Abstract
Nornicotine is a secondary tobacco alkaloid that is produced by the N-demethylation of nicotine. Nornicotine production and accumulation in tobacco are undesirable because nornicotine serves as the precursor in the synthesis of the well characterized carcinogen N′-nitrosonornicotine during the curing and processing of tobacco. Although nornicotine is typically a minor alkaloid in tobacco plants, in many tobacco populations a high percentage of individuals can be found that convert a substantial proportion of the nicotine to nornicotine during leaf senescence and curing. We used a microarray-based strategy to identify genes that are differentially regulated between closely related tobacco lines that accumulate either nicotine (nonconverters) or nornicotine (converters) as the predominant alkaloid in the cured leaf. These experiments led to the identification of a small number of closely related cytochrome P450 genes, designated the CYP82E2 family, whose collective transcript levels were consistently higher in converter versus nonconverter tobacco lines. RNA interference-induced silencing of the CYP82E2 gene family suppressed the synthesis of nornicotine in strong converter plants to levels similar to that observed in nonconverter individuals. Although each of the six identified members of the P450 family share >90% nucleotide sequence identity, sense expression of three selected isoforms revealed that only one (CYP82E4v1) was involved in the conversion of nicotine to nornicotine. Yeast expression analysis revealed that CYP82E4v1 functions as a nicotine demethylase. Identification of the gene(s) responsible for nicotine demethylation provides a potentially powerful tool toward efforts to minimize nornicotine levels, and thereby N′-nitrosonornicotine formation, in tobacco products.
Keywords: N′-nitrosonornicotine, N-demethylation, tobacco, alkaloid, tobacco-specific nitrosamines
The four major alkaloids produced in Nicotiana tabacum are nicotine, nornicotine, anabasine, and anatabine. In most commercial varieties, nicotine is the predominant alkaloid representing 90-95% of the total alkaloid content, whereas the remaining three alkaloids account for a relatively small percentage (5-10%) of the alkaloid pool. During curing and processing of tobacco products, a portion of the leaf alkaloids undergoes nitrosation, leading to the formation of tobacco-specific nitrosamines (TSNAs) (1, 2). Numerous studies have documented the carcinogenic properties of TSNAs (1, 3, 4). Although seven distinct TSNAs have been detected in tobacco, N′-nitrosonornicotine (NNN) and 4-methylnitrosoamino-1-(3-pyridyl)-1-butanone (NNK) are considered the most detrimental given their potency and abundance in tobacco products (1, 3, 5).
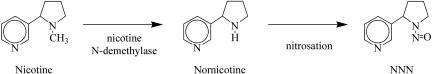
The primary biochemical mechanism of NNN formation is the N-nitrosation of nornicotine, an alkaloid produced through the N-demethylation of nicotine by the enzyme nicotine N-demethylase (Fig. 1) (6). Although nornicotine typically represents <5% of the total alkaloid content in cultivated tobacco, nornicotine levels can dramatically increase by a mechanism termed “conversion” in which plants that accumulate nicotine as their principal alkaloid give rise to progeny that metabolize a large portion (as high as 95%) of leaf nicotine to nornicotine. In individuals that have genetically converted (termed “converters”), N-demethylation of nicotine to nornicotine primarily occurs during senescence and curing (7). For reasons yet to be determined, the frequency of conversion is much higher in burley than in flue-cured tobaccos and occurs at rates as high as 20% per generation in some burley cultivars.
Fig. 1.
Structures of nicotine, nornicotine, and NNN.
Investigations aimed at understanding the genetic mechanisms underlying the conversion of nicotine to nornicotine began more than half a century ago. The pioneering work of Griffith et al. (8) followed by reports by Burk and Jeffrey (9) and Mann et al. (10) demonstrated that the high nornicotine phenotype in converter tobacco plants is controlled by a single dominant genetic locus. Because nicotine N-demethylase, the enzyme that mediates the final step in nornicotine biosynthesis (Fig. 1), clearly plays a pivotal role in the conversion process, several studies have been conducted to characterize this protein (11-14). Although the inability to purify active nicotine N-demethylase from crude extracts has impeded the isolation and identification of the enzyme, experimental evidence from these studies suggested that the enzyme may be a cytochrome P450 (P450) monooxygenase. Whether the dominant converter locus revealed by genetic studies represents the nicotine N- demethylase gene itself, or alternatively, encodes an upstream regulator of the processes that ultimately give rise to the metabolism of nicotine to nornicotine in the leaf, is unknown.
Identification of the gene(s) involved in the conversion of nicotine to nornicotine could lead to new approaches to help minimize nornicotine accumulation in the tobacco leaf. Maintaining low nornicotine levels is not only desirable because of its well characterized role as the precursor of NNN, but also because the results of a recent study suggest that nornicotine per se may be responsible for unwanted health effects. Dickerson and Janda (15) demonstrated that nornicotine can induce aberrant glycation of proteins and showed the increased accumulation of modified proteins in the blood plasma of smokers. Furthermore, the same report provided evidence that nornicotine can react covalently with commonly used steroid drugs, such as prednisone, potentially altering both the efficacy and toxicity of these drugs.
Here, we report the identification of a closely related family of tobacco P450 genes, at least one member of which encodes an enzyme with nicotine demethylase activity. The inhibition of gene expression of this P450 family was effective in suppressing nornicotine production in strong converter tobacco genotypes to levels typically observed in nonconverter individuals.
Materials and Methods
Plant Materials. All plant materials used in this study were provided by Earl Wernsman (North Carolina State University). Isogenic doubled haploid burley lines DH 91-1307-46(NC) and DH91-1307-46(Con) (nonconverter and converter, respectively) were used in generating the cDNA libraries that served as the source for creating the EST databases. For microarray hybridizations, RNAs were isolated from the full-sib doubled haploid burley lines DH98-326-3 (nonconverter) and DH98-326-1 (converter), and DH98-325-5 (nonconverter) and DH98-325-6 (converter). Near-isogenic flue-cured lines SC58(cTcT) (nonconverter) and SC58(CTCT) (converter) were also used in microarray assays. SC58(CTCT) was developed through the introgression of the single dominant converter locus (CT) found in the tobacco progenitor species Nicotiana tomentosiformis (10) into cultivar SC58.
All plants were maintained in growth chambers or greenhouses by using standard potting soil and fertilizer. For the microarray studies, individual leaves were excised, and their petioles were inserted into a solution of 0.1% ethephon or 1% sodium bicarbonate for 5-7 h, treatments that have been shown to accelerate the metabolism of nicotine to nornicotine (16, 17). Treated leaves were subsequently placed in small plastic storage bags after being lightly sprayed with water (to maintain high humidity) and cured for 3 days at 30°C in the dark. Leaves of the transgenic plant materials were dipped in a solution of 0.2% ethephon, dried, and cured in plastic storage bags for 7 days at room temperature in the dark before alkaloid analysis.
cDNA Libraries and ESