Abstract
Many nuclear transport pathways are mediated by importin β-related transport receptors. Here, we identify human importin (Imp) 4b as well as mouse Imp4a, Imp9a and Imp9b as novel family members. Imp4a mediates import of the ribosomal protein (rp) S3a, while Imp9a and Imp9b import rpS7, rpL18a and apparently numerous other substrates. Ribosomal proteins, histones and many other nuclear import substrates are very basic proteins that aggregate easily with cytoplasmic polyanions such as RNA. Imp9 effectively prevents such precipitation of, for example, rpS7 and rpL18a by covering their basic domains. The same applies to Imp4, Imp5, Imp7 and Impβ and their respective basic import substrates. The Impβ–Imp7 heterodimer appears specialized for the most basic proteins, such as rpL4, rpL6 and histone H1, and is necessary and sufficient to keep them soluble in a cytoplasmic environment prior to rRNA or DNA binding, respectively. Thus, just as heat shock proteins function as chaperones for exposed hydrophobic patches, importins act as chaperones for exposed basic domains, and we suggest that this represents a major and general cellular function of importins.
Keywords: chaperone/histone/importin/ribosomal protein
Introduction
The nuclear envelope (NE) divides eukaryotic cells into a nuclear and a cytoplasmic compartment, uncouples transcription from translation and thereby necessitates nucleo-cytoplasmic transport (reviewed in Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999). tRNAs, mRNAs and rRNAs, for example, need to be exported from the nucleus to the cytoplasm, where they mediate translation. Conversely, virtually all nuclear functions are dependent on proteins which are imported from the cytoplasm. The nuclear compartment already represents the final destination for many import substrates, such as histones or polymerases. Numerous other proteins, however, pass through nuclei only transiently. Ribosomal proteins, for example, are first imported, assemble in the nucleoli with rRNAs, and finally become exported as ribosomal subunits to the cytoplasm (for a review see Venema and Tollervey, 1999).
Macromolecular transport between the nucleus and cytoplasm proceeds through nuclear pore complexes (NPCs) and is normally receptor mediated. Impβ-type transport receptors account for most, but not all, nuclear transport pathways (for reviews see Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Conti and Izaurralde, 2001). They constitute a diverse protein superfamily and occur in two forms, import mediators (importins) and exportins. They circulate between nucleus and cytoplasm, recognize cargo molecules and transfer them from one side of the NE to the other. Substrate loading and release is guided by a gradient of RanGTP across the NE, whereby a high nuclear RanGTP concentration favours cargo loading onto exportins and substrate displacement from importins, while cytoplasmic conditions with low RanGTP levels release substrates from exportins but allow importin–cargo complexes to form.
Nuclear import is a highly selective process that involves the specific recognition of appropriate import signals by suitable receptors. Apart from a few exceptions, nuclear import signals generally are basic and often part of DNA- or RNA-binding sites. In some cases, just a short peptide sequence is recognized. The nuclear localization signal (NLS) from the SV40 large T antigen, for example, comprises five basic residues in a total length of eight (Kalderon et al., 1984; Makkerh et al., 1996). The ribosomal protein L23a illustrates another scenario that probably also applies to the majority of ribosomal and other very basic nucleic acid-binding proteins. In this case, the importins recognize a domain of at least 43 residues and a net charge of +18 (Jäkel and Görlich, 1998). Such a signal is certainly longer than necessary if only a nuclear destination of the protein was to be ‘indicated’. It therefore has been suggested previously that importins not only direct their cargoes to the nuclear compartment, but might also serve a second purpose, namely by shielding very basic cargoes against undesired interactions during transit to the nucleus (Jäkel and Görlich, 1998; Jäkel et al., 1999).
Here we show that numerous basic proteins indeed aggregate in an importin-free cytosol, apparently through multivalent ionic interactions with cytoplasmic polyanions such as tRNA. We identify several importins, namely Imp4a, Imp4b, Imp9a and Imp9b, and show that these not only mediate active transport through NPCs, but also effectively suppress the aggregation of their basic import cargoes in polyanionic environments. Previously described importins such as Impβ, Imp5, Imp7 and the Impβ–Imp7 (Impβ/7) heterodimer also possess such activity. The anti-aggregation activity of importins involves shielding of basic patches on the cargo and requires a precise match between cargo and receptor. A single, abundantly expressed importin might be, in principle, sufficient to accomplish all required nuclear import tasks. However, it is hard to see how a single receptor could possibly shield each of the thousands of different RNA- or DNA-binding domains that are present in the vast number of different nuclear import substrates. The stringent demands on the anti-aggregation activity of importins could therefore explain why for the safe cargo-transfer to the nucleus, mammalian cells employ not just one, but at least 15 distinct importins and 11 heterodimeric import receptor complexes.
Results
Identification of importins 4a, 4b, 9a and 9b
By searching the database, we identified several expressed sequence tags (ESTs) that ultimately allowed the identification of mouse Imp4a, Imp9a and Imp9b, and human Imp4b. Full-length Imp4 cDNAs were isolated by several rounds of 3′ and 5′ RACE using mouse or human cDNA as a starting material (see Materials and methods). The 5′ and 3′ ends of Imp9a and Imp9b could be derived from available ESTs in the database and so sufficient sequence information was already available to amplify the complete open reading frames by RT–PCR (see Materials and methods).
Imp4 is a 119 kDa protein that groups within the Impβ superfamily (Fornerod et al., 1997; Görlich et al., 1997) together with Impβ (Chi et al., 1995; Görlich et al., 1995; Imamoto et al., 1995a), transportin (Pollard et al., 1996) and Imp5 (Deane et al., 1997; Jäkel and Görlich, 1998). Imp4 orthologues are evident in nearly all eukaryotic organisms analysed so far, including Xenopus, Drosophila, Arabidopsis, Schizosaccharomyces pombe and Saccharomyces cerevisiae (Yrb4p/Kap123; Rout et al., 1997; Schlenstedt et al., 1997). However, Imp4 could not be identified in Caenorhabditis elegans. Orthologous nuclear transport receptors are typically >98% identical amongst mammals. Mouse Imp4a and human Imp4b share only ∼80% identical residues and for the time being we would consider them as distinct proteins, in particular as functional differences are evident (see below).
Imp9 has a predicted mass of 130 kDa and appears to have orthologues in the majority of higher eukaryotes, including human, Xenopus, Drosophila, fish (Danio rerio), Arabidopsis and S.cerevisiae (HRC1004/Kap114; see Morehouse et al., 1999; Pemberton et al., 1999; Muhlhausser et al., 2001), but it appears to be absent from the C.elegans genome. Mouse contains two Imp9 forms, Imp9a and Imp9b, which differ by only four residues. It is unclear at present if Imp9a and Imp9b originate from distinct genes or represent allelic variants.
Imp9 mediates import of the ribosomal protein S7
Imp4 and Imp9 are clearly related in sequence to established nuclear transport receptors; however, it was unclear whether they would also function in nuclear transport and which cargoes they would carry. To clarify this point, we produced them in a recombinant form and carried out a thorough biochemical analysis.
