Abstract
The vir genes of Agrobacterium tumefaciens tumor-inducing (Ti) plasmids direct the transfer of oncogenic portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells (T-DNA) into plant cells and are coordinately induced by plant-released phenolic chemical signals. We have used DNA microarrays, representing all genes of the octopine- and nopaline-type Ti plasmids, to identify all Ti-plasmid-encoded genes in the vir regulons of both plasmids. Acetosyringone (AS) induced the expression of all known members of the vir regulons, as well as a small number of additional genes. Unexpectedly, AS also caused a modest induction of virtually every Ti plasmid gene. This suggested that the copy number of the Ti plasmid might increase in response to AS, a hypothesis confirmed by DNA dot blotting. VirA and VirG were the only Vir proteins required for this copy number increase. Promoter resections and primer extension analysis of the repABC promoter region showed that expression of the promoter closest to repA (promoter P4) was induced by AS. We also identified a sequence resembling a consensus VirG-binding motif ≈70 nucleotides upstream from the P4 transcription start site. Mutating this sequence blocked the AS-induced copy number increase of a RepABC-dependent miniplasmid, indicating that phospho-VirG increases copy number solely by enhancing repABC expression.
Keywords: agrobacterium, microarrays, transcriptional profiling
Many microbes that associate with plant or animal hosts coordinately regulate large sets of genes required for these associations and induce expression of these genes in response to host-released chemical signals. For example, Sinorhizobium meliloti induces expression of the nod regulon in response to flavonoids released from host legumes (1), whereas Vibrio cholerae induces expression of several toxins and adhesins in response to bile salts released into the gut of human or animal hosts (2). This phenomenon is also well illustrated by the plant pathogen Agrobacterium tumefaciens. The entire set of vir genes, whose products transfer fragments of oncogenic portion of the tumor-inducing (Ti) plasmid that is transferred to plant cells (T-DNA) from the large Ti plasmid to the nuclei of host cells, is induced by chemical signals released from host plants (3). The primary signal molecules that stimulate expression of the vir regulon are phenolic compounds [including acetosyringone (AS), which is widely used in many laboratories as a vir gene inducer], particular monosaccharides, and extracellular acidity (4).
The expression of vir genes is controlled by the periplasmic sugar-binding protein ChvE, the transmembrane histidine protein kinase VirA, and the cytoplasmic response regulator VirG (4). Phospho-VirG activates expression of at least 14 vir operons, containing a total of 30 genes (3, 5, 6) on the octopine-type Ti plasmid, whereas far fewer VirG-inducible genes have been identified on the nopaline-type Ti plasmid (7). The products of some vir genes play direct roles in T-DNA transfer, whereas the products of other vir genes are not required for tumorigenesis and probably play other roles during the initial stages of infection (8). To date, all genes known to be in the vir regulon are located on Ti plasmids. It is quite plausible that additional members of this regulon remain to be described. It is also quite possible that VirG-regulated genes may exist on replicons other than the Ti plasmid (9).
Ti plasmids are generally referred to by the opines they can catabolize, and the two best studied are often referred to as the nopaline-type plasmid originally found in strain C58 and the octopine-type Ti plasmid, which is found in virtually identical forms in strains such as A6, B6, 15955, Ach5, and R10. All known Ti plasmids contain (i) one or more T-DNAs, (ii) the vir genes, which direct the transfer of T-DNA into plant cells, (iii) a set of tra and trb genes, which direct the conjugation of the Ti plasmid, (iv) a large number of genes that direct the uptake and catabolism of nutrients called opines that are released from plant tumors, and (v) a repABC operon, which directs vegetative plasmid replication and partitioning (8). Similar repABC operons are widespread among the plasmids and some chromosomes of α-proteobacteria, but their regulation is largely unexplored (10-12).
In this study, we have used DNA microarrays to identify all members of the vir regulons that lie on the two most widely studied Ti plasmids. These microarrays contained all ORFs of the two Ti plasmids as well as ≈20 chromosomal genes thought to be constitutively expressed and several members of the putative SOS and heat-shock regulons. It seemed conceivable that either or both of these regulons might be AS-inducible, because phenolics are somewhat toxic at the concentrations used (13), and the T-DNA is processed to a single-stranded form (14) that might possibly cause SOS induction (15). As described below, these transcriptional profiling experiments confirmed the induction of virtually all known members of the vir regulons on the Ti plasmids and identified a small number of previously undescribed members. They also led to the discovery that VirA and VirG directly activate expression of the repABC operon, causing an increased copy number of this plasmid in response to host-released chemical signals.
