullough P.A., Hughson,F.M., Skehel,J.J. and Wiley,D.C. (1994) Structure of influenza haemagglutinin at the pH of membrane fusion. Nature, 371, 37–43. [DOI] [PubMed] [Google Scholar]
- Carson M. (1987) Ribbon models of macromolecules. J. Mol. Graph., 5, 103–106. [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chappell J.D., Gunn,V.L., Wetzel,J.D., Baer,G.S. and Dermody,T.S. (1997) Mutations in type 3 reovirus that determine binding to sialic acid are contained in the fibrous tail domain of viral attachment protein σ1. J. Virol., 71, 1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.D., Barton,E.S., Smith,T.H., Baer,G.S., Duong,D.T., Nibert,M.L. and Dermody,T.S. (1998) Cleavage susceptibility of reovirus attachment protein σ1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the σ1 neck. J. Virol., 72, 8205–8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.D., Duong,J.L., Wright,B.W. and Dermody,T.S. (2000) Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol., 74, 8472–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly J.L., Rodgers,S.E., Clarke,P., Ballard,D.W., Kerr,L.D., Tyler,K.L. and Dermody,T.S. (2000) Reovirus-induced apoptosis requires activation of transcription factor NF-κB. J. Virol., 74, 2981–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBiasi R.L., Edelstein,C.L., Sherry,B. and Tyler,K.L. (2001) Calpain inhibition protects against virus-induced apoptotic myocardial injury. J. Virol., 75, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden K.A., Wang,G., Yeager,M., Nibert,M.L., Coombs,K.M., Furlong,D.B., Fields,B.N. and Baker,T.S. (1993) Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol., 122, 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R., Horne,D., Cashdollar,L.W., Joklik,W.K. and Lee,P.W.K. (1990) Identification of conserved domains in the cell attachment proteins of the three serotypes of reovirus. Virology, 174, 399–409. [DOI] [PubMed] [Google Scholar]
- Fraser R.D., Furlong,D.B., Trus,B.L., Nibert,M.L., Fields,B.N. and Steven,A.C. (1990) Molecular structure of the cell-attachment protein of reovirus: correlation of computer-processed electron micrographs with sequence-based predictions. J. Virol., 64, 2990–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong D.B., Nibert,M.L. and Fields,B.N. (1988) σ1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol., 62, 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. (2001) The familiar and the unexpected in structures of icosahedral viruses. Curr. Opin. Struct. Biol., 11, 195–199. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zhou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kaye K.M., Spriggs,D.R., Bassel-Duby,R., Fields,B.N. and Tyler,K.L. (1986) Genetic basis for altered pathogenesis of an immune-selected antigenic variant of reovirus type 3 (Dearing). J. Virol., 59, 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) Halloween—masks and bones. In Bailey,S., Hubbard,R. and Waller,D. (eds), From First Map To Final Model. SERC Daresbury Laboratory, Warrington, UK, pp. 59–66.