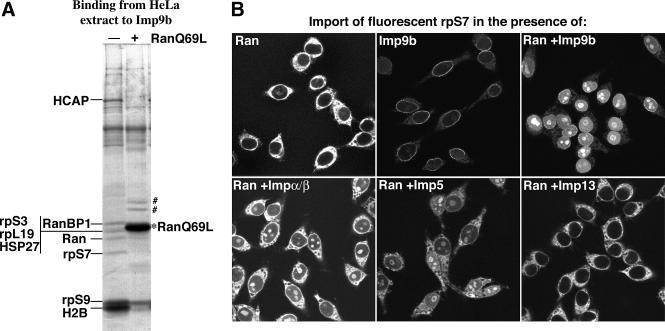
To identify Imp9-specific transport substrates, we performed affinity chromatography on immobilized Imp9 and retrieved the following putative cargoes from a HeLa cell extract: the hepatocellular carcinoma-associated protein, the ribosomal proteins S3, S7, S9 and L19, the heat shock protein HSP27 and the core histone H2B (Figure 1A). They were all displaced from Imp9 by RanGTP, suggesting that Imp9 binds them in a cytoplasmic environment, releases them in the nucleus and thus functions as their import receptor. Of these interacting proteins, we chose rpS7 for a further, detailed analysis.
Fig. 1. (A) Identification of potential Imp9-specific transport substrates. Immobilized Imp9b was used to retrieve interacting proteins from a cytosolic HeLa extract. Binding was performed in the absence or presence of RanQ69L GTP (5 µM) to mimic a cytoplasmic and nuclear environment, respectively. Analysis of bound proteins was by SDS–PAGE, followed by Coomassie Blue staining. The hepatocellular carcinoma-associated protein (HCAP), the ribosomal proteins S3, S7, S9 and L19 as well as the heat shock protein HSP27 and the core histone H2B were identified by mass spectrometry as putative import substrates. ‘#’ indicates two bands derived from the added RanQ69L. (B) Imp9 mediates import of rpS7. Import of fluorescently labelled rpS7 (1 µM) into nuclei of permeabilized cells was performed for 15 min with the indicated combinations of nuclear transport receptors (1 µM each). The panels show confocal sections of the rpS7 distribution after import and fixation. Nuclear import of rpS7 was efficient with Imp9b or the Impα/β heterodimer and turned out to be strictly Ran and energy dependent. As expected for a ribosomal protein, rpS7 accumulated strongly in the nucleoli, whenever import was efficient.
We next tested the capacity of various nuclear transport receptors to import rpS7. Imp9b indeed mediated the nuclear import of rpS7 efficiently and, as expected for a ribosomal protein, rpS7 accumulated predominantly in the nucleoli (Figure 1B). Import was equally efficient with Imp9a (not shown). Some import was also observed in the presence of the Impα/β complex (Görlich et al., 1994, 1995; Chi et al., 1995; Imamoto et al., 1995b) and to a lesser extent with Imp5 (Deane et al., 1997; Jäkel and Görlich, 1998), while import by Imp13 (Mingot et al., 2001) was not significant. Imp9-dependent rpS7 import showed a strong Ran and GTP dependence, indicating that substrate release from the receptor is rate limiting for the overall import reaction.
In the absence of import receptor, rpS7 showed a very prominent cytoplasmic staining, indicating a severe aggregation of rpS7 with certain cytoplasmic structures (Figure 1B). The cytoplasmic aggregation could be suppressed fully by Imp9. This effect occurred even in the absence of Ran and was thus not a consequence of import, but rather points to a cytoplasmic function for Imp9. The anti-aggregation activity towards rpS7 was specific for Imp9 and not observed with other tested import receptors. We take this as a strong indication that Imp9 represents the physiological import receptor for rpS7, and we will now focus on this aggregation phenomenon.
The aggregation problem of basic proteins
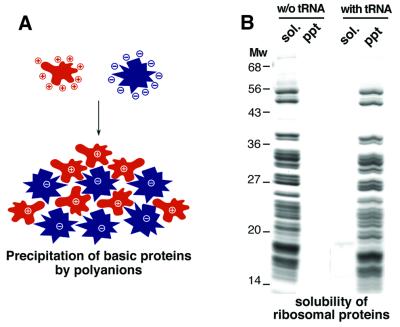
It is a common experience that basic proteins, such as histones and ribosomal proteins, easily precipitate upon contact with polyanions. This problem is evident, for example, with the heavy precipitates that form when a (high-salt) nuclear extract is dialysed back to physiological ionic strength. These extracts largely consist of (polyanionic) nucleic acids and (polycationic) nucleic acid-binding proteins, and it is easy to imagine how large aggregates form through multivalent ionic interactions (see Figure 2A). A more defined illustration of the effect is shown in Figure 2B. Purified, RNA-free total ribosomal proteins are perfectly soluble in water and a variety of buffers, but the addition of polyanions such as tRNA triggers the formation of large aggregates and quantitative precipitation (Figure 2B). The aggregation does not occur at high ionic strength (not shown), indicating that aggregate formation is indeed driven by ionic interactions.
Fig. 2. (A) A schematic illustration of the ‘basic chaperone’ problem. Basic proteins (in red) can form large aggregates through multivalent ionic interactions with polyanions (in blue) such as nucleic acids. (B) A practical demonstration of the basic chaperone problem. Total ribosomal proteins (TP80) were purified from HeLa ribosomes as described in Materials and methods. The pure TP80 preparation (90 µg/ml protein) is perfectly soluble in the absence of polyanions, but the ribosomal proteins collectively precipitate upon addition of tRNA (30 µg/ml). sol. = soluble fraction; ppt = precipitate.
Imp9 prevents aggregation of rpS7 by shielding basic patches of the protein
It is clear that an aggregated ribosomal protein would neither become imported into nuclei nor assemble with rRNA to form ribosomal subunits. This consideration implies that cells must employ efficient anti-aggregation mechanisms to avoid such a problem. What could these be? Ribosomal proteins become complexed in the cytoplasm with importins. Importins in turn are very acidic proteins with net charges that should suffice to neutralize the positive charge of most of their respective import substrates (–59 in the case of Imp9a or –68 in the case of Imp7). That importins exhibit a general anti-aggregation activity towards their substrates therefore appeared a tempting speculation, which we wanted to test directly and systematically.
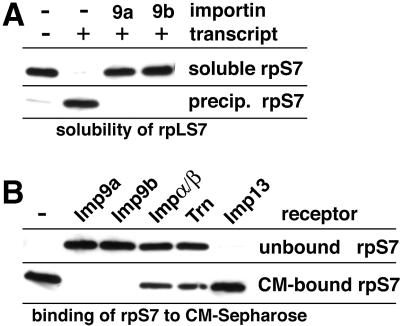
rpS7 is perfectly soluble in a pure form, but quantitatively precipitates upon addition of an in vitro transcribed RNA that was used to mimic the aggregation problem in the cytosol (Figure 3A). Strikingly, the addition of a slight molar excess of Imp9a or Imp9b suppressed this aggregation completely. The effect was highly specific and could be abrogated by RanGTP, which antagonizes the importin–cargo interaction (not shown, but see below).
