Abstract
To assess the mechanisms of repression of the erythroid-specific carbonic anhydrase II (CAII) locus we used chromatin immunoprecipitation and show that an NCoR–histone deacetylase (HDAC)3 complex is recruited by the nuclear receptor v-ErbA to the intronic HS2 enhancer turning it into a potent silencer. Furthermore we demonstrate that efficient CAII silencing requires binding of a MeCP2-targeted HDAC-containing corepressor complex to the hypermethylated CpG-island at the promoter. Activation of transcription by either AZAdC or thyroid hormone results in loss of one of the two corepressor complexes. Thyroid hormone further replaces the enhancer-bound NCoR–corepressor complex by the TRAP220 coactivator. Treatment with the HDAC inhibitor trichostatin A (TSA) causes activation of CAII transcription and histone H3 and H4 hyperacetylation at the enhancer, apparently without affecting binding of the two corepressor complexes. Unexpectedly, histone H3 and H4 at the fully repressed promoter are already hyperacetylated despite the close apposition of the MeCP2-targeted HDAC complex. Acetylation of histone H4, but not H3, at the promoter is moderately increased following TSA treatment. Our data suggest that the hyperacetylated but repressed CAII promoter is (partially) remodeled and primed for activation in v-ErbA-transformed cells.
Keywords: corepressor/erythroleukemia/HDAC/methylation/v-ErbA
Introduction
Gene-specific regulation lies at the heart of cell differentiation in higher eukaryotes. Many genes in the human genome appear to be transiently active during differentiation and in small subsets of cells. This exquisite regulation is brought about by the selective assembly and disassembly of cell-type-specific transcription factor complexes, the enhanceosomes (reviewed in Merika and Thanos, 2001). Specificity is further determined by epigenetic events such as DNA methylation as well as by local remodeling of chromatin to permit or preclude binding of regulatory factors to nucleosomal DNA. In contrast to our biochemical understanding of the factors involved in gene activation and repression, in vivo data about gene regulation mediated by DNA-bound transcription factors and associated cofactor complexes remain scarce.
Nuclear hormone receptors provide a unique model to study the mechanisms of transcriptional activation as well as repression. Transcriptional activation by liganded nuclear hormone receptors is accomplished by recruitment of coactivator proteins, which contain intrinsic acetyltransferase activity. Coactivators that interact with nuclear receptors include p160-related factors and the acetyltransferases p300/CBP and PCAF (reviewed in Glass and Rosenfeld, 2000). A multiprotein complex, TRAP– Mediator (also referred to as ARC–DRIP–CRSP; reviewed by Malik and Roeder, 2000) has recently been shown to interact with liganded nuclear receptors (Fondell et al., 1996; Näär et al., 1999; Ranchez et al., 1999). A single subunit, TRAP220, is thought to target the complex to the thyroid hormone receptor (TR) and other liganded nuclear hormone receptors through a LXXLL motif (Yuan et al., 1998; Ito et al., 1999). Chromatin immunoprecipitation (ChIP) experiments suggest that assembly of enhanceosome and coactivator complexes is ordered and dynamic (Agalioti et al., 2000; Shang et al., 2000).
Transcriptional repression by unliganded nuclear hormone receptors is thought to be brought about by recruitment of multi-subunit corepressor complexes containing intrinsic histone deacetylase (HDAC) activity. Most of our current knowledge on repression and corepressor proteins stems from in vitro binding studies. Two-hybrid analysis led to the identification of the nuclear receptor corepressor, NCoR (Horlein et al., 1995), and its related family member, SMRT (Chen and Evans, 1995). The first in vivo proof for NCoR function in nuclear receptor action came from studies analyzing NCoR–/– mice which indicated that NCoR was required for active repression by nuclear receptors and other repressors (Jepsen et al., 2000). The biochemical identification of multiple NCoR-containing complexes (Guenther et al., 2000; Li et al., 2000; Underhill et al., 2000; Jones et al., 2001) posed the question as to which of these complexes are involved in nuclear hormone receptor functioning in vivo.
Recent studies indicate that methylation is another important mechanism of gene silencing in cancer (Costello et al., 2000). Although numerous studies suggested the involvement of methyl-binding proteins such as MeCP2 in gene silencing by methylation, direct in vivo evidence is currently lacking. The methyl-CpG binding protein MeCP2 is the founding member of a family of proteins that contain homologous methyl-CpG-binding domains (MBDs) (reviewed by Wade, 2001). The recent discovery that MBD family members reside in distinct complexes containing HDACs and other unique as well as common subunits provided a link between DNA methylation, histone deacetylation and gene silencing.
Our current understanding of how corepressor and coactivator complexes associate with nuclear receptors in vivo on chromatin and exert their regulatory function is far from complete. To unravel these questions, we used the chicken erythroleukemia cell line, HD3, which is transformed by the avian erythroblastosis virus (AEV). AEV encodes two co-operating oncoproteins: v-ErbA, a mutated thyroid hormone receptor (Sap et al., 1986) and v-ErbB, a constitutively active EGF–TGF-α receptor tyrosine kinase (Downward et al., 1984). Notwithstand ing extensive studies in the past decade, the molecular mechanisms by which v-ErbA deploys its oncogenic potential remain rather vague. It seems clear however that v-ErbA contributes to the leukemogenic phenotype by silencing erythroid-specific genes such as the lysozyme gene and the carbonic anhydrase II gene (CAII) (Zenke et al., 1988; Baniahmad et al., 1990; Pain et al., 1990). We recently identified a v-ErbA responsive element (VRE) in the hypersensitive 2 (HS2) enhancer in intron 2 of the CAII gene. The VRE is adjacent to two GATA-1 sites (Ciana et al., 1998; Braliou et al., 2001). Mutation of the VRE unleashed the enhancer function resulting in a very marked increase in transcription (Braliou et al., 2001; Rietveld et al., 2001).
In this study, we used in vitro and in vivo approaches to assess the identity of the proteins associated with the CAII promoter and HS2 enhancer in its silent state. Using ChIP experiments, we show that v-ErbA and GATA-1 cohabitate the HS2 enhancer and that recruitment of a NCoR– HDAC3 corepressor complex by v-ErbA turns the erythroid enhancer into a silencer. Efficient silencing of the CAII gene further involves binding of a MeCP2-targeted HDAC-containing corepressor complex to the promoter. Dissociation of one of the corepressor complexes through the addition of either the methylation inhibitor AZAdC or thyroid hormone suffices to activate CAII transcription. Furthermore, we show that the HDAC inhibitor trichostatin A (TSA) activates CAII transcription and induces histone hyperacetylation at the HS2 enhancer without affecting corepressor binding. The repressed CAII promoter is hyperacetylated despite the presence of HDAC-containing complexes, suggesting that histone H3 and H4 at repressed promoter are not necessarily hypoacetylated.
