Abstract
The c-myb proto-oncogene product (c-Myb) is a transcriptional activator. Vertebrate c-Myb is a key regulator of the G1/S transition in cell cycle, while Drosophila Myb (dMyb) is important for the G2/M transition. Here we report that dMyb induces expression of cyclin B, a critical regulator of the G2/M transition, in Drosophila eye imaginal disc. In the wild-type eye disc, dmyb mRNA was expressed in the stripes both anterior and posterior to the morphogenetic furrow. Ectopic expression of C-terminal-truncated dMyb in the eye disc caused ectopic expression of cyclin B and the rough eye phenotype. This rough eye phenotype correlated with prolonged M phase, caused by overexpression of cyclin B. Cyclin B expression was lost in dmyb-deficient clones. In Schneider cells, the activity of the cyclin B promoter was dramatically reduced by loss of dMyb using the RNA interference method. Mutations of the multiple AACNG sequences in the cyclin B promoter also abolished the promoter activity. These results indicate that dMyb regulates the G2/M transition by inducing cyclin B expression via binding to its promoter.
Keywords: cell cycle/cyclin B/Drosophila Myb/G2/M progression/transcription
Introduction
The c-myb proto-oncogene is the cellular progenitor of the v-myb oncogenes carried by the chicken retroviruses AMV and E26, which can transform myelomonocytic hematopoietic cells (Roussel et al., 1979; Klempnauer et al., 1982). The level of c-myb expression is predominantly, although not exclusively, high in immature hematopoietic cells and its expression is turned off during terminal differentiation (Gonda et al., 1984). c-myb-deficient mice showed a defect in definitive hematopoiesis in fetal liver, indicating that c-myb plays a role in the proliferation of immature hematopoietic cells (Mucenski et al., 1991). Furthermore, analysis of transgenic mice expressing dominant-negative forms of the c-myb gene product (c-Myb) indicated that c-myb is essential for the development of T cells (Badiani et al., 1994; Allen et al., 1999). The mammalian myb gene family contains two other members, A-myb and B-myb, in addition to c-myb (Nomura et al., 1988). A-myb is highly expressed in a limited range of cell types, including male germ cells and female breast ductal epithelium (Trauth et al., 1994). Consistent with this expression pattern, A-myb-deficient males are infertile due to a block in spermatogenesis and null A-myb females show underdevelopment of breast tissue following pregnancy (Toscani et al., 1997). The cell-type specificity of B-myb expression is broader than those of c-myb and A-myb in both adult tissues and embryos (Nomura et al., 1988; Sitzmann et al., 1996); B-myb is essential for inner cell mass (ICM) formation in the early stages of development (Tanaka et al., 1999). The myb gene is well conserved not only in vertebrate but also in other species. Drosophila melanogaster has one myb gene (dmyb), which is required in diverse cellular lineages throughout the course of Drosophila development (Peters et al., 1987; Katzen and Bishop, 1996; Katzen et al., 1998). Thus, the myb gene family plays an important role in the proliferation of various types of cells in many species.
c-Myb is a transcriptional activator that recognizes a specific DNA sequence, 5′-AACNG-3′ (Biedenkapp et al., 1988; Ness et al., 1989; Sakura et al., 1989; Weston and Bishop, 1989; Tanikawa et al., 1993). c-Myb has three functional domains that are responsible for DNA binding, transcriptional activation and negative regulation (Sakura et al., 1989). The DNA-binding domain in the N-terminal region of c-Myb consists of three imperfect tandem repeats of 51–52 amino acids, each containing a helix–turn–helix variation motif (Ogata et al., 1994). This DNA-binding domain is well conserved among many Myb proteins in other species, including dmyb gene product (dMyb). The transcriptional activation domain of c-Myb is rich in acidic amino acids and binds to the transcriptional co-activator CREB-binding protein (CBP; Dai et al., 1996; Oelgeschlager et al., 1996). The Drosophila homolog of CBP (dCBP) also binds to the transcriptional activation domain of dMyb (Hou et al., 1997). However, the molecular mechanism of c-Myb-induced transcriptional activation remains unknown. Deletion of the negative regulatory domain (NRD), located in the C-terminal portion of the molecule, increases both trans-activation and transformation capacity, implying that this domain normally represses c-Myb activity (Sakura et al., 1989; Hu et al., 1991; Dubendorff et al., 1992; Kanei-Ishii et al., 1992).
It is generally believed that vertebrate Myb proteins play an important role in cell cycle regulation, specifically at the G1/S transition. During IL-2-promoted G1 progression of T cells, expression of c-myb is transiently induced, with maximal levels occurring at the midpoint of G1 (Stern and Smith, 1986). Transcription of a group of target genes, which encode the regulators required for the G1/S transition and include Cdc2 and c-Myc, is directly activated by c-Myb (Evans et al., 1990; Nakagoshi et al., 1992; Ku et al., 1993). A-myb and B-myb are also expressed during the late G1-to-S phase transition in vascular smooth muscle cells, fibroblasts and hematopoietic cells (Reiss et al., 1991; Marhamati et al., 1997). Cell cycle-dependent expression of B-myb is controlled via the E2F-binding sites in the B-myb promoter region (Lam and Watson, 1993) and the trans-activating capacity of both A-Myb and B-Myb is positively regulated by phosphorylation by cyclin A/Cdk2 (Ziebold and Klempnauer, 1997; Saville and Watson, 1998). In contrast to the vertebrate myb genes, dmyb is required for the G2/M transition, although there remains a possibility that dmyb is also involved in the regulation of G1/S transition (Katzen et al., 1998). The phenotypes of dmyb mutants can be partially suppressed by ectopic expression of either cdc2 or string (Cdc25; Katzen et al., 1998), both of which have been shown to promote the G2/M transition. However, the mechanism by which dMyb controls the G2/M transition remains unknown.
The Drosophila compound eye is a powerful system for the study of cell cycle control during development. The Drosophila compound eye, which is composed of an orderly array of ∼800 unit eyes (ommatidia), develops from a columnar epithelium, the eye imaginal disc. During the third larval instar, differentiation initiates in the posterior region of the eye disc and progresses anteriorly as a wave marked by a physical constriction in the apical surface of the epithelium called the morphogenetic furrow (MF). Ahead of the MF, cells are undifferentiated and progress through the cell cycle asynchronously. Cells arrest in G1, beginning just anterior to the MF and this G1 arrest is mediated in part by roughex (Thomas et al., 1994), a Drosophila cyclin-dependent kinase inhibitor (Foley et al., 1999). G1 cells either enter a final, synchronous S phase behind the MF or differentiate into retinal neurons. A complex of Cdk1 (Cdc2) and cyclin B is the main regulator of the G2/M transition for the synchronized cells behind the MF and multiple factors control its activity (Harper and Elledge, 1996; King et al., 1996; Lew and Kornbluth, 1996). However, the destruction of cyclin B via the polyubiquitylation pathway (Glotzer et al., 1991) is normally required for the inactivation of the cyclin B–Cdk1 complex and for the exit from mitosis (Murray, 1995). Thus, the eye disc provides a system where cell-cycle progression during development can be directly visualized as a continuum from the anterior edge of the MF extending posteriorly (Ready et al., 1976; Thomas et al., 1994).
In this paper, we studied the function of dMyb in cell-cycle regulation of the Drosophila eye imaginal disc. We show that dMyb controls the G2/M transition by directly activating the cyclin B promoter.
