Abstract
Menin, the product of the Men1 gene mutated in familial multiple endocrine neoplasia type 1 (MEN1), regulates transcription in differentiated cells. Menin associates with and modulates the histone methyltransferase activity of a nuclear protein complex to activate gene expression. However, menin-dependent histone methyltransferase activity in endocrine cells has not been demonstrated, and the mechanism of endocrine tumor suppression by menin remains unclear. Here, we show that menin-dependent histone methylation maintains the in vivo expression of cyclin-dependent kinase (CDK) inhibitors to prevent pancreatic islet tumors. In vivo expression of CDK inhibitors, including p27 and p18, and other cell cycle regulators is disrupted in mouse islet tumors lacking menin. Chromatin immunoprecipitation studies reveal that menin directly associates with regions of the p27 and p18 promoters and increases methylation of lysine 4 (Lys-4) in histone H3 associated with these promoters. Moreover, H3 Lys-4 methylation associated with p27 and p18 is reduced in islet tumors from Men1 mutant mice. Thus, H3 Lys-4 methylation is a crucial function of menin in islet tumor suppression. These studies suggest an epigenetic mechanism of tumor suppression: by promoting histone modifications, menin maintains transcription at multiple loci encoding cell cycle regulators essential for endocrine growth control.
Keywords: islet of Langerhans, Men1, multiple endocrine neoplasia, tumor suppressor, diabetes mellitus
Mutation of the Men1 tumor suppressor gene, which encodes the protein menin, promotes pathogenesis of type 1 multiple endocrine neoplasia (MEN1) syndrome and sporadic neuroendocrine tumors in humans (1, 2). The molecular basis for the endocrine tumor bias in MEN1 is unknown. Targeted heterozygous Men1 inactivation in mice produces a spectrum of endocrine tumors similar to those observed in human patients with MEN1 syndrome (3–5). For example, Men1+/- mice develop pancreatic islet tumors, the most common enteroendocrine tumor in human MEN1 syndrome, as well as tumors in the anterior pituitary, adrenal cortex, and the stomach. In some cases, loss of chromosome 19, which harbors Men1 in mice, was detected in islet tumors (5), consistent with loss of heterozygosity and the postulated tumor suppressor function of menin. Homozygous Men1 loss in mice leads to embryonic lethality by midgestation [embryonic day (E) 11.5–13.5] and is found associated with a variety of developmental defects, including undergrowth and defective organogenesis of the neural tube, heart, and liver (6). Thus, menin may have general roles in cell growth and development.
Menin localizes to the nucleus and regulates gene transcription (7). Recent biochemical studies demonstrate that menin associates with a nuclear protein complex that includes the trithorax group (TrxG) members MLL, MLL2, and Ash2 (8–10). This complex promotes the methylation of histone H3 at lysine 4 (Lys-4), consistent with the hypothesized role of MLL as a histone methyltransferase (HMTase; ref. 11). Site-specific histone modification is a major epigenetic mechanism for maintaining stable gene transcription in terminally differentiated cells (12, 13). Thus, menin may function as a tumor suppressor by regulating histone methylation in promoters of specific target genes that govern neuroendocrine cell growth and differentiation. However, menin-dependent HMTase activity at endocrine cell promoters has not yet been demonstrated. Thus, it is unclear whether dysregulated gene expression promotes endocrine tumors, a defining phenotype of MEN1. Moreover, it has not yet been demonstrated that menin regulates histone H3 methylation in vivo to suppress endocrine tumor formation, for example, in mouse MEN1 models.
Clues to identifying relevant genes in endocrine cells that might be regulated by menin-dependent HMTase activity come from recent studies revealing the roles of cyclin-dependent kinases (CDKs) and their inhibitors in regulating endocrine cell growth. Mice homozygous for null alleles of p18INK4C (which encodes a member of the Ink4 family of CDK inhibitors, hereafter called p18) or for p27Kip1 (which encodes a member of the Cip/Kip family of CDK inhibitors, hereafter called p27) had generalized organomegaly but did not develop spontaneous endocrine tumors until 10 months of age or later, when they developed pituitary adenomas (14–17). However, simultaneous loss of p18 and p27 was sufficient to produce a specific spectrum of endocrine tumors similar to that seen in human MEN1 and MEN2 syndromes, including tumors in the pituitary, parathyroid, thyroid, endocrine pancreas, stomach, and duodenum (18). p18 and p27 are known to inhibit functions of cell cycle regulators like CDK2 and CDK4 (19). Thus, these studies suggested that Men1 inactivation might disrupt expression of p18, p27, and other established cell cycle regulators in endocrine cells. A recent study showed that menin can regulate p18 and p27 expression by fibroblasts in vitro, but menin-dependent changes in levels of histone methylation were not reported (10). Thus, the molecular basis for menin-dependent control of cell cycle regulators remained unclear. Here, we report that menin is required to regulate histone methylation in genes encoding specific cell cycle regulators, thereby maintaining the expression of these genes in endocrine cells, and that this property is the basis for endocrine tumor suppression by menin in a mouse model of MEN1.
Materials and Methods
Generation, Genotyping, and Physiologic Assessment of Men1+/- Mice. Generation, breeding, and genotyping of mice harboring a null allele of the Men1 gene on the C57BL genetic background have been described (8). Blood glucose levels during random feeding and glucose challenge were measured as described (20, 21). Serum glucose values are represented as the mean from at least eight animals per genotype per time point, with standard error of the mean indicated by a bar. Statistical significance was established here and elsewhere by using a two-tailed t test.