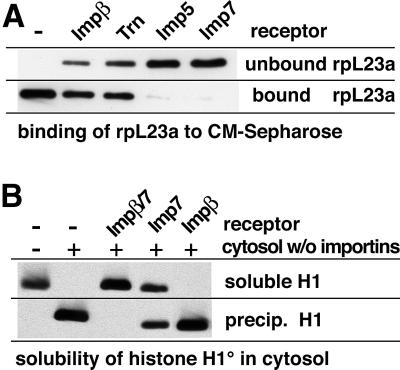
Fig. 3. (A) Imp9 suppresses aggregation of the rpS7 with polyanions. rpS7 (0.5 µM) was incubated with the indicated combinations of in vitro transcribed RNA (60 µg/µl) and Imp9a or 9b (0.75 µM each). Soluble fractions and precipitates were separated by centrifugation and analysed for rpS7 content by western blotting (for details see Materials and methods). Addition of RNA caused a quantitative precipitation of rpS7, which in turn was fully suppressed by the presence of the importins. (B) Imp9 shields basic patches in rpS7 against ionic interactions. rpS7 (0.5 µM) was incubated with various nuclear transport receptors (0.75 µM each) and subsequently subjected to binding to the cation exchanger CM-Sepharose. rpS7 alone bound tightly to CM. This binding was fully suppressed by Imp9a or 9b, indicating an efficient shielding of the exposed basic patches of S7. Imp13, which does not support rpS7 import, was also totally negative in this shielding/anti-aggregation assay. Transportin (Trn) and the Impα/β heterodimer, which are functional but suboptimal import receptors for rpS7, had a partial effect.
rpS7 contains a very basic patch (residues 98–120, charge +12, pI 12.9) and therefore binds tightly to cation exchangers such as carboxymethyl (CM)-Sepharose (Figure 3B). Pre-incubation of rpS7 with Imp9a or Imp9b prevented this binding completely, indicating that Imp9 shields the basic patch against ionic interactions, which in turn is a plausible mechanism to suppress aggregation. Shielding of rpS7 was weak with transportin or the Impα/β heterodimer, and not detectable with Imp13, which also has no import activity towards rpS7. These differential effects correlate well with the already mentioned observation that only Imp9 could suppress the aggregation of rpS7 with the cytoplasmic remnants of the permeabilized cells (Figure 1B). They also represent a specificity control and indicate that shielding of basic patches requires a precise match between receptor and substrate. The observation that the Impα/β heterodimer can mediate rpS7 import (Figure 1B), but not protect rpS7 against undesired cytoplasmic interactions (Figures 1B and 3B), further suggests that the requirements for suppression of aggregation can be more stringent than for nuclear import per se.
rpL18a requires importins 9 or β for import and suppression of cytoplasmic aggregation
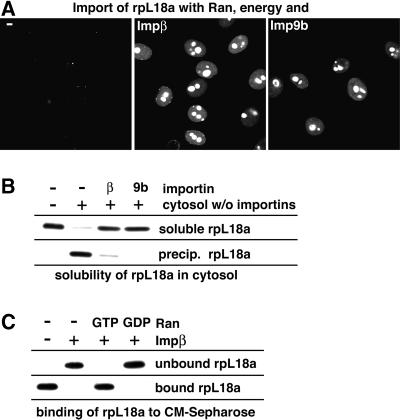
Our next example is rpL18a. Its import pathway had not been elucidated so far, so we tested the available transport receptors for their ability to import rpL18a and found Impβ and Imp9b to be the most efficient receptor species (Figure 4A; Table I). Although the net charge of rpL18a is only modest (+6), it contains 38 basic residues and might therefore face aggregation problems similar to those of other ribosomal proteins. So far, we mimicked the aggregation problem of basic proteins by addition of RNA or binding to an anionic surface. To come closer to the physiological situation, we added the ribosomal protein this time to a total cytoplasmic HeLa extract, which had been selectively depleted of nuclear transport receptors (see Materials and methods). Figure 4B shows that rpL18a readily precipitates in this importin-free cytoplasmic environment. The aggregation was fully suppressed by prior re-addition of Imp9b or Impβ (Figure 4B).
Fig. 4. (A) Nuclear import of the Alexa 488-labelled ribosomal protein L18a (0.5 µM) was performed with Ran and an energy-regenerating system. The presence of either Impβ or Imp9b (0.75 µM each) resulted in efficient nuclear import and nucleolar accumulation of rpL18a. (B) rpL18a (0.5 µM) precipitates in importin-free cytosol, but remained soluble when the cytosol was replenished with 0.75 µM Impβ or Imp9 (for details see Materials and methods). (C) Impβ prevents rpL18a binding to CM-Sepharose (for details see Figure 3B and Materials and methods). This shielding of rpL18a against interactions with polyanionic surfaces was highly specific and could be abrogated by 2.5 µM RanQ69LGTP, which displaces rpL18a from Impβ. The shielding was unaffected by 2.5 µM RanGDP, which leaves the Impβ–rpL18a complex intact.
Table I. Comparison of the effects of importins in terms of nuclear import and suppression of ionic aggregation of some basic cargoes.
| Cargo | Most efficient import receptors | Most efficient as chaperones |
|---|---|---|
| rpS7 | Imp9a, Imp9b≥Impα/β>Imp5 | Imp9b, Imp9a |
| rpL18a | Impβ, Imp9 | Imp9, Impβ |
| rpS3a | Imp4a, Imp4b, Impβ/7, Imp5>Impα/β | Imp4a |
| rpL23a | Imp5, Imp7, Impβ, transportin | Imp5, Imp7 |
| rpL6 | Impβ/7>Imp9, Imp7, Impβ | Impβ/7 |
| rpL4 | Impβ/7>Imp9, Impβ (in molar excess) | Impβ/7 |
| UBC9a | Imp13 | Imp13 |
| Histone H1 | Impβ/7 | Impβ/7>Imp7 |
aSee Mingot et al. (2001) and our unpublished data.
rpL18a also binds tightly to the polyanionic cation exchanger CM-Sepharose. Impβ (Figure 4C) and Imp9 (not shown) quantitatively suppressed this binding, indicating a shielding of the basic patch by the importins. Again, this effect was highly specific and did not occur when rpL18a was displaced by RanGTP from its cognate importin. RanGDP, which does not interfere with the Impβ–rpL18a interaction, had no effect.