Materials and Methods
Design of Oligonucleotides for PCR Amplification. Strains and plasmids used in this study are described in Table 2, which is published as supporting information on the PNAS web site. Oligonucleotides were designed by using the primer3 program (16). We used default settings except for the following parameters: product size range, 501-900 (most preferred) or 300-500 (acceptable) and %GC, 40-60; maximum polyX, 3; and CG clamp, 1. The primers were synthesized by IDT Technologies (Coralville, IA) and are listed in Table 3, which is published as supporting information on the PNAS web site. PCR reactions were carried out by using genomic DNA from derivatives of strains R10 or C58 as templates and Platinum TaqDNA polymerase (Invitrogen). PCR products were purified by using 96-well multiscreen filter plates (Millipore).
Spotting PCR Products onto Glass Slides. Purified PCR products were transferred to 384-well plates, and water was removed by evaporation. DNA fragments were resuspended in 3×SSC buffer supplemented with 1.5 M betaine (17) and spotted on glass slides coated with aminosilane (CMT-GAPSII, Corning). Each DNA sample was spotted three times per slide by using a MicroGrid II DNA Arrayer (Matrix Science, Boston). DNA was crosslinked to the slides using heat and UV irradiation (18).
Bacterial Culturing and RNA Extraction. Strains were cultured for 24 h in acidified AB minimal medium (19) in the presence or absence of 100 μM AS. The initial OD600 of each culture was 0.25, and cultures were diluted as needed to keep the OD600 at or below 0.5. Two milliliters of the resulting cultures were mixed with 4 ml of RNA Protect Bacteria Reagent (Qiagen, Valencia, CA) and processed as recommended by the manufacturer. RNA was purified by using RNeasy mini columns (Qiagen). Before elution, RNA was treated with DNase I to remove contaminating DNA. Integrity of RNA was monitored by agarose gel electrophoresis.
cDNA Synthesis and Hybridization. Fluorescent cDNA probes were synthesized, purified, and used to probe microarrays following a published procedure (18). Briefly, RNA samples were treated with Superscript II reverse transcriptase (Invitrogen) in the presence of dNTPs (Invitrogen), aminoallyl-dUTP (Sigma), and random hexamers (Invitrogen). The resulting cDNA was purified with PCR purification columns (Qiagen), coupled to either Cy3 or Cy5 dyes by incubation with Cy3- or Cy5-NHS (Amersham Pharmacia Biosciences) at pH 9.3, and purified again with PCR purification columns. cDNA concentrations and efficiency of dye incorporation were measured by using a NanoDrop ND-1000 fluorescence spectrophotometer (NanoDrop Technologies, Wilmington, DE). Glass-slide microarrays were incubated in prehybridization buffer (1% BSA/5 × SSC/0.1% SDS) at 42°C for 2 h to decrease nonspecific binding of the probes. The slides were then hybridized with the probes in hybridization buffer (5 × SSC/0.1% SDS/50% form-amide) at 42°C overnight and washed sequentially with a buffer containing 1 × SSC and 0.2% SDS, at 50°C, then a buffer containing 0.1 × SSC and 0.2% SDS, at 22°C and then washed twice with a buffer containing 0.1 × SSC buffer at 22°C. Slides were analyzed by using an Axon GenePix 400B scanner (Axon Instruments, Union City, CA). Tagged image file format (TIFF) images (16 bit) were analyzed by using the scanalyze 2.50 program (http://rana.lbl.gov/EisenSoftware.htm). A normalization coefficient was computed by averaging the signal intensity ratio of 21 chromosomally encoded genes on each array. Four replicate experiments (with independent bacterial culturing, RNA preparation, cDNA probe synthesis, dye coupling, and hybridizations) were performed with dyes swapped in two trials for each strain.
DNA Dot Blotting. Strains were cultured for 24 h in the presence or absence of 100 μM AS and harvested at midexponential phase (OD600 = 0.4). Cells were resuspended in TE buffer (10 mM Tris·HCl/1 mM EDTA, pH 8.0) and lysed by incubation at 55°C with 1.25% SDS/0.6 M NaCl/0.6 mg/ml proteinase K for 30 min. The lysate was extracted with phenol-chloroform, and the nucleic acids were ethanol-precipitated and resuspended in TE buffer supplemented with RNase (20 μg/ml). DNA was purified, and 3-fold serial dilutions were spotted onto Zeta-Probe Blotting membranes (Bio-Rad), using a Minifold I Dot Blot System (Schleicher & Schuell). Each filter was probed with radiolabeled PCR products containing the virD5, bapA, kdpD, or rpoD genes to detect the Ti plasmid, pAtC58, the linear chromosome, and the circular chromosome, respectively. Radioactivity was quantified by using a Storm B840 Phosphorimager and imagequant software (Molecular Dynamics).