Results
Effect of overexpression of C-truncated dMyb in eye imaginal disc
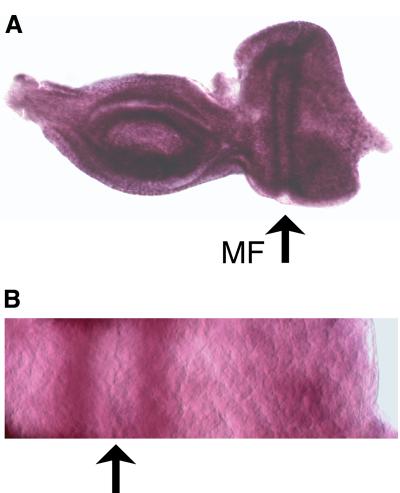
To study the role of dmyb in cell-cycle regulation, we first analyzed the expression of dmyb in the eye imaginal disc by in situ hybridization (Figure 1). In the wild-type eye disc, dmyb mRNA was strongly expressed in the stripes both anterior and posterior to the MF (Figure 1A; note that strong non-specific signals at dorsal and ventral sides are due to the disc edge, which was not extended). A higher magnification image of the part of the eye disc containing the MF clearly indicates the spatial relationship between the dmyb-positive stripes and the MF (Figure 1B).
Fig. 1. Expression of dmyb mRNA in the eye imaginal disc. (A) In situ hybridization was performed using a dmyb-specific anti-sense RNA probe. The dmyb mRNA was detected at both stripes anterior and posterior to the MF in the wild-type eye imaginal disc. Anterior is to the left, dorsal is up. (B) High magnification of the eye disc containing the MF and the dmyb-positive stripes.
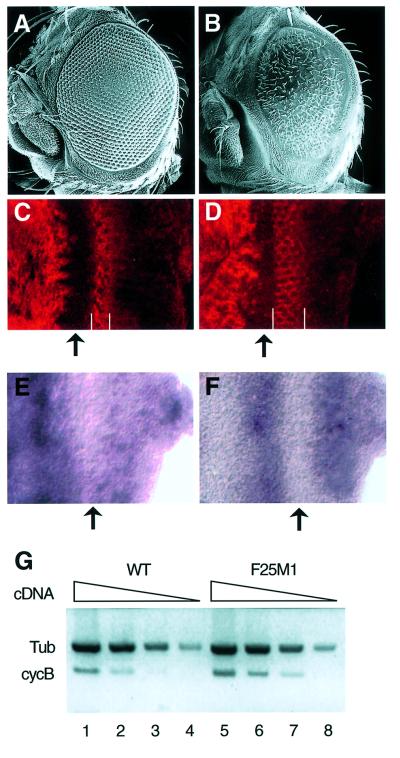
To investigate the effect of overexpression of dmyb in Drosophila, we generated transgenic flies carrying a transgene encoding wild-type dMyb expressed from the eye-specific expression vector, Glass multimer reporter (pGMR). The pGMR P-element vector contains a pentamer of the Glass-binding site derived from the Drosophila Rh1 promoter (Hay et al., 1994). Since Glass is expressed in the morphogenetic furrow (MF) and the whole region posterior to the MF of the third instar eye imaginal disc (Moses and Rubin, 1991), expression of dMyb from this transgene was expected in the MF and the posterior region of the eye disc. Forty independent transgenic lines expressing wild-type dMyb were generated, but none of them exhibited any morphological abnormality in the adult eye (data not shown). The C-terminal portion of vertebrate c-Myb negatively regulates c-Myb activity (Sakura et al., 1989; Hu et al., 1991; Dubendorff et al., 1992; Kanei-Ishii et al., 1992). However, it remains unclear whether dMyb also contains a NRD in its C-terminal region, since a dMyb protein lacking the C-terminal 241 amino acids activates a promoter containing tandem repeats of the Myb-binding site to almost the same extent as wild-type dMyb in co-transfection assays using Drosophila cultured cells (Hou et al., 1997). In spite of this, it might be possible that the C-terminal portion of dMyb negatively regulates the dMyb activity in the eye disc. Therefore, we also generated transgenic flies carrying a transgene encoding a C-terminal-truncated dMyb lacking the C-terminal 241 amino acids (dMybΔC) expressed from the pGMR vector. In contrast to the transgenic flies overexpressing wild-type dMyb, 32 independent transgenic lines expressing the C-truncated dMyb showed a variety of dominant morphological disorders of the adult compound eye (rough eye phenotypes), probably due to the position effect (Figure 2A and B). These results suggest that the C-truncated dMyb has a stronger activity than wild-type dMyb in eye imaginal disc cells. In the present study, we used two of the transgenic lines, GMR–dMybΔC-F25 and GMR– dMybΔC-F54, which exhibited severe and mild rough eye phenotypes, respectively. As expected, in the transgenic flies (GMR–dMybΔC-F25), dmyb mRNA was ectopically expressed in the MF and the whole posterior region to the MF in the developing eye imaginal disc (data not shown).
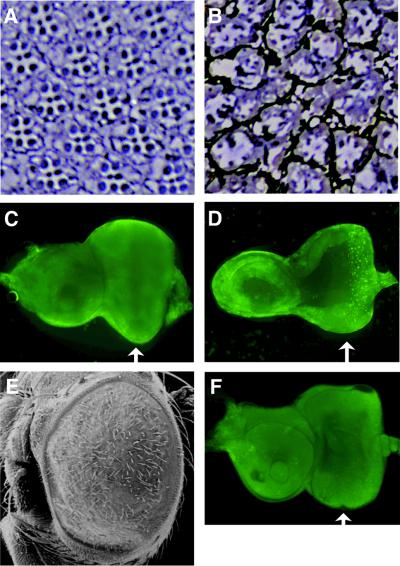
Fig. 2. Rough eye phenotype and ectopic expression of cyclin B in the transgenic flies expressing C-truncated dMyb. (A and B) Scanning electron micrographs of adult eyes. Normal compound eye of wild type (A) and severe rough eye of GMR–dMybΔC-F25 (B) flies. (C and D) Immuno-staining of the eye imaginal disc with anti-cyclin B antibody. High magnification of the eye disc is indicated. Eye discs were prepared from wild-type (C) and GMR–dMybΔC-F25 (D) flies. The widths of the cyclin B-expressing cells are indicated by white bars. Anterior is to the left, dorsal is up. (E and F) Expression of cycB mRNA in the eye imaginal disc. In situ hybridization was performed using a cycB-specific anti-sense RNA probe. High magnification of the eye disc is indicated. The cycB mRNA was detected at the whole region anterior to the MF and in the whole region posterior to the MF in the wild-type eye imaginal disc (E), whereas a strong cycB mRNA signal was detected in the broad stripe posterior to the MF in the GMR–dMybΔC-F25 disc (F). Anterior is to the left, dorsal is up. (G) RT–PCR analysis of the cyclin B mRNA. Poly(A)+ RNAs were prepared from the wild-type and GMR–dMybΔC-F25 eye discs and the cDNAs corresponding to the cyclin B and β1-tubulin genes were sythesized. Using various amounts of the first-strand synthesis reaction mixture (5, 0.5, 0.1 and 0.02 µl for lanes 1–4 and 5–8, respectively), the DNA fragments were then amplified. Quantification of the PCR products were performed by Southern blot analysis.