Imp4a functions as an import receptor and cytoplasmic chaperone towards rpS3a
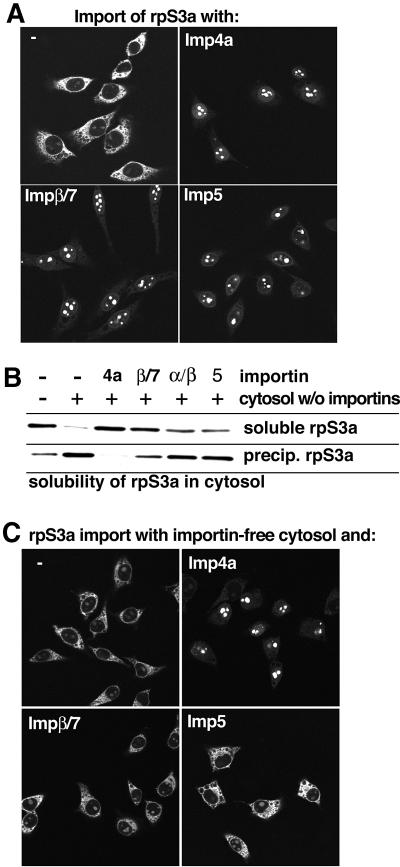
By using affinity chromatography with immobilized Imp4a (not shown), we identified the ribosomal protein S3a as a potential import substrate from a HeLa cell extract. Indeed, Imp4a mediated efficient rpS3a import (Figure 5A). It also suppressed the otherwise strong cytoplasmic aggregation of rpS3a with the cytoplasmic remnants of the permeabilized cells (Figure 5A) and prevented the rpS3a precipitation in the importin-free cytosol (Figure 5B).
Fig. 5. Imp4 is a functional nuclear import receptor and a cytoplasmic chaperone for the ribosomal protein S3a. (A) Nuclear import of Alexa 488-labelled rpS3a (0.5 µM) was performed with Ran, an energy-regenerating system and the indicated transport receptors (1.5 µM each). Imp4a, Imp5 and the Impβ/7 heterodimer were efficient in rpS3a import. Under these conditions, all three receptor species suppressed aggregation of rpS3a with the cytoplasmic remnants of the permeabilized cells. (B) Imp4a is necessary and sufficient to suppress precipitation of rpS3a in a cytoplasmic environment. The anti-precipitation assays contained 0.5 µM rpS3a in either buffer or a cytoplasmic HeLa extract depleted of importins. Note that rpS3a readily precipitates in a cytoplasmic environment when importins are absent. This aggregation was completely suppressed by re-addition of Imp4a (1.5 µM), while Imp5 (1.5 µM), and the Impα/β (1.5 µM) and Impβ/7 (0.75 µM) heterodimers show only a weak anti-precipitation activity towards this substrate. (C) Nuclear import of rpS3a was performed exactly as in (A), but importin-free cytosol (see Materials and methods) was included in the reaction. Under these conditions, only Imp4a was efficient as a chaperone and import mediator.
Imp5 and the Impβ/7 heterodimer were also efficient import receptors for rpS3a and strongly reduced rpS3a aggregation with the cytoplasmic remnants (Figure 5A). However, the two receptor species were rather inefficient when it came to preventing the rpS3a precipitation in the importin-depleted cytosol (Figure 5B). This clear discrepancy between the two assays indicated that the permeabilized cells had lost some cytosolic components, which otherwise aggregate with rpS3a.
Indeed, when the import assays were repeated in the presence of the importin-depleted cytosol (Figure 5C), only Imp4a, but neither Imp5 nor the Impβ/7 dimer, showed efficient import and chaperone activity towards rpS3a. These data confirm Imp4a as the physiological import mediator and cytoplasmic chaperone for rpS3a. In addition, they emphasize that suppression of precipitation relies on a precise recognition of the aggregation-prone cargo.
As a side point, it needs to be mentioned that Imp4b is also an import receptor for rpS3a, but a poor chaperone for this substrate, which suggests that Imp4a and 4b are not functionally identical.
Importins 5 and 7 also function as chaperones for exposed basic domains
We previously reported that rpL23a could be imported efficiently by transportin, Impβ, Imp5 or Imp7 (Jäkel and Görlich, 1998). The primary interaction site for all four receptors is the so-called BIB domain, which represents the most basic part of rpL23a (pI ∼12). However, there are also numerous basic residues outside this domain. Figure 6A demonstrates that rpL23a binds strongly to CM-Sepharose in the absence of cognate importins, while Imp5 or Imp7 abolish this binding completely. Interestingly, Impβ and transportin in this case have only a partial effect. We assume that the difference is that transportin and Impβ bind the BIB domain only, while Imp5 and Imp7 are also capable of shielding basic residues outside this domain.
Fig. 6. (A) The ribosomal protein L23a (0.5 µM) was pre-incubated with various nuclear transport receptors (0.75 µM) and subjected to binding to CM-Sepharose. Imp5 and Imp7 efficiently shielded rpL23a against ionic interactions and prevented binding to the cation exchanger, while Impβ and transportin had only a partial effect. (B) Histone H1 was incubated with the indicated combinations of importin-free cytosol, Impβ and Imp7. H1 is perfectly soluble in a pure form, but quantitatively precipitates in a cytosol depleted of importins. Re-addition of a physiological concentration of the Impβ/7 heterodimer (the functional import receptor for H1) suppressed the aggregation completely. Imp7 alone had a partial effect; Impβ had no effect at all.
The Impβ/7 heterodimer is specialized for import and shielding of the most basic proteins
The import substrates considered so far have been moderately basic proteins. rpL18a, for example, has a total of 39 basic residues and a net charge +6, while the net charges of rpS7 and rpL23a amount to +16 and +28, respectively. More extreme cases are histone H1 (net charge +58), rpL6 (+47) and the most basic ribosomal protein, rpL4, which contains a total of 98 basic residues and has a net charge of +74.
Figure 6B shows that pure H1 is perfectly soluble, but quantitatively precipitates in the importin-free cytosol. We have shown previously that H1 is imported by the Impβ/7 heterodimer (Jäkel et al., 1999) and, strikingly, this receptor species is the only one that can suppress the cytoplasmic aggregation of H1 (Figure 6B). Imp7 alone has a partial effect, while Impβ alone has no effect at all.
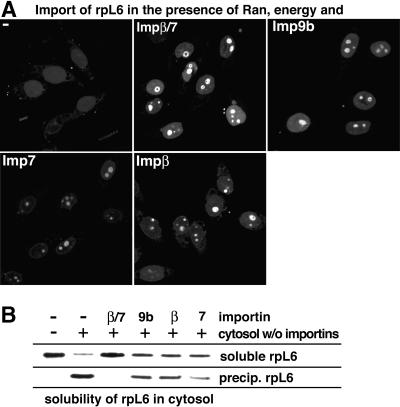
rpL6 can be imported by several import receptor species such as Imp9, Imp7 or Impβ, but the Impβ/7 heterodimer is the most efficient (Figure 7A). The Impβ/7 heterodimer is also the only receptor species that can fully suppress the rpL6 aggregation in an otherwise importin-free cytosol (Figure 7B).
Fig. 7. (A) Efficient nuclear import of rpL6 (0.5 µM) occurred with the Impβ/7 heterodimer (0.75 µM) and to a lesser extent with either Impβ, Imp7 or Imp9b. (B) rpL6 (0.5 µM) was incubated with the indicated combinations of importin-depleted cytosol and transport receptors. rpL6 aggregated in the depleted cytosol, but remained soluble when 0.75 µM Impβ/7 heterodimer was re-added. Impβ, Imp7 or Imp9b (1.5 µM each) could only partially suppress aggregation.