Primer Extension Analysis, Plasmid Copy Number Estimation, and S1 Nuclease Protection Assays. The transcription start site of the AS-inducible promoter was identified by primer extension analysis and oligonucleotide TP29 (5′-AGCTCATATCAGTTTCCTGTCAGTT-3′) (11). The copy number of a repABC-dependent plasmid was estimated by using a published procedure (11), except that the strain contained pHC090, which expresses VirA and VirG, and was treated with AS for 24 h. The copy number of the plasmids was determined by gel electrophoresis after digestion with KpnI. Nuclease S1 protection assays to quantitate mRNA levels were carried out by using 5′ radiolabeled oligonucleotides, as described in Table 2 (20).
Results
Identification of the AS Stimulons of Two Ti Plasmids. In an effort to identify the complete set of vir genes of the Ti plasmids pTiA6NC and pTiC58, we constructed microarrays containing all known or predicted Ti plasmid genes (21, 22). These microarrays also contained 36 additional genes encoded on circular and linear chromosomes. Of these, eight were judged as likely to be part of the heat-shock regulon, and seven were thought likely to be part of the SOS regulon. The remaining 21 genes were chosen as probable constitutive controls whose expression was unlikely to be affected by AS.
Strain A348(pCC113)(pSM243cd) was used to identify the octopine Ti plasmid vir regulon, whereas strain C58(pCC113)(pSM243cd) was used to identify that of the nopaline Ti plasmid. Plasmid pCC113 contains the virG gene of pTiBo542 (the so-called “supervir” plasmid) and was provided to enhance the efficiency of vir gene induction (23). Plasmid pSM243cd contains a virB1-lacZ fusion and facilitates the monitoring of vir gene induction (24). Both strains were cultivated in acidified AB minimal medium containing or lacking 100 μM AS for 24 h, assayed for β-galactosidase activity, and used to purify total RNA for transcriptional profiling analysis. AS slightly impaired the growth rate of this strain, as we have seen in the past (data not shown). The results are summarized in Fig. 2 and Tables 4 and 5, which are published as supporting information on the PNAS web site.
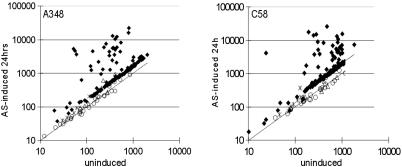
Fig. 2.
Transcription profile of AS-induced and uninduced cultures of A348(pCC113)(pSM243cd) (Left) and C58(pCC113)(pSM243cd) (Right). Ti plasmid-encoded genes are indicated by using diamonds, whereas genes thought to be members of the putative SOS or heat-shock regulons are indicated by using stars and triangles, respectively, and other non-Ti plasmid-encoded genes are indicated by using circles.
A number of observations were made. First, all known members of the AS stimulon of pTiA6NC (virB, -C, -D, -E, -F, -G, -H, -J, -K, -P, and -R) showed an induction ratio of at least 2.5-fold (Table 4). The virA gene was induced only 2.2-fold, confirming other data that this gene is induced rather weakly (25). Two strongly inducible genes were found in this plasmid, designated ysf and ysg, both of which are small genes and do not resemble any sequenced genes in public databases. Unexpectedly, traM was induced 2.6-fold. This gene encodes an antiactivator of the LuxR-type regulator TraR (26-28), suggesting that phospho-VirG may block TraR function via TraM.
In pTiC58, the virA, -B, -C, -D, -E, -G, and tzs operons were already known to be induced by AS (8), and all were observed in this study to be induced at least 2.5-fold (Table 5). Three additional genes (virH, virF, and virK) were suspected to be part of this stimulon, because orthologous genes of the octopine-type plasmid are AS inducible (5, 24), and all three were observed to be strongly induced. The traM gene was moderately induced (2.5-fold), like the orthologous gene of pTiA6.
Several previously uncharacterized genes were found to be AS-stimulated, including atu6155, atu6157, atu6158, atu6162, atu6163R, and virE0. Of these, atu6155 and atu6157 lie in a probable operon with virK (Fig. 1). The atu6158, atu6162, atu6163R, and tzs genes appear to be expressed as monocistronic transcripts. An ORF designated atu6163 (22) is complementary to atu6163R and was not detectably transcribed (see below). The virE0 gene may lie in an operon with virE1, virE2, and virE3, although the intergenic region between it and virE1 is rather large (115 nucleotides). Atu6162 strongly resembles several conserved hypothetical genes found in other members of the Rhizobiaceae. Atu6158 resembles genes related to capsule biosynthesis. The other three inducible genes do not resemble any gene in the public database. We searched the DNA sequences of both Ti plasmids for sequences similar to the consensus VirG-binding site (29). One or more of these sequences was found directly upstream of virtually every AS-induced operon (Tables 6 and 7, which are published as supporting information on the PNAS web site).
Fig. 1.