Results
Association of a corepressor complex with v-ErbA in vitro and in vivo
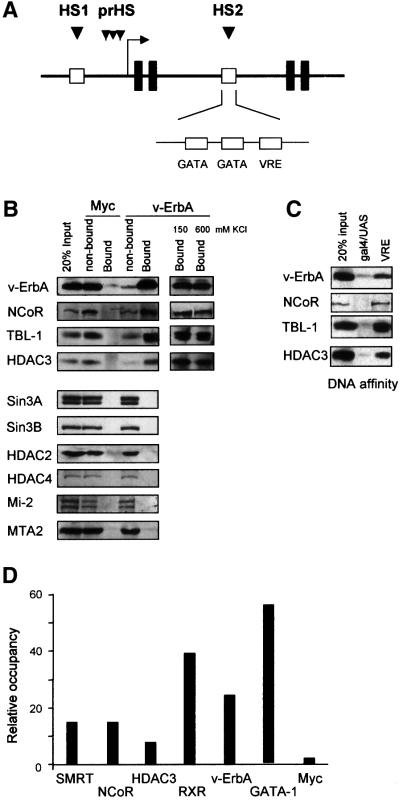
Recently we have shown that the HS2 region within the CAII gene contains a binding site for v-ErbA as well as two functional GATA-1 binding sites (Figure 1A) (Ciana et al., 1998; Braliou et al., 2001). In this study, we set out to unravel the factors and mechanism involved in repression of the CAII gene by v-ErbA in HD3 cells. To identify corepressor components associated with v-ErbA in HD3 cells, immunoprecipitations were performed using an antibody directed against the gag moiety of v-ErbA (1G10). Western blot analysis of the precipitates revealed that the 270 kDa NCoR protein coprecipitated with v-ErbA (Figure 1B). The histone deacetylase HDAC3 and the transducin β-like 1 protein (TBL-1) also efficiently co-immunoprecipitated with v-ErbA. The interaction between NCoR–HDAC3–TBL-1 complex and v-ErbA resisted stringent washing with buffers containing up to 600 mM KCl and 0.25% nonident P-40 (NP-40).
Fig. 1. v-ErbA recruits a specific corepressor complex. (A) Schematic depiction of the CAII genomic locus. Black boxes indicate exons 1–4. DNase I hypersensitive sites at the promoter (prHS) are indicated, as well as HS1 and HS2 (open boxes). At the HS2 region, protected regions in in vivo footprint experiments are indicated, these are two putative GATA-1 binding sites and a VRE. (B) Coupled co-immunoprecipitation–western blot experiment with v-ErbA antibody (1G10) showing association of the NCoR–HDAC3–TBL-1 proteins with v-ErbA but not with a control myc antibody. (C) DNA-affinity assay showing specific association of NCoR–TBL-1–HDAC3 with v-ErbA. DNA-affinity experiments were performed with specific oligomerized VRE and Gal4 oligonucleotides. (D) ChIP experiment performed with the indicated antibodies. The recovered DNA was quantified in real-time PCR with a primer set for the CAII HS2 region. The signal was compared with an unrelated control region, located 15 kb upstream of the HS2 and 7 kb upstream of the start site. This control signal was arbitrarily set at 1 and HS2 signal was calculated compared to this.
In contrast to a number of other studies (Heinzel et al., 1997; Lee et al., 2000; Underhill et al., 2000) we found no association of v-ErbA with Sin3A/B, HDAC2 or the class II histone deacetylase HDAC4 (Huang et al., 2000; Kao et al., 2000) (Figure 1B). Furthermore, association between v-ErbA and components of the NuRD complex such as the ATPase Mi-2 or the metastasis-associated protein MTA2 (Wade et al., 1999; Zhang et al., 1999) could not be detected (Figure 1B).
To obtain additional evidence for the association between v-ErbA and the NCoR–SMRT complex, a DNA affinity resin bearing oligomerized oligonucleotides containing the HS2–VRE was used. Figure 1C shows that VRE-bound v-ErbA associated with NCoR, HDAC3 and TBL-1, whereas these proteins are not bound to the unrelated Gal4 DNA binding site. Notably, components of the Sin3 and NuRD complexes could not be detected in these experiments (data not shown).
The outcome of our biochemical analysis in HD3 cells reveals that v-ErbA recruits a corepressor complex containing HDAC3. To show that this corepressor complex is targeted to the HS2 enhancer in vivo, we conducted ChIP experiments. Chromatin was purified from proliferating HD3 cells and ChIP assays were performed using the 1G10 antibody that is specific for the gag moiety of v-ErbA. This analysis revealed association of v-ErbA with the HS2 enhancer, but not with an unrelated control region, in vivo in HD3 cells (Figure 1D). An antibody directed against the heterodimeric partner RXR of v-ErbA efficiently precipitated the HS2 region indicating that RXR co-occupies the HS2 with v-ErbA. Antibodies against the GATA-1 transcription factor specifically precipitated the HS2 enhancer in HD3 cells that do not express CAII. The HS2 region was also efficiently precipitated with antibodies directed against NCoR, its close relative SMRT, and HDAC3 (Figure 1D), indicating the in vivo association of a v-ErbA–NCoR–HDAC3 complex with the HS2 region in HD3 cells. An unrelated anti-myc antibody did not specifically precipitate the HS2 regulatory region.
The ChIP experiments along with the previously published in vivo footprinting data (Ciana et al., 1998) show that v-ErbA as well as GATA-1 are bound to the HS2 enhancer in vivo in proliferating HD3 cells that do not transcribe CAII. These data corroborate and extend the notion that in proliferating HD3 cells, v-ErbA nullifies the positive transcriptional activity of GATA factors (Ciana et al., 1998; Braliou et al., 2001). Our findings thus provide evidence for the in vivo association of an NCoR–corepressor complex to a presumed endogenous target gene by an unliganded nuclear hormone receptor.