- Kostrewa D. et al. (2001) X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. EMBO J., 20, 4391–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.W., Hayes,E.C. and Joklik,W.K. (1981) Protein σ1 is the reovirus cell attachment protein. Virology, 108, 156–163. [DOI] [PubMed] [Google Scholar]
- Lescar J., Roussel,A., Wien,M.W., Navaza,J., Fuller,S.D., Wengler,G. and Rey,F.A. (2001) The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell, 105, 137–148. [DOI] [PubMed] [Google Scholar]
- Martin-Padura I. et al. (1998) Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol., 142, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison L.A., Sidman,R.L. and Fields,B.N. (1991) Direct spread of reovirus from the intestinal lumen to the central nervous system through vagal autonomic nerve fibers. Proc. Natl Acad. Sci. USA, 88, 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert M.L., Dermody,T.S. and Fields,B.N. (1990) Structure of the reovirus cell-attachment protein: a model for the domain organization of σ1. J. Virol., 64, 2976–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert M.L., Chappell,J.D. and Dermody,T.S. (1995) Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved σ1 protein. J. Virol., 69, 5057–5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Oberhaus S.M., Smith,R.L., Clayton,G.H., Dermody,T.S. and Tyler,K.L. (1997) Reovirus infection and tissue injury in the mouse central nervous system are associated with apoptosis. J. Virol., 71, 2100–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olland A.M., Jané-Valbuena,J., Schiff,L.A., Nibert,M.L. and Harrison,S.C. (2001) Structure of the reovirus outer capsid and dsRNA-binding protein σ3 at 1.8 Å resolution. EMBO J., 20, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pletnev S.V., Zhang,W., Mukhopadhyay,S., Fisher,B.R., Hernandez,R., Brown,D.T., Baker,T.S., Rossmann,M.G. and Kuhn,R.J. (2001) Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell, 105, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch K.M., Nibert,M.L. and Harrison,S.C. (2000) Structure of the reovirus core at 3.6 Å resolution. Nature, 404, 960–967. [DOI] [PubMed] [Google Scholar]
- Rey F.A., Heinz,F.X., Mandl,C., Kunz,C. and Harrison,S.C. (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature, 375, 291–298. [DOI] [PubMed] [Google Scholar]
- Rodgers S.E., Barton,E.S., Oberhaus,S.M., Pike,B., Gibson,C.A., Tyler,K.L. and Dermody,T.S. (1997) Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J. Virol., 71, 2540–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok R.W., Barge,A., Albiges-Rizo,C. and Dayan,S. (1990) Structure of adenovirus fibre. II. Morphology of single fibres. J. Mol. Biol., 215, 589–596. [DOI] [PubMed] [Google Scholar]
- Ruigrok R.W., Barge,A., Mittal,S.K. and Jacrot,B. (1994) The fibre of bovine adenovirus type 3 is very long but bent. J. Gen. Virol., 75, 2069–2073. [DOI] [PubMed] [Google Scholar]
- Spriggs D.R., Bronson,R.T. and Fields,B.N. (1983) Hemagglutinin variants of reovirus type 3 have altered central nervous system tropism. Science, 220, 505–507. [DOI] [PubMed] [Google Scholar]
- Strauss J.H. and Strauss,E.G. (2001) Virus evolution: how does an enveloped virus make a regular structure? Cell, 105, 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler K.L. and Fields,B.N. (1996) Reoviruses. In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Vol. 2. Lippincott-Raven, Philadelphia, PA, pp. 1597–1623.
- Tyler K.L., McPhee,D.A. and Fields,B.N. (1986) Distinct pathways of viral spread in the host determined by reovirus S1 gene segment. Science, 233, 770–774. [DOI] [PubMed] [Google Scholar]
- Tyler K.L., Squier,M.K., Rodgers,S.E., Schneider,S.E., Oberhaus,S.M., Grdina,T.A., Cohen,J.J. and Dermody,T.S. (1995) Differences in the capacity of reovirus strains to induce apoptosis are determined by the viral attachment protein σ1. J. Virol., 69, 6972–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanRaaij M.J., Mitraki,A., Lavigne,G. and Cusack,S. (1999) A triple β-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature, 401, 935–938. [DOI] [PubMed] [Google Scholar]
- Weiner H.L., Drayna,D., Averill,D.R.,Jr and Fields,B.N. (1977) Molecular basis of reovirus virulence: role of the S1 gene. Proc. Natl Acad. Sci. USA, 74, 5744–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H.L., Powers,M.L. and Fields,B.N. (1980) Absolute linkage of virulence and central nervous system cell tropism of reoviruses to viral hemagglutinin. J. Infect. Dis., 141, 609–616. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W., Dessen,A., Calder,L.J., Harrison,S.C., Skehel,J.J. and Wiley,D.C. (1999) Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol., 16, 3–9. [DOI] [PubMed] [Google Scholar]
- Xia D., Henry,L.J., Gerard,R.D. and Deisenhofer,J. (1994) Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure, 2, 1259–1270. [DOI] [PubMed] [Google Scholar]
- Zhang C. and Kim,S.-H. (2000) A comprehensive analysis of the Greek key motifs in protein β-barrels and β-sandwiches. Proteins, 40, 409–419. [DOI] [PubMed] [Google Scholar]