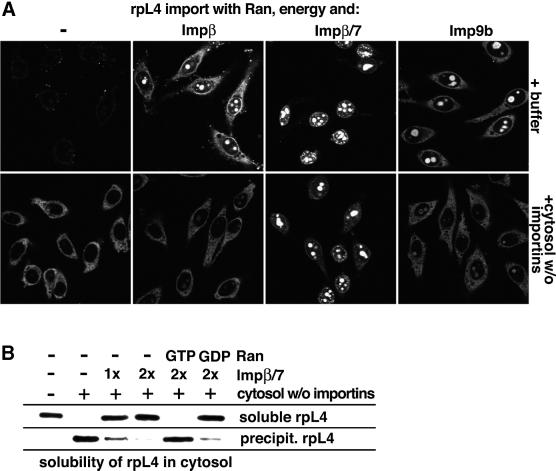
We finally investigated our most basic import substrate, rpL4. Its import was also most efficient with the Impβ/7 heterodimer (see Figure 8A and below). This heterodimer already has a net charge of approximately –130 and thus appears particularly suitable for compensating the charge of very basic substrates. However, even this receptor species was required in a 3-fold molar excess over rpL4 to achieve optimal import. rpL4 can, in principle, also be imported by Impβ and Imp9, provided these receptors are offered in an ∼6-fold excess over the substrate (Figure 8A). Under these conditions, nearly as much rpL4 arrived in the nucleoli as when import was performed with the Impβ/7 heterodimer. When the import buffer was replaced, however, by the importin-free cytosol, then Imp9- and Impβ-mediated import of rpL4 became diminished to zero (Figure 8A), probably because the shielding by Imp9 and Impβ is not strong enough to overcome the non-specific ionic interactions under these conditions. In contrast, rpL4 import by the Impβ/7 heterodimer remained unaffected, emphasizing that this receptor species represents the physiological import receptor for rpL4 and that this receptor species can suppress all undesired interactions of rpL4 which might arise in a cytoplasmic environment.
Fig. 8. (A) Import of 0.5 µM rpL4 into the nuclei of permeabilized cells was performed in the presence of Ran and an energy-regenerating system. Nuclear/nucleolar accumulation of rpL4 occurred with either 3 µM Impβ or 3 µM Imp9b, but import was most efficient with 1.5 µM Impβ/7 heterodimer (upper panels). The inclusion of importin-free cytosol in the assay completely abolished Impβ- and Imp9-mediated rpL4 import, but left import by the Impβ/7 heterodimer unaffected (lower panels). In the absence of cytosol and import receptors, most aggregates were deposited outside the cells and remained out of the focal plane. (B) rpL4 is the most positively charged ribosomal protein and relies on the Impβ/7 heterodimer to escape aggregation in a cytoplasmic extract. However, in contrast to rpL6, a higher receptor/cargo ratio was necessary to prevent aggregation. The RanGTP and RanGDP controls demonstrate the specificity of the chaperone function of Impβ/7.
As now expected, rpL4 quantitatively aggregated in an importin-free cytosol, but not when sufficient Impβ/7 heterodimer was present (Figure 8B). To suppress aggregation fully, an at least 2-fold molar excess of the Impβ/7 receptor over rpL4 was required. This concentration (1.2 µM) is nevertheless still below the physiological concentrations of Impβ and Imp7 in the cytoplasm, which we estimate at ∼2 µM each.
RanGTP-dependent release of rpL4 from the heterodimer again results in an aggregation (Figure 8B). This is not only a stringent specificity control of the effect, but also makes the point that the chaperone function of importins is largely confined to the cytoplasm (see Discussion). It predicts that basic proteins such as rpL4 might be transferred from importins onto nuclear chaperones that keep them soluble on their way from nuclear pores until their final incorporation into ribosomal subunits.
Discussion
Nucleo-cytoplasmic transport comprises a great diversity of distinct pathways that accommodate the import of ∼10 000 different proteins and the export of proteins, RNAs and ribonucleoprotein particles (Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Cokol et al., 2000). Here, we have allocated functions to the importins 4a, 9a and 9b and also elucidated the nuclear import pathways of the ribosomal proteins S3a, S7, L4, L6 and L18a. This kind of import substrate not only needs to reach nuclei, but is also highly basic and faces severe aggregation problems in the presence of polyanionic macromolecules such as nucleic acids. These proteins indeed quantitatively precipitate in a cytoplasmic environment unless their basic patches are shielded against undesired interactions. Depletion and re-addition experiments indicate that importins are the only cytoplasmic factors that are capable of keeping proteins such as ribosomal proteins S7, S3a, L4, L6 and L18a or histone H1 soluble in a cytoplasmic environment. Crucially, we have shown that importins are effective at a physiological, i.e. low micromolar, concentration.
The basic chaperone problem was first noticed and studied systematically in the attempts to assemble nucleosomes from purified core histones and DNA (for a review see Philpott et al., 2000). When mixed under physiological ionic conditions, these components precipitated but formed no nucleosomes, suggesting that chromatin formation must be catalysed in vivo by some specific factor. The first identified chromatin assembly factor is nucleoplasmin, a highly abundant nuclear protein from Xenopus oocytes (reviewed in Dingwall and Laskey, 1990). It binds core histones, suppresses undesired aggregations and thereby allows the histone transfer onto DNA to occur in a controlled manner. The term chaperone was coined originally to describe such nucleoplasmin-like anti-aggregation activity. It is now also widely used for other factors with anti-aggregation activity, such as HSPs, and applies perfectly to the importins.
An aggregation occurs typically through multivalent interactions, which can be of hydrophobic or ionic nature. Importins prevent ‘ionic’ aggregation by shielding basic protein domains. This is evident from the observation that recognition by cognate import receptors prevents ribosomal proteins and the linker histone from binding to a cation exchanger (Figures 3B, 4C, 6A and data not shown for H1, L6 and L4). Consistent with this, importins normally bind the most basic part of their import substrate. The primary importin-binding site of rpL23a, for example, has a pI of 12.2 and a net charge of +16 (BIB domain; Jäkel and Görlich, 1998). The situation is somewhat more complex for rpS7. The primary Imp9b-binding site maps to residues 98–120, but its affinity increases when placed into the original protein context, indicating that a folded motif and not just a linear sequence is recognized (not shown). However, the primary interaction site is again far more basic (pI ∼12.9) than the rest of the molecule (pI ∼9.1).
HSPs bind folding and assembly intermediates with exposed hydrophobic regions. They suppress aggregation and thereby increase the probability of proper folding and assembly (reviewed in Agashe and Hartl, 2000). Genetic experiments with Escherichia coli demonstrated that in vivo depletion of certain HSP combinations causes the majority of proteins to aggregate (Deuerling et al., 1999). ‘Chaperoning’ of basic proteins against ionic aggregation is also not an exotic requirement, instead, the problem applies to a significant proportion of cellular proteins. Most proteins that enter nuclei also bind nucleic acids and are therefore equipped with basic nucleic acid-binding sites. This applies to linker and core histones, ribosomal proteins, constituents of the signal recognition particle and mRNA-binding splicing factors with SR domains, to list only some major classes. In HeLa cells, these might together account for ∼10–20% of the total cellular protein, and most of the DNA- and RNA-binding proteins we have tested do face problems with aggregation. An impressive illustration of this fact is the behaviour of the mixture of total ribosomal protein (TP80), which collectively precipitates upon an ‘unprotected’ contact with tRNA (Figure 2B). In all cases tested, the precipitation of individual proteins could be suppressed by binding of the optimal import receptor.