MeCP2-instigated repression at the CAII promoter
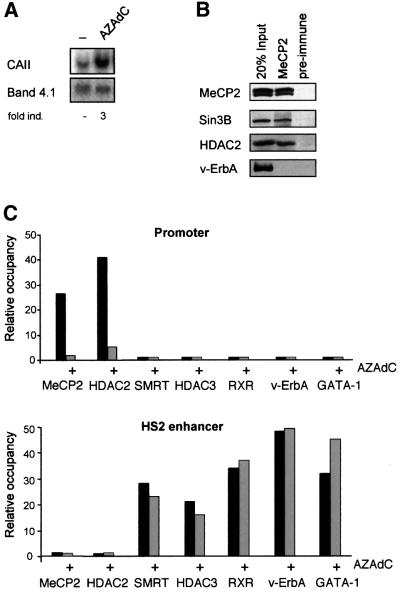
Previous as well as unpublished experiments (Ciana et al., 1998; data not shown) suggested that apart from repression instigated by v-ErbA, additional mechanism(s) involving HDACs contribute to CAII gene silencing. When carefully inspecting the CAII promoter region, we found that it is rather GC rich (82%) and that the CpG:GpC ratio in this region is 1.0, meeting the criteria for a CpG island (Gardiner-Garden and Frommer, 1987). Bisulfite sequencing revealed that the CAII promoter contains a high frequency (56 ± 8%) of methylated CpG dinucleotides in the region between –400 and –200 (data not shown). Analyzing different clones (n = 15) we found that the methylation pattern was heterogeneous and disperse, and a consistent pattern of methylated CpG residues could not be observed. Direct involvement of methylation in CAII repression could be shown by treatment of HD3 cells with the methylation inhibitor AZAdC for 48 h, which efficiently activated CAII transcription (Figure 2A). Follow ing treatment, the CAII promoter region was significantly demethylated (∼50%) compared with untreated cells (data not shown).
Fig. 2. The hypermethylated CAII promoter CpG island binds the MeCP2–corepressor complex in vivo. (A) Northern blot showing activation of CAII expression by treatment with the methylation inhibitor AZAdC (48 h, 500 nM) in HD3 cells. Band 4.1 serves as loading control. (B) Coupled co-immunoprecipitation–western blotting experiment with MeCP2 antibody showing association of the Sin3B–HDAC2 proteins with MeCP2, but not with preimmune serum. (C) ChIP experiment performed with indicated antibodies on chromatin from untreated HD3 cells and from cells treated for 48 h with AZAdC (500 nM). Recovered DNA was quantified using real-time PCR with control-, HS2- and promoter spanning primer sets. The background control region 7 kb upstream of the CAII promoter is used as control for which the PCR signal is arbitrarily set to 1 and signals were calculated compared to this.
Gene silencing by methylation reportedly involves binding of methyl-CpG binding proteins that recruit HDAC activity (Jones et al., 1998; Nan et al., 1998). Hence, the role of methyl-binding proteins in CAII repression was assessed. The methyl-binding protein MeCP2 appeared to be expressed at relatively high levels in HD3 cells. In co-immunoprecipitation experiments with MeCP2 antibody, HDAC2 and Sin3B proteins copurified with the MeCP2 protein, indicating that MeCP2 recruited a Sin3–HDAC complex from HD3 cell extracts (Figure 2B). Furthermore, a Sin3B antibody efficiently co-immunoprecipitated MeCP2 from HD3 cells (data not shown).
ChIP assays were used to assess whether MeCP2 binds to the methylated promoter region in vivo and recruits a HDAC-containing complex to the CpG island in the CAII promoter. Antibodies against MeCP2 or HDAC2 specifically immunoprecipitated the CAII CpG island when using chromatin preparations derived from HD3 cells (Figure 2C). Treatment with AZAdC dislodged MeCP2 and HDAC2 from the promoter without significantly affecting binding of the factors bound to the HS2 enhancer (Figure 2C). Thus, the hypermethylated promoter recruits a MeCP2–HDAC2-containing corepressor complex that contributes to CAII repression in proliferating HD3 cells.
CAII activation by thyroid hormone treatment
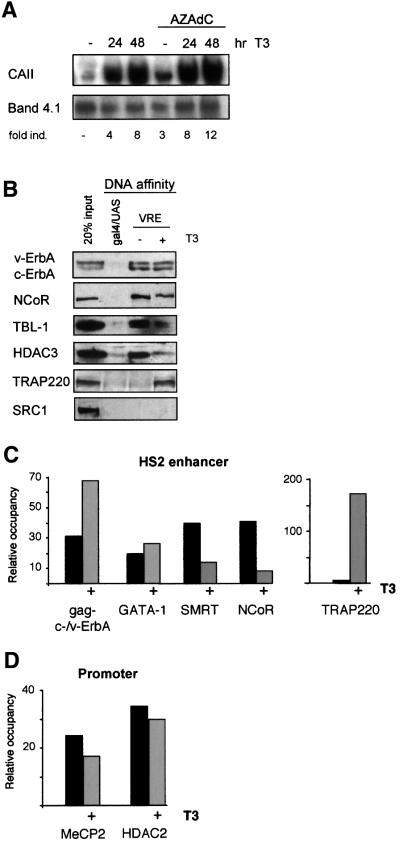
The generally accepted ‘on–off’ model for repression and activation by nuclear receptors implies that ligand binding causes a conformational change in the ligand-binding domain resulting in dissociation of corepressor and subsequent association of coactivator proteins (reviewed in Glass and Rosenfeld, 2000). To visualize the ‘on–off’ switch, we turned to a HD3-derived cell line, HD3-V3, which expresses a ligand-responsive TR fused to the retroviral gag moiety (gag-c-ErbA) at levels equivalent to that of v-ErbA. Our previous studies had shown that addition of thyroid hormone (T3) triggers differentiation of these erythroblasts with a concomitant activation of CAII transcription (Disela et al., 1991). HD3-V3 cells grown in hormone-depleted medium express low levels of CAII mRNA and addition of T3 boosts the levels of CAII mRNA notwithstanding the continued presence of the ligand-unresponsive, constitutive repressor v-ErbA in these cells (Ciana et al., 1998; Figure 3A).
Fig. 3. Thyroid hormone-induced CAII activation induces cofactor exchange at the HS2 enhancer region. (A) Northern blot showing activation of CAII transcription in HD3-V3 cells treated with T3 (300 nM), AZAdC (500 nM) or both. Band 4.1 served as a control. (B) DNA-affinity pull-down experiment with oligomerized VRE oligonucleotides in extracts from HD3-V3 cells either untreated or stimulated for 24 h with 300 nM T3. Western blot analysis shows the association/release of indicated corepressor/coactivator subunits with respect to c-/v-ErbA bound to the VRE. (C) HS2 enhancer ChIP experiment with soluble chromatin isolated from untreated and T3-treated (24 h, 300 nM) HD3-V3 cells using antibodies directed against the indicated proteins. HS2 PCR-signals were quantified as in Figure 1D. (D) as (C), but primer sets that amplify the promoter region were used to quantify the recovered DNA.
To identify factors recruited to the HS2 enhancer upon T3 activation, DNA-affinity resins were used to isolate interacting factors in nuclear extracts from HD3-V3 cells prepared before and after ligand treatment. Western blot analysis revealed the binding of roughly equal amounts of gag-c-ErbA and v-ErbA to the DNA beads using T3-treated and untreated extracts. Furthermore, the NCoR/HDAC3/TBL-1 corepressor proteins co-purified efficiently via gag-c-/v-ErbA from extracts prepared from untreated cells (Figure 3B). Using extracts prepared from HD3-V3 cells grown in the presence of T3, binding of NCoR, HDAC3 and TBL-1 to the DNA-affinity resin was significantly diminished. In contrast, the coactivator protein TRAP220, a component of the TRAP–Mediator complex (Yuan et al., 1998) could be recovered. Sur prisingly, SRC-1, another coactivator reported to interact with liganded TR (Onate et al., 1995), did not bind to ligand-activated gag-c-ErbA although it could readily be detected in HD3-V3 extracts. Similar analyses using HD3 cell extracts (lacking a ligand responsive TR receptor) revealed that v-ErbA was unable to recruit either SRC-1 or TRAP220 (data not shown), in line with the fact that v-ErbA is unable to bind ligand.