Histological Analysis. Immunohistochemical analyses were performed as described (20, 21). The antisera and detection methods used here are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Isolation of Pancreatic Islet RNA, Protein, and RT-PCR Analysis. We isolated pancreatic islets by intraductal collagenase perfusion using standard methods (22). RNA was isolated from the purified islets and reverse-transcribed into cDNA by using a Retroscript Kit (Ambion, Austin, TX). RT-PCR conditions are described in Supporting Materials and Methods.
Chromatin Immunoprecipitation (ChIP) Assay. ChIP assays were performed on pancreatic islets and MIN6 cells by using a kit from Upstate Biotechnology (Lake Placid, NY). Briefly, DNA was crosslinked to protein with formaldehyde. Cellular lysates were obtained by scraping, followed by pulsed ultrasonication to shear cellular DNA. We performed overnight immunoprecipitations with the following antibodies: anti-menin (goat, 1:100 Santa Cruz Biotechnology), anti-dimethylated histone H3 Lys-4 (1:300 Upstate Biotechnology), or anti-trimethylated histone H3 Lys-4 (1:300, Upstate Biotechnology). On the next day, the crosslinks were reversed, and bound DNA was purified by phenol:chloroform extraction. We performed PCR using primers specific for p18, p27, p15, p21, and cdk4 proximal promoter sequences. Primer sequences are available upon request.
Transfections and Western Blot Studies. MIN6 murine insulinoma cells were transfected with DNA encoding wild-type menin, with DNA encoding alleles of Men1 that specify the menin missense forms A242V, L22R, and T344R (8), or with vector DNA lacking Men1 sequences. In some experiments, these cells were simultaneously transfected with an small interfering RNA (siRNA) specific for menin or a scrambled siRNA control (23). After transfection, cells were grown for 48 h before the lysates were processed for Western blotting. Blots were developed as described (24). Probes for Western blotting are listed in Supporting Materials and Methods.
Luciferase Reporter Assays. Luciferase assays were performed according to the manufacturer's specifications (Promega). Additional details are in refs. 25 and 26 and in Supporting Materials and Methods.
Results
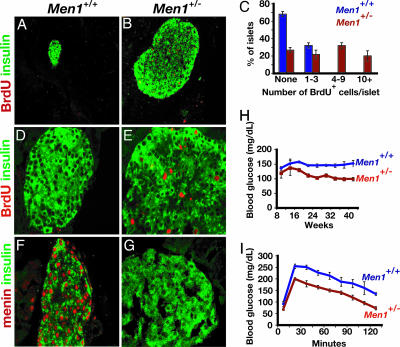
Menin Controls Pancreatic Islet Growth and Glucose Regulation. The strategy for generating mice harboring null alleles of Men1 used in this study has been reported (8), but the endocrine cell phenotypes in adult mutants from these lines have not been described. Based on our ability to purify and assess fates of islet cells, we focused our studies on pancreatic islets. Mice heterozygous for the Men1-null allele (Men1+/-) were indistinguishable from wild-type control littermates during the first several months of postnatal life. By 7 months of age, we observed several islet phenotypes in Men1+/- mice (Fig. 1). A majority of pancreatic islets appeared enlarged and hyperplastic in Men1+/- pancreata (Fig. 1 A and B) in all animals examined (n > 20). Immunostaining revealed that islet enlargement resulted from an increase in numbers of insulin+ β cells (Fig. 1 A, B, and D–G, and data not shown). Consistent with these results, we found increased Men1+/- pancreatic islet cell incorporation of bromodeoxyuridine (BrdUrd), a marker of DNA synthesis during the cell cycle S phase (Fig. 1 A–E). Islet and pancreatic architecture was not detectably altered in Men1+/- mice (data not shown). Immunohistochemistry revealed a significant reduction of menin protein in the β cells of these islets by 7 months (Fig. 1 F and G). Serial islet isolation and analyses of mRNA by RT-PCR demonstrated that levels of Men1 mRNA from Men1+/- mice were reduced before the appearance of hyperplastic islets (Fig. 2A). Reduction of islet Men1 mRNA likely reflected haploinsufficiency at earlier ages (6 weeks; Fig. 2A), but, by 18 weeks and later, we observed further reductions of islet Men1 mRNA, coinciding with the loss of detectable nuclear menin in β cells. These islet phenotypes mimic those reported in patients with MEN1 syndrome and in Men1+/- mice that developed insulin-producing tumors after loss of the other Men1 allele (3, 5, 7, 27–29). Thus, this in vivo model provided an opportunity to elucidate the basis for islet tumor formation after loss of menin expression.
Fig. 1.
Pancreatic β cell hyperplasia, increased BrdUrd incorporation, loss of menin expression, and hypoglycemia in Men1+/- mice. (A, B, D, and E) Immunostaining of BrdUrd (red) and insulin (green) in pancreatic tissue from 9-month-old Men1+/+ (A and D) and Men1+/- mice (B and E). (C) Quantification of BrdUrd+ cells in islets (n = 100) from Men1+/+ and Men1+/- mice. (F and G) Immunohistochemical detection of nuclear menin (red) and insulin (green) in pancreas from Men1+/+ (F) and Men1+/- mice (G). (H) Blood glucose level during random feeding in mice with the indicated age and genotype (wild-type or Men1+/-). n = 8 or more animals for each genotype. After 20 weeks of age, hypoglycemia in Men1+/- mice remained statistically significant at P < 0.01. (I) Glucose tolerance testing of 28-week-old wild-type (n = 11), and Men1+/- littermates (n = 10) after 14-h overnight fast. Data in C, H, and I are presented as average ± SEM. [Original magnification: ×16 (A and B) and ×63 (D–G).]