Since it has been demonstrated that dMyb plays an important role in the G2/M cell cycle transition, we investigated the expression of regulators of cell-cycle control. We found ectopic expression of cyclin B, which is a key regulator of the G2/M transition (Knoblich and Lehner, 1993), in the transgenic flies expressing the C-truncated dMyb. Consistent with earlier reports (Thomas et al., 1994), imaginal disc cells in the developing wild-type eye became synchronized at the G1 phase of the cell cycle within the MF and cyclin B was strongly expressed in the stripe posterior to the MF (Figure 2C). The width of the cyclin B-expressing region in the GMR–dMybΔC-F25 eye imaginal discs was broader than in wild-type control discs (Figure 2C and D). Since the level of cyclin B is tightly regulated by protein degradation, we used in situ hybridization to confirm that ectopic expression of cyclin B in the GMR–dMybΔC-F25 eye imaginal discs was caused at the transcriptional level. In the wild-type eye disc, the cyclin B (cycB) mRNA was expressed in the whole region anterior to the MF at a high level and the whole region posterior to the MF at a low level (Figure 2E), suggesting that the high expression of cyclin B protein in the stripe posterior to the MF of the wild-type eye disc may be due to protein stabilization. In contrast, cycB mRNA was strongly expressed in the broad stripe posterior to the MF of GMR–dMybΔC-F25 eye imaginal discs (Figure 2F). To confirm the upregulation of the cycB gene in the GMR–dMybΔC-F25 eye imaginal discs, we performed RT–PCR analysis using a series of decreasing amount of RNA prepared from the wild-type and the GMR–dMybΔC-F25 eye imaginal discs (Figure 2G). The results indicated that the level of cycB mRNA in the GMR–dMybΔC-F25 eye imaginal discs was ∼2.2-fold higher than that in wild-type discs. Since we used the whole eye disc as a source of RNA, the degree of increase in cycB mRNA levels in the region posterior to the MF may be higher than 2.2-fold. Thus, overexpression of C-truncated dMyb in the whole region posterior to the MF of eye imaginal discs caused the rough eye phenotype and ectopic expression of cycB mRNA in the stripe posterior to the MF. No induction of cycB mRNA in the posterior region other than in the stripe suggests that activation of the cycB promoter requires not only dMyb but also other transcription factor(s) expressed in the stripe posterior to the MF.
Correlation between the delayed exit from M phase and the rough eye phenotype
The kinase activity of cyclin B–Cdk1 complex promotes mitosis, whereas the destruction of cyclin B and loss of kinase activity is associated with and required for exit from mitosis (Sigrist et al., 1995). Therefore, ectopic expression of cyclin B in the posterior region of GMR–dMybΔC eye imaginal discs might be expected to slow down progression through M phase. To investigate whether the M-phase population in the posterior region of GMR–dMybΔC eye imaginal discs is in fact higher than that in wild-type discs, we performed immunostaining using an antibody specific for phosphorylated histone H3 (Figure 3A and B). Phosphorylation of histone H3 is induced at M phase and phosphorylated histone H3 can be used as a marker of M-phase cells. The entire region in the anterior and posterior portion of MF was investigated. In order to count the stained cells that are located both at the apical and basal surface, discs were visualized with a fluorescent microscope at the lower magnification. The number of cells stained by anti-phospho-histone H3 antibody in the whole region posterior to the MF of the GMR–dMybΔC-F25 and the wild-type eye discs were 92.7 ± 10.6 and 58.7 ± 6.6 (average of 11 discs), respectively (Table I). Thus, the population of M-phase cells in the posterior region of GMR–dMybΔC-F25 eye discs was 58% larger than the corresponding population in wild-type discs. In contrast, the number of cells stained by anti-phospho-histone H3 antibody in the whole region anterior to the MF of the GMR–dMybΔC-F25 and the wild-type eye discs were 44.3 ± 6.5 and 48.9 ± 5.5 (average of 11 discs), respectively, indicating that the populations of M-phase cells were similar sizes in the anterior region of GMR–dMybΔC-F25 and wild-type eye discs.
Fig. 3. Prolonged M phase correlates with the rough eye phenotype in the GMR–dMybΔC flies. (A and B) Increased population of M-phase cells in the GMR–dMybΔC-F25 eye imaginal disc. Eye discs prepared from wild-type (A) and GMR–dMybΔC-F25 (B) flies were immuno-stained with anti-phospho-histone H3 antibody. (C and D) No extra division cycles in the posterior region of the GMR–dMybΔC-F25 eye imaginal disc. BrdU incorporation was examined using eye discs prepared from wild-type (C) and GMR–dMybΔC-F25 (D) flies. (E and F) Rescue of the mild rough eye phenotype of the GMR–dMybΔC-F54 flies by cycB mutation. Scanning electron micrographs of mild rough eye of GMR–dMybΔF54 flies (E) and restored eye of the GMR– dMybΔC-F54 flies carrying one copy of the cycB1 allele (F).
Table I. Number of phospho-histone H3 positive cells.
| Genotype (No. of discs examined) | Anterior to the MF (mean ± SE) | Posterior to the MF (mean ± SE) |
|---|---|---|
| Wild type (n = 11) | 48.9 ± 5.5 | 58.7 ± 6.6 |
| F25/+ (n = 11) | 44.3 ± 6.5 | 92.7 ± 10.6 |
| F25/stg4 (n = 15) | 40.1 ± 5.9 | 71.7 ± 7.2 |
| cycB1/+;F25/+ (n = 15) | 47.9 ± 7.1 | 89.1 ± 9.1 |
| F25/GMR–p35 (n = 15) | 50.5 ± 4.8 | 99.1 ± 8.3 |
| F54/+ (n = 15) | 50.1 ± 4.7 | 65.4 ± 6.1 |
| F54/stg4 (n = 15) | 43.7 ± 6.8 | 59.9 ± 8.2 |
| cycB1/+;F54/+ (n = 15) | 48.1 ± 7.1 | 60.3 ± 4.9 |
An increase in the number of cells that are phospho-histone H3 positive may be caused by either of two mechanisms: the progression through M phase is slowed down or cells progress through extra division cycles instead of exiting the cell cycle after M2. To examine this, we investigated the S-phase cells that incorporate bromodeoxyuracil (BrdU; Figure 3C and D). The number of S-phase cells and their distribution in the posterior region of eye disc were similar for the wild-type and GMR–dMybΔC eyes, indicating that cells in the GMR– dMybΔC eye discs do not progress through extra division cycles.
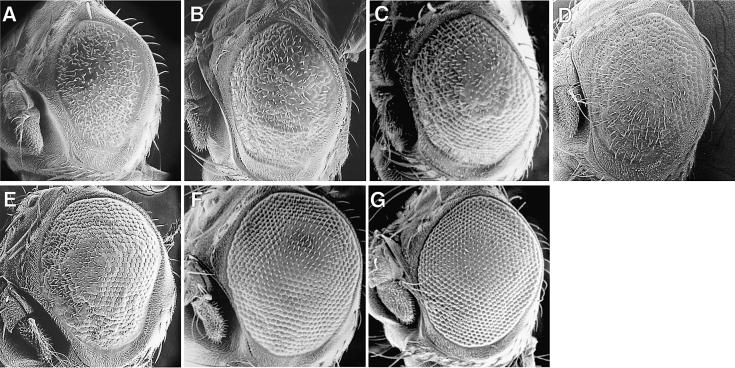
We next asked whether ectopic expression of cyclin B really correlates with the rough eye phenotype. In order to investigate this, we crossed transgenic flies expressing the C-truncated dMyb with a cyclin B mutant, cycB1 (Jacobs et al., 1998). In two independent transgenic lines expressing the C-truncated dMyb (F25 and F54), the mild rough eye phenotype of the GMR–dMybΔC-F54 flies was clearly restored by crossing with a cycB1 mutant (Figure 3E and F). However, the more severe rough eye phenotype of the GMR–dMybΔC-F25 transgenic line was not restored by crossing with a cycB1 mutant (data not shown). These results correlated with the number of M-phase cells in the posterior region of eye discs that were stained with anti-phospho-histone H3 antibody (Table I). To examine further the relationship between the delayed progression through M phase and the rough eye phenotype, we crossed the GMR–dMybΔC-F25 or GMR–dMybΔC-F54 flies with the string Cdc25 mutant (Figure 4B and F). The Cdc25 tyrosine phosphatase activates Cdk1 by dephosphorylation. The severe rough eye phenotype of the GMR– dMybΔC-F25 flies was partially restored by crossing with the string mutant, while the mild rough eye phenotype of the GMR-dMybΔC-F54 flies was clearly restored. Further, we also investigated whether rough eye phenotype observed in the GMR–dMybΔC flies was rescued by reducing the dosage of cdc2 and dCBP (Figure 4C, D and G). Loss of one copy of cdc2 or dCBP partly or almost completely restored the rough eye phenotype of the GMR–dMybΔC flies. Rescue of the rough eye phenotype by loss of one copy of stg, cdc2 or dCBP is well correlated with the number of M-phase cells in the posterior region of eye discs (Table I). These results suggest that the rough eye phenotype observed in the GMR–dMybΔC flies correlates, at least in part, with the delayed progression through M phase, which was caused by ectopic expression of cyclin B.