To provide proof for the ligand-dependent recruitment of TRAP220, ChIP experiments were performed with chromatin preparations from untreated and T3-treated HD3-V3 cells. Ligand addition caused a 2-fold increase in the occupancy of the HS2 by gag-c-/v-ErbA (Figure 3C). The relative occupancy of the HS2 by GATA-1 was not affected by ligand treatment of the cells. A 4- to 5-fold decrease in the binding of NCoR and SMRT to the HS2 region was observed after the addition of ligand. Note that v-ErbA, expressed to similar levels as gag-c-ErbA, is unable to respond to ligand and will sustain low level recruitment of NCoR–SMRT complexes even in the presence of T3. Using a TRAP220-specific antibody, a strong increase in TRAP220 occupancy of the HS2 enhancer (up to 150-fold) could be observed in T3-treated compared with untreated cells (Figure 3C). Consistent with the in vitro data we could not find evidence for the recruitment of the p160-coactivators SRC-1 or AIB1 to ligand-bound gag-c-ErbA in ChIP experiments (data not shown).
An intriguing question is whether the MeCP2–HDAC2 complex that binds the hypermethylated CpG island remained associated upon ligand addition. ChIP experiments did not reveal significant differences in the association of MeCP2 and HDAC2 with the CpG island before or after ligand administration (Figure 3D). This suggests that the MeCP2–HDAC2 complex at the promoter might continue to repress transcription in the presence of T3, an interpretation that is supported by northern blot analyses; addition of T3 plus AZAdC activated CAII transcription in an additive manner (Figure 3A). Thus, T3 and AZAdC activate CAII transcription via distinct and independent pathways.
Taken together, the data show that T3 addition trig gered the ‘on–off’ switch, i.e. dislodging the NCoR– SMRT-containing corepressor complex and recruiting a TRAP220-containing coactivator complex to the enhancer.
The repressed CAII promoter is hyperacetylated despite corepressor binding
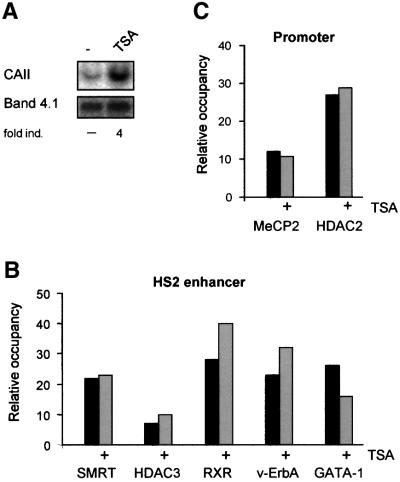
The HDAC inhibitor TSA efficiently activates transcription from the CAII gene in HD3 cells (Figure 4A; Ciana et al., 1998). To investigate whether CAII activation by TSA affects binding of corepressor complexes at the enhancer and promoter, ChIP assays were performed using chromatin isolated from untreated and TSA-treated HD3 cells. Figure 4B shows that TSA did not significantly affect binding of GATA-1, v-ErbA, RXR or the NCoR– SMRT complex to the HS2 enhancer region. Furthermore, TSA did not affect promoter methylation and binding of MeCP2–HDAC2 to the promoter region (Figure 4C; data not shown).
Fig. 4. Corepressor binding after TSA treatment. (A) Northern blot showing activation of CAII expression by treatment of HD3 cells with TSA (24 h, 100 nM). Band 4.1 served as a loading control. (B) ChIP experiments with indicated antibodies, using chromatin isolated from untreated and TSA-treated HD3 cells. The recovered DNA was quantified in real-time PCR as in Figure 1D. (C) as (B), but primer sets that amplify the promoter region were used to quantify the recovered DNA.
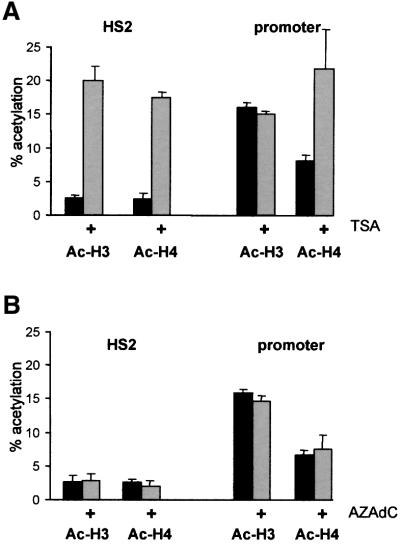
To assess the pattern of histone acetylation–deacetyl ation in the CAII locus, ChIP assays were performed with antibodies directed against the acetylated tails of histones H3 and H4. Nucleosomes at the HS2 enhancer were hypoacetylated in proliferating HD3 cells and TSA treatment caused a 7- to 8-fold increase in H3- and H4-acetylation consistent with the inactivation of the HDAC activity associated with the NCoR–SMRT complex (Figure 5A). Note that the percentage of histone acetylation is plotted in Figure 5A. The relative concentration of CAII promoter fragments present in CsCl-purified chromatin before immunoprecipitation was arbitrarily set to 100%. Surprisingly, histone H3 and H4 associated with the start site region appear to be hyperacetylated in proliferating HD3 cells where the CAII promoter is repressed. Acetylation of histones in the promoter region is generally accepted as a hallmark of an active promoter. TSA treatment did not increase further the acetylated state of histone H3 associated with the start site whereas acetylation of histone H4 was elevated 2-fold (Figure 5A). Thus, the CAII promoter appears to be hyperacetylated despite the presence of a HDAC-containing MeCP2 complex in close proximity to the start site of transcription.
Fig. 5. The promoter of the repressed CAII gene is hyperacetylated. (A) ChIP experiment with acetylated H3 and H4 antibodies. The ChIP input signal was set at 100% and all other signals were calculated compared to this signal. (B) as (A), but chromatin from untreated and AZAdC-treated HD3 cells was used.
To investigate whether dissociation of the MeCP2– HDAC2 complex from the promoter affected the acetylation state of nucleosomes at the promoter and at the enhancer, HD3 cells were treated with the methylation inhibitor AZAdC. The MeCP2–HDAC2 complex dissociated from the promoter following AZAdC treatment (Figure 2C) but this did not significantly alter the level of acetylation of histone H3 and H4, neither at the promoter nor at the enhancer (Figure 5B). These results suggest that increasing histone acetylation at the enhancer is not a prerequisite for derepression of the CAII gene.
