Abstract
Mice lacking transcription factor interferon consensus sequence binding protein (ICSBP) develop a syndrome similar to human chronic myeloid leukemia and are immunodeficient. In order to define the molecular mechanisms responsible for the cellular defects of ICSBP–/– mice, we used bone marrow-derived macrophages (BMM) to identify genes deregulated in the absence of ICSBP. Here, we report that disabled-2 (Dab2), a signal phosphoprotein, is transcriptionally up-regulated and accumulates in the cytoskeleton/membrane fraction of ICSBP–/– BMM. Moreover, our results revealed Dab2 as a novel IFN-γ-response gene. Both ICSBP and the Ets-transcription factor PU.1 bind to the Dab2 promoter, whereby ICSBP represses PU.1-induced Dab2 promoter transactivation in vitro. Notably, repression of Dab2 expression by ICSBP is also found in myeloid progenitors. Overexpression of Dab2 leads to accelerated cell adhesion and spreading, accompanied by enhanced actin fiber formation. Furthermore, cell adhesion induces transient Dab2 phosphorylation and its translocation to the cytoskeletal/membrane fraction. Our results identify a novel role of Dab2 as an inducer of cell adhesion and spreading, and strongly suggest that the up-regulation of Dab2 contributes to the hematopoietic defect seen in ICSBP–/– mice.
Keywords: adhesion/Dab2/ICSBP/IFN-γ/myelopoiesis
Introduction
Progression of pluripotent hematopoietic stem cells to mature, terminally differentiated effector cells of the hemato-lymphoid system proceeds through a process of sequential changes in the gene expression pattern, driven by multiple extrinsic and intrinsic signals. Transcription factors are shared mediators of both extrinsic and intrinsic signals and control gene activity directly. Both general transcription factors, indispensable for development of all hematopoietic cells, as well as lineage specific factors have been described (Tenen et al., 1997; Orkin, 2000).
Analysis of mice deficient for interferon consensus sequence binding protein (ICSBP), a member of the interferon regulatory factor (IRF) family of transcription factors, has provided evidence for a critical role of ICSBP in hematopoiesis, in particular as a regulatory switch in myeloid differentiation. ICSBP deficiency in mice results in a complex phenotype, dominated by immunodeficiency and a myelo-lymphoproliferative syndrome that resembles chronic myeloid leukemia (CML) (Holtschke et al., 1996; Fehr et al., 1997).
Several lines of evidence support the notion of ICSBP involvement in human CML, a disease caused by the Bcr–Abl fusion protein (reviewed in Deininger et al., 2000). Patients with CML have strongly reduced levels of ICSBP, which can be restored after interferon (IFN)-α therapy (Schmidt et al., 1998). Furthermore, ICSBP is significantly reduced in mice with a Bcr–Abl-induced CML-like disease, and forced overexpression of ICSBP inhibits the myeloproliferative syndrome in that system (Hao and Ren, 2000).
The hematopoietic precursor cells from ICSBP–/– mice, similar to those from CML patients, reveal gross alterations in their response to growth factors and altered adhesion. The most remarkable characteristic of the ICSBP–/– bone marrow progenitors is their altered response to CSF-1, suggesting a critical role of ICSBP in the proliferation and differentiation of the myeloid cell lineage (Scheller et al., 1999). This notion was confirmed and further extended by Tamura et al. (2000), who showed that ICSBP directed macrophage differentiation of a myeloid progenitor cell line established from ICSBP–/– mice.
Recently, we have found that in the absence of ICSBP, CSF-1R signaling is attenuated (Kallies et al., 2002), as seen from a rapid termination of MAP kinase phosphorylation and reduced cell growth. This coincides with enhanced accumulation of c-Cbl, which is known to down-regulate CSF-1R signaling by its ubiquitin-ligase activity. Our results indicate that c-Cbl is proteolytically degraded and that this proteolytic activity is reduced in ICSBP–/– bone marrow-derived macrophages (BMM). However, the primary target genes directly deregulated by the lack of the transcription factor ICSBP remain to be found. In view of the profound effects of the ICSBP deficiency on myeloid differentiation and proliferation, the identification of ICSBP target genes should contribute to our understanding of normal myelopoiesis and could reveal insights into novel mechanisms underlying myeloproliferative diseases.
Here we have identified disabled-2 (dab2) as a gene differentially transcribed in myeloid cells from ICSBP–/– and ICSBP+/+ mice. Mouse Dab2 is a mitogen-responsive phosphoprotein with sequence homology to Dab proteins of humans as well as Caenorhabditis elegans and Drosophila melanogaster (Xu et al., 1995; Albertsen et al., 1996). Dab2 contains a phosphotyrosine-interacting domain, a C-terminal proline-rich domain and a potential actin-binding motif, KKEK (Xu et al., 1995). It has been reported that Dab2 interaction with Grb2 reduces the binding between Grb2 and Sos and thus could modulate growth factor/Ras pathways (Xu et al., 1998). Recently, Dab2 was characterized as a critical link between transforming growth factor (TGF)-β receptors and the Smad proteins (Hocevar et al., 2001). Results presented here identify Dab2 as a novel ICSBP down-regulated target of the IFN-γ pathway and show for the first time that Dab2 induces macrophage adhesion and spreading.
Results
Dab2 expression is enhanced in BMM from ICSBP–/– mice
In order to identify the molecular mechanisms responsible for altered myeloid differentiation, we used BMM from ICSBP+/+ and ICSBP–/– mice to search for genes differentially expressed in the absence of ICSBP. cDNA expression was compared by expression arrays, which allow the comparison of expression levels of almost 1200 different genes in parallel. After normalizing the expressed signals to a set of housekeeping genes, only six genes (1.3% of the positive signals) were differentially expressed by >2-fold in ICSBP–/– BMM (data not shown). The low numbers of differentially expressed genes confirmed the high comparability of both BMM populations. The most pronounced difference in expression was observed for dab2, with a 3.2-fold increase in mRNA levels in ICSBP–/– BMM (Figure 1A). This result was confirmed also by semiquantitative RT–PCR (Figure 1B). Dab2 is a mitogen-responsive phosphoprotein with signal transduction capability (Xu et al., 1995), and therefore an interesting candidate gene, potentially involved in molecular pathways leading to altered cellular properties of ICSBP–/– BMM.
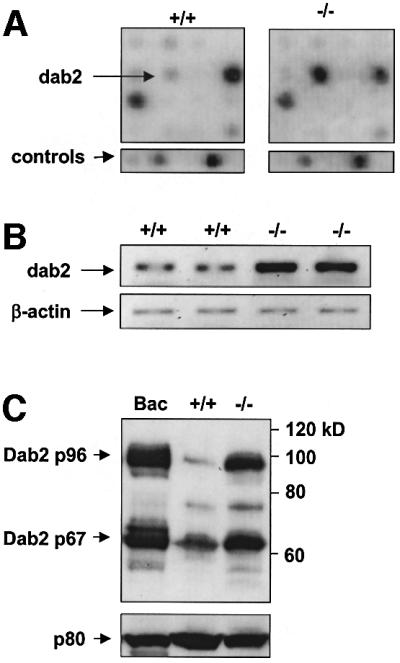
Fig. 1. Dab2 is overexpressed in ICSBP–/– BMM. (A) The hybridization patterns from ICSBP+/+ and ICSBP–/– BMM from a section of the cDNA expression arrays are shown. The arrows indicate the position of the Dab2 cDNA and of two control genes (β-actin and a 40S ribosomal protein). The arrays show a strong hybridization signal of Dab2 in ICSBP–/– but a weak signal in ICSBP+/+ BMM. (B) Semiquantitative RT–PCR analysis demonstrating the mRNA expression levels of the Dab2 gene in BMM from ICSBP+/+ and ICSBP–/– mice. (C) Western blot analysis of total protein extracts showing the different protein expression levels of Dab2 in ICSBP+/+ and ICSBP–/– BMM. Bac1.2F5 cells, expressing p96 and p67 (Xu et al., 1995), served as a control to identify the Dab2 isoforms expressed in BMM. The Dab2 proteins were detected by M2 antiserum. Equal protein loading was confirmed by the protein p80 detected by ICSBP antiserum.