Fig. 4. Restoration of the rough eye phenotype of the GMR–dMybΔC flies by Cdc25, cdc2 and dCBP mutation. Scanning electron micrographs of severe rough eye of GMR–dMybΔC-F25 flies (A) and mild rough eye of GMR–dMybΔF54 flies (E). The partially or almost completely restored eye caused by loss of one copy of the string (B and F), cdc2 (C and G) and nejire (dCBP) allele (D).
Apoptosis in the eye imaginal disc expressing the C-truncated dMyb
To examine further the molecular basis of the rough eye phenotype in the GMR–dMybΔC-F25 flies, retinal sections were examined by microscopy (Figure 5A and B). A wild-type compound eye contains ∼800 ommatidia, each of which contains eight photoreceptor cells (Tomlinson, 1988; Ready, 1989). Six of the photoreceptor cells, R1–R6, extend the full depth of the ommatidia and contain large rhabdomeres positioned along the periphery of the ommatidia. The rhabdomeres of the R7 and R8 cells are smaller and occupy the central distal and proximal portions of the ommatidia, respectively. Consequently, only seven of the eight photoreceptor cells are present in any single cross section. In the GMR–dMybΔC-F25 flies, the ommatidia were found to contain abnormal numbers and positioning of ommatidial cells. GMR–dMybΔC-F25 ommatidia were characterized by a variable number of photoreceptor and outer photoreceptor cells in each ommatidium. The exact number of cells was difficult to score, as most of the ommatidia were markedly misshapen. In addition, most of the rhabdomeres in these ommatidia were found to be immature in size. Since programmed cell death plays a key role in the development of the architecture of the adult eye (Bangs and White, 2000), we investigated whether apoptosis occurs in the GMR– dMybΔC-F25 third instar eye imaginal disc (Figure 5C and D). Acridine Orange staining of the GMR–dMybΔC-F25 eye disc demonstrated that cell death is induced in the region posterior to the MF, whereas cell death was not evident in the wild-type eye discs. To examine whether the cell death in GMR–dMybΔC-F25 eye disc is blocked by the apoptosis inhibitor p35, we crossed transgenic flies expressing the C-truncated dMyb with the transgenic flies expressing p35 (GMR–p35). In the transgenic lines expressing both the C-truncated dMyb and p35, apoptosis was not observed, but the rough eye phenotype was still observed (Figure 5E and F). Consistent with this, the eye discs of those transgenic flies had the increased number of M-phase cells that were stained with anti-phospho-histone H3 antibody (Table I). These results suggest that both abnormal cell cycle regulation and increased cell death in the eye disc caused the observed loss of ommatidial cells in the GMR–dMybΔC-F25 retina.
Fig. 5. Decreased number of photoreceptors and induced apoptosis in the GMR–dMybΔC flies. (A and B) Eye sections from wild-type [white; (A)] and GMR–dMybΔC-F25 (B) flies. (C and D) Apoptotic cells were stained with Acridine Orange in wild-type (C) and GMR–dMybΔC-F25 (D) eye discs. Arrows indicate the position of the MF. (E) Scanning electron micrographs of eye of GMR–dMybΔF25 flies expressing p35 ectopically. (F) Apoptotic cells were stained with Acridine Orange in eye discs of GMR–dMybΔF25 flies expressing p35 ectopically.
dMyb directly activates cyclin B transcription
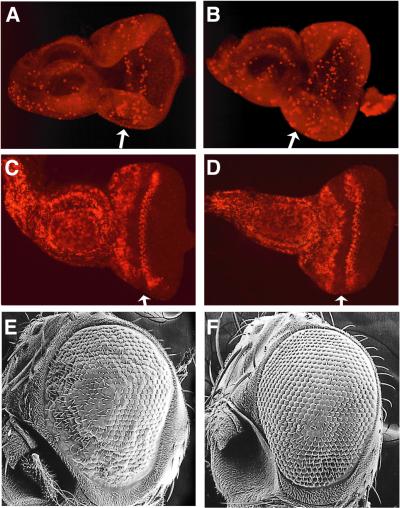
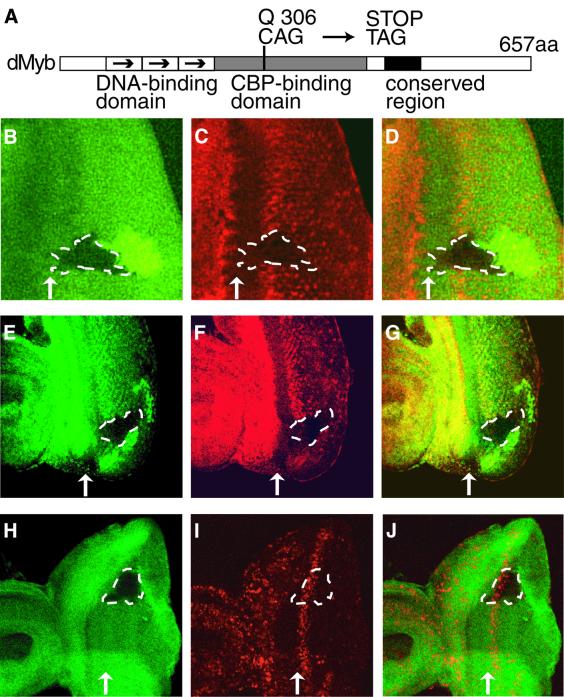
We investigated whether dMyb is required for cyclin B expression using mosaic analysis. We first isolated the dmyb mutant by ethyl methanesulfonate (EMS) treatment; the isolated allele had a nonsense mutation at Q306 in the transcriptional activation domain and the dMyb mutant protein encoded by this allele probably cannot bind to co-activator dCBP or binds with a decreased affinity compared with the wild type (Figure 6A). In the dmyb mutant clones, which crossed the stripe of the cyclin B-expressing region posterior to the MF, the level of cyclin B expression was dramatically decreased (Figure 6B–D). A decrease in cyclin B expression was similarly observed in the clone lacking dCBP, which is an essential co-activator of dMyb (Figure 6E–G). To further confirm that loss of cyclin B accumulation in the dmyb mutant clones is not due to cells remaining at G1 phase in the MF, we investigated BrdU incorporation in the dmyb mutant clones. In the dmyb mutant clones that were crossed with the MF, the cells in the region posterior to the MF normally incorporated BrdU (Figure 6H–J), indicating that S phase of the second mitotic wave occurred on schedule in the dmyb mutant clones. Thus, loss of dmyb leads to abrogation of cyclin B accumulation.