Genetic maps of the vir regions of the octopine- (A) and nopaline-type (B) Ti plasmids. Genes shown in red are members of the vir regulon, whereas genes shown in black are IS elements, and genes shown in gray encode uncharacterized proteins. Horizontal arrows indicate known or suspected operons.
The third conclusion from these studies is that recA and uvrA, which are likely to lie in the SOS regulon, appeared to be slightly induced by AS in both strains (1.7-fold for recA, 1.5-fold for uvrA in A348, 2.7-fold for recA, and 1.5-fold for uvrA in C58), whereas other putative SOS genes were not induced. Among likely heat-shock genes, dnaK was slightly induced in both strains (1.7-fold in A348 and 1.4-fold in C58), whereas the groESL operon was weakly induced in the A348 derivative but not induced in the C58 strain.
The fourth and possibly most exciting finding is that virtually all genes of both Ti plasmids appear slightly induced by AS, when normalized against the 21 chromosomal genes that we judged likely to be constitutively expressed (Fig. 2). Induction of non-vir genes of pTiA6 was 1.5- to 1.7-fold, whereas induction of pTiC58 genes was 1.8- to 2.0-fold (Fig. 2). This finding is consistent with an older report that the copy number of the Ti plasmid was increased upon cocultivation with tobacco protoplasts (30) and is considered further below.
Confirmation of Induction Using S1 Nuclease Protection Assays. To confirm the induction of all new members of the two vir regulons, we tested induction of a total of 33 genes by nuclease S1 protection assays (20). In these assays, a 5′-radiolabeled oligonucleotide is protected from nuclease digestion by hybridization to the complementary mRNA, thus allowing quantitation of that mRNA. All new members of the two AS stimulons described above were confirmed to be AS-inducible in these assays (Fig. 3 and Fig. 10, which is published as supporting information on the PNAS web site). We also used these assays to test induction of several members of the heat-shock and SOS regulons by AS. The genes shown to be slightly induced in microarray experiments seemed to be slightly induced again in this assay when normalized against the rpoA and fusA genes, which were judged likely to be constitutively expressed. In future studies, it will be interesting to test whether expression of the vir regulon is required for this modest induction of these genes.
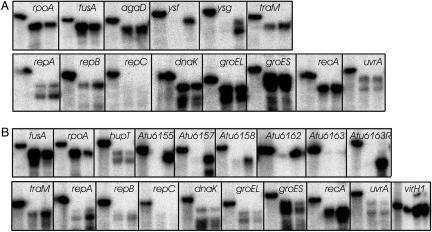
Fig. 3.
S1 nuclease protection assays for A348 (A) and C58 (B). In each panel, the left lane shows the radio-labeled oligonucleotide in the absence of nuclease digestion, while the middle and right lanes show nuclease-resistant oligonucleotides after hybridization with mRNA from uninduced or AS-induced cultures, respectively. The rpoA and fusA genes are chromosomally encoded and judged likely to be constitutively expressed, whereas agaD and hupT are Ti plasmid-encoded and serve as controls for Ti plasmid genes that are not directly VirG-regulated.
Our observation that all Ti plasmid genes are slightly induced by AS prompted us to test the expression of the repA, repB, and repC genes using nuclease protection assays. The repA and repB genes of both plasmids were significantly induced by AS (Fig. 3). In contrast, expression of repC was not detectable. We have shown elsewhere that all three genes are expressed as an operon (10), and that repC expression is down-regulated by a small antisense RNA molecule transcribed from the gene that lies between repB and repC (12). This antisense RNA molecule probably accounts for the lack of detectable repC expression. Looking back at the microarray data, expression of the repABC operon may have been elevated by AS slightly more than that of other Ti plasmid genes, although in all cases, induction was <2.5-fold. This apparently poor induction (Table 4) is probably due to the weak expression of these genes, which might have masked their true level of induction.
The Copy Number of the Ti Plasmid Is Increased by AS. Under normal laboratory growth conditions, the Ti plasmid copy number is approximately one per chromosome (31). Because the repABC operon is induced by AS, and because induction of this operon by another stimulus increases Ti plasmid copy number (10), we hypothesized that AS might increase the copy number of this plasmid. To test this, we used DNA dot blots to compare the relative abundance of the four replicons of strain A348(pCC113)(pSM243cd) and C58(pCC113)(pSM243cd) with or without AS induction. The copy number of both Ti plasmids was increased ≈4-fold by AS induction when compared with the circular or linear chromosomes (Fig. 4). Very similar results were obtained by using a strain containing the octopine-type virG gene rather than the “supervir” virG gene of pCC113 (data not shown). Interestingly, the copy number of the cryptic plasmid pAtC58 also appeared to increase by AS, although this increase was <2-fold (Fig. 4).
Fig. 4.