Proteins such as rpL4 or H1 are safe from aggregation probably only after their final incorporation into ribosomal subunits or chromatin, respectively. One should therefore assume that they rarely occur naked in the cell and become shielded immediately after synthesis or possibly even co-translationally. To achieve this, it appears crucial that importins are extremely abundant. The importins β, 4, 5, 7 and 9 as well as transportin 1 and SR 2 reach cellular concentrations of ∼1–2 µM each (data not shown) and so the total import receptor concentration in the cytosol might amount to 10 µM or >1 mg/ml. Importins are thus concentrated similarly to abundant HSPs. This should ensure a high probability of encountering an appropriate importin during synthesis or very soon thereafter, and provide a sufficient capacity to accommodate the great number and amounts of basic chaperone substrates.
HSPs such as HSP70 actively use an ATPase cycle to control substrate binding and release (Bukau and Horwich, 1998). Likewise, substrate binding to importins is coupled to RanGTP cycles, which ensures a stable interaction in the cytoplasm and favours RanGTP-driven release in the nucleus (Rexach and Blobel, 1995; Chi et al., 1996; Görlich et al., 1996). What happens after nuclear import? It appears unlikely that aggregation-prone substrates are dropped unselectively by the importins immediately after nuclear entry. For some receptor–substrate pairs, the presence of RanGTP is not sufficient for release of a substrate; instead, an appropriate binding site for the cargo is also required (Pemberton et al., 1999). Thus, in these cases, the importins might deposit their cargo molecules directly on their final destinations. Alternatively, the basic cargo could be handed over to a nuclear chaperone such as CAF-1 (Smith and Stillman, 1989) or nucleoplasmin (Dingwall and Laskey, 1990) in the case of core histones, or transferred to some of the highly acidic nucleolar proteins, such as NO29 or NO38 (Schmidt-Zachmann et al., 1987; Zirwes et al., 1997), in the case of ribo somal proteins. This aspect certainly deserves further investigation.
For import alone, one common tag and one cognate receptor should suffice to localize any substrate to the nucleus and so a frequently asked question is why are there so many different nuclear import receptor species. We suggest here that the answer to the problem lies in the chaperone task of importins. The charge distribution of their cargoes is determined by their function and is constrained, for example, by the specific nucleic acid motifs to which they finally bind. The great number of differently shaped RNA- and DNA-binding sites obviously cannot be shielded by a single type of receptor. This circumstance might explain why mammalian cells employ ∼15 single receptors (Impβ, 4, 5, 7, 8, 9a, 9b, 11, 13, transportin 1 and 2, SR1 and SR2) and another ∼11 Impβ complexes (including Impα1–α7, Imp7, Imp8, Xripα and snurportin 1) for this purpose (see Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Strom and Weis, 2001). Indeed, in many cases, the perfect chaperone function appears a more stringent demand than just the mediation of import (see also Table I). rpS7, for example, can also be imported, but not effectively shielded by the Impα/β complex, while only Imp9 is highly efficient in both tasks. rpL4 is another illustrative example. In a defined system, i.e. in the absence of polyanionic aggregants, it can be imported efficiently by either Imp9 or the Impβ/7 heterodimer. In the presence of a cytoplasmic extract, however, import is only observed with the Impβ/7 heterodimer, which correlates perfectly with the capacity of this receptor species to suppress aggregation quantitatively in such extracts. Finally, only Imp4a, but not Imp5 or the Impβ/7 dimer, efficiently protects rpS3a against cytoplasmic aggregation, even though all three import receptor species can, in principle, mediate import of the protein.
The nuclear transport system comprises >30 nucleoporins, the RanGTPase system, numerous nuclear transport receptors and the NE. The transport system per se should be beneficial for the cell only when it is complete and operational. On the other hand, such a complex syndicate of components could hardly have arisen all at once, which in turn implies that some elements of the present nuclear transport system might have been beneficial even without mediating transport. The anti-aggregation activity of importins should have been useful even before nuclear pores were functional and might thus represent the most ancient part of the nuclear transport machinery.
Materials and methods
Cloning of new importins
Imp9 was identified by searching the NCBI database for homologues of the yeast protein HRC1004/YGL241w (Kap114p, ACC P53067). The primary sequences of the various identified homologous human ESTs (HUMNK548, ACC D29439, ACC R58833, ACC R72848, ACC J08686, ACC AA446641, etc.) were used to design primers to clone the entire corresponding human cDNA by RT–PCR. In addition, we found an EST from D.melanogaster and mouse with homology to Imp9. The corresponding clones (LD45303, ACC AI513074, IMAGp998A133793Q2, IMAGp998B184710Q2 and IMAGp998M154710Q2) were obtained from the respective suppliers (Research Genetics and RZPD) and the entire sequence was determined.
Imp4 was identified by searching the NCBI database for vertebrate homologues to S.cerevisiae Yrb4p/Kap123. We identified several ESTs from rat and human (H34644, K201471, AA035137, AA204944, AA342016 and AA342017) and determined the complete coding sequence by 3′ and 5′ RACE using human mRNA. This information was then used to identify mouse ESTs that code for the region around the start and stop codons of Imp4. We then isolated the corresponding human and mouse cDNAs by RT–PCR.
Plasmids for recombinant expression in E.coli
The coding sequences for rpL6 and rpL18a were each amplified by PCR from HeLa cDNA and cloned into the NcoI–BamHI sites of the 2z60 vector (described in Görlich et al., 1997), which allows bacterial expression as a fusion with an N-terminal zz tag and a C-terminal His6 tag rpS3a was cloned as an NcoI–BamHI fragment into the 2zChis2cys vector, a pQE80 derivative that provides an N-terminal zz tag, a C-terminal His6 tag and two C-terminal cysteines for labelling (see below). rpL4 was amplified with a 5′ BamHI and a 3′ SalI site and cloned into the BamHI–SalI sites of the vector pQE80 (Qiagen) for expression with an N-terminal His6 tag. The replacement histone H1o was amplified from pWA312 (gift of Dr Werner Albig, Göttingen) and cloned with an extra N-terminal cysteine into the NcoI–BamHI sites of pQE60, which allows expression with a C-terminal His6 tag.
Imp9a and Imp9b were amplified from mouse cDNA (3T3 cells) and each cloned into the KpnI–HindIII sites of pQE80 for expression with an N-terminal His6 tag or into zzTev80 to allow expression with an N-terminal zz tag. To improve expression, the codon usage at the extreme 5′ end was optimized. Mouse Imp4a was cloned from mouse cDNA into the SalI–HindIII site of pQE30; human Imp4b was cloned from HeLa cDNA into the KpnI–HindIII sites of either zzTev60 (for expression with N-terminal zz tag) or pQE80 (for expression with an N-terminal His6 tag).