Fig. 6. Loss of cyclin B expression in dmyb-deficient clones. (A) Schematic representation of dMyb mutant. The functional domains and the position of the dmybel1 mutation are indicated. (B–G) Decreased cyclin B expression in dmyb (B–D) or dCBP (E–G) mutant clones in the eye imaginal disc. dmybel1 (B) and nej3 (E) homozygous clones visualized by the lack of β-galactosidase and Myc marker staining (green), respectively, are outlined with dotted lines. Cyclin B expression was monitored in the same disc by staining for the cyclin B protein (red in C and F). The right-hand panels (D and G) show the two single staining patterns superimposed. Anterior is to the left, dorsal is up. At least 10 clones were examined and similar results were obtained with all the clones examined. (H–J) BrdU incorporation of the cells lacking dmyb. dmybel1 (H) homozygous clones visualized by the lack of β-galactosidase marker staining (green) are outlined with dotted lines. BrdU incorporation was monitored in the same disc (red in I and J). The right-hand panel (J) show the two single staining patterns superimposed. Anterior is to the left, dorsal is up. At least 10 clones were examined and similar results were obtained with all the clones examined.
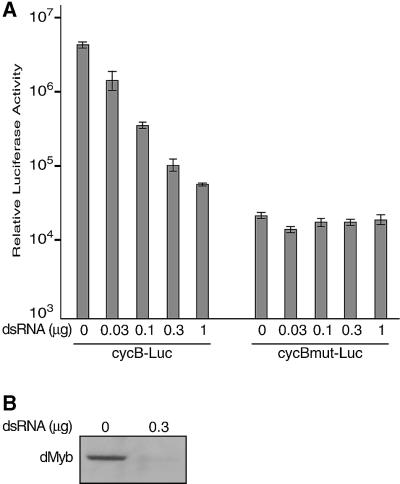
We next used a luciferase reporter assay to investigate whether dMyb directly activated transcription from the cyclin B promoter (Figure 7). Co-transfection of the wild-type dMyb expression vector into Drosophila Schneider cells with the reporter plasmid pcycB–Luc, from which luciferase is expressed under the control of the cyclin B promoter, increased luciferase expression only a few fold (data not shown). The dMyb protein lacking the C-terminal 241 amino acids, which was ectopically expressed in the GMR–dMybΔC flies, also enhanced luciferase expresson from the cycB promoter by a few fold (data not shown). We speculated that this low degree of activation might be due to the presence of an excess amount of the dMyb protein in Schneider cells. Therefore, we tried to decrease the level of endogenous dMyb by using the RNA interference (RNAi) method (Caplen et al., 2000). Co-transfection of the increasing amounts of 700 bp double-stranded RNA of the dmyb cDNA with the reporter plasmid pcycB–Luc suppressed luciferase expression in a dose-dependent manner. As a control, we used the reporter plasmid pcycBmut–Luc, which contains mutations in the seven AACNG sites in the cycB promoter. The level of the luciferase expression from this mutant reporter was much lower than that of pcycB–Luc containing the wild-type cycB promoter, indicating that these dMyb-recognition sequences are important to induce cycB transcription. As expected, the dMyb double-stranded RNA did not affect the luciferase expression from the reporter plasmid pcycBmut–Luc. To confirm that the dmyb double-stranded RNA really decreased the level of endogenous dMyb protein, we performed western blotting. The Schneider cell lysates gave rise to a single band of ∼90 kDa, whereas this band was not detected in the lysates prepared from the Schneider cells transfected with the dmyb double-stranded RNA. Thus, the level of endogenous dMyb proteins was specifically decreased by the RNAi method. These results demonstrate that dMyb activates the cycB promoter and regulates G2/M transition.
Fig. 7. dMyb directly activates the cycB promoter. (A) Co-transfection assays. Increasing amounts of the 700 bp double-stranded RNA corresponding to the dMyb cDNA were transfected into Schneider cells together with a luciferase reporter construct containing the cyclin B promoter. The cycB–Luc or cycBmut–Luc reporters contained the wild-type or mutant cycB promoter in which the seven AACNG sequences were mutated. Luciferase assays were carried out and data are shown as the average with standard deviation of three independent experiments. (B) Western blotting of dMyb. Total cell lysates were prepared from the Schneider cells transfected with the dMyb double-stranded RNA or the control non-transfected cells. Lysates were separated by SDS–PAGE and western blotting was performed using the anti-dMyb antibody.
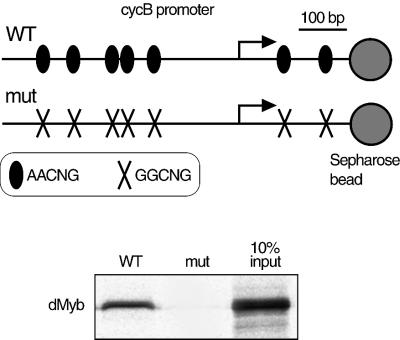
We next tested whether dMyb could directly bind to the cyclin B promoter (Figure 8). The 733 bp DNA fragments containing the cyclin B promoter (from nucleotides +236 to –497; the transcription start site at +1) were attached to streptavidin–Sepharose beads and mixed with the full-length dMyb protein translated in vitro. dMyb formed a complex with this DNA fragment containing seven Myb recognition sequences, 5′-AACNG-3′ (Tanikawa et al., 1993). In contrast, dMyb did not bind to the 733 bp DNA fragment in which the all the seven dMyb recognition sequences were mutated. These results indicate that dMyb directly binds to the AACNG sequences in the cyclin B promoter.
Fig. 8. dMyb directly binds to the cyclin B promoter. Two 733 bp DNA fragments containing the cycB promoter region (nucleotides +236 to –497; the transcription start site at +1) are shown at the top of figure. The seven sites containing the dMyb recognition sequence (AACNG) are indicated. The DNA fragments were immobilized on Sepharose beads and the full-length dMyb protein translated in vitro was incubated with the DNA–Sepharose resin. After washing, the proteins that bound to the cyclin B promoter–Sepharose resin were analyzed by SDS–PAGE. dMyb was detected by autoradiography.
Discussion
Expression of dmyb mRNA was detected in the stripes anterior and posterior to the MF of the wild-type eye imaginal disc (Figure 1). In the developing eye imaginal disc, cells become synchronized at G1 phase of the cell cycle within the MF, where roughex prevents cells in G1 from entering S phase prematurely (Thomas et al., 1994). The expression pattern of dmyb mRNA in eye imaginal discs suggests that dMyb plays a role in the G2/M transition, as does a previous report on the wing phenotype of dmyb mutants (Katzen et al., 1998). Expression of cyclin B was lost in the dmyb-deficient clones and dMyb directly activates the cycB promoter in co-transfection assays. These results indicate that dMyb regulates the G2/M transition, at least partly, by inducing cyclin B expression. In the GMR–dMybΔC eye disc, dMybΔC was ectopically expressed in the whole region posterior to the MF, whereas cycB mRNA was induced in the broad stripe posterior to the MF only. Induction of cycB mRNA may require not only dMyb but also an uncharacterized factor, which is expressed in the stripe posterior to the MF.
Ectopic expression throughout the entire region posterior to the MF of the eye disc of a dMyb mutant, which lacks the C-terminal 241 amino acids, caused the rough eye phenotype, whereas similar ectopic expression of wild-type dMyb did not have this effect. In co-transfection assays using Schneider cells, the C-truncated dMyb mutant stimulated transcription from an artificial promoter containing tandem repeats of the Myb binding site to almost the same extent as wild-type dMyb (Hou et al., 1997). In the case of the luciferase reporter that contained the cyclin B promoter, overexpression of wild-type dMyb only slightly enhanced the luciferase expression and the endogenous dMyb appeared to be sufficient for the transcription from the cyclin B promoter. It is likely that the amounts of dMyb required to induce the maximum level of transcription may be different depending on the promoter context. Another possibility is that the role of the C-terminal portion of dMyb may also be different depending on the promoter context; the C-truncated dMyb may have a stronger trans-activating capacity than wild-type dMyb for the cyclin B promoter but not for the artificial promoter containing tandem repeats of the Myb binding site.