Dab2 is expressed as two isoforms, p96 and p67, in vivo (Xu et al., 1995); both of them were also present in ICSBP+/+ and ICSBP–/– BMM (Figure 1C). However, a striking accumulation of both isoforms was found in ICSBP–/– cells. Thus, our results demonstrate a significant up-regulation of Dab2 at both RNA and protein levels in ICSBP–/– BMM.
ICSBP represses PU.1-induced transactivation of the Dab2 promoter
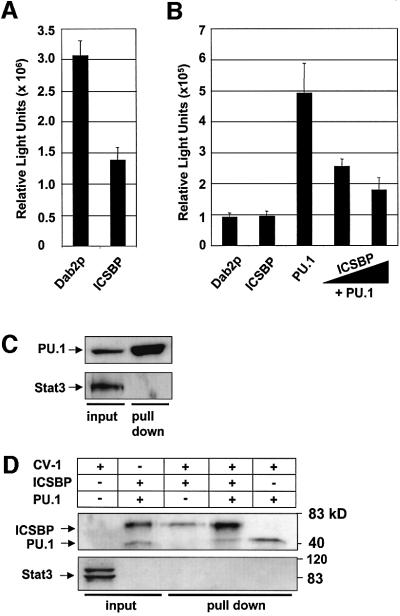
To analyze whether ICSBP regulates Dab2 transcription directly, a luciferase reporter plasmid under the transcriptional control of the Dab2 promoter (pGL3-Dab2p) was transfected into two cell lines, K562 and CV-1, both lacking endogenous ICSBP expression (Rosenbauer et al., 1999; data not shown). As demonstrated in Figure 2A, transcription of the reporter gene driven by Dab2 regulatory sequences was repressed by the co-transfection of ICSBP expression vector (pcDNA-ICSBP) in K562 cells. Consequently, ICSBP acts as a negative transcriptional regulator of the Dab2 promoter.
Fig. 2. ICSBP down-regulates PU.1 induced transactivation of the Dab2 promoter. (A) K562 cells were transfected with pGL3-Basic containing a 2 kb fragment of the mouse Dab2 promoter together with pcDNA or pcDNA-ICSBP and the reference vector pRL-TK. Luciferase reporter gene activities were measured as described in Materials and methods. (B) CV-1 cells were transiently transfected with a Dab2 promoter driven luciferase reporter gene (300 ng) along with the following expression vectors: PU.1 200 ng; ICSBP 200 ng or 600 ng. Empty vector DNA was added so that each reaction contained a total of 800 ng of expression plasmid. (C) Endogenously expressed PU.1 binds to the Dab2 regulatory sequence. K562 nuclear extracts were incubated with the immobilized Dab2 promoter DNA, and the bound fraction was analyzed by western blotting using antibodies as indicated. (D) ICSBP and PU.1 bind to the Dab2 promoter. Magnetic beads containing the 2 kb mouse Dab2 promoter were incubated with nuclear extracts of CV-1 cells, which were supplemented with 10 µl of 35S-labeled ICSBP, PU.1, or both ICSBP and PU.1. The beads were washed and the eluates were subjected to SDS–PAGE and autoradiography. 35S-labeled protein (1 µl) was used as an input control. Probing of the membrane with an anti-Stat3 antibody confirmed the specificity of the employed assay.
In CV-1 cells only very low levels of Dab2-directed luciferase expression could be detected, which was not affected by the co-transfection of the ICSBP expression vector. These data suggest that other co-factors, which are required for Dab2 promoter activation, might be absent in CV-1 cells. In contrast to K562 cells, CV-1 cells do not express the hematopoietic-specific Ets-transcription factor PU.1 (Behre et al., 1999). Since PU.1 is a well-defined binding partner of ICSBP (Brass et al., 1996), we analyzed the effect of PU.1 on the Dab2 promoter by co-transfecting pGL3-Dab2p along with a PU.1 expression vector into CV-1 cells. PU.1 transactivated the Dab2 promoter >5-fold, indicating that PU.1 is a potent positive-regulator of Dab2. Interestingly, when increasing amounts of ICSBP were co-transfected together with constant amounts of pGL3-Dab2p and PU.1, a repression of PU.1-induced Dab2 promoter transactivation was observed (Figure 2B). Together, these results indicate that ICSBP down-regulates the PU.1-dependent transcription of the Dab2 promoter.
To test whether ICSBP and PU.1 can physically interact with the Dab2 promoter, we employed a DNA affinity binding assay that has previously been used as a sensitive method for the analysis of protein recruitment to regulatory DNA elements (Wang et al., 2000). The Dab2 promoter DNA was conjugated to magnetic beads and incubated with nuclear extracts from CV-1 cells, which were supplemented with [35S]methionine-labeled ICSBP, PU.1, or both ICSBP and PU.1. The proteins bound to the immobilized DNA were separated from the unbound fraction, resolved by SDS–PAGE and visualized by autoradiography. Although both ICSBP and PU.1 were recruited to the Dab2 promoter individually, ICSBP binding increased >6-fold in the presence of PU.1 (Figure 2D). The specificity of the assay was tested by probing the membrane with an antibody against Stat3 as a negative control. Although Stat3 was highly expressed in CV-1 cells, it was not recruited to the conjugated beads. Furthermore, endogenous PU.1 expressed by K562 cells was also found to bind strongly to the Dab2 promoter (Figure 2C). Thus, the employed DNA affinity binding assay revealed specific binding of both PU.1 and ICSBP to the Dab2 regulatory DNA, and indicated that ICSBP binding is supported by PU.1.
ICSBP suppresses Dab2 expression in myeloid progenitors
The role of ICSBP as a transcriptional repressor of Dab2 was directly confirmed by its enforced expression in two mouse myeloid progenitor cell lines. IC34L cells, a line derived from ICSBP–/– mice, infected with either a control retroviral vector or a vector expressing a hydroxytamoxifen (TAM)-inducible ICSBP, express Dab2 but no endogenous ICSBP. After ICSBP induction by TAM, Dab2 expression was no longer detectable in these cells, whereas TAM induction of control infected IC34L cells had no effect on Dab2 levels (Figure 3A). Similar results were obtained with FDC-P1Mac11 cells, a line that expresses Dab2 but only very limited amounts of ICSBP (Figure 3B). After transfection with an ICSBP expression plasmid, Dab2 expression was significantly reduced. Further evidence supporting a direct regulation of Dab2 gene expression by ICSBP in myeloid progenitor cells was revealed by DNA affinity binding. As shown in Figure 3C, both ICSBP and PU.1 were specifically recruited to the Dab2 promoter when incubated with nuclear extracts of IC34L cells.