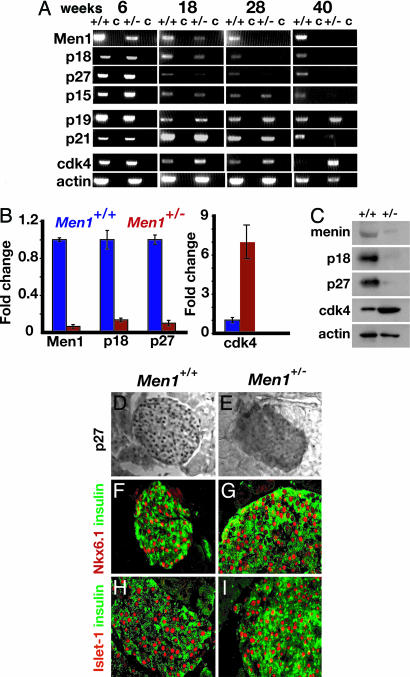
Fig. 2.
Disrupted expression of genes encoding cell cycle regulators in Men1+/- pancreatic islets. (A) Conventional RT-PCR analysis of mRNA isolated from Ficoll-gradient-purified pancreatic islets for the indicated markers. Levels of mRNA specifying p19 and actin (a loading control) are not detectably altered in Men1+/- mice. (B) Quantitative RT-PCR on islets from 40-week-old Men+/+ and Men1+/- mice for the indicated markers. (C) Western blot comparing expression of the indicated proteins in purified islets from 40-week-old Men1+/+ and Men1+/- mice. Actin serves as a loading control. (D and E) Immunohistochemical detection of nuclear p27Kip1 (black dots) reveals reduced expression in pancreatic islets of Men1+/- mice. (F and G) Nuclear expression of the transcription factor Nkx6.1 (red) by insulin+ β cells (green) is maintained in Men1+/- mice. (H and I) Nuclear expression of the transcription factor Islet-1 (red) by insulin+ β cells (green) is maintained in Men1+/- mice. [Original magnification: ×40 (D and E) and ×63 (F–I).]
Men1+/- mice had mild but significant hypoglycemia during random feeding and after overnight fasting by 6–7 months (Fig. 1 H and I). During glucose challenge, Men1+/- mice had reduced blood glucose levels compared with matched wild-type littermate controls. Thus, heterozygosity for the Men1-null allele in our animal model resulted in a dominant transmission of phenotypes at high penetrance, including loss of detectable menin expression in islet β cells, islet hyperplasia, and systemic hypoglycemia.
Menin Is Required to Maintain Islet Expression of Multiple Genes Encoding CDK Inhibitors. Prior studies showed that menin is a transcriptional regulator (8–10, 30–34), but little is known about the molecular targets of menin activity in endocrine cells, and how these targets might mediate the tumor suppressor activity of menin. Recent reports showed that mutations disrupting genes encoding cell cycle regulators, including CDK4 and the CDK inhibitors p27 and p18, are sufficient to promote hyperplasia of pancreatic islets and other endocrine organs (17, 18, 35). Thus, gene loci encoding cell cycle regulators in endocrine cells are candidate targets of menin. Consistent with this possibility, p27, p18, and other members of the Cip/Kip and Ink4 family of CDK inhibitors are expressed in postnatal mouse islets (Fig. 2A).
To test the possibility that menin regulates islet expression of cell cycle regulators, we measured gene expression by RT-PCR and immunohistochemistry in the whole pancreas and in islets isolated from Men1+/- and control littermate mice at regular intervals for 1 year. By 18 weeks, islet expression of p18 and p27 was reduced in Men1+/- mice (Fig. 2A), a reduction that became more pronounced as these mice aged (Fig. 2 A and B). Consistent with these findings, Western blots revealed reduced levels of p27, p18, and menin protein in Men1+/- mouse islets at 40 weeks (Fig. 2C), and analysis of islets in sectioned pancreata revealed reduction of nuclear p27 in Men1+/- islets (Fig. 2 D and E). By 40 weeks, in Men1+/- mice, we also observed reduction of p15 and p21Cip1/Waf1, two additional CDK inhibitors (Fig. 2A). Islet expression of the Cip/Kip member p57Kip2 was undetectable in these studies (data not shown). By contrast, expression of factors characteristic of differentiated β cells like insulin, Nkx6.1, Islet-1, Pdx1, and the Ink4 family member p19, was maintained in Men1+/- mice (Figs. 1 and 2 A and F–I, and data not shown). We also observed increased levels of CDK4 mRNA, starting at 18 weeks (Fig. 2 A and B), corroborated by Western blots showing increased CDK4 protein (Fig. 2C). Thus, reductions in islet menin expression preceded or accompanied the observed changes in islet p15, p18, p21, p27, and CDK4 expression (Fig. 2 and data not shown). Collectively, our data suggest that menin activity maintains the expression of specific genes encoding cell cycle regulators in adult pancreatic islet cells.
Menin Induces p27 and p18 Expression and Suppresses Growth of β Cells. To investigate the mechanisms underlying menin regulation of genes like p27 and p18 in differentiated endocrine cells like β cells, we tested whether menin was sufficient to induce expression of these putative target genes in β cells. MIN6 cells are a well established line derived from mouse pancreatic β cell tumors (36). Western blots revealed that MIN6 cells expressed nearly undetectable levels of menin (Fig. 4A, which is published as supporting information on the PNAS web site). Transient transfection of MIN6 cells with Men1 increased levels of menin protein and mRNA (Fig. 4 A and B). Western blots also demonstrated that increased levels of p27 and p18 protein accumulated in MIN6 cells after Men1 transfection (Fig. 4A). Consistent with these results, we observed increased expression of menin, p18, and p27 mRNA after MIN6 transfection with Men1 (Fig. 4B). Cotransfection of a Men1-specific siRNA with Men1-encoding DNA strongly inhibited expression of menin, p27, and p18 protein (Fig. 4 A and B).