We have observed that the dmyb-deficient clones are not tiny, suggesting that the dmyb-deficient cells keep proliferating for a while to produce clones of reasonable size. This may suggest that loss of dMyb function slows down cell-cycle progression through mitosis but does not stop the cell cycle. In fact, we observed the presence of M-phase cells stained with anti-phospho-histone H3 antibody in the dmyb-deficient clone (data not shown), suggesting that loss of dMyb does not block the G2/M transition. This is consistent with the report that loss of either cyclin A or cyclin B does not block the cell-cycle progression in Drosophila embryos and that loss of both cyclins blocks the cell-cycle progression (Knoblich and Lehner, 1993). Probably, dMyb is required for expression of cyclin B but not cyclin A. However, we cannot exclude the possibility that the dMyb protein is relatively stable and persists in mutant cells for several generations after mitotic recombination events so that mutant cells can continue to divide for a while. Further analyses are required for understanding the precise role of dMyb in cell-cycle regulation.
Our results suggest that overexpression of the C-truncated dMyb retains the cells in M phase and that the abnormal cell-cycle regulation causes apoptosis. These two events result in the rough eye phenotype. Since R2–5 and R8 are already determined by the time that the GMR promoter is activated, these cells presumably are not affected by overexpression of dMyb. This suggests that R1, 6 and 7 and later cell types are specifically affected in these discs. One possibility is that, since mitosis may be prolonged, cells are unable to respond to differentiation signals in the appropriate temporal window and this may lead in part cell death.
Materials and methods
Isolation of dmyb mutants
We established ∼700 X-linked recessive lethal lines by feeding flies with 25 mM EMS. To screen for dmyb mutants, the females were mated with male transgenic flies, which were generated by P element-mediated transformation and carried a 13 kb XhoI genomic fragment containing the dmyb open reading frame (ORF) on an autosomal chromosome. We isolated two hemizygous lethal strains, el1 and el2507, lethality of which was rescued by the autosomal dmyb gene. Sequence analysis indicated that the dmybel1 allele had a nonsense mutation at Q306 in the dmyb ORF caused by a C to T transition at nucleotide 1521 (nucleotide numbering system is the same as in DDBJ/EMBL/GenBank accession No. X05939). At 25°C, ∼60% of dmybel1 hemizygotes were embryonic lethal and the remainder were lethal at the third larval or early pupal stage. A shift in temperature to 30°C resulted in embryonic or first larval lethality of all hemizygotes, demonstrating that the dmybel1 mutation is temperature-sensitive.
Generation of transgenic flies expressing wild-type and C-truncated dMyb
To generate a truncated dMyb protein lacking 241 amino acids from its C-terminus, a stop codon was introduced into the dmyb cDNA using a PCR-based method. Wild-type or C-truncated dmyb cDNA was inserted into the pGMR1 vector (Hay et al., 1994). Forty independent lines were generated by P element-mediated transformation using wild-type dMyb and 32 independent lines were generated using the C-truncated dMyb. None of the wild-type dMyb transgenic lines exhibited any morphological abnormality in the adult eye. In contrast, the transgenic lines expressing the C-truncated dMyb showed a variety of dominant rough-eye phenotypes; two of these, GMR–dMybΔC-F25 and GMR–dMybΔC- F54, were used in the present study.
Fly stocks and histology
The CycB1 and cdc2E10 mutants were provided by C.F.Lehner (Stern et al., 1993; Jacobs et al., 1998). The stg4 mutant was obtained from Bloomington Stock Center. The dCBP mutant, nej, was described previously (Akimaru et al., 1997). The transgenic flies expressing p35 (GMR–p35) was obtained from G.M.Rubin (Hay et al., 1994). For analysis by scanning electron microscopy, adult eyes were dehydrated, critical-point dried as described by Kimmel et al. (1990) and examined using a JEOL 6100 electron microscope. Whole-mount in situ hybridization using dmyb- or cycB-specific probes was carried out as described by Tautz and Pfeifle (1989). Tangential eye sections were prepared as described by Tomlinson and Ready (1987). For Acridine Orange staining, eye discs from late third instar larvae were dissected in phosphate-buffered saline (PBS) with 2 mg/ml Acridine Orange, washed with PBS and mounted directly. M-phase cells were stained with rabbit anti-phospho-histone H3 (Upstate) in a ratio of 1:200. A secondary antibody coupled to rhodamine (Cappel) was used at 1:100. Immunofluorescent staining was visualized using a Zeiss Axioplan2 fluorescent microscope.
RT–PCR assay
The RT–PCR assay was carried out as described by Knoblich et al. (1994). The third instar eye discs from GMR–dMybΔC-F25 or wild-type larvae were dissected and mRNA was prepared using the Quickprep Micro mRNA purification kit (Pharmacia). First-strand cDNA was prepared from 50 ng of purified mRNA using the first-strand cDNA synthesis kit (Pharmacia). Various amounts of the first-strand synthesis reaction mixture were used as the template. PCR was carried out with the primer for amplification of a 200 bp cyclin B fragment and the primer for amplification of a 300 bp β1-tubulin fragment (Knoblich et al., 1994). Quantification of the PCR products was performed by Southern blot analysis. PCR products were separated by agarose gel electrophoresis, transfered to a nylon membrane and hybridized with a 32P-labeled PCR primer for cyclin B and β1-tubulin. Signals were visualized by BAS-2500 system (Fuji).
Clonal analysis
Somatic clones of mutant cells were generated using the FLP recombin ation target/Flippase (FRT/FLP)-mediated recombination system (Xu and Rubin, 1993). First instar larvae were heat-shocked for 1 h at 37°C to induce clones. The relevant genotypes used were: for dmyb clones, w dmybel1 FRT18A/ w arm–lacZ FRT18A, hsp70–flp/+; for nej clones, w nej3 FRT18A/ w hsp70–πMyc FRT18A, hsp70–flp/+. After the induction of dmyb clones, the dmyb mutant larvae were grown at 30°C until dissection at the third larval stage. Eye imaginal discs were dissected from late third stage larvae, fixed and stained with the appropriate antibodies to mark clones and detect cyclin B expression as described by Thomas et al. (1988). Detection of BrdU incorporation was performed as described by Johnston and Schubiger (1996). Only the nej mutant larvae were heat-shocked prior to dissection as previously described (Akimaru et al. 1997). The antibodies and dilutions were used were: mouse anti-β-galactosidase (Promega), 1:500; mouse anti-c-Myc (Santa Cruz), 1:100; rabbit anti-CycB (gift from C.Lehner), 1:2000; mouse anti-BrdU (Santa Cruz), 1:30. Secondary antibodies coupled to fluorescein isothiocynate and rhodamine (Cappel) were used at 1:100 dilution. Immunofluorescent staining was visualized using a Zeiss 510 laser confocal microscope 510.
Co-transfection assay
The 733 bp fragment containing the Drosophila cycB promoter was prepared from pKScycB (Dalby and Glover 1992) by PCR using the BglII site-containing primer (5′-CCTTCAGATCTATTGTACATATAATGCCAGTCTT) and the HindIII site-containing primer (5′-CCTATAAGCTTTTCTATCTGTACAAACACAATTA) and inserted into the pGL3-basic vector (Promega) to generate the reporter plasmid pcycB0.7–Luc. To generate the reporter plasmid pcycBmut0.7–Luc, all seven dMyb recognition sequences containing the AACNG sequence were mutated to GGCNG by a PCR-based method. To produce the dmyb double-stranded RNA, the 700 bp EcoRI–EcoRV fragment of the dmyb cDNA was inserted into pGEM3Zf(+). The plasmid was digested with EcoRI or HindIII and RNA was synthesized using SP6 or T7 RNA polymerase. RNA was extracted with phenol–chloroform and precipitated three times with ethanol. The equal amount of the complementary RNA fragments were annealed by incubation at 65°C for 10 min followed by slow cooling to room temperature. A mixture of 0.5 µg of the luciferase reporter pcycB0.7–Luc, 0–1 µg of dmyb double-stranded RNA and 0.1 µg of the internal control plasmid pCMV–Luc was transfected into Schneider cells. The total amount of DNA was adjusted to 2.6 µg by adding the control plasmid pAct5c0 lacking the cDNA insert. Luciferase assays were then performed.