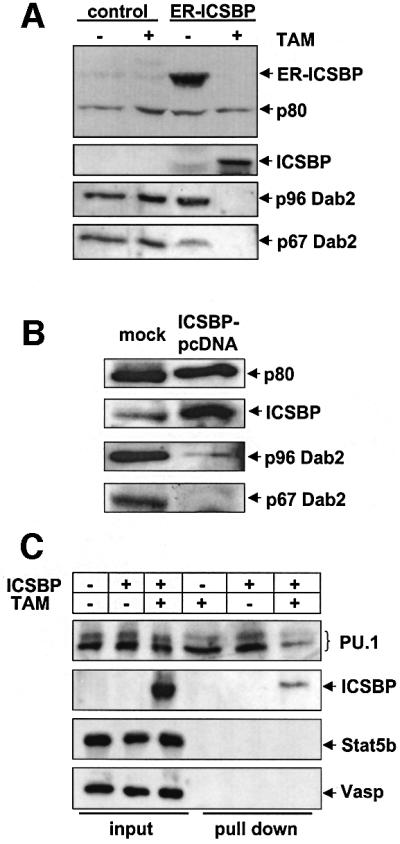
Fig. 3. ICSBP regulates Dab2 expression in myeloid progenitor cells. (A) TAM-inducible ICSBP represses Dab2 expression in IC34L cells. The IC34L precursor cell line, isolated from ICSBP–/– mouse, was transfected with a TAM-inducible estrogen (ER)–ICSBP fusion construct or with control vector by retroviral infection as described in Materials and methods. Cells were treated with TAM for 16 h and total cell extracts were analyzed by western blotting using an anti-Dab2 antibody and ICSBP antiserum. (B) Overexpression of ICSBP in the bipotential myeloid precursor cell line FDC-P1Mac11 down-regulates endogenous Dab2 expression. FDC-P1Mac11 cells were either mock-transfected or transiently transfected with pcDNA-ICSBP. After 24 h, total protein was extracted and analyzed by western blotting using a Dab2 antiserum (M2) and ICSBP antiserum. (C) ICSBP and PU.1 bind to the Dab2 promoter in myeloid progenitor cells. The Dab2 promoter DNA was immobilized by magnetic beads and incubated with nuclear extracts of IC34L cells transfected with the TAM-inducible ER–ICSBP fusion protein or a control vector. Proteins recruited to the Dab2 promoter were analyzed by western blotting using antibodies as indicated.
The fact that ICSBP represses Dab2 expression in myeloid progenitors indicates that Dab2 gene regulation by ICSBP is not limited to more mature monocytic cells, e.g. BMM, but also plays a role in myeloid progenitors.
dab2 is a novel IFN-γ-responsive gene
The expression of ICSBP is induced by IFN-γ and also, to a lesser extent, by LPS (Wang et al., 2000). Whether dab2 expression is controlled by IFN-γ was tested in the experiment shown in Figure 4. As expected, Dab2 transcription, as well as protein expression, was repressed in ICSBP+/+ BMM stimulated with IFN-γ. In contrast, no reduction in dab2 mRNA and protein levels was observed in ICSBP–/– cells in response to IFN-γ. Furthermore, ICSBP recruitment to the dab2 promoter was strongly increased after IFN-γ induction, as revealed by DNA affinity binding (Figure 4C). In contrast, no effect of IFN-γ on PU.1 binding to the dab2 promoter was observed. Taken together, our results show that dab2 is a novel downstream target of the IFN-γ pathway, transcriptionally regulated by ICSBP.
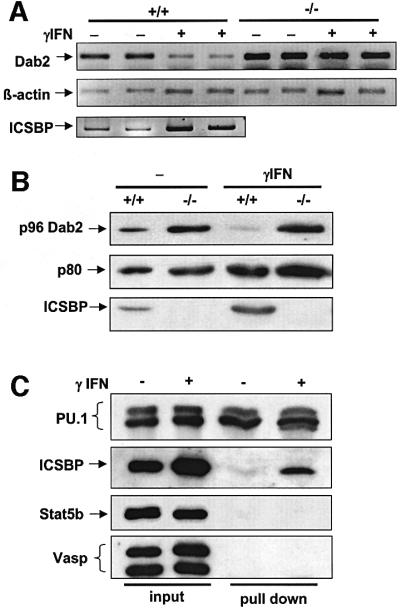
Fig. 4. Dab2 is a novel IFN-γ-responsive protein. (A and B) BMM from ICSBP+/+ and ICSBP–/– mice were cultivated for 19 h in the absence (–) or presence (+) of 200 U/ml IFN-γ. (A) Total RNA was isolated and subjected to semiquantitative RT–PCR analysis using primers for Dab2, β-actin and ICSBP. (B) Total cell extracts were analyzed by western blotting with an anti-Dab2 antibody and an anti-ICSBP antiserum. (C) IFN-γ induces ICSBP binding to the Dab2 promoter. Nuclear extracts of the macrophage-like cell line Bac1.2F5, either untreated (–) or treated with IFN-γ (200 U/ml) (+) for 16 h, were incubated with immobilized Dab2 promoter DNA. The bound fraction was analyzed by western blotting using antibodies as indicated.
Dab2 is phosphorylated, and accumulates in the cytoskeleton/membrane fraction of ICSBP–/– BMM
Mitogen stimulation, such as treatment with CSF-1 or tetradecanoylphorbol-13-acetate (TPA), leads to serine-phosphorylation of Dab2 by PKC (Xu et al., 1995). This modification seems to be important for at least some Dab2 functions, since only the phosphorylated protein can inhibit AP-1 activity (Tseng et al., 1999). As shown by others, phosphorylated Dab2 can easily be detected by its retarded electrophoretic mobility in gels compared with the unphosphorylated protein (Xu et al., 1995). To investigate whether the increase of Dab2 protein expression in ICSBP–/– BMM is also accompanied by its phosphorylation, we analyzed the phosphorylation of p96 after CSF-1 stimulation. We found that the slower migrating phospho-p96 (pp96) appears 5 min after stimulation of both ICSBP+/+ and ICSBP–/– BMM (Figure 5A).
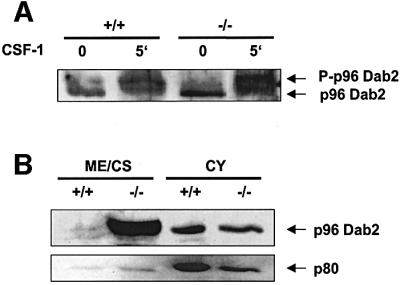
Fig. 5. Phosphorylation and subcellular distribution of Dab2 in BMM. (A) Phosphorylation of p96 Dab2 in ICSBP+/+ and ICSBP–/– BMM following CSF-1 stimulation. Cells were deprived of CSF-1 for 16 h, left untreated or were stimulated with 150 ng/ml CSF-1 at 37°C for 5 min. Total cell extracts were analyzed by western blotting with a Dab2 antiserum (M15). To obtain a better separation of p96 and its phosphorylated form (P-p96), only 30% of the ICSBP–/– protein extract, compared with the ICSBP+/+ extract, was loaded on the gel. (B) Subcellular distribution of p96 Dab2 in BMM from ICSBP+/+ and ICSBP–/– mice. Membrane/cytoskeleton (ME/CS) and cytosolic (CY) fractions (25 µg each) were loaded on SDS–PAGE and subjected to western blotting using M15 Dab2 and ICSBP antisera.
It has been reported previously that Dab2 is found mainly in the cytosol but appears in the particulate fraction after stimulation with TPA (Tseng et al., 1999). We therefore analyzed the subcellular distribution of Dab2 in asynchronously growing BMM by separating the cytosolic from the cytoskeleton/membrane-associated proteins. Comparable amounts of Dab2 were present in the cytosol of both ICSBP+/+ and ICSBP–/– cells (Figure 5B). However, a strongly enhanced accumulation of Dab2 was seen in the cytoskeleton/membrane fraction of ICSBP–/– BMM.