DNA dot blots of strains A348(pCC113)(pSM243cd) (Upper) and C58(pCC113)(pSM243cd) (Lower) cultured in the presence or absence of 100 μM AS. Genomic DNA samples were serially diluted in 3-fold increments and spotted onto nylon membranes. Membranes were hybridized with 32P-end-labeled probes specific for each replicon, which were synthesized by PCR on rpoD, kdpD, bapA, and virD5 genes by using oligonucleotides described in Table 3 and radiolabeled by treatment with EcoRI and the Klenow fragment of DNA polymerase in the presence of [α-32P]-dATP.
AS-Enhanced Ti Plasmid Copy Number Requires VirA and VirG. In an effort to determine how AS enhances the Ti plasmid copy number, we tested a collection of strains having mutations in various vir operons for defects in this phenomenon. Mutations in virA or virG abolished the copy number increase, whereas mutations in virB, virD, virE, virH, virC, virK, virL, virM, virP, or virR had little or no effect (Table 1). Because the only vir genes required for enhanced copy number were virA and virG themselves, we hypothesized that phospho-VirG might directly activate the repABC operon, and that this activation might cause the observed copy number increase.
Table 1. AS-elevated copy numbers of Ti plasmids with vir mutations.
| vir mutants | Copy number change |
|---|---|
| A348 (wild type) | 3.31 |
| 226MX (virA) | 0.95 |
| 27MX (virB) | 2.17 |
| 365MX (virC) | 2.56 |
| 304MX (virD) | 1.93 |
| 358MX (virE) | 2.04 |
| 363MX (virG) | 0.99 |
| 219MX (virH) | 4.05 |
| VIK27 (virK) | 2.29 |
| VIK22 (virL) | 3.04 |
| VIK21 (virM) | 2.98 |
| VIK40 (virP) | 3.16 |
| VIK41 (virR) | 3.29 |
To determine whether phospho-VirG regulates repABC transcription, we introduced a plasmid that expresses virA and virG derived from the octopine-type Ti plasmid (pHC090) into Ti plasmid-less strains with various plasmid-borne rep-lacZ fusions. In plasmids containing the promoters P1-P4 of repABC operon (11), AS induced the expression of repA, repB, and repC (data not shown). Importantly, these fusions were constructed by using a plasmid vector whose copy number is not affected by AS.
To further localize the sites required for AS-inducible expression of the rep operon, we constructed a set of nested deletions of the region containing promoters P1-P4, each fused to lacZ. Most fusions showed AS-induced levels of β-galactosidase activity (Fig. 5). However, plasmids pHC3FS and pHC4FS did not show any induction, whereas induction of pKP9FS was somewhat weaker than that of other plasmids. Full induction required a region containing 130 nucleotides upstream of the repA translation start site, whereas no induction was detected with a plasmid having only 113 nucleotides of upstream DNA. Consistent with the resection data, a DNA sequence weakly similar to the vir box consensus motif was found between nucleotides -123 and -110 (Fig. 6).
Fig. 5.
Localization of the AS inducible rep promoter. Each plasmid contains repC-lacZ translational fusions and various amounts of rep promoter DNA or rep promoter-lacZ transcriptional fusions and were introduced into strain NTL4(pHC090), a Ti plasmid-less strain with a plasmid expressing virA and virG from their native promoters. Base positions at the endpoints of resections refer to distances from the repA gene translation start site. The resulting strains were cultured overnight in the presence or absence of 100 μM AS in acidified AB minimal medium (pH 5.5) and tested for β-galactosidase-specific activity (33).
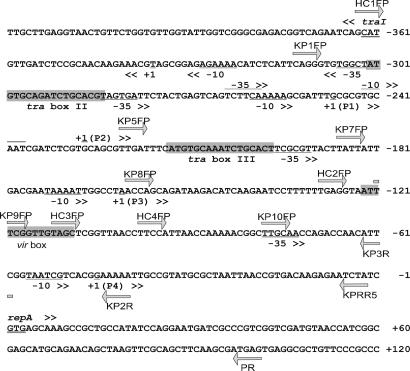
Fig. 6.
DNA sequence of traI-repA intergenic region. Arrows indicate the 5′ ends of oligonucleotides used for PCR amplification. Nested deletions of the 5′ end of this region were made by using reverse primer PR and various forward primers, as indicated, whereas nested deletions of the 3′ end were made by using forward primer KP1FP and various reverse primers, as indicated. The locations of promoters P1-P4, two tra boxes, and the putative vir box are indicated. The translation start sites of the divergent repA and traI genes are double-underlined.