Taken together, the repressed CAII promoter is hyperacetylated despite the association of HDAC2 protein at the CpG island. Furthermore, dissociation of the MeCP2 complex did not alter the extent of acetylation at the promoter. In contrast, TSA caused extensive histone acetylation at the HS2 enhancer in accordance with the inactivation of HDAC3 present at this transcription control region.
Discussion
The CAII locus has been extensively studied in our lab and this resulted in the identification and characterization of the DNase hypersensitive region HS2 in the CAII locus (Ciana et al., 1998). Here we show that v-ErbA represses transcription by recruitment of a specific corepressor complex to the HS2 enhancer in vivo and in vitro. Furthermore, the promoter binds a methylation-dependent MeCP2–corepressor complex. Recruitment of multiple specific HDAC-containing complexes efficiently silences transcription of CAII. Thus, this study provides in vivo evidence for a role in corepressor complexes in the regulation of a natural chromosomal target gene.
Repression instigated at the HS2 enhancer
To unravel the molecular mechanisms involved in repression by v-ErbA, we have used in vitro as well as in vivo approaches to identify the (co)factors involved in CAII repression. Using immunoprecipitation and DNA affinity resins, we show that v-ErbA recruits a corepressor complex containing NCoR–SMRT, HDAC3 and TBL-1. TBL-1 is related to the Tup1 and Groucho corepressors with respect to its WD40 domain that has the potential to interact with histone H3 in vitro. TBL-1 might therefore serve to bring HDAC3 to chromatin thereby facilitating histone deacetylation (Guenther et al., 2000). Several NCoR-containing corepressor complexes have been identified which possess HDAC activity but display distinct protein subunit compositions (Guenther et al., 2000; Li et al., 2000; Underhill et al., 2000; Urnov et al., 2000; Wen et al., 2000; Jones et al., 2001). A number of studies implicated the Sin3–HDAC complex in repression by NCoR–SMRT (Heinzel et al., 1997; Lee et al., 2000). However, this and other studies (Urnov et al., 2000) did not provide evidence for the recruitment of a Sin3–HDAC complex by nuclear hormone receptors either in vitro or in vivo. Nonetheless, Sin3–HDAC complexes are present in HD3 erythroblasts and are recruited by MeCP2. Furthermore we have shown that neither the class II HDAC4 (Huang et al., 2000) nor subunits of the Mi2– NURD complex (Wade et al., 1999; Zhang et al., 1999) are recruited by v-ErbA.
ChIP experiments established that GATA-1 and v-ErbA–RXR are recruited to the HS2 enhancer in vivo in proliferating erythroblasts that express CAII at very low levels. Furthermore, a NCoR–HDAC3-containing corepressor complex is in turn recruited by v-ErbA to the HS2 enhancer corroborating and extending the notion that the enhancer activity of the HS2 governed by two adjacent GATA-1 sites is nullified by v-ErbA.
MeCP2- instigated repression at the CAII promoter region
Experiments involving AZAdC revealed that DNA methylation also contributes to repression of CAII transcription. AZAdC activated CAII expression and caused (partial) demethylation of the hypermethylated promoter CpG island. The methylation pattern within the CAII CpG island is disperse, which appears to be a feature common to repressed genes in a number of different cell lines (De Smet et al., 1999; Müller et al., 2000; Danam et al., 2001). Interestingly, preliminary studies indicate that the CAII promoter is hypomethylated in primary erythroid cells suggesting that methylation may be due to transformation by AEV or extensive culturing of the cells. Methylation of genes appears to be confined to tissue-specific genes but not housekeeping genes in various cultured cell lines (Antequera et al., 1990). The CAII gene is an erythroid-specific gene that may not be required in cell culture and hence may have become methylated during extensive culturing of the cells.
The mechanism by which methylation represses transcription has long been unclear but recent data suggested the involvement of MBDs that reside in distinct complexes. The MBDs all share a high homology in the methyl-binding domain with the founding member of this family, MeCP2 (reviewed by Bird and Wolffe, 1999; Ballestar and Wolffe, 2001). We have shown that MeCP2 is expressed at relatively high levels in HD3 cells. In our immunoprecipitation experiments, MeCP2 coprecipitated Sin3B and HDAC2 and the involvement of MeCP2– HDAC2 in CAII promoter binding in vivo was established using ChIP. As a single methylated CpG is able to bind MeCP2 (Nan et al., 1993), it is likely that multiple MeCP2 proteins bind to the promoter region and recruit HDAC-containing complexes.
Dynamic regulation at the CAII locus
Our data provide evidence for the involvement of minimally two distinct corepressor complexes in repression of the CAII gene. Furthermore we found that dislodging of only one of these complexes suffices to activate CAII transcription. In this study, CAII gene transcription has been initiated in different ways, either via the recruitment of coactivators (activation) or by release or inactivation of corepressors (derepression). Dissociation of the v-ErbA-bound corepressor complex from the enhancer could be accomplished by treatment of HD3-V3 cells with T3. We provide in vivo evidence that T3 activation caused a release of the NCoR–SMRT-containing complex from gag-c-ErbA bound to the HS2 enhancer in vivo. T3-activated gag-c-ErbA was shown to recruit the coactivator TRAP220 to the HS2 enhancer. TRAP220 anchors the TRAP complex, which is composed of >25 distinct polypeptides, to liganded TR (Fondell et al., 1996; reviewed in Ito and Roeder, 2001). We did not find evidence for a role of the p160 coactivators SRC-1 and AIB1 in ligand activation instigated by gag-c-ErbA after 24 h of ligand administration, suggesting that p160 and TRAP coactivators do not bind simultaneously. However, the p160 coactivators might bind gag-c-ErbA shortly after hormone treatment as suggested by a model in which coactivators are sequentially recruited (Glass and Rosenfeld, 2000). Ligand administration did not affect association of the MeCP2–corepressor complex with the promoter, indicating that the HS2 enhancer is able to partially overcome repression instigated at the CpG island.