Antibody preparation and immunoblotting
The anti-dMyb antibody was raised against the bacterially expressed N-terminal 300 amino acids of dMyb–GST fusion protein. The antibody was purified using a protein G column (Pharmacia). Three days after transfection of the dmyb double-stranded RNA, total cell lysates from 3 × 106 cells were used for western blotting.
Promoter-binding assay
Biotin-labeled 733 bp DNA fragments containing the wild-type or mutated cyclin B promoter (nucleotides +236 to –497; the transcription start site at +1) were synthesized by PCR using the primers (5′-GGCACAAAAACAAACCGAACACAG or 5′-GGCACAAAAACAGGCCGAACACAG and 5′-biotin-CTATCTGTACAAACACAATTAAC). PCR products were bound to streptavidin–Sepharose 4B in buffer A (20 mM Tris–HCl pH 8.0, 0.5 M NaCl) by incubating for 15 min at room temperature. Immobilized DNA fragments (0.2 µg) were incubated with in vitro translated full-length dMyb in buffer B [20 mM Tris–HCl pH 7.5, 50 mM NaCl, 4.8 µg poly(dG-dC), 1 mM dithiothreitol (DTT), 0.1% bovine serum albumin (BSA)] for 30 min at 25°C. After washing with buffer B five times, bound proteins were analyzed by SDS–PAGE followed by autoradiography.
Acknowledgments
Acknowledgements
We thank M.Nonaka for help with the isolation of dmyb mutants, D.M.Glover for the cycB promoter clone, S.Luschnig for the Ubi–GFP strain, Y.Hiromi for the arm–lacZ strain, C.Lehner for the anti-cyclin B antibody, CycB1 and cdc2E10 mutants, G.M.Rubin for the GMR–p35 strain and Yoshiki Ohno for assistance with scanning electron microscopy.
References
- Akimaru H., Chen,Y., Dai,P., Hou,D.X., Nonaka,M., Smolik,S.M., Armstrong,S., Goodman,R.H. and Ishii,S. (1997) Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature, 386, 735–738. [DOI] [PubMed] [Google Scholar]
- Allen R.D. III, Bender,T.P. and Siu,G. (1999) c-Myb is essential for early T cell development. Genes Dev., 13, 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiani P., Corbella,P., Kioussis,D., Marvel,J. and Weston,K. (1994) Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev., 8, 770–782. [DOI] [PubMed] [Google Scholar]
- Bangs P. and White,K. (2000). Regulation and execution of apoptosis during Drosophila development. Dev. Dyn., 218, 68–79. [DOI] [PubMed] [Google Scholar]
- Biedenkapp H., Borgmeyer,U., Sippel,A.E. and Klempnauer,K.-H. (1988) Viral myb oncogene encodes a sequence-specific DNA binding activity. Nature, 335, 835–837. [DOI] [PubMed] [Google Scholar]
- Caplen N.J., Fleenor,J., Fire,A. and Morgan,R.A. (2000) dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene, 252, 95–105. [DOI] [PubMed] [Google Scholar]
- Dai P., Akimaru,H., Tanaka,Y., Hou,D.X., Yasukawa,T., Kanei-Ishii,C., Takahashi,T. and Ishii,S. (1996) CBP as a transcriptional coactivator of c-Myb. Genes Dev., 10, 528–540. [DOI] [PubMed] [Google Scholar]
- Dalby B. and Glover,D.M. (1992) 3′ non-translated sequences in Drosophila cyclin B transcripts direct posterior pole accumulation late in oogenesis and peri-nuclear association in syncytial embryos. Development, 115, 989–997. [DOI] [PubMed] [Google Scholar]
- Dubendorff J.W., Whittaker,L.J., Eltman,J.T. and Lipsick,J.S. (1992) Carboxyl-terminal elements of c-Myb negatively regulate transcriptional activation in cis and in trans. Genes Dev., 6, 2524–2535. [DOI] [PubMed] [Google Scholar]
- Evans J.T., Moore,T.L., Kuehl,W.M., Bender,T. and Ting,J.P.-Y. (1990) Functional analysis of c-Myb protein in T-lymphocytic cell lines shows that it trans-activates the c-myc promoter. Mol. Cell. Biol., 10, 5747–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E., O’Farrell,P.H. and Sprenger,F. (1999) Rux is a cyclin-dependent kinase inhibitor (CKI) specific for mitotic cyclin–Cdk complexes. Curr. Biol., 9, 1392–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray,A.W. and Kirschner,M.W. (1991) Cyclin is degraded by the ubiquitin pathway. Nature, 349, 132–138. [DOI] [PubMed] [Google Scholar]
- Gonda T.J. and Metcalf,D. (1984) Expression of myb, myc and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature, 310, 249–251. [DOI] [PubMed] [Google Scholar]
- Harper J.W. and Elledge,S.J. (1996) Cdk inhibitors in development and cancer. Curr. Opin. Genet. Dev., 6, 56–64. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wolff,T. and Rubin,G.M. (1994) Expression of baculovirus p35 prevents cell death in Dorosophila. Development, 120, 2121–2129. [DOI] [PubMed] [Google Scholar]
- Hou D.X., Akimaru,H. and Ishii,S. (1997) Trans-activation by the Drosophila myb gene product requires a Drosophila homologue of CBP. FEBS Lett., 413, 60–64. [DOI] [PubMed] [Google Scholar]
- Hu Y., Ramsay,R.G., Kanei-Ishii,C., Ishii,S. and Gonda,T.J. (1991) Transformation by carboxyl-deleted Myb reflects increased transactivating capacity and disruption of a negative regulatory domain. Oncogene, 6, 1549–1553. [PubMed] [Google Scholar]
- Jacobs H.W., Knoblich,J.A. and Lehner,C.F. (1998) Drosophila Cyclin B3 is required for female fertility and is dispensable for mitosis like Cyclin B. Genes Dev., 12, 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.A. and Schubiger,G. (1996) Ectopic expression of wingless in imaginal discs interferes with decapentaplegic expression and alters cell determination. Development, 122, 3519–3529. [DOI] [PubMed] [Google Scholar]
- Kanei-Ishii C., MacMillan,E.M., Nomura,T., Sarai,A., Ramsay,R.G., Aimoto,S., Ishii,S. and Gonda,T.J. (1992) Transactivation and transformation by Myb are negatively regulated by a leucine-zipper structure. Proc. Natl Acad. Sci. USA, 89, 3088–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen A.L. and Bishop,J.M. (1996) myb provides an essential function during Drosophila development. Proc. Natl Acad. Sci. USA, 93, 13955–13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen A.L., Jackson,J., Harmon,B.P., Fung,S.M., Ramsay,G. and Bishop,J.M. (1998) Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes Dev., 12, 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel B.E., Heberlein,U. and Rubin,G.M. (1990) The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev., 4, 712–727. [DOI] [PubMed] [Google Scholar]
- King R.W., Deshaies,R.J., Peters,J.M. and Kirschner,M.W. (1996) How proteolysis drives the cell cycle. Science, 274, 1652–1659. [DOI] [PubMed] [Google Scholar]
- Klempnauer K.-H., Gonda,T.J. and Bishop,J.M. (1982) Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell, 31, 453–463. [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. and Lehner,C.F. (1993) Synergistic action of Drosophila cyclins A and B during the G2–M transition. EMBO J., 12, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A., Sauer,K., Jones,L., Richardson,H., Saint,R. and Lehner,C.F. (1994) Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell, 77, 107–120. [DOI] [PubMed] [Google Scholar]