To determine whether menin regulates MIN6 cell growth, we monitored rates of cell accumulation. We transfected MIN6 cells with DNA encoding IRES:GFP, or menin:IRES:GFP. As an additional control, we cotransfected cells with menin:IRES:GFP and a Men1-specific siRNA. We then measured proliferation and BrdUrd incorporation on GFP+ cells purified by FACS (see Materials and Methods). We observed that menin:IRES:GFP transfection (compared with transfection with control DNA encoding IRES:GFP) impaired MIN6 cell accumulation (Fig. 4C). Cotransfection of MIN6 cells with menin:IRES:GFP and a Men1-specific siRNA restored MIN6 cell growth (Fig. 4C). Cotransfection with menin: IRES:GFP DNA and a control siRNA did not restore MIN6 cell growth (data not shown). To assess effects of Men1 on proliferation further, we quantified BrdUrd incorporation in MIN6 cells. Nuclear labeling by BrdUrd after transfection with menin:IRES:GFP DNA was reduced compared with control MIN6 cells cotransfected with menin:IRES:GFP and a Men1-specific siRNA, or compared with MIN6 cells transfected with control DNA encoding IRES:GFP (Fig. 4D). These in vitro studies correlate well with our in vivo findings in Men1+/- islets and provide evidence that menin regulates the expression of p27 and p18, and growth of β cells.
If menin regulates growth by maintaining expression of cell cycle regulators like p18 and p27, we postulated that a subset of tumor-derived mutant menin proteins might not induce expression of these putative target loci in MIN6 cells. We analyzed levels of menin, p18, and p27 in MIN6 cells after transfection with wild-type Men1, or with patient-derived Men1 missense mutant alleles that specify amino acid substitutions, designated A242V, and L22R. In prior studies (8), the A242V mutant form lacked histone methyltransferase-reconstituting activity, distinct from the L22R form, which retained this activity. MIN6 cells expressed equivalent levels of wild-type or mutant menin after transfection (Fig. 4E). However, the level of p18 and p27 protein induced by the A242V mutant form was reduced compared with those induced by the wild-type or L22R forms of menin (Fig. 4E). Compared with controls, growth of MIN6 cells transfected with the A242V allele was unperturbed, whereas cells transfected with the L22R allele grew more slowly (Fig. 4F). Consistent with this finding, we found that the percentage of BrdUrd incorporation was greater in MIN6 cells transfected with the A242V allele than in cells transfected with the L22R allele (48 ± 1 versus 32 ± 3; n = 3, P < 0.01). Collectively, these data support the conclusion that menin maintains expression of CDK inhibitors to suppress tumor formation in pancreatic islets.
Menin Associates with Regulatory Elements in the Mouse p27 and p18 Promoters. To determine whether menin directly regulates the expression of p27 or p18, we performed ChIP assays with MIN6 cells, and with pancreatic islets isolated from Men1+/- and Men1+/+ mice. Endogenous menin levels in MIN6 cells were nearly undetectable (Fig. 4). Thus, anti-menin ChIP studies in MIN6 cells required transfection of these cells with Men1-encoding DNA (Fig. 5 B and D, which is published as supporting information on the PNAS web site, compare + and - Men1 transfection controls). This requirement for Men1 transfection facilitated comparisons of wild-type and mutant menin activity in MIN6 cells.
Anti-menin ChIP revealed that menin associates with specific regions of the mouse p18 and p27 promoters (Fig. 5 A–D). For these studies, we used a set of primers directed to overlapping portions of the 1,000-bp immediately 5′ of the p18 or p27 transcription start sites (Fig. 5 A and C). Menin associates with sequences between -750 and -300 in the p18 promoter (enriched by p18-specific primer pairs 3 and 4; Fig. 5A), and with sequences between -725 and -350 in the p27 promoter (enriched by p27-specific primer pairs 3 and 4; Fig. 5C). Similar results were obtained from ChIP studies with pancreatic islets. Menin associated with specific regions proximal to the p18 and p27 promoters, and this association was lost in islets from Men1+/- mice (Fig. 3C). Thus, menin binds to specific promoter-proximal regions of the p27 and p18 loci, arguing that menin directly regulates the expression of p27 and p18. Anti-menin antibodies did not coprecipitate sequences in p18 or p27 that flanked these regions (Fig. 5 B and D) or sequences from p15, p21Cip1/Waf1, and CDK4 controls (Fig. 6, which is published as supporting information on the PNAS web site), suggesting that regulation of p15, p21Cip1/Waf1, or CDK4 expression by menin in islets (see Fig. 2A) may not be direct.
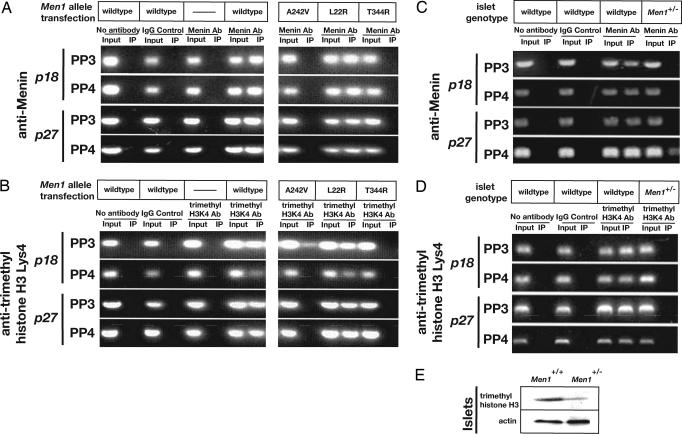
Fig. 3.