AZAdC treatment caused demethylation and release of the promoter-bound MeCP2–corepressor complex but binding of the enhancer-bound v-ErbA–corepressor complex was not affected. Interestingly, although AZAdC induced CAII transcription, acetylation of histone H3 and H4 associated with the promoter was not affected. Thus, dissociation of a closely positioned HDAC-enzymatic activity did not increase H3 and H4 histone acetylation at the CAII promoter. It might be that promoter-associated HDAC2 specifically targets histones H2A and H2B without affecting H3 and H4. Another possibility is that specific lysine residues are targeted by HDAC2. In this study antibodies directed against the di-acetylated H3 (K9/K14) and tetra-acetylated H4 tails that do not accurately discriminate between (de)acetylation of specific lysine residues were used. It will be of interest to use antibodies that can detect (de)acetylation of specific lysine residues at the histone tails. Deckert and Struhl (2001) already found that histone acetylation is differentially affected by repressor proteins. Furthermore, experiments in yeast using deletion strains for the HDA1 and RPD3 histone deacetylases show that these deacetylases target specific lysine residues in the H3 and H4 tails (Vogelauer et al., 2000; Wu et al., 2001). In cells from RETT syndrome patients that express a truncated MeCP2 protein in the absence of wild-type MeCP2 protein, an increase in the mono-acetylated histone isoform H4 K16 is observed, indicating that the MeCP2-associated corepressor complex targets specific lysine residues (Wan et al., 2001). Moreover, recent data show that lysine or arginine methylation of histone tails might be important in transcriptional regulation (Nielsen et al., 2001; Noma et al., 2001). The MeCP2- and v-ErbA-targeted corepressor complexes might contain methylase proteins that regulate CAII transcription by histone methylation as has been shown for the Rb protein (Nielsen et al., 2001).
How does TSA induce CAII transcription?
Whereas activation of CAII by AZAdC or thyroid hormone treatment coincides with the release of one corepressor complex from the CAII locus, addition of TSA did not alter the binding of these complexes to the locus. TSA treatment induced the anticipated hyperacetylation of histones H3 and H4 at the HS2 enhancer probably due to the inhibition of the activity of the HDAC3-containing corepressor complex. Remarkably, the promoter appears to be hyperacetylated even in its repressed state in untreated proliferating cells, suggesting that the promoter chromatin may be ‘open’ and primed for transcription. Similar phenomena have been reported for a number of genes in different organisms. For example, in the mouse yolk sac, the inactive β-globin minor promoter is hyperacetylated (Forsberg et al., 2000) and the promoter of the mouse MMTV-LTR was reported to have a relatively high level of acetylation when the gene is inactive (Sheldon et al., 2001). Hyperacetylated histones are also associated with a selected number of other yeast promoters, suggesting that inactive promoters are not necessarily hypoacetylated (Deckert and Struhl, 2001). If histone acetylation is not the trigger for transcription activation at the represssed CAII promoter by TSA, the question arises as to what other mechanisms could be involved. An explanation might be that TSA affects the acetylation state of non-histone components, such as basal transcription factors. Recent data suggest that the activity of the basal transcription factors TFIIE and TFIIF may be regulated by acetylation (Imhof et al., 1997). TSA treatment may also result in acetylation of GATA-1 associated with the HS2 enhancer and activation of this important erythroid-specific transcription factor (Boyes et al., 1998). Interestingly, at the yeast PHO8 promoter, histone acetylation provides a signal for the remodeling of chromatin by SWI–SNF (Reinke et al., 2001). PHO8 promoter acetylation is transient, since acetylation levels at the repressed and activated PHO8 promoter appear to be similar. Thus, histone acetylation might precede activation of a gene, ‘priming’ it for expression. The acetylated state of the CAII promoter might reflect this intermediate state of the promoter, which is still repressed but primed for activation.
Detailed analysis of the mechanisms by which HDAC inhibitors such as TSA activate transcription will be required to provide more insights for effective use of these inhibitors for cancer treatment.
Materials and methods
ChIP and real-time PCR
Chromatin isolation was performed according to Orlando et al. (1997). Approximately 3 × 108 HD3 cells were grown in 100 cm2 dishes, cross-linked by the addition of formaldehyde (1% final concentration), sonicated and chromatin was isolated by CsCl gradients. The equivalent of ∼107 cells were used for one immunoprecipitation reaction. For chromatin immunoprecipitations, 200 µl chromatin samples were diluted 2-fold in IP buffer (1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0, 150 mM NaCl and protease inhibitors) and precleared with 70 µl of a 50% protein A/G–Sepharose slurry (Santa Cruz) containing 10 µg sonicated salmon sperm DNA and 1 mg/ml bovine serum albumin (BSA) for 1 h with agitation at 4°C. Precleared chromatin was incubated with antibodies overnight at 4°C after which 50 µl of a 50% protein A/G–Sepharose slurry was added and immunocomplexes were recovered. Immunoprecipitates were washed five times subsequently with low salt buffer (as IP buffer), twice with high salt buffer (1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0, 500 mM NaCl and protease inhibitors) and twice with TE buffer (10 mM Tris, 1 mM EDTA). The immunocomplexes were eluted twice from the beads by adding 200 µl 1% SDS in 0.1 M NaHCO3 at room temperature for 15 min. For the reversal of cross-links, 16 µl 5 M NaCl was added and the samples were incubated at 65°C for 4 h, after which the samples were phenol– chloroform extracted and precipitated with 20 µg glycogen as carrier. Samples were dissolved in 50 µl TE.
The precipitated DNA was quantified by real-time PCR with the GeneAmp 5700 Sequence Detection System (PE Biosystems) using the SYBR Green I kit (PE Biosystems). Unless indicated otherwise, a PCR with a background control primer set (NC3/4) was performed which was used as control and was arbitrarily set to 1. All other PCR signals were calculated compared to this. Furthermore, total DNA from the IP input was included in the PCR and all PCR signals were corrected for this. Oligonucleotides used for ChIP analysis: CAII HS2: HS7, 5′-TCTGGAACATCCTTGCTA-3′; 3P, 5′-AGCGGATGATGTAGAGAT-3′; background control: NC3, 5′-GCAGACACTGGCAGGTTTC-3′; NC4, 5′-TATGAGCCTTAGCCTTAG-3′; promoter: 7A, 5′-CGTGCCCCGCGCACGGAG-3′; 8, 5′-GGCGGGGGGGCAAGAGGCG-3′.
Cell culture
Two derivatives of the AEV-transformed cell line HD3, namely HD3-EpoR and HD3-V3, expressing, respectively, the murine erythropoietin receptor or a gag-chicken TR fusion, were used. Before T3 treatment, HD3-V3 cells were grown for 48 h in medium containing stripped serum; 300 nM T3 was added to the medium where indicated.
Northern blot
Total RNA was extracted using the guanidium–CsCl method; CAII and Band 4.1 mRNA levels were detected by northern blot analysis as previously described (Zenke et al., 1990).