The downstream end of this AS-inducible promoter was defined by using pKP2R and pKP3R, because the first was inducible, whereas the latter was not (Fig. 5B). This indicates that all sequences required for this inducible promoter lie at least 41 nucleotides upstream of repA. The region between 130 and 41 nucleotides upstream of repA contains just one known promoter, designated P4 (11). To confirm that VirA and VirG up-regulate this promoter, we carried out primer extension assays by using mRNA from AS-induced or uninduced cultures of strain NTL4(pHC090)(pKPRR5). The results showed increased transcription from promoter P4 upon AS induction (Fig. 7), demonstrating that this promoter is AS-inducible. This promoter was previously demonstrated to be autorepressed by the RepA and RepB proteins (11).
Fig. 7.
Primer extension assay using mRNA from strain NTL4(pHC090)(pKPRR5) after overnight incubation in the presence or absence of 100 μM AS. Two radiolabeled oligonucleotide primers were added, one of which hybridizes to repA mRNA, and the other of which hybridizes to the lacZα gene (of pHC090), which serves as a constitutive control.
Phospho-VirG Enhances the Ti Plasmid Copy Number Solely by Activating Promoter P4. The results described above show that phospho-VirG activates transcription of P4 but do not prove that this activation is the sole cause of the AS-induced increase in Ti plasmid replication. To test this, we reconstituted this system using a minimal repABC-dependent replicon (pKP23) and a second plasmid expressing VirA and VirG (pHC090). The copy number of pKP23 increased ≈4-fold upon AS induction (Fig. 8). This increase required VirA and/or VirG, because it was abolished in a strain that lacked pHC090 (data not shown). We identified a DNA sequence, centered 116 nucleotides upstream of repA, that resembles the consensus VirG-binding site (Fig. 8). This region is required for AS inducibility of the repABC operon. We disrupted this putative VirG-binding site by altering three conserved nucleotides and found that the copy number of the resulting plasmid, pHC122, was not enhanced by AS (Fig. 8). We also altered five nucleotides in this sequence to create a consensus VirG-binding site. The copy number of the resulting plasmid, pHC121, was enhanced by AS slightly more than the wild type (Fig. 8). These data indicate that phospho-VirG binding to a site upstream of P4 is required for enhanced copy number, which in turn indicates that phospho-VirG enhances copy number solely via its effects on repABC transcription.
Fig. 8.
Increase in copy number of repABC-dependent plasmids by phosphoVirG. Strain NTL4(pHC090)(pKP23) was incubated overnight in the presence or absence of 100 μM AS. Purified plasmid DNA was digested with KpnI, which linearizes the minireplicons (upper band of each lane) and cuts pHC090 at two sites, generating two lower bands of each lane. The two pHC090-derived fragments were used to normalize the band intensities of repABC-dependent plasmids. Plasmids pHC121 and pHC122 are derivatives of pKP23 with the indicated alterations in the vir box motif.
Discussion
One of the goals of this study was to attempt to identify all Ti plasmid-encoded members of the vir regulons of pTiA6NC and pTiC58. The core set of vir genes that are essential for tumorigenesis are highly conserved in sequence and order between these plasmids (8, 21, 22), and we confirmed older data that each of these genes is induced by AS. In addition to these genes, both plasmids have a large number of members of this regulon that are not essential for tumorigenesis and are not conserved. For example, in pTiA6NC, virM, -L, -K, -D3, -D5, -E3, -P, and -R are inducible by AS but not essential for tumorigenesis (8). Of these, virM, -L, -P, and -R are not conserved between the two Ti plasmids. We also identified two new members of the pTiA6NC vir regulon (ysf and ysg) that are linked to other vir genes. The roles of these genes in tumorigenesis have not been determined, but neither is conserved in pTiC58, suggesting that they are not essential for tumorigenesis.
A larger number of AS-inducible genes were discovered in the nopaline-type Ti plasmid, partly because such genes had not previously been sought as rigorously as in the octopine-type plasmid. These include Atu6155, Atu6157, Atu6158, Atu6162, and Atu6163R, in addition to three other operons that were anticipated to be induced, because their orthologs on pTiA6NC are inducible (virH, virF, and virK). That so many members of this regulon are not found in the octopine-type Ti plasmid suggests that wound-released chemical signals elicit slightly different responses from strains carrying these different plasmids, suggesting they have slightly different strategies for colonizing host plants.
Our finding that traM is induced by AS was of considerable interest, because TraM interacts with TraR to block expression of the tra regulon (26, 27). For several years, we and others have wondered why tra regulatory machinery includes such an antiactivator, because very few other quorum-sensing systems are controlled by analogous antiactivator proteins (32). Induction of TraM by AS predicts that vir gene induction blocks tra gene expression. Although traM induction by AS is not high, it may well suffice to block low levels of activated TraR. Such a mechanism to block simultaneous expression of the vir and tra systems might well be useful, because they are both type IV secretion systems and might well interfere with each other's function. The idea that vir gene expression blocks tra gene expression rather than the converse could be interpreted to suggest that plant transformation takes precedence over interbacterial conjugation.