All constructs were verified by DNA sequencing.
Recombinant expression and protein purification
The ribosomal proteins L4, L6 and L18a, and histone H1o were expressed in E.coli BLR/pREP4 and purified on Ni-NTA–agarose (Qiagen) under 6 M guanidine hydrochloride. Nickel-eluted rpL6 and rpL18a were gel filtrated on NAP5 columns (Pharmacia), and rpL4 on Superdex 200 (Pharmacia). Gel filtration was under 50 mM Tris–HCl pH 7.5, 100 mM NaCl. Ni-eluted histone H1o was purified further on SP-Sepharose (Pharmacia) and then applied to an NAP5 column equilibrated with 50 mM Tris–HCl pH 7.5, 100 mM NaCl. Purification of ribosomal proteins L23a (expressed from p2zL23a) and S7 (expressed from p2zS7 and p6zS7Cys) has been described previously (Jäkel and Görlich, 1998).
Imp9a, Imp9b, Imp4a and Imp4b were expressed in BLR/Rep4 and purified on Ni-NTA–agarose followed by gel filtration on a Superdex 200 column equilibrated in 50 mM Tris–HCl pH 7.5, 50 mM NaCl, 300 mM sucrose, 1 mM magnesium acetate, 1 mM dithiothreitol (DTT).
Preparation of total ribosomal proteins
The extraction of ribosomal proteins from HeLa cell ribosomes (TP80) was based on the TP50 protocol (see Nierhaus, 1990). A total of 700 A260 units (2 ml) of purified HeLa ribosomes were adjusted to a final concentration of 30 mM magnesium acetate and 66% (v/v) acetic acid, incubated for 45 min at 4°C and then centrifuged at 10 000 g for 30 min. The ribosomal proteins stay in the supernatant and were precipitated with 5 vols of acetone. The pellet was dried under vacuum and then dissolved in 1 ml of 20 mM Tris–HCl pH 7.5, 6 M urea, 4 mM magnesium acetate, 400 mM ammonium chloride, 0.2 mM EDTA, 5 mM β-mercaptoethanol. The solution was dialysed twice against 1 l of the same buffer and finally four times against 1 l of 50 mM Tris–HCl pH 7.5. SDS–PAGE analysis was used to verify that the vast majority of the ribosomal proteins were recovered quantitatively by the purification procedure. One A230 unit of the TP80 preparation was estimated to correspond to 220 µg of ribosomal protein (Nierhaus, 1990).
Protein labelling
Fluorescent import substrates were prepared by labelling endogenous cysteines (for rpL4, rpL6 and rpL18a) or engineered cysteine residues (for H1, rpS3a and rpS7) with Alexa 488 maleimide (1:1 stoichiometry to protein). Free fluorophore was removed by gel filtration on NAP5 columns (Pharmacia).
Import assays
HeLa cells were grown on 12 mm coverslips and permeabilized with 60 µg/ml digitonin for 3 min. Import reactions were performed at room temperature for 15 min essentially as described previously (Jäkel et al., 1998). Transport buffer was 20 mM HEPES–KOH pH 7.5, 110 mM potassium acetate, 5 mM magnesium acetate, 0.5 mM EGTA, 250 mM sucrose. The Ran mix contained 3 µM RanGDP, 0.3 µM NTF2, 0.2 µM RanBP1 and 0.2 µM RNA1p. The energy-regenerating system contained 10 mM creatine phosphate, 0.5 mM ATP and GTP, and 50 µg/ml creatine kinase. Where indicated, importin-depleted cytosol was included at 10 mg/ml protein concentration. Fixation was with 2% paraformaldehyde.
Binding experiments
For the binding experiment of Figure 1A, zz-tagged Imp9a was immobilized to ∼2 µg/µl IgG–Sepharose (Pharmacia). A 20 µl aliquot of immobilized protein was incubated for 3 h with 0.8 ml of hypotonic HeLa extract adjusted to 20 mM Tris–HCl pH 7.5, 100 mM NaCl and 3 mM MgCl2. Bound proteins were eluted with 1.5 M MgCl2, precipitated with 95% isopropanol and analysed by SDS–PAGE followed by Coomassie Blue staining. Proteins in the Coomassie Blue-stained bands were identified by mass spectrometry after a tryptic digest. Identification of Imp4 substrates was performed the same way, except that zz-tagged Imp4b was used.
Anti-aggregation assays
Various importins were tested for their ability to prevent aggregation of several basic nuclear import substrates with polyanions. Three sources of polyanions were used in this study. (i) purified tRNA from calf thymus (30 µg/ml final concentration in assay); (ii) a large RNA generated by run-on in vitro transcription (T7) from pBluescript SKII (60 µg/ml final RNA concentration); and (iii) HeLa cytosol depleted of importins (at ∼10 mg/ml final protein concentration). All anti-aggregation assays were performed with 50 mM Tris–HCl pH 7.5, 100 mM NaCl. The procedure was as follows: the precipitation substrate (0.5 µM final concentration) was pre-incubated with the individual importins (see figure legends for concentrations) for 15 min on ice. Then RNA or cytosol depleted of importins was added as precipitants and the reaction incubated for another 15 min on ice. To separate soluble and precipitated material, the samples were centrifuged to sediment particles larger than 30S. The pelleted (precipitated) material was solubilized in SDS sample buffer. Equal fractions (2.5%) of supernatant and pellet diluted in sample buffer were analysed by SDS–PAGE and western blotting to detect the precipitation substrate. Ribosomal proteins S3a, S7, L18a and L23a were detected by their z tags, rpL4 and histone H1o with an anti-His6 tag antibody. Two controls were included in each experiment. A reaction without precipitant demonstrated the solubility of the substrate in the absence of polyanions. Another incubation in the presence of the precipitant but without an importin demonstrates the precipitation tendency of the substrates.
The quantitative and selective transport receptor depletion from cytosol will be described elsewhere (K.Ribbeck and D.Görlich, submitted).
Shielding assay
Patches of exposed basic residues mediate the unspecific interaction of basic proteins with polyanions. The shielding assay was employed to demonstrate the ability of importins to cover these patches, thus preventing an interaction of their import substrates with anionic surfaces. To mimic this situation, we tested the capacity of various importins to shield ribosomal proteins against binding to the cation exchanger CM-Sepharose (Pharmacia). The assays were performed at 4°C in 50 mM Tris–HCl pH 7.5, 100 mM NaCl as follows: 0.5 µM of the ribosomal protein was pre-incubated in the absence or presence of 0.75 µM import receptor. After 15 min, 100 µl of the mixture was added to 20 µl of CM-Sepharose. After 20 min binding with mild shaking, unbound material was separated from the CM-Sepharose by centrifugation through a Mobicol microcolumn and diluted in SDS sample buffer. CM-Sepharose-bound material was eluted with 20 mM NaOH, 4% SDS, 0.5 M sucrose. The eluate was adjusted to 62.5 mM Tris–HCl and 20 mM DTT. Equal fractions (3.7%) of unbound and bound material were analysed by SDS–PAGE followed by western blotting and detection of the ribosomal proteins via their fused z tags.