Together, these results indicate that loss of ICSBP in BMM leads to enhanced expression of Dab2, which becomes phosphorylated after CSF-1 stimulation and accumulates in the membrane/cytoskeletal fraction.
Enhanced expression of Dab2 augments spreading and adhesion of macrophages
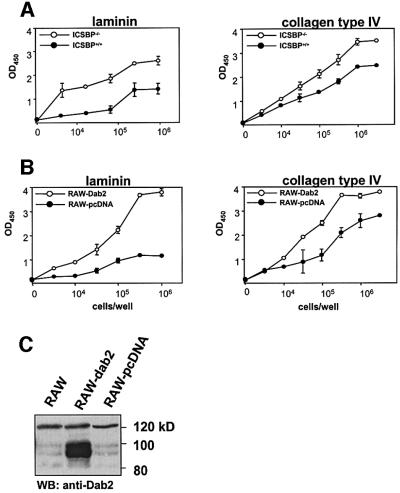
We reported previously that myeloid progenitors from ICSBP–/– mice have altered adhesion properties to extracellular matrix (ECM) components (Scheller et al., 1999). Sheng et al. (2000) suggested a role of Dab2 in cell positioning control and in mediating the exigency for basement membrane attachment of epithelial cells. Therefore, we have compared the adhesion of BMM from ICSBP–/– and ICSBP+/+ mice to laminin and collagen IV using an adhesion assay. These two compounds are the main components of the basement membrane and other ECM. As seen in Figure 6A, an increased adhesion of ICSBP–/– BMM to both substrates was observed.
Fig. 6. Overexpression of Dab2 leads to enhanced adhesion of macrophages to laminin and collagen type IV. (A) BMM from ICSBP+/+ and ICSBP–/– mice, and (B) RAW cells stably transfected with a pcDNA vector containing the complete Dab2-cDNA or the empty pcDNA were used in an adhesion assay. Indicated numbers of enzymatically labeled cells were plated on 96-well plates coated with laminin or collagen type IV. After 15 min incubation, non-adherent cells were removed by washing, and the adherent cells were stained and processed according to the manufacturer’s protocol. The graphs show the results of a representative experiment; three experiments were performed in triplicate. (C) Total cell extracts of RAW cells stably transfected with pcDNA-Dab2, untransfected or transfected with the empty vector were subjected to western blotting with a Dab2 antibody. Extracts of 5 × 105 cells per lane were loaded on the gel.
To investigate whether the enhanced adhesion of ICSBP–/– BMM to laminin or collagen IV could be linked directly to Dab2 overexpression, the murine macrophage cell line RAW 264.1 was transfected with a Dab2 expression vector, and four stably transfected lines overexpressing Dab2 were established. Their independent origin was confirmed by Southern blots (data not shown). Only a slight expression of the endogenous Dab2 gene was detected in non-transfected RAW cells, as well as in cells stably transfected with the empty plasmid (Figure 6C).
The adhesion of Dab2-overexpressing RAW cells to laminin and collagen IV was markedly enhanced, similar to what is seen in ICSBP–/– BMM (Figure 6B). Thus Dab2 is a potent inducer of cell adhesion and it is very likely that its deregulated expression is, at least in part, directly responsible for the altered adhesion properties of ICSBP–/– myeloid cells.
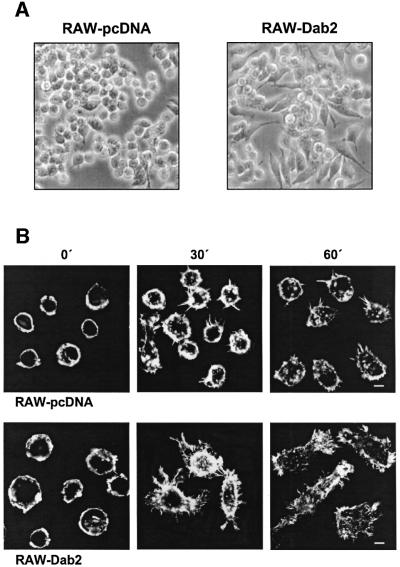
In addition to the enhanced adhesion, we noted an accelerated spreading of ICSBP–/– BMM. This phenomenon was even more prominent in Dab2-overexpressing RAW cells (Figure 7A). We therefore compared the adhesion-induced morphological changes of Dab2 and control transfected RAW cells. Cell spreading and the adhesion-induced reorganization of the actin cytoskeleton was monitored by plating the cells on laminin-coated glass coverslips, followed by rhodamin-labeled phalloidin staining at different time points (Figure 7B). When maintained in suspension, both types of cells had a rounded, contracted shape and displayed a cortical ring of actin filaments. Notably, intensification of adhesion-induced lamellae and filopodia formation was more prominent in cells overexpressing Dab2. Furthermore, cell spreading and formation of lamellae and filopodia started earlier and was clearly visible as early as 30 min after plating the cells.
Fig. 7. Overexpression of Dab2 leads to accelerated spreading and formation of pseudopodia in macrophages. (A) Phase-contrast microscopy of RAW cells transfected with Dab2-pcDNA or the empty vector, and plated on the plastic surface of tissue culture plates. (B) Confocal immunofluorescence microscopy of actin cytoskeletal organization of RAW cells transfected with Dab2-pcDNA or the empty vector. Cells were trypsinized and plated on laminin-coated coverslips, cultured for 30 and 60 min, fixed and processed for F-actin staining using rhodamine-conjugated phalloidin. Scale bar = 5 µm.
Taken together, we found that Dab2 overexpression augments macrophage adhesion and spreading and is associated with reorganization of the actin cytoskeleton.
Dab2 is phoshorylated and translocates to cytoskeleton/membrane upon macrophage adhesion
Adhesion is known to induce phosphorylation and translocation of several cytosolic signal proteins to components of the cytoskeleton and cell membrane (Machesky and Hall, 1997). To further examine the role of Dab2 in adhesion-induced signaling pathways, we examined whether Dab2 becomes phosphorylated during adhesion of cells to components of ECM. Serine phosphorylation was shown to be required for at least some functions of Dab2 in epithelial cells (Tseng et al., 1999). RAW cells overexpressing Dab2 were seeded on plates coated with laminin, or were kept in suspension. The appearance of phosphorylated forms of p96 Dab2 was monitored by western blotting (Figure 8A). The adhesion-induced phosphorylation of Dab2 was transient; it was clearly observed after 30 min and lasted for ∼2 h. Dab2 phosphorylation was observed also in RAW cells after their adhesion on other substrates, fibronectin and poly-d-lysine. Similar results were also obtained with BMM (Figure 8B and C).
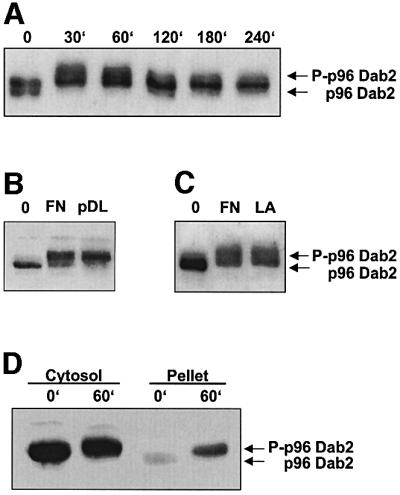
Fig. 8. Dab2 is transiently phosphorylated and translocates into the cytoskeleton/membrane fraction in response to adhesion. (A) RAW macrophages overexpressing Dab2 were trypsinized and plated on laminin-coated culture dishes or were kept in suspension. At the time points indicated, total cell extracts of adherent cells and cells kept in suspension were analyzed by western blotting. RAW cells overexpressing Dab2 (B) or ICSBP–/– BMM (C) were treated as described in (A), and plated on culture dishes coated with fibronectin (FN), poly-d-lysine (pDL) or laminin (LA). After 30 min, total cell extracts of adherent cells and cells kept in suspension were prepared, and subjected to SDS–PAGE and western blotting with the anti-Dab2 antibody. (D) RAW cells overexpressing Dab2 were trypsinized and plated on culture dishes coated with fibronectin, or kept in suspension. After 60 min, cytosolic and particulate fractions were prepared as described in Materials and methods, and were analyzed by western blotting with anti-Dab2 antibody.