Antibodies
The following antibodies were used: 1G10 antibody against v-ErbA, Myc antibody 1-9E10.2 (ascites hybridoma cells ATCC CRL-1729). The polyclonal antibodies NCoR (C-20), HDAC1 (H51), HDAC2 (H54), Sin3A (AK11), Sin3B (AK12), SRC1 (M-341), TRAP220 (M-255) and RXR (ΔN197) were from Santa Cruz. The polyclonal antibodies against ac-H3 (06-599) and ac-H4 (06-598) were obtained from Upstate Biotechnology. Other antibodies used: polyclonal antibody against NCoR (provided by J.Torchia), polyclonal antibodies against MeCP2 (provided by A.Wolffe), monoclonal antibody against GATA-1 (provided by D.Engel), polyclonal antibody against HDAC4 (provided by T.Kouzarides), polyclonal antibody against MTA2 (provided by R.Brouwer), HDAC3 (provided by J.Wong). TBL-1 rabbit polyclonal antibody was raised against amino acids 1–251 of human TBL-1. The CHD4/Mi-2 antibodies are described in Xue et al. (1998). SMRT rabbit polyclonal antibody has been described in Li et al. (2000).
DNA-affinity binding assay
Double-stranded oligos Gal4: 5′-GATCGGAGGACAGTACTCCG-3′, VRE: 5′-TCGACCCAGCAAGGTCACAGCAGGGCTTTTTTTC-3′. Oligos were multimerized and end-labeled using the large Klenow fragment of Escherichia coli DNA polymerase and biotin-11-dCTP (Gibco-BRL), followed by purification on a spin column. Double-stranded oligo (100 µg) was incubated with 1 mg streptavidin–agarose beads (Sigma) in WB1000 (1 M NaCl, 20 mM Tris pH 8.0, 1 mM EDTA, 10% glycerol) buffer for 30 min at room temperature. Beads were washed twice with WB1000 and three times with WB150 [150 mM NaCl, 20 mM Tris pH 8.0, 1 mM EDTA, 10% glycerol, 1× protease inhibitor mix, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 µg/µl BSA]. Diluted nuclear extracts (150 mM NaCl) were incubated with 10 µg bio-oligo coupled to streptavidine–agarose beads for 2 h at 4°C on a rotating wheel. Bound proteins were washed five times with buffer WB150, boiled in 1× sample buffer and subjected to immunoblotting.
Immunoprecipitations
Nuclear extracts from HD3 or HD3-V3 cells were prepared according to Dignam et al. (1983). Diluted nuclear extracts (200 mM NaCl) were pre-cleared with protein A–agarose beads, the protein extract was then incubated with either anti-v-ErbA, anti-myc or anti-MeCP2 for 4 h at 4°C on a rotating wheel. Bound proteins were washed five times in buffer D (200 mM NaCl, 0.25% NP-40, 10 mM HEPES, 1 mM EDTA, protease inhibitors, 1 mM PMSF, 1 µg/µl BSA), boiled in 1× sample buffer and subjected to immunoblotting.
Acknowledgments
Acknowledgements
We thank C.Logie, M.Lohrum and S.Mandrup for reading the manuscript and members of the laboratory for suggestions and continued discussions. We are grateful to J.Wong, J.Torchia, A.Wolffe, T.Kouzarides, D.Engel and R.Brouwer for kindly providing antibodies. The project was supported by an EU-Biomed II grant and KWF grant to E.C.
References
- Agalioti T., Lomvardas,S., Parekh,B., Yie,J., Maniatis,T. and Thanos,D. (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell, 103, 667–678. [DOI] [PubMed] [Google Scholar]
- Antequera F., Boyes,J. and Bird,A. (1990) High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell, 62, 503–514. [DOI] [PubMed] [Google Scholar]
- Ballestar E. and Wolffe,A.P. (2001) Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem., 268, 1–6. [DOI] [PubMed] [Google Scholar]
- Baniahmad A., Steiner,C., Kohne,A.C. and Renkawitz,R. (1990) Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell, 61, 505–514. [DOI] [PubMed] [Google Scholar]
- Bird A.P. and Wolffe,A.P. (1999) Methylation-induced repression—belts, braces and chromatin. Cell, 99, 451–454. [DOI] [PubMed] [Google Scholar]
- Boyes J., Byfield,P., Nakatani,Y. and Ogryzko,V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- Braliou G.G., Ciana,P., Klaassen,W., Gandrillon,O. and Stunnenberg,H.G. (2001) The v-ErbA oncoprotein quenches the activity of an erythroid-specific enhancer. Oncogene, 20, 775–787. [DOI] [PubMed] [Google Scholar]
- Chen J.D. and Evans,R.M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature, 377, 454–457. [DOI] [PubMed] [Google Scholar]
- Ciana P., Braliou,G.G., Demay,F.G., von Lindern,M., Barettino,D., Beug,H. and Stunnenberg,H.G. (1998) Leukemic transformation by the v-ErbA oncoprotein entails constitutive binding to and repression of an erythroid enhancer in vivo. EMBO J., 17, 7382–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello J.F. et al. (2000) Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature Genet., 24, 132–138. [DOI] [PubMed] [Google Scholar]
- Danam R.P., Howell,S.R., Remack,J.S. and Brent,T.P. (2001) Hetero geneous methylation of the O(6)-methylguanine–DNA methyltrans ferase promoter in immortalized IMR90 cell lines. Int. J. Oncol., 18, 187–193. [DOI] [PubMed] [Google Scholar]
- Deckert J. and Struhl,K. (2001) Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol., 21, 2726–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet C., Lurquin,C., Lethe,B., Martelange,V. and Boon,T. (1999) DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol. Cell. Biol., 19, 7327–7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disela C., Glineur,C., Bugge,T., Sap,J., Stengl,G., Dodgson,J., Stunnenberg,H., Beug,H. and Zenke,M. (1991) v-erbA over expression is required to extinguish c-erbA function in erythroid cell differentiation and regulation of the erbA target gene CAII. Genes Dev., 5, 2033–2047. [DOI] [PubMed] [Google Scholar]
- Downward J., Yarden,Y., Mayes,E., Scrace,G., Totty,N., Stockwell,P., Ullrich,A., Schlessinger,J. and Waterfield,M.D. (1984) Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature, 307, 521–527. [DOI] [PubMed] [Google Scholar]
- Fondell J.D., Ge,H. and Roeder,R.G. (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA, 93, 8329–8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg E.C., Downs,K.M., Christensen,H.M., Im,H., Nuzzi,P.A. and Bresnick,E.H. (2000) Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA, 97, 14494–14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M. and Frommer,M. (1987) CpG islands in vertebrate genomes. J. Mol. Biol., 196, 261–282. [DOI] [PubMed] [Google Scholar]
- Glass C.K. and Rosenfeld,M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- Guenther M.G., Lane,W.S., Fischle,W., Verdin,E., Lazar,M.A. and Shiekhattar,R. (2000) A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev., 14, 1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Heinzel T. et al. (1997) A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature, 387, 43–48. [DOI] [PubMed] [Google Scholar]
- Horlein A.J. et al. (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature, 377, 397–404. [DOI] [PubMed] [Google Scholar]