DDBJ/EMBL/Genbank accession numbers
The nucleotide sequences reported in this manuscript are listed under the following accession numbers: human importin 9, AF410465; mouse importin 9a, AF273672; mouse importin 9b, AF273673; Drosophila importin 9, AF245516; human importin 4b, AF411122; mouse importin 4a, AF123388.
Acknowledgments
Acknowledgements
We wish to thank S.Kostka and R.Kraft for peptide sequencing, Petra Rübmann for excellent technical help, Gerd Lipowski for initial work on Imp4 and Imp9, and Martin Pool as well as the members of our laboratory for critical reading of the manuscript and stimulating discussions. This work was supported by an EMBO long-term fellowship (to J.M.M.), the Fonds der Chemischen Industrie and grants from the DFG (SFB 352) and the HFSPO (RG0198/1998M).
Note added in proof
While this paper was in press, Muhlhausser et al. (2001) also reported on importin 9 and identified histones H2A and H2b as cargoes.
References
- Agashe V.R. and Hartl,F.U. (2000) Roles of molecular chaperones in cytoplasmic protein folding. Semin. Cell. Dev. Biol., 11, 15–25. [DOI] [PubMed] [Google Scholar]
- Bukau B. and Horwich,A.L. (1998) The Hsp70 and Hsp60 chaperone machines. Cell, 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Chi N.C., Adam,E.J. and Adam,S.A. (1995) Sequence and characterization of cytoplasmic nuclear protein import factor p97. J. Cell Biol., 130, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N.C., Adam,E.J.H., Visser,G.D. and Adam,S.A. (1996) RanBP1 stabilises the interaction of Ran with p97 in nuclear protein import. J. Cell Biol., 135, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M., Nair,R. and Rost,B. (2000) Finding nuclear localization signals. EMBO rep., 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E. and Izaurralde,E. (2001) Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol., 13, 310–319. [DOI] [PubMed] [Google Scholar]
- Deane R., Schafer,W., Zimmermann,H.P., Müller,L., Görlich,D., Prehn,S., Ponstingl,H. and Bischoff,F.R. (1997) Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-β but interacts differently with RanBP1. Mol. Cell. Biol., 17, 5087–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E., Schulze-Specking,A., Tomoyasu,T., Mogk,A. and Bukau,B. (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature, 400, 693–696. [DOI] [PubMed] [Google Scholar]
- Dingwall C. and Laskey,R.A. (1990) Nucleoplasmin: the archetypal molecular chaperone. Semin. Cell Biol., 1, 11–17. [PubMed] [Google Scholar]
- Fornerod M., van Deursen,J., van Baal,S., Reynolds,A., Davis,D., Murti,K.G., Fransen,J. and Grosveld,G. (1997) The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J., 16, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Görlich D., Prehn,S., Laskey,R.A. and Hartmann,E. (1994) Isolation of a protein that is essential for the first step of nuclear protein import. Cell, 79, 767–778. [DOI] [PubMed] [Google Scholar]
- Görlich D., Kostka,S., Kraft,R., Dingwall,C., Laskey,R.A., Hartmann,E. and Prehn,S. (1995) Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol., 5, 383–392. [DOI] [PubMed] [Google Scholar]
- Görlich D., Pante,N., Kutay,U., Aebi,U. and Bischoff,F.R. (1996) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J., 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Dabrowski,M., Bischoff,F.R., Kutay,U., Bork,P., Hartmann,E., Prehn,S. and Izaurralde,E. (1997) A novel class of RanGTP binding proteins. J. Cell Biol., 138, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N., Shimamoto,T., Kose,S., Takao,T., Tachibana,T., Matsubae,M., Sekimoto,T., Shimonishi,Y. and Yoneda,Y. (1995a) The nuclear pore targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett., 368, 415–419. [DOI] [PubMed] [Google Scholar]
- Imamoto N., Tachibana,T., Matsubae,M. and Yoneda,Y. (1995b) A karyophilic protein forms a stable complex with cytoplasmic components prior to nuclear pore binding. J. Biol. Chem., 270, 8559–8565. [DOI] [PubMed] [Google Scholar]
- Jäkel S. and Görlich,D. (1998) Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J., 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S., Albig,W., Kutay,U., Bischoff,F.R., Schwamborn,K., Doenecke,D. and Görlich,D. (1999) The importin β/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J., 18, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Richardson,W.D., Markham,A.F. and Smith,A.E. (1984) Sequence requirements for nuclear location of simian virus 40 large T antigen. Nature, 311, 33–38. [DOI] [PubMed] [Google Scholar]
- Makkerh J.P.S., Dingwall,C. and Laskey,R.A. (1996) Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr. Biol., 6, 1025–1027. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Mingot J.M., Kostka,S., Kraft,R., Hartmann,E. and Gorlich,D. (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J., 20, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morehouse H., Buratowski,R.M., Silver,P.A. and Buratowski,S. (1999) The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl Acad. Sci. USA, 96, 12542–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlhausser P., Muller,E.C., Otto,A. and Kutay,U. (2001) Multiple pathways contribute to nuclear import of core histones. EMBO rep., 2, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Nierhaus K. (1990) Reconstitution of ribosomes. In Spedding,G. (ed.), Ribosomes and Protein Synthesis. Oxford University Press, New York, NY, pp. 161–190.
- Pemberton L.F., Rosenblum,J.S. and Blobel,G. (1999) Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol., 145, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott A., Krude,T. and Laskey,R.A. (2000) Nuclear chaperones. Semin. Cell. Dev. Biol., 11, 7–14. [DOI] [PubMed] [Google Scholar]
- Pollard V.W., Michael,W.M., Nakielny,S., Siomi,M.C., Wang,F. and Dreyfuss,G. (1996) A novel receptor-mediated nuclear protein import pathway. Cell, 86, 985–994. [DOI] [PubMed] [Google Scholar]
- Rexach M. and Blobel,G. (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell, 83, 683–692. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Blobel,G. and Aitchison,J.D. (1997) A distinct nuclear import pathway used by ribosomal proteins. Cell, 89, 715–725. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G., Smirnova,E., Deane,R., Solsbacher,J., Kutay,U., Görlich,D., Ponstingl,H. and Bischoff,F.R. (1997) Yrb4p, a yeast ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J., 16, 6237–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Zachmann M.S., Hugle-Dorr,B. and Franke,W.W. (1987) A constitutive nucleolar protein identified as a member of the nucleoplasmin family. EMBO J., 6, 1881–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. and Stillman,B. (1989) Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell, 58, 15–25. [DOI] [PubMed] [Google Scholar]
- Ström A.C. and Weis,K. (2001) Importin-β-like nuclear transport receptors. Genome Biol., 2, 3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- Zirwes R.F., Schmidt-Zachmann,M.S. and Franke,W.W. (1997) Identification of a small, very acidic constitutive nucleolar protein (NO29) as a member of the nucleoplasmin family. Proc. Natl Acad. Sci. USA, 94, 11387–11392. [DOI] [PMC free article] [PubMed] [Google Scholar]