We next investigated whether adhesion induces the translocation of cytosolic Dab2 to the particulate fraction, i.e. cytoskeleton/membrane. Dab2-overexpressing cells were plated on laminin-coated plates or were kept in suspension. Figure 8D shows that phosphorylated forms of Dab2 accumulated in the particulate fraction of adherent cells, indicating a translocation and cytoskeleton association of Dab2. In conclusion, the results presented demonstrate that cell adhesion induces transient Dab2 phosphorylation and translocation to the cytoskeleton fraction.
Discussion
The molecular basis of the hematopoietic disorder in mice deficient for transcription factor ICSBP is not known. In order to illuminate their deregulated transcription program, we have searched for differentially expressed genes in BMM from ICSBP–/– and ICSBP+/+ mice. Here, we report that the gene encoding the signal phosphoprotein Dab2 is transcriptionally up-regulated in ICSBP-deficient BMM. Protein analyses confirmed the overexpression of Dab2 and showed that, in the absence of ICSBP, Dab2 accumulates in the membrane/cytoskeleton fraction. The functional significance of Dab2 in these cells is supported by its phosphorylation after CSF-1 stimulation. Further more, we provide direct evidence that Dab2 expression is controlled by IFN-γ via ICSBP.
The loss of Dab-2 gene expression has been associated with oncogenic processes in some tissues (Fulop et al., 1998; Fazili et al., 1999). However, very little is known about the transcriptional regulation of the Dab2 gene. Morrisey et al. (2000) were able to show that Dab2 mRNA is up-regulated by GATA-6 in embryonic stem cells and in the visceral endoderm. Here, the mode of transcriptional regulation of Dab2 by ICSBP was investigated using co-transfection experiments and DNA affinity binding. The results revealed that Dab2 transcription is positively controlled by PU.1, and that ICSBP specifically represses PU.1-induced Dab2 transactivation. Both PU.1 and ICSBP are hematopoietic-restricted transcription factors critical for the development and key functions of hematopoietic cells (Holtschke et al., 1996; Tenen et al., 1997). We have shown that ICSBP represses the transactivation of Dab2 via its recruitment to the Dab2-regulatory DNA region, indicating that Dab2 is a direct downstream target of ICSBP in myeloid cells. Interestingly, although PU.1 is not required for ICSBP–Dab2 promoter interaction, we found an increased ICSBP binding in the presence of PU.1. Cross-talk between PU.1 and ICSBP has been noted before as a critical event in the regulation of several genes. ICSBP and PU.1 can form a ternary complex with a composite IRF/Ets motif found in regulatory DNA elements of several promoters (Eisenbeis et al., 1995). A computer search did not reveal any composite IRF/Ets motif within the 2 kb Dab2 promoter, but did identify several single Ets sites. Interestingly, recent reports have shown that ICSBP can bind to single Ets sites, probably through protein– protein interactions (Wang et al., 2000; Grazia Cappiello et al., 2001). Here, we reveal Dab2 as a novel downstream target of the IFN-γ pathway. The link between Dab2 and IFN-γ is interesting, since IFN-γ is a well-characterized macrophage differentiation and activation signal (Stark et al., 1998). In contrast to the ICSBP+/+ controls, the ICSBP–/– BMM failed to down-regulate Dab2 expression after IFN-γ stimulation at both RNA and protein levels. Thus the presence of ICSBP appears to be critical for IFN-γ-induced Dab2 suppression in macrophages.
The biological function of Dab2 has not yet been fully elucidated. It is conceivable, owing to Dab2 ubiquitous expression and the diversity of reported activities, that Dab2 is involved in several signaling pathways. It has been suggested, but not proven, that Dab2 functions as a repressor of Ras signaling (Xu et al., 1998). More recently, Hocevar et al. (2001) demonstrated the role of Dab2 in TGF-β signaling using genetic complementation. Dab2 was shown to interact with Smad2 and Smad3, and to promote TGF-β receptor signaling. In light of this finding, it is tempting to speculate that Dab2 mediates a cross-talk between IFN-γ and TGF-β receptor pathways. Several of the phenotypic changes observed in ICSBP–/– mice are compatible with an enhanced TGF-β signaling.
An involvement of Dab2 in cytoskeleton regulatory pathways has been suggested previously. Enforced expression of Dab2 was shown to restore the attachment of epithelial cells to the basal membrane and thus support a role of Dab2 in integrin signaling (Sheng et al., 2000). Another example of Dab2 interactions with components of the cytoskeleton was reported by Morris and Cooper (2001), who found that Dab2 associates with the clathrin adaptor protein AP-2 and regulates endocytosis of cell surface receptors. Moreover, Dab1, a neurone-specific relative of Dab2, was shown to play a role in cell positioning and in modulating cell adhesion and migration (Sheldon et al., 1997; Gotthardt et al., 2000). Never theless, a direct involvement of Dab2 in the regulation of cytoskeleton-based functions, such as cell adhesion and spreading, had not been reported.
We have noted that altered adhesion and spreading is a prominent feature of BMM from ICSBP–/– mice. Since we identified Dab2 as a major target of ICSBP in these cells, we tested whether these cellular alterations can be linked directly to the enhanced Dab2 expression. Our results demonstrate that an enforced expression of Dab2 in RAW macrophages leads to a similar phenotype, and provide evidence for a role of Dab2 in macrophage adhesion and spreading. Both, Dab2-overexpressing ICSBP–/– BMM and RAW macrophages adhere more strongly and spread faster on laminin and collagen IV. Adhesion to these substrates is mainly stimulated through integrin receptors (Giancotti and Ruoslahti, 1999). However, similar to on laminin and collagen IV, the adhesion of Dab2-overexpressing cells on poly-d-lysine was also accelerated (data not shown). Since adhesion to poly-d-lysine does not induce specific integrin signaling pathways (Morino et al., 1995), a more general role of Dab2 in adhesion and spreading can be proposed. This view is further supported by the finding that Dab2 is rapidly and transiently phosphorylated upon adhesion on all substrates tested, including poly-d-lysine. We found that adhesion-induced phosphorylation of Dab2 correlates with its mobilization to the cytoskeletal/membrane subcellular fraction. The pathway by which phosphorylated Dab2 becomes associated with this fraction is unknown. However, Dab2 has a potential actin-binding motif, KKEK (Xu et al., 1995), that could facilitate a direct interaction of Dab2 with the cytoskeleton.
The molecular mechanism by which Dab2 affects cell adhesion is not yet clear, but the results presented here suggest that Dab2 is an adhesion-responsive phosphoprotein that may stimulate the reorganization of the cytoskeleton, and thereby induces cell adhesion and spreading. Alternatively, Dab2 expression and subsequent phosphorylation could also have a regulatory function in adhesion-induced signal transduction events, which in turn lead to a reorganization of the cytoskeleton.