- Ku D.H., Wen,S.C., Engelhard,A., Nicolaides,N.C., Lipson,K.E., Marino,T.A. and Calabretta,B. (1993) c-myb transactivates cdc2 expression via Myb binding sites in the 5′-flanking region of the human cdc2 gene. J. Biol. Chem., 268, 2255–2259. [PubMed] [Google Scholar]
- Lam E.W. and Watson,R.J. (1993) An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J., 12, 2705–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J. and Kornbluth,S. (1996) Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Curr. Opin. Cell Biol., 8, 795–804. [DOI] [PubMed] [Google Scholar]
- Marhamati D.J, Bellas,R.E., Arsura,M., Kypreos,K.E. and Sonenshein,G.E. (1997) A-myb is expressed in bovine vascular smooth muscle cells during the late G1-to-S phase transition and cooperates with c-myc to mediate progression to S phase. Mol. Cell. Biol., 17, 2448–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses K. and Rubin,G.M. (1991) Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev., 5, 583–593. [DOI] [PubMed] [Google Scholar]
- Mucenski M.L., McLain,K., Kier,A.B., Swerdlow,S.H., Schereiner,C.M., Miller,T.A., Pietryga,D.W., Scott,W.J. and Potter,S.S. (1991) A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell, 65, 677–689. [DOI] [PubMed] [Google Scholar]
- Murray A. (1995) Cyclin ubiquitination the destructive end of mitosis. Cell, 81, 149–152. [DOI] [PubMed] [Google Scholar]
- Nakagoshi H., Kanei-Ishii,C., Sawazaki,T., Mizuguchi,G. and Ishii,S. (1992) Transcriptional activation of the c-myc gene by the c-myb and B-myb gene products. Oncogene, 7, 1233–1240. [PubMed] [Google Scholar]
- Ness S.A., Marknell,A. and Graf,T. (1989) The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell, 59, 1115–1125. [DOI] [PubMed] [Google Scholar]
- Nomura N., Takahashi,M., Matsui,M., Ishii,S., Date,T., Sasamoto,S. and Ishizaki,R. (1988) Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res., 16, 11075–11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschlager M., Janknecht,R., Krieg,J., Schreek,S. and Luscher,B. (1996) Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J., 15, 2771–2780. [PMC free article] [PubMed] [Google Scholar]
- Ogata K., Morikawa,S., Nakamura,H., Sekikawa,A., Inoue,T., Kanai,H., Sarai,A., Ishii,S. and Nishimura,Y. (1994) Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell, 79, 639–648. [DOI] [PubMed] [Google Scholar]
- Peters C.W., Sippel,A.E., Vingron,M. and Klempnauer,K.-H. (1987) Drosophila and vertebrate myb proteins share two conserved regions, one of which functions as a DNA-binding domain. EMBO J., 6, 3085–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready D.F. (1989) A multifaceted approach to neural development. Trends Neurosci., 12, 102–110. [DOI] [PubMed] [Google Scholar]
- Ready D.F., Hanson,T.E. and Benzer,S. (1976) Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol., 53, 217–240. [DOI] [PubMed] [Google Scholar]
- Reiss K., Travali,S., Calabretta,B. and Baserga,R. (1991) Growth regulated expression of B-myb in fibroblasts and hematopoietic cells. J. Cell Physiol., 148, 338–343. [DOI] [PubMed] [Google Scholar]
- Roussel M., Saule,S., Lagrou,C., Beug,H., Graf,T. and Stehelin,D. (1979) Three new types of viral oncogene of cellular origin specific for haematopoietic cell transformation. Nature, 281, 452–455. [DOI] [PubMed] [Google Scholar]
- Sakura H., Kanei-Ishii,C., Nagase,T., Nakagoshi,H., Gonda,T.J. and Ishii,S. (1989) Delineation of the three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc. Natl Acad. Sci. USA, 86, 5758–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville M.K. and Watson,R.J. (1998) The cell-cycle regulated transcrip tion factor B-Myb is phosphorylated by cyclin A/Cdk2 at sites that enhance its transactivation properties. Oncogene, 17, 2679–2689. [DOI] [PubMed] [Google Scholar]
- Sigrist S., Jacobs,H., Stratmann,R. and Lehner,C.F. (1995) Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3. EMBO J., 14, 4827–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitzmann J., Noben-Trauth,K., Kamano,H. and Klempnauer,K.-H. (1996) Expression of B-Myb during mouse embryogenesis. Oncogene, 12, 1889–1894. [PubMed] [Google Scholar]
- Stern J.B. and Smith,K.A. (1986) Interleukin-2 induction of T-cell G1 progression and c-myb expression. Science, 233, 203–206. [DOI] [PubMed] [Google Scholar]
- Stern B., Ried,G., Clegg,N., Grigliatti,T.A. and Lehner,C.F. (1993) Genetic analysis of the Drosophila cdc2 homolog. Development, 117, 219–232. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Patestos,N.P., Maekawa,T. and Ishii,S. (1999) B-myb is required for inner cell mass formation at an early stage of development. J. Biol. Chem., 274, 28067–28070. [DOI] [PubMed] [Google Scholar]
- Tanikawa J., Yasukawa,T., Enari,M., Ogata,K., Nishimura,Y., Ishii,S. and Sarai,A. (1993) Recognition of specific DNA sequences by the c-myb proto-oncogene product: role of three repeat units in the DNA-binding domain. Proc. Natl Acad. Sci. USA, 90, 9320–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. and Pfeifle,C.A. (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma, 98, 81–85. [DOI] [PubMed] [Google Scholar]
- Thomas B.J., Gunning,D.A., Cho,J. and Zipursky,L. (1994) Cell cycle progression in the developing Drosophila eye: roughex encodes a novel protein required for the establishment of G1. Cell, 77, 1003–1014. [DOI] [PubMed] [Google Scholar]
- Thomas J.B., Crews,S.T. and Goodman,C.S. (1988) Molecular genetics of single-minded locus: a gene involved in the development of Drosophila nervous system. Cell, 52, 133–141. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. (1988) Cellular interactions in the developing Drosophila eye. Development, 104, 183–193. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. and Ready,D.F. (1987) Neuronal differentiation in the Drosophila ommatidium. Dev. Biol., 120, 366–376. [DOI] [PubMed] [Google Scholar]
- Toscani A., Mettus,R.V., Coupland,R., Simpkins,H., Letvin,J., Orth,J., Hatton,K.S. and Reddy,E.P. (1997) Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature, 386, 713–717. [DOI] [PubMed] [Google Scholar]
- Trauth K., Mutschler,B., Jenkins,N.A., Gilbert,D.J., Copelend,N.G. and Klempnauer,K.-H. (1994) Mouse A-myb encodes a trans-activator and is expressed in mitotically active cells of the developing central nervous system, adult testis and B lymphocytes. EMBO J., 13, 5994–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston K. and Bishop,J.M. (1989) Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell, 58, 85–93. [DOI] [PubMed] [Google Scholar]
- Xu T. and Rubin,G.M. (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development, 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Ziebold U. and Klempnauer,K.-H. (1997) Linking Myb to the cell cycle: cyclin-dependent phosphorylation and regulation of A-Myb activity. Oncogene, 15, 1011–1019. [DOI] [PubMed] [Google Scholar]