Menin promotes methylation of histone H3 associated with chromatin at the p18 and p27 genes. MIN6 cells were transfected with DNA specifying wild-type menin (wild-type), or mutant missense alleles of Men1 derived from human tumors that encode the amino acid substitutions A242V, L22R, or T344R. (A) Anti-menin ChIP was performed by using PP3 and PP4 specific for regions in the p18 promoter, or PP3 and PP4 specific for regions in the p27 promoter. Menin produced from alleles that retain histone methyltransferase activity (wild-type, L22R) associate with these promoters. (B) ChIP analysis using antibodies specific for trimethylated histone H3 Lys-4. (C and D) ChIP analysis using antibodies specific to menin and trimethylated histone H3 Lys-4 performed on isolated islets from Men1+/+ and Men1+/- mice. (E) Comparison of trimethylated histone H3 Lys-4 levels in Men1+/+ and Men1+/- islets by Western blotting. Actin protein served as a loading control.
We performed transient transfection studies to confirm that the promoter-proximal elements in p27 and p18 that associate with menin in ChIP studies mediate menin-dependent transcription (Fig. 5 E–H). Compared with luciferase expression directed from a minimal promoter (pGL), which showed no detectable menin-dependence, expression directed from a synthetic transgene comprised of 1.5 kb of p18 promoter-proximal DNA linked to luciferase or from a synthetic transgene comprised of 1.6 kb of p27 promoter-proximal DNA linked to luciferase was clearly increased by menin (Fig. 5 F and H). Dissection of the p18 regulatory region identified a minimal menin-responsive element in the region from -854 to -517 bp 5′ of an established p18 transcriptional start site (p18 Luc20; Fig. 5 E and F). Dissection of the p27 regulatory region using a similar strategy identified a menin-responsive element in the region from -936 to -502 bp 5′ of an established p27 transcriptional start site (p27 Luc55; Fig. 5 G and H). Further parsing of this p27 region into smaller elements led to reduced luciferase expression (Fig. 5H). Thus, defined promoter-proximal cis-regulatory elements immediately 5′ of known transcription start sites in p27 and p18 were sufficient for directing menin-dependent transcription. In some in vitro contexts, menin can bind directly to DNA (37). To determine whether menin might associate with a common specific sequence in p27 and p18, we performed comparisons of DNA sequences in p27 Luc55 and p18 Luc20 (see Supporting Materials and Methods). This analysis (data not shown) did not reveal evidence for specific sequences of four or more consecutive bases common to the menin-responsive elements in p27 and p18.
If association with target chromatin sequences is important for menin activity, we postulated that a subset of tumor-derived mutant menin protein would be unable to associate with cis-regulatory sequences in the p27 and p18 promoters. We performed anti-menin ChIP studies after transfection with patient-derived Men1 missense mutant alleles that specify one of three amino acid substitutions: A242V, T344R, and L22R. Both A242V and T344R forms lack HMTase-reconstituting activity (8), and both failed to permit ChIP enrichment of specific sequences from the p27 and p18 promoters. By contrast, both L22R and wild-type menin, which retain HMTase-reconstituting activity, permitted ChIP enrichment of these sequences (Fig. 3A). These results suggest that the association of menin with p27 and p18 cis-regulatory chromatin is involved in the tumor suppressor function of menin.
Menin Promotes Methylation of Histone H3 Lys-4 Associated with the p27 and p18 Loci. Menin associates with a nuclear protein complex that includes the TrxG members MLL, MLL2, and Ash2 (8–10). Association of menin with this complex promotes the methylation of histone H3 at specific residues like Lys-4, consistent with the hypothesized role of MLL as a methyltransferase (11). Methylation of H3 Lys-4 has been correlated with activated expression of Hox c8 (11) and other genes (13). Thus, we used ChIP analysis to test whether menin enhances methylation of H3 Lys-4 associated with the p27 and p18 promoter regions. ChIP assays using antibodies specific for tri-methyl H3 Lys-4 revealed a menin-dependent increase of H3 Lys-4 methylation in MIN6 cells associated with both the p27 and p18 promoters (Fig. 3B). Consistent with prior analyses demonstrating association of menin protein with these promoter sites, the PCR primers used in these trimethyl H3 Lys-4 ChIP studies spanned regions of the p27 and p18 promoters that associate with menin. Similar results were obtained in studies of pancreatic islets. H3 Lys-4 methylation associated with both the p27 and p18 promoters was readily detected in islets isolated from Men1+/+ mice, but severely reduced or undetectable in Men1+/- islet tumors lacking menin (Fig. 3D). These results suggest that menin regulates H3 Lys-4 methylation at the p27 and p18 promoters. Consistent with these data, Western blotting analysis revealed a reduction of total trimethyl-histone H3 Lys-4 levels in these islet tumors (Fig. 3D).
If menin directs histone methyltransferase activity in cis-regulatory chromatin regions governing expression of genes like p27 and p18, then we predicted that some tumor-derived mutant alleles of Men1 would fail to enrich H3 Lys-4 methylation associated with these genes. As shown in Fig. 3B, MIN6 cells transfected with the wild-type or patient-derived L22R mutant form of menin had elevated levels of trimethyl H3 Lys-4 in association with p27 and p18 chromatin. In contrast, mutations in Men1 producing the A242V and T344R forms completely failed to enrich trimethyl H3 Lys-4 in p27 and in p18 (Fig. 3B). These results are consistent with our ChIP studies demonstrating that L22R associates with the p27 and p18 promoters (Fig. 3A) and our analyses showing increased expression of p27 and p18 protein in MIN6 cells transfected with the L22R allele of Men1 (Fig. 4E). Collectively, these data indicate that mutation of Men1 disrupts menin HMTase complex activity at the p27 and p18 loci, impairing appropriate expression of these key endocrine cell cycle regulators.