The role of Dab2 in myeloid cells has not been studied previously. Interestingly, we found that Dab2 is expressed in myeloid progenitor cells derived from ICSBP–/– mice, and in the bipotential myeloid precursor cell line FDC-P1Mac11. In both progenitor cell lines, Dab2 expression was markedly down-regulated after enforced expression of ICSBP. Thus our finding that ICSBP suppresses Dab2 expression in BMM and myeloid progenitors may be essential for elucidating the role of ICSBP in myelopoiesis. Previous analyses of ICSBP–/– mice have demonstrated that ICSBP can act as a molecular switch in directing myeloid differentiation (Holtschke et al., 1996; Scheller et al., 1999; Tamura et al., 2000).
Congruently with the previously described negative effect of Dab2 on cell proliferation (Fulop et al., 1998), we found that both ICSBP–/– BMM (Kallies et al., 2002) and Dab2-transfected RAW cells (data not shown) proliferate slower than control cells. This observation is in good agreement with our previously published results showing that the strongly reduced number of macrophage colonies generated from ICSBP–/– myeloid progenitors in vitro is compensated by an increased granulocytic proliferation (Scheller et al., 1999). Furthermore, it also reflects the situation in ICSBP–/– mice that develop predominantly granulocytic leukemia (Holtschke et al., 1996).
Here we show that in addition to its anti-proliferative effect, Dab2 augments adhesion and spreading, two fundamental features critical for the development and function of myeloid cells. It is well established that adhesive interactions between hematopoietic progenitors and stromal ligands of bone marrow play an important role in the regulation of hematopoiesis (Verfaillie, 1998). Thus, it is conceivable, that up-regulated Dab2 expression contributes largely to the hematopoietic alterations of ICSBP–/– mice.
Materials and methods
Mice and cells
ICSBP mutant mice were described before (Holtschke et al., 1996). All experiments were carried out in 6- to 7-week-old mice on a C57Bl/6 X 129/Sv F2 background. BMM were generated as described previously (Meraz et al., 1996) with some modifications. Bone marrow cells were plated on 100 mm non-tissue culture plates (Falcon) and cultivated in complete Dulbecco’s modified Eagle’s medium (DMEM) in the presence of 20 ng/ml recombinant murine CSF-1 for 7 days. Human K562 cells, the murine macrophage line RAW 264.1 and the murine myeloid-progenitor line FDC-P1Mac11 were maintained in complete RPMI 1640 medium, the latter in the presence of 10 ng/ml IL-3 (Tebu). Monkey kidney CV-1 cells were cultured in complete DMEM. The murine monocytic line Bac1.2F5 was maintained in complete DMEM supplemented with 25% conditioned L929 cell supernatant as a source of CSF-1. The IC34L myeloid progenitor cell line was isolated from ICSBP–/– mice suffering from an acute myeloblastic leukemia induced by murine leukemia virus infection. The cells were maintained in α-MEM supplemented with IL3 and 20% horse serum; expression of the myeloid markers CD11b and GR1 can be induced by stimulation with granulocyte-macrophage colony-stimulating factor.
Reagents and antibodies
Purified recombinant murine IFN-γ and purified recombinant murine CSF-1 were from R&D Systems. Polyclonal rabbit antisera against Dab2 (M2 and M15) (Xu et al., 1995), against ICSBP (Rosenbauer et al., 1999) and against Vasp (Smolenski et al., 1998) have been described previously. Rabbit polyclonal antibodies against PU.1, Stat3 and Stat5b, goat polyclonal antibody against Dab2, and horseradish peroxidase-conjugated secondary IgGs were from Santa Cruz Biotechnology.
cDNA arrays
Atlas™ mouse 1.2 cDNA expression arrays were purchased from Clontech Laboratories (Palo Alto, CA). Cytoplasmic RNA from BMM was extracted with RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA labeling, hybridization and washing of the cDNA Atlas array membranes were carried out according to the instructions accompanying the macroarrays. Array exposures on the phosphoimager were normalized by equalizing the intensity of the signals from a set of housekeeping genes provided on the arrays. Arrays were analyzed with Atlas Image 1.01a software (Clontech Laboratories).
Synthetic oligonucleotides and RT–PCR analysis
The sequences of primers used for RT–PCR are as follows. Dab2: upper strand, 5′-GGAGCATGTAGACCATCATG-3′, lower strand, 5′-AAGGATTTCCGAAAGGGCTC-3′; β-actin: upper strand, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′, lower strand, 5′-TAAAACGCAGCTCAGTAACAGTCC-3′; ICSBP: upper strand, 5′-GGCCTGGGCAGTTTTTAAAG-3′, lower strand, 5′-AAAAGGGTCTCTGGTGTGAG-3′. T7, T3, RV3 and GL2 primers were purchased from Promega.
RT–PCR analysis was performed as described previously (Schmidt et al., 1998). For the Dab2 and ICSBP RT–PCRs 26 cycles, for β-actin 19 cycles of the following parameters were used: 24 s, 94°C; 22 s, 60°C; 36 s, 72°C.
Isolation of the Dab2 promoter region
A murine genomic 129 PAC library (Resourcenzentrum, Berlin, Germany) was screened using full-length Dab2 cDNA as a probe. A vector with a ∼2 kb PstI insert containing the Dab2 genomic region 5′ of the first exon (including 50 bp of exon 1) was isolated and sequenced. The sequence is available at DDBJ/EMBL/GenBank under accession number AJ295277.
Bead assay for detection of DNA-binding proteins and in vitro labeling of proteins
A DNA binding assay was performed similar to as described previously (Wang et al., 2000). In brief, the biotinylated 2 kb Dab2 promoter fragment (see above) was synthesized from pGL3-Dab2p by PCR using a biotinylated RV3 and a standard GL2 primer. Purified PCR product (30 pmol) was conjugated to magnetic beads (Dynabeads M280 streptavidin, Dynal) according to the manufacturer’s protocol. The conjugated beads were suspended in 10 µl of TGEDN buffer [120 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.1 M NaCl, 1 mM dithiothreitol (DTT), 0.1% Triton X-100, 10% glycerol] and incubated with 500 µg of nuclear extracts of CV-1, K562, IC34L or Bac1.2F5 cells, which were prepared according to Schreiber et al. (1990). After addition of 20 µg of herring sperm DNA (Sigma), the reaction was rotated at 4°C for 2 h. The bound fraction was washed five times in TGEDN buffer, eluted in TGEDN buffer supplemented with 0.5% SDS and 1 M NaCl, and analyzed on a 10% SDS–PAGE. Nuclear extract (50 µg) was loaded as input controls. Western blots were performed with antibodies as indicated.
ICSBP and PU.1 were labeled with [35S]methionine and in vitro translated using pcDNA-based T7 expression vectors, the Promega TNT T7 coupled transcription/translation system and [35S]methionine (NEN). 35S signals were quantified on a PhosphorImager using ImageQuant software (Molecular Dynamics).
Luciferase assay and plasmids
K562 cells were transfected by electroporation and CV-1 cells were transfected by LipofectAMINE reagent (Gibco-BRL). For K562 transfection, 15 µg of pGL3 or pGL3-Dab2p and 15 µg of pcDNA or pcDNA-ICSBP were used. In addition, cells were transfected with 1 µg of the internal control pRL-TK. After 14 h, relative light units from the pGL3 or pGL3-Dab2p vectors were measured and standardized relative to the expression of pRL-TK. CV-1 cells were transfected with 0.6 µg of pGL3 or pGL3-Dab2p and pcDNA, pcDNA-PU.1 or pcDNA-ICSBP as indicated. In addition, cells were transfected with 0.05 µg of the internal control pRL-Null and relative light units were standardized relative to the expression of pRL-Null. Experiments were performed twice in triplicate. Vectors pRL-Null, pRL-TK, pcDNA and pGL3-basic were purchased from Promega. The 2 kb Dab2 promoter (see above) was cloned into the SmaI site of pGL3-basic to create pGL3-Dab2p. The ICSBP expression vector was as described previously (Rosenbauer et al., 1999), the PU.1 expression plasmids were a gift from D.Tenen. A TAM-inducible ICSBP protein was generated by ligating the mouse ICSBP coding region to a mutated hormone-binding domain of the murine estrogen receptor (Littlewood et al., 1995). This was inserted into a SFEV retroviral vector expressing eGFP (Baum et al., 1995) and used to infect IC34L cells. FDC-P1Mac11 cells were electroporated at 400 V and 500 µF with 15 µg of pcDNA-ICSBP.