Perhaps the most unexpected finding of this study is that the repABC operon was induced by phospho-VirG. That we did not detect this induction directly from the microarrays was probably due to their extremely low expression of the intact operon, due to negative autoregulation by RepA and RepB (11). This induction was clearly detected by using nuclease protection assays. A series of rep-lacZ fusions and primer extension assays clearly showed that promoter P4 is directly activated by phopho-VirG. Induced expression of repABC enhanced Ti plasmid copy number and thereby indirectly enhanced the expression of all Ti plasmid-encoded genes. Increased expression of all vir genes, as well as increased copies of the T-DNA, could lead to increased production of T-strands and of the T-strand export machinery, which could lead to enhanced levels of tumorigenesis (10).
We have previously shown (10) that the quorum-sensing regulator TraR and its cognate autoinducer also activate transcription of repABC, causing an enhanced copy number of this plasmid. In that case, induction occurs at the P1, P2, and P3 promoters. Taken together with the present study, we believe that when A. tumefaciens grows saprophytically, its Ti plasmid replicates at rates just high enough to ensure vertical transmission. In contrast, when the bacterium detects either host-released signals or quorum signals, the plasmid's copy number significantly increases by activation of repABC operon (Fig. 9). This seems appropriate, because minimizing the plasmid copy number during saprophytic growth would lessen the metabolic burden to the host bacterium. In contrast, during plant colonization, the Ti plasmid carries out numerous important functions. Increased gene expression (due to increased gene dosage) could well facilitate such activities as tumorigenesis and opine utilization.
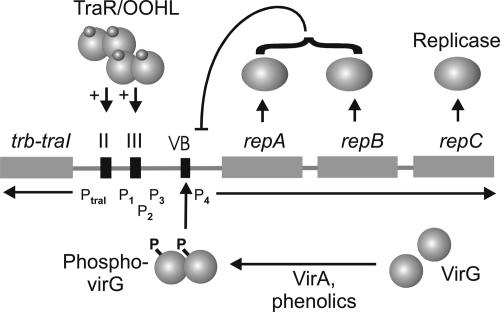
Fig. 9.
Summary of the regulation of the repABC operon. According to this model, wound-released signals act through VirA to phosphorylate VirG, which activates promoter P4, whereas 3-oxo-octanoylhomoserine lactone-TraR complexes activate promoters P1-P3 (10). All four promoters are autorepressed by RepA/RepB complexes, which bind to an operator directly downstream of promoter P4 (11).
At present, there is no convincing evidence that VirG regulates any gene localized on either of the two chromosomes or on the cryptic plasmid. Because many natural isolates of A. tumefaciens lack Ti plasmids, it would make sense that chromosomal genes not rely on VirA or VirG for their regulation. However, our finding that the copy number of pAtC58 (also referred to as the cryptic plasmid) may also be slightly enhanced, if confirmed, would suggest that at least these two plasmids share common regulatory elements. The use of whole-genome microarrays will facilitate the search for VirG-regulated genes on other replicons.
Supplementary Material
Acknowledgments
We are grateful to members of our laboratory for helpful discussions and suggestions and to Derek Wood and Eugene Nester for permission to describe the consensus vir box. We are also grateful to Paul Debbie and the Center for Gene Expression Profiling (Cornell University) for printing microarray slides. This work was supported by the Monsanto Company and the National Institutes of Health (Grant GM41892).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ti, tumor-inducing; AS, acetosyringone; T-DNA, portion of the Ti plasmid that is transferred to plant cells.
References
- 1.Barnett, M. J., Toman, C. J., Fisher, R. F. & Long, S. R. (2004) Proc. Natl. Acad. Sci. USA 101, 16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hung, D. T. & Mekalanos, J. J. (2005) Proc. Natl. Acad. Sci. USA 102, 3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson, T. M. & Das., A. (1998) in The Rhizobeaceae: Molecular Biology of Model Plant-Associated Bacteria, eds. Spaink, H. P., Kondorosi, A. & Hooykaas, P. J. J. (Kluwer, Dordrecht, The Netherlands), pp. 265-279.