Discussion
These studies reveal an epigenetic mechanism for tumor suppression by menin in pancreatic islets. One of our principal findings is that menin binds in vivo to specific regions of the p27 and p18 promoters in pancreatic endocrine cells. Prior studies (17, 18) have demonstrated that simultaneous loss of p27 and p18 function in mice leads to a spectrum of tumors similar to that seen in patients with type 1 MEN syndrome, suggesting that these cyclin-dependent kinase inhibitors serve as essential growth regulators of neuroendocrine tissues. Thus, our studies show that menin governs expression of at least two established regulators of endocrine cell proliferation. p18 and p27 are known to inhibit functions of CDK4 and other cyclin-dependent kinases (19). For pancreatic islets, this view is consistent with recent work showing that regulation of CDK4 is required for islet growth control (33, 35). Replacement of the endogenous CDK4 gene with an Ink4-resistant activated CDK4 allele resulted in mice with hyperplastic islets, comprised chiefly of insulin-producing β cells (35). Our data suggest that menin is required to control CDK4 expression in islets; however, results from ChIP studies suggest that menin effects on islet expression of CDK4 and of CDK inhibitors like p15 and p21 is indirect. Thus, further studies are required to reveal how menin controls expression of targets like p15, p21, and CDK4.
We found that menin associates with specific regions of the p27 and p18 proximal promoters. Prior studies showed that these regions contained or were adjacent to positive transcriptional regulatory elements (26, 27, 38). Here, we used luciferase-based assays to show that DNA encompassing these regions was sufficient for stimulating menin-dependent transcription. Based on data suggesting that menin forms a complex with MLL2 and regulates its histone methyltransferase activity (8, 9), we speculate that menin promotes histone methylation at 5′ promoter sites, and might function as a transcriptional coactivator at promoters near the transcriptional start site of specific targets like p27 and p18. Consistent with this view are data from previous studies that demonstrate the direct association of menin with transcriptional activators and repressors (30–34). MLL and MLL2, TrxG members that associate with menin, have also been recently shown to bind promoter-proximal cis-regulatory elements to maintain expression of gene targets like p27 and p18 in terminally differentiated cells (10, 11, 39), including fibroblasts. Our analysis did not identify specific DNA sequence motifs common to regions in p27 and p18 that might mediate menin-dependent transcription, consistent with at least one prior study (37) that showed that menin can bind DNA in a sequence-independent manner. Thus, additional studies are needed to identify how menin, or other factors associated with menin, might localize to the p27 and p18 promoters.
The second principal finding in our study is that menin promotes methylation of histone H3 Lys-4 in vivo at p27 and p18, genes crucial for islet growth control and tumor suppression. We showed that loss of menin expression by islet tumors in Men1+/- mice prevented Lys-4 methylation of histone H3 associated with p27 and p18. Thus histone H3 Lys-4 methylation by menin is a possible mechanism for controlling expression of p27 and p18 by pancreatic endocrine cells in vivo. A subset of tumor-derived mutant forms of menin failed to associate with p27 or p18 cis-regulatory sequences or stimulate H3 Lys-4 methylation in cultured islet cell lines. These studies therefore provide evidence that some mutant forms of menin lack at least two activities (binding to specific promoters, and stimulating histone H3 Lys-4 methylation) that regulate tumorigenesis in islet cells. Both activities may be eliminated by mutations like A242V and T344R, but it remains unknown whether Men1 mutations that affect promoter binding by menin invariably impair menin-dependent H3 methylation. Moreover, our work also reveals that some patient-derived mutant alleles of Men1, like L22R, specify forms of menin that retain both activities; thus, it is unclear whether dysregulated p27 or p18 expression serves as the basis for tumor pathogenesis in these cases.
Could menin regulate other epigenetic modifications that control neuroendocrine gene expression? Prior studies suggest that H3 Lys-4 methylation antagonizes CpG methylation (13), which is thought to be an important means for repressing promoter activity of p27 and p18 in some contexts (40). Histone H3 Lys-4 methylation also disrupts binding of the nucleosome remodeling and deactylase (NuRD) repressor complex to H3 tails (41, 42). Further studies may show whether menin-induced H3 Lys-4 methylation can alter promoter methylation or prevent histone deactylation at target loci like p27 and p18. The association of menin with TrxG proteins like MLL (10) and the well established antagonism betweeen TrxG and polycomb group (PcG) proteins in maintenance of steady-state gene expression (39) raise the possibility that PcG proteins may regulate growth of islets and other neuroendocrine cells.
We propose that menin is required for specific methylation of histone H3 Lys-4 to ensure stable expression of target genes like p27 and p18 in endocrine cells. Thus, our data may also begin to explain the specific bias for endocrine tumor formation in MEN1, a hallmark feature of this familial tumor syndrome. Growth of mouse endocrine organs is sensitive to simultaneous loss of p27 and p18 activity (17, 18), suggesting that Ink4 family members like p18 and Cip/Kip members like p27 may have partially overlapping functions in maintaining growth control of diverse neuroendocrine cells. Thus, we speculate that the bias toward endocrine tumor pathogenesis in MEN1 results from a requirement for menin to maintain expression of CDK inhibitors in adult endocrine tissues. Understanding the molecular mechanisms underlying growth control by menin in endocrine cells may improve our ability to provide diagnoses, prognoses, and treatment for patients with MEN1 and diabetes.