Protein extracts and western blotting
Cells were washed with phosphate-buffered saline (PBS), trypsinized, washed with medium and PBS and pelleted. For western blot analyses, cells were lysed on ice in RIPA buffer (50 mM Tris–HCl pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 50 mM NaCl, 10 mM EDTA) supplemented with 1 mg/ml antipain, 1 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin, 1 mM Na3(VO)4, and 1 mM phenylmethylsulfonyl fluoride and incubated for 30 min on ice. Samples were than centrifuged at 20 000 g for 10 min and the supernatant was boiled with 2× SDS sample buffer. Protein corresponding to 5 × 105 cells was separated on an SDS–PAGE, transferred to nitrocellulose membranes and immunoblottings were performed with antibodies as indicated.
Subcellular fractionation
BMM were fractionated as described previously (Wang et al., 1994) with minor modifications. All steps were performed at 4°C. Cells were harvested by trypsin as described above, suspended in 5 vol. of buffer C (10 mM HEPES–KOH pH 7.6, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1 mM EDTA, 1 mM EGTA) supplemented with proteinase inhibitors (see above) and incubated for 1 h before passing 20 times through a 22-gauge needle. Lysates were centrifuged in a microcentrifuge for 1 min at 20 000 g. The supernatants were further spun at 100 000 g for 1 h to obtain the cytosolic fractions; the resulting pellets were extracted in RIPA buffer supplemented with proteinase inhibitors (see above) and were designated as cytoskeletal plus membrane fractions. Alternatively, cells were suspended in extraction buffer (10 mM PIPES pH 6.8, 250 mM sucrose, 3 mM MgCl2, 120 mM KCl, 1 mM EGTA, 0.15% Triton X-100) and incubated for 30 min on ice before passing 20 times through a 22-gauge needle. Lysates were centrifuged at 20 000 g for 10 min. The supernatant was designated as cytosolic fraction, the pellet was extracted with RIPA buffer and designated as particulate, i.e. cytoskeleton/membrane fraction.
Preparation of ECM-coated plates and immunofluorescence microscopy
Mouse type IV collagen (Sigma) and laminin (Tebu) were diluted in PBS and added to glass coverslips (Nunc) at 4 µg/cm2. After incubation overnight at 4°C the coverslips were air dried for at least 45 min at room temperature, rinsed twice with PBS, blocked by incubation with 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature, and used for adhesion assays. The tissue culture dishes or 96-well culture plate were coated in the same way. Rat fibronectin and poly-d-lysine (Tebu) were diluted in PBS and used at 20 µg/ml.
For immunofluorescence studies, RAW cells from subconfluent cultures were replated onto laminin-coated coverslips at a density of 2.5 × 105 cells/well in serum-free medium containing 1% BSA and allowed to spread at 37°C for the time periods indicated. The samples were washed twice with PBS and fixed in freshly prepared 2% paraformaldehyde in PBS pH 7.0, for 20 min at room temperature. Cells were then permeabilized in 0.5% NP-40 in PBS for 3 min. Following two more PBS washes, the coverslips were blocked with 1% BSA in PBS for 30 min and stained with rhodamine–phalloidin (Molecular Probes) at the concentration of one unit/coverslip for 20 min, washed in PBS and water, and mounted with Ultramount (Dako). Images of representative fields were obtained using a Zeiss LSM 510 confocal laser-scanning microscope and the accompanying LSM 510 software.
Adhesion assay
Cell adhesion was measured using the Fast Quant Assay (Amplificon) according to the manufacturer’s protocol. In brief, cells were trypsinized, suspended in 1 ml of medium at 107 cells/ml, and labeled by adding Fast Quant enzymatic label solution. The cells were washed in medium, centrifuged and resuspended in serum-free medium containing 1% BSA. After diluting the cells to the desired concentration, 100 µl of suspension per well were added to ECM-coated 96-well tissue culture plates and allowed to adhere at 37°C for the time periods indicated. After incubation, non-adherent cells were removed by washing. Two hundred microliters of the color reaction substrate were added to each well and the plate was incubated for 5 min in the dark. Stop solution (50 µl) was added to each well and the optical density was measured at 450 nm using a microplate reader.
Generation of Dab2-overexpressing RAW cell lines
RAW 264.1 cells were transfected with a pcDNA expression plasmid containing the full-length cDNA of the 96 kDa Dab2 using Effectene Transfection Reagent (Qiagen) according to the manufacturer’s protocol. Four independent Dab2-overexpressing cell lines were generated. Their independent origin was confirmed by Southern blotting using the pcDNA expression vector as a probe.
Acknowledgments
Acknowledgements
We thank M.Wietstruk for excellent technical assistance, and Dr B.Wiesner for his advice with confocal microscopy. We are particularly grateful to Dr D.G.Tenen for the PU.1 expression constructs and for giving F.R. the opportunity to perform some of the described experiments in his laboratory. This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (SFB 506), and from the Fonds der Chemischen Industrie.