- 4.Brencic, A., Augert, E. R. & Winans, S. C. (2005) Mol. Microbiol. 57, 1522-1531. [DOI] [PubMed] [Google Scholar]
- 5.Kalogeraki, V. S. & Winans, S. C. (1998) J. Bacteriol. 180, 5660-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalogeraki, V. S., Zhu, J., Stryker, J. L. & Winans, S. C. (2000) J. Bacteriol. 182, 1774-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogowsky, P. M., Close, T. J., Chimera, J. A., Shaw, J. J. & Kado, C. I. (1987) J. Bacteriol. 169, 5101-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu, J., Oger, P. M., Schrammeijer, B., Hooykaas, P. J., Farrand, S. K. & Winans, S. C. (2000) J. Bacteriol. 182, 3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engstrom, P., Zambryski, P., Montagu, M. V. & Stachel., S. (1987) J. Mol. Biol. 197, 635-645. [DOI] [PubMed] [Google Scholar]
- 10.Pappas, K. M. & Winans, S. C. (2003) Mol. Microbiol. 48, 1059-1073. [DOI] [PubMed] [Google Scholar]
- 11.Pappas, K. M. & Winans, S. C. (2003) Mol. Microbiol. 49, 441-455. [DOI] [PubMed] [Google Scholar]
- 12.Chai, Y. & Winans, S. C. (2005) Mol. Microbiol. 56, 1574-1585. [DOI] [PubMed] [Google Scholar]
- 13.Bais, H. P., Prithiviraj, B., Jha, A. K., Ausubel, F. M. & Vivanco, J. M. (2005) Nature 434, 217-221. [DOI] [PubMed] [Google Scholar]
- 14.Tzfira, T., Rhee, Y., Chen, M. H., Kunik, T. & Citovsky, V. (2000) Annu. Rev. Microbiol. 54, 187-219. [DOI] [PubMed] [Google Scholar]
- 15.Shinagawa, H. (1996) EXS 77, 221-235. [DOI] [PubMed] [Google Scholar]
- 16.Rozen, S. & Skaletsky, H. (2000) Methods Mol. Biol. 132, 365-386. [DOI] [PubMed] [Google Scholar]
- 17.Diehl, F., Grahlmann, S., Beier, M. & Hoheisel, J. D. (2001) Nucleic Acids Res. 29, E38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hegde, P., Qi, R., Abernathy, K., Gay, C., Dharap, S., Gaspard, R., Hughes, J. E., Snesrud, E., Lee, N. & Quackenbush, J. (2000) Biotechniques 29, 548-562. [DOI] [PubMed] [Google Scholar]
- 19.Cangelosi, G. A., Best, E. A., Martinetti, G. & Nester, E. W. (1991) Methods Enzymol. 204, 384-397. [DOI] [PubMed] [Google Scholar]
- 20.Zhu, J. & Winans, S. C. (1998) Mol. Microbiol. 27, 289-297. [DOI] [PubMed] [Google Scholar]
- 21.Goodner, B., Hinkle, G., Gattung, S., Miller, N., Blanchard, M., Qurollo, B., Goldman, B. S., Cao, Y., Askenazi, M., Halling, C., et al. (2001) Science 294, 2323-2328. [DOI] [PubMed] [Google Scholar]
- 22.Wood, D. W., Setubal, J. C., Kaul, R., Monks, D. E., Kitajima, J. P., Okura, V. K., Zhou, Y., Chen, L., Wood, G. E., Almeida, N. F., Jr., et al. (2001) Science 294, 2317-2323. [DOI] [PubMed] [Google Scholar]
- 23.Chen, C. Y., Wang, L. & Winans, S. C. (1991) Mol. Gen. Genet. 230, 302-309. [DOI] [PubMed] [Google Scholar]
- 24.Stachel, S. E. & Zambryski, P. C. (1986) Cell 46, 325-333. [DOI] [PubMed] [Google Scholar]
- 25.Winans, S. C., Kerstetter, R. A. & Nester, E. W. (1988) J. Bacteriol. 170, 4047-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen, G., Malenkos, J. W., Cha, M. R., Fuqua, C. & Chen, L. (2004) Mol. Microbiol. 52, 1641-1651. [DOI] [PubMed] [Google Scholar]
- 27.Qin, Y., Smyth, A. J., Su, S. & Farrand, S. K. (2004) Mol. Microbiol. 53, 1471-1485. [DOI] [PubMed] [Google Scholar]
- 28.Swiderska, A., Berndtson, A. K., Cha, M. R., Li, L., Beaudoin, G. M., 3rd, Zhu, J. & Fuqua, C. (2001) J. Biol. Chem. 276, 49449-49458. [DOI] [PubMed] [Google Scholar]
- 29.Heath, J. D., Charles, T. C. & Nester, E. W. (1995) in Two-Component Signal Transduction., eds. Hoch, J. A. & Silhavy, T. J. (Am. Soc. Microbiol. Press., Washington, DC), pp. 367-385.
- 30.Veluthambi, K., Ream, W. & Gelvin, S. B. (1988) J. Bacteriol. 170, 1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallie, D. R., Zaitlin, D., Perry, K. L. & Kado, C. I. (1984) J. Bacteriol. 157, 739-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He, X., Chang, W., Pierce, D. L., Seib, L. O., Wagner, J. & Fuqua, C. (2003) J. Bacteriol. 185, 809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Laboratory Press, Plainview, NY).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.