Supplementary Material
Acknowledgments
We thank Dr. M. Fontaine for assistance with islet isolation, Dr. A. Bhushan (Hillblom Islet Center, University of California, Los Angeles) and N. Smart for advice on p27 immunohistochemistry, J. Heit and Drs. A. Hoffman and T. Li for advice on histone ChIP methods, R. Gupta for technical advice on performing islet ChIP, and B. Burgering (Center for Biomedical Genetics, University Medical Center, Utrecht, The Netherlands) and A. Brunet (Department of Genetics, Stanford University School of Medicine) for p27 reporter constructs. S. K. Karnik was supported by a Stanford Cancer Council award, a gift from the Verto Institute, and a Ruth Kirschstein postdoctoral fellowship (National Institutes of Health). This work was supported by grants from the Biomedical Scholars Program of the Pew Charitable Trusts, and gifts through the Verto Institute from Dr. Raymond Sackler and Beverly Sackler to M.M. and S. K. Kim, and a gift from the Stephen and Caroline Kaufer Fund for Neuroendocrine Tumor Research (to S. K. Kim).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MEN, multiple endocrine neoplasia; TrxG, trithorax group; Lys-4, lysine 4; HMTase, histone methyltransferase; CDK, cyclin-dependent kinase; ChIP, chromatin immunoprecipitation; siRNA, small interfering RNA; PP, primer pair.
References
- 1.Larsson, C., Skoseid, B., Oberg, K., Nakamura, Y. & Nordenskjold, M. (1988) Nature 332, 85-87. [DOI] [PubMed] [Google Scholar]
- 2.Chandrasekharappa, S. C., Guru, S. C., Manickam, P., Olufemi, S. E., Collins, F. S., Emmert-Buck, M. R., Debelenko, L. V., Zhuang, Z., Lubensky, I. A., Liotta, L. A., et al. (1997) Science 276, 404-407. [DOI] [PubMed] [Google Scholar]
- 3.Crabtree, J. S., Scacheri, P. C., Ward, J. M., Garrett-Beal, L., Emmert-Buck, M. R., Edgemon, K. A., Lorang, D., Libutti, S. K., Chandrasekharappa, S. C., Marx, S. J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biondi, C., Gartside, M., Tonks, I., Paterson, C., Hayward, N. K. & Kay, G. F. (2002) Genesis 32, 150-151. [DOI] [PubMed] [Google Scholar]
- 5.Bertolino, P., Tong, W. M., Galendo, D., Wang, Z. Q. & Zhang, C. X. (2003) Mol. Endocrinol. 17, 1880-1892. [DOI] [PubMed] [Google Scholar]
- 6.Bertolino, P., Radovanovic, I., Casse, H., Aguzzi, A., Wang, Z. Q. & Zhang, C. X. (2003) Mech. Dev. 120, 549-560. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal, S. K., Lee Burns, A., Sukhodolets, K. E., Kennedy, P. A., Obungu, V. H., Hickman, A. B., Mullendore, M. E., Whitten, I., Skarulis, M. C., Simonds, W. F., et al. (2004) Ann. N.Y. Acad. Sci. 1014, 189-198. [DOI] [PubMed] [Google Scholar]
- 8.Hughes, C. M., Rozenblatt-Rosen, O., Milne, T. A., Copeland, T. D., Levine, S. S., Lee, J. C., Hayes, D. N., Shanmugam, K. S., Bhattacharjee, A., Biondi, C., et al. (2004) Mol. Cell 13, 587-597. [DOI] [PubMed] [Google Scholar]
- 9.Yokoyama, A., Wang, Z., Wysocka, J., Sanyal, M., Aufiero, D. J., Kitabayashi, I., Herr, W. & Cleary, M. L. (2004) Mol. Cell. Biol. 24, 5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milne, T. A., Hughes, C. M., Lloyd, R. M., Yang, Z., Rozenblatt-Rosen, O., Dou, Y., Schnepp, R. W., Krankel C., LiVolsi, V. A., Gibbs, D., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 749-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milne, T. A., Briggs, S. D., Brock, H. W., Martin, M. E., Gibbs, D., Allis, C. D. & Hess, J. L. (2002) Mol. Cell 10, 1107-1117. [DOI] [PubMed] [Google Scholar]
- 12.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074-1080. [DOI] [PubMed] [Google Scholar]
- 13.Lachner, M. & Jenuwein, T. (2002) Curr. Opin. Cell Biol. 14, 286-298. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama, K., Ishida, N., Shirane, M., Inomata, A., Inoue, T., Shishido, N., Horii, I., Loh, D. Y. & Nakayama, K. (1996) Cell 85, 707-720. [DOI] [PubMed] [Google Scholar]
- 15.Kiyokawa, H., Kineman, R. D., Manova-Todorova, K. O., Soares, V. C., Hoffman, E. S., Ono, M., Khanam, D., Hayday, A. C., Frohman, L. A. & Koff, A. (1996) Cell 85, 721-732. [DOI] [PubMed] [Google Scholar]
- 16.Fero, M. L., Rivkin, M., Tasch, M., Porter, P., Carow, C. E., Firpo, E., Polyak, K., Tsai, L. H., Broudy, V., Perlmutter, R. M., et al. (1996) Cell 85, 733-744. [DOI] [PubMed] [Google Scholar]