References
- Albertsen H.M., Smith,S.A., Melis,R., Williams,B., Holik,P., Stevens,J. and White,R. (1996) Sequence, genomic structure and chromosomal assignment of human DOC-2. Genomics, 33, 207–213. [DOI] [PubMed] [Google Scholar]
- Baum C., Hegewisch-Becker,S., Eckert,H.G., Stocking,C. and Ostertag,W. (1995) Novel retroviral vectors for efficient expression of the multidrug resistance (mdr-1) gene in early hematopoietic cells. J. Virol., 69, 7541–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behre G., Whitmarsh,A.J., Coghlan,M.P., Hoang,T., Carpenter,C.L., Zhang,D.E., Davis,R.J. and Tenen,D.G. (1999) c-Jun is a JNK-independent coactivator of the PU.1 transcription factor. J. Biol. Chem., 274, 4939–4946. [DOI] [PubMed] [Google Scholar]
- Brass A.L., Kehrli,E., Eisenbeis,C.F., Storb,U. and Singh,H. (1996) Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev., 10, 2335–2347. [DOI] [PubMed] [Google Scholar]
- Deininger M.W.N., Goldmann,J.M. and Melo,J.V. (2000). The molecular biology of chronic myeloid leukemia. Blood, 96, 3343–3356. [PubMed] [Google Scholar]
- Eisenbeis C.F., Singh,H. and Storb,U. (1995) Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev., 9, 1377–1387. [DOI] [PubMed] [Google Scholar]
- Fazili Z., Sun,W., Mittelstaedt,S., Cohen,C. and Xu,X.-X. (1999) Disabled-2 inactivation is an early step in ovarian tumorigenicity. Oncogene, 18, 3104–3113. [DOI] [PubMed] [Google Scholar]
- Fehr T., Schoedon,G., Odermatt,B., Holtschke,T., Schneemann,M., Bachmann,M.F., Mak,T.W., Horak,I. and Zinkernagel,R.M. (1997) Crucial role of interferon consensus sequence binding protein, but neither of interferon regulatory factor 1 nor of nitric oxide synthesis for protection against murine listeriosis. J. Exp. Med., 185, 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop V., Colitti,C.V., Genest,D., Berkowitz,R.S., Yiu,G.K., Ng,S.-W., Szepesi,J. and Mok,S.C. (1998) DOC-2/hDab2, a candidate tumor suppressor gene involved in the development of gestational trophoblastic diseases. Oncogene, 17, 419–424. [DOI] [PubMed] [Google Scholar]
- Giancotti F.G. and Ruoslahti,E. (1999) Integrin signaling. Science, 285, 1028–1032. [DOI] [PubMed] [Google Scholar]
- Gotthardt M., Trommsdorff,M., Nevitt,M.F., Shelton,J., Richardson,J.A., Stockinger,W., Nimpf,J. and Herz,J. (2000) Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J. Biol. Chem., 275, 25616–25624. [DOI] [PubMed] [Google Scholar]
- Grazia Cappiello M., Sutterwala,F.S., Trinchieri,G., Mosser,D.M. and Ma,X. (2001) Suppression of IL-12 transcription in macrophages following Fc gamma receptor ligation. J. Immunol., 166, 4498–4506. [DOI] [PubMed] [Google Scholar]
- Hao S.X. and Ren,R. (2000) Expression of interferon consensus sequence binding protein (ICSBP) is downregulated in Bcr–Abl-induced murine chronic myelogenous leukemia-like disease and forced coexpression of ICSBP inhibits Bcr–Abl-induced myeloproliferative disorder. Mol. Cell. Biol., 20, 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocevar B.A., Smine,A., Xu,X.-X. and Howe,P.H. (2001) The adator molecule Disabled-2 links the transforming growth factor β receptor to the Smad pathway. EMBO J., 20, 2789–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtschke T. et al. (1996) Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell, 87, 307–317. [DOI] [PubMed] [Google Scholar]
- Kallies A., Rosenbauer,F., Scheller,M., Knobeloch,K.-P. and Horak,I. (2002) Accumulation of c-Cbl and rapid termination of CSF-1 receptor signaling in ICSBP-deficient bone marrow-derived macrophages. Blood, 99, in press. [DOI] [PubMed] [Google Scholar]
- Littlewood T.D., Hancock,D.C., Danielian,P.S., Parker,M.G. and Evan,G.I. (1995) A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res., 23, 1686–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L.M. and Hall,A. (1997) Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J. Cell Biol., 138, 913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraz M.A. et al. (1996) Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell, 84, 431–442. [DOI] [PubMed] [Google Scholar]
- Morino N. et al. (1995) Matrix/integrin interaction activates the mitogen activated protein kinase, p44erk-1 and p42erk-2. J. Biol. Chem., 270, 269–273. [DOI] [PubMed] [Google Scholar]
- Morris S.M. and Cooper,J.A. (2001) Disabled-2 colocalizes with the LDLR in clathrin-coated pits and interacts with AP-2. Traffic, 2, 111–123. [DOI] [PubMed] [Google Scholar]
- Morrisey E.E., Musco,S., Chen,M.Y.Z., Lu,M.M., Leiden,J.M. and Parmacek,M.S. (2000) The gene encoding the mitogen-responsive phosphoprotein Dab2 is differentially regulated by GATA-6 and GATA-4 in the visceral endoderm. J. Biol. Chem., 275, 19949–19954. [DOI] [PubMed] [Google Scholar]
- Orkin S.H. (2000) Diversification of haematopoietic stem cells to specific lineages. Nature Rev. Genet., 1, 57–64. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F., Waring,J.F., Foerster,J., Wietstruk,M., Philipp,D. and Horak,I. (1999) Interferon consensus sequence binding protein and interferon regulatory factor-4/Pip form a complex that represses the expression of the interferon-stimulated gene-15 in macrophages. Blood, 94, 4274–4281. [PubMed] [Google Scholar]
- Scheller M., Foerster,J., Heyworth,C.M., Waring,J.F., Lohler,J., Gilmore,G.L., Shadduck,R.K., Dexter,T.M. and Horak,I. (1999) Altered development and cytokine responses of myeloid progenitors in the absence of transcription factor, interferon consensus sequence binding protein. Blood, 94, 3764–3771. [PubMed] [Google Scholar]
- Schmidt M. et al. (1998) Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood, 91, 22–29. [PubMed] [Google Scholar]
- Schreiber E., Harshman,K., Kemler,I., Malipiero,U., Schaffner,W. and Fontana,A. (1990) Astrocytes and glioblastoma cells express novel octamer-DNA binding proteins distinct from the ubiquitous Oct-1 and B cell type Oct-2 proteins. Nucleic Acids Res., 18, 5495–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon M. et al. (1997) Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature, 389, 730–733. [DOI] [PubMed] [Google Scholar]
- Sheng Z., He,J., Tuppen,J.A., Sun,W., Fazili,Z., Smith,E.R., Dong,F.B. and Xu,X.-X. (2000) Structure, sequence and promoter analysis of human Disabled-2 gene (DAB2). Genomics, 70, 381–386. [DOI] [PubMed] [Google Scholar]
- Smolenski A., Bachmann,C., Reinhard,K., Hönig-Liedl,P., Jarchau,T., Hoschuetzky,H. and Walter,U. (1998) Analysis and regulation of vasodilator-stimulated phosphoprotein serine 239 phosphorylation in vitro and in intact cells using a phosphospecific monoclonal antibody. J. Biol. Chem., 273, 20029–20035. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998). How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Tamura T., Nagamura-Inoue,T., Shmeltzer,Z., Kuwata,T. and Ozato,K. (2000) ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity, 13, 155–165. [DOI] [PubMed] [Google Scholar]
- Tenen D.G., Hromas,R., Licht,J.D. and Zhang,D.-E. (1997) Transcription factors, normal myeloid development and leukemia. Blood, 90, 489–519. [PubMed] [Google Scholar]
- Tseng C.-P., Ely,B.D., Pong,R.-C., Wang,Z., Zhou,J. and Hsieh,J.-T. (1999) The role of DOC-2/DAB2 protein phosphorylation in the inhibition of AP-1 activity. J. Biol. Chem., 274, 31981–31986. [DOI] [PubMed] [Google Scholar]
- Verfaillie C.M. (1998) Adhesion receptors as regulators of the hematopoietic process. Blood, 92, 2609–2612. [PubMed] [Google Scholar]
- Wang I.-M., Contursi,C., Masumi,A., Ma,X., Trinchieri,G. and Ozato,K. (2000) An IFN-γ-inducible transcription factor, IFN consensus sequence binding protein (ICSBP), stimulates IL-12 p40 expression in macrophages. J. Immunol., 165, 271–279. [DOI] [PubMed] [Google Scholar]
- Wang X., Sato,R., Brown,M.S., Hua,X. and Goldstein,J.L. (1994) SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell, 77, 53–62. [DOI] [PubMed] [Google Scholar]
- Xu X.X., Yang,W., Jackowski,S. and Rock,C.O. (1995) Cloning of a novel phosphoprotein regulated by colony-stimulating factor 1 shares a domain with the Drosophila disabled gene product. J. Biol. Chem., 270, 14184–14191. [DOI] [PubMed] [Google Scholar]
- Xu X.X., Yi,T., Tang,B. and Lambeth,J.D. (1998) Disabled-2 (Dab2) is an SH3 domain-binding partner of Grb2. Oncogene, 16, 1561–1569. [DOI] [PubMed] [Google Scholar]