- Huang E.Y., Zhang,J., Miska,E.A., Guenther,M.G., Kouzarides,T. and Lazar,M.A. (2000) Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev., 14, 45–54. [PMC free article] [PubMed] [Google Scholar]
- Imhof A., Yang,X.J., Ogryzko,V.V., Nakatani,Y., Wolffe,A.P. and Ge,H. (1997) Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol., 7, 689–692. [DOI] [PubMed] [Google Scholar]
- Ito M. and Roeder,R.G. (2001) The TRAP–SMCC–Mediator complex and thyroid hormone receptor function. Trends Endocrinol. Metab., 12, 127–134. [DOI] [PubMed] [Google Scholar]
- Ito M. et al. (1999) Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell, 3, 361–370. [DOI] [PubMed] [Google Scholar]
- Jepsen K. et al. (2000) Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell, 102, 753–763. [DOI] [PubMed] [Google Scholar]
- Jones P.L., Veenstra,G.J., Wade,P.A., Vermaak,D., Kass,S.U., Landsberger, N., Strouboulis,J. and Wolffe,A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- Jones P.L., Sachs,L.M., Rouse,N., Wade,P.A. and Shi,Y.B. (2001) Multiple N-CoR complexes contain distinct histone deacetylases. J. Biol. Chem., 276, 8807–8811. [DOI] [PubMed] [Google Scholar]
- Kao H.Y., Downes,M., Ordentlich,P. and Evans,R.M. (2000) Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev., 14, 55–66. [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Kim,J.H., Lee,Y.C., Cheong,J. and Lee,J.W. (2000) Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-κB and serum response factor. J. Biol. Chem., 275, 12470–12474. [DOI] [PubMed] [Google Scholar]
- Li J., Wang,J., Wang,J., Nawaz,Z., Liu,J.M., Qin,J. and Wong,J. (2000) Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J., 19, 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S. and Roeder,R.G. (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci., 25, 277–283. [DOI] [PubMed] [Google Scholar]
- Merika M. and Thanos,D. (2001) Enhanceosomes. Curr. Opin. Genet Dev., 11, 205–208. [DOI] [PubMed] [Google Scholar]
- Müller C., Readhead,C., Diederichs,S., Idos,G., Yang,R., Tidow,N., Serve,H., Berdel,W.E. and Koeffler,H.P. (2000) Methylation of the cyclin A1 promoter correlates with gene silencing in somatic cell lines, while tissue-specific expression of cyclin A1 is methylation independent. Mol. Cell. Biol., 20, 3316–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näär A.M., Beaurang,P.A., Zhou,S., Abraham,S., Solomon,W. and Tjian,R. (1999) Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature, 398, 828–832. [DOI] [PubMed] [Google Scholar]
- Nan X., Meehan,R.R. and Bird,A. (1993) Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res., 21, 4886–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- Nielsen S.J. et al. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature, 412, 561–565. [DOI] [PubMed] [Google Scholar]
- Noma K., Allis,C.D. and Grewal,S.I. (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science, 293, 1150–1155. [DOI] [PubMed] [Google Scholar]
- Onate S.A., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1995) Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science, 270, 1354–1357. [DOI] [PubMed] [Google Scholar]
- Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Pain B., Melet,F., Jurdic,P. and Samarut,J. (1990) The carbonic anhydrase II gene, a gene regulated by thyroid hormone and erythropoietin, is repressed by the v-erbA oncogene in erythrocytic cells. New Biol., 2, 284–294. [PubMed] [Google Scholar]
- Ranchez C., Lemon,B.D., Suldan,Z., Bromleigh,V., Gamble,M., Naar, A.M., Erdjument-Bromage,H., Tempst,P. and Freedman,L.P. (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature, 398, 824–828. [DOI] [PubMed] [Google Scholar]
- Reinke H., Gregory,P.D. and Horz,W. (2001) A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell, 7, 529–538. [DOI] [PubMed] [Google Scholar]
- Rietveld L.E.G., Caldenhoven,E. and Stunnenberg,H.G. (2001) Avian erythroleukemia: a model for corepressor function in cancer. Oncogene, 20, 3100–3109. [DOI] [PubMed] [Google Scholar]
- Sap J., Munoz,A., Damm,K., Goldberg,Y., Ghysdael,J., Leutz,A., Beug,H. and Vennstrom,B. (1986) The c-erb-A protein is a high-affinity receptor for thyroid hormone. Nature, 324, 635–640. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Sheldon L.A., Becker,M. and Smith,C.L. (2001) Steroid hormone receptor-mediated histone deacetylation and transcription at the mouse mammary tumor virus promoter. J. Biol. Chem., 276, 32423–32426. [DOI] [PubMed] [Google Scholar]
- Underhill C., Qutob,M.S., Yee,S.P. and Torchia,J. (2000) A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem., 275, 40463–40470. [DOI] [PubMed] [Google Scholar]
- Urnov F.D., Yee,J., Sachs,L., Collingwood,T.N., Bauer,A., Beug,H., Shi,Y.B. and Wolffe,A.P. (2000) Targeting of N-CoR and histone deacetylase 3 by the oncoprotein v-erbA yields a chromatin infrastructure-dependent transcriptional repression pathway. EMBO J., 19, 4074–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu,J., Suka,N. and Grunstein,M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Wade P.A. (2001) Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene, 20, 3166–3173. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Gegonne,A., Jones,P.L., Ballestar,E., Aubry,F. and Wolffe,A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- Wan M., Zhao,K., Lee,S.S. and Francke,U. (2001) MECP2 truncating mutations cause histone H4 hyperacetylation in Rett syndrome. Hum. Mol. Genet., 10, 1085–1092. [DOI] [PubMed] [Google Scholar]
- Wen Y.D., Perissi,V., Staszewski,L.M., Yang,W.M., Krones,A., Glass,C.K., Rosenfeld,M.G. and Seto,E. (2000) The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl Acad. Sci. USA, 97, 7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Suka,N., Carlson,M. and Grunstein,M. (2001) TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell., 7, 117–126. [DOI] [PubMed] [Google Scholar]
- Xue Y., Wong,J., Moreno,G.T., Young,M.K., Cote,J. and Wang,W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell, 2, 851–861. [DOI] [PubMed] [Google Scholar]
- Yuan C.X., Ito,M., Fondell,J.D., Fu,Z.Y. and Roeder,R.G. (1998) The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl Acad. Sci. USA, 95, 7939–7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke M. et al. (1988) v-erbA specifically suppresses transcription of the avian erythrocyte anion transporter (band 3) gene. Cell, 52, 107–119. [DOI] [PubMed] [Google Scholar]
- Zenke M., Munoz,A., Sap,J., Vennstrom,B. and Beug,H. (1990) v-erbA oncogene activation entails the loss of hormone-dependent regulator activity of c-erbA. Cell, 61, 1035–1049. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ng,H.H., Erdjument-Bromage,H., Tempst,P., Bird,A. and Reinberg,D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]