- 17.Franklin, D. S., Godfrey, V. L., Lee, H., Kovalev, G. I., Schoonhoven, R., Chen-Kiang, S., Su, L. & Xiong, Y. (1998) Genes Dev. 12, 2899-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin, D. S., Godfrey, V. L., O'Brien, D. A., Deng, C. & Xiong, Y. (2000) Mol. Cell. Biol. 20, 6147-615820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer, P. M., Endicott, J. & Meijer, L. (2003) Prog. Cell Cycle Res. 5, 235-248. [PubMed] [Google Scholar]
- 20.Kim, S. K., Hebrok, M., Li, E., Oh, S. P., Schrewe, H., Harmon, E. B., Lee, J. S. & Melton, D. A. (2000) Genes Dev. 14, 1866-1871. [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S. K., Selleri, L., Lee, J. S., Zhang, A. Y., Gu, X., Jacobs, Y. & Cleary, M. L. (2002) Nat. Genet. 30, 430-435. [DOI] [PubMed] [Google Scholar]
- 22.Beilhack, G. F., Scheffold, Y. C., Weissman, I. L., Taylor, C., Jerabek, L., Burge, M. J., Masek, M. A. & Shizuru, J. A. (2003) Diabetes 52, 59-68. [DOI] [PubMed] [Google Scholar]
- 23.Scacheri, P. C., Rozenblatt-Rosen, O., Caplen, N. J., Wolfsberg, T. G., Umayam, L., Lee, J. C., Hughes, C. M., Shanmugam, K. S., Bhattacharjee, A., Meyerson, M. & Collins, F. S. (2004) Proc. Natl. Acad Sci. USA 101, 1892-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karnik, S. K., Brooke, B. S., Bayes-Genis, A., Sorensen, L., Wythe, J. D., Schwartz, R. S., Keating, M. T. & Li, D. Y. (2003) Development (Cambridge, U.K.) 130, 411-423. [DOI] [PubMed] [Google Scholar]
- 25.Phelps, D. E., Hsiao, K., Li, Y., Hu, N., Franklin, D. S., Westphal, E., Lee, E. Y. & Xiong, X. (1998) Mol. Cell. Biol. 18, 2334-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medema, R. H., Kops, G. J. P., Bos, J. L. & Burgering, B. M. T. (2000) Nature 404, 782-784. [DOI] [PubMed] [Google Scholar]
- 27.Crabtree, J. S., Scacheri, P. C., Ward, J. M., McNally, S. R., Swain, G. P., Montagna, C., Hager, J. H., Hanahan, D., Edlund, H., Magnuson, M. A., et al. (2003) Mol. Cell. Biol. 23, 6075-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biondi, C. A., Gartside, M. G., Waring, P., Loffler, K. A., Stark, M. S., Magnuson, M. A., Kay, G. F. & Hayward, N. K. (2004) Mol. Cell. Biol. 24, 3125-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertolino, P., Tong, W. M., Herrera, P. L., Casse, H., Zhang, C. X. & Wang, Z. Q. (2003) Cancer Res. 63, 4836-4841. [PubMed] [Google Scholar]
- 30.Agarwal, S. K., Guru, S. C., Heppner, C., Erdos, M. R., Collins, R. M., Park, S. Y., Saggar, S., Chandrasekharappa, S. C., Collins, F. S., Spiegel, A. M., et al. (1999) Cell 96, 143-152. [DOI] [PubMed] [Google Scholar]
- 31.Guru, S. C., Goldsmith, P. K., Burns, A. L., Marx, S. J., Spiegel, A. M., Collins, F. S. & Chandrasekharappa, S. C. (1998) Proc. Natl. Acad. Sci. USA 95, 1630-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sukhodolets, K. E., Hickman, A. B., Agarwal, S. K., Sukhodolets, M. V., Obungu, V. H., Novotny, E. A., Crabtree, J. S., Chandrasekharappa, S. C., Collins, F. S., Spiegel, A. M., et al. (2003) Mol. Cell. Biol. 23, 493-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin, S., Mao, H., Schnepp, R. W., Sykes, S. M., Silva, A. C., D'Andrea, A. D. & Hua, X. (2003) Cancer Res. 63, 4204-4210. [PubMed] [Google Scholar]
- 34.Lin, S. Y. & Elledge, S. J. (2003) Cell 113, 881-889. [DOI] [PubMed] [Google Scholar]
- 35.Rane, S. G., Dubus, P., Mettus, R. V., Galbreath, E. J., Boden, G., Reddy, E. P. & Barbacid, M. (1999) Nat. Genet. 22, 44-52. [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki, J., Araki, K., Yamato, E., Ikegami, H., Asano, T., Shibasaki, Y., Oka, Y. & Yamamura, K. (1990) Endocrinology 127, 126-132. [DOI] [PubMed] [Google Scholar]
- 37.La, P., Silva, A. C., Hou, Z., Wang, H., Schnepp, R. W., Yan, N., Shi, Y. & Hua, X. (2004) J. Biol. Chem. 279, 49045-49054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blais, A., Monte, D., Pouliot, F. & Labrie, C. (2002) J. Biol. Chem. 277, 31679-31693. [DOI] [PubMed] [Google Scholar]
- 39.Ringrose, L. & Paro, R. (2004) Annu. Rev. Genet. 38, 413-443. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Aguilera, A., Delgado, J., Camacho, F. I., Sanchez-Beato, M., Sanchez, L., Montalban, C., Fresno, M. F., Martin, C., Piris, M. A. & Garcia, J. F. (2004) Blood 103, 2351-2357. [DOI] [PubMed] [Google Scholar]
- 41.Zegerman, P., Canas, B., Pappin, D. & Kouzarides, T. (2002) J. Biol. Chem. 277, 11621-11624. [DOI] [PubMed] [Google Scholar]
- 42.Nishioka, K., Chuikov, S., Sarma, K., Erdjument-Bromage, H., Allis, C. D., Tempst, P. & Reinberg, D. (2002) Genes Dev. 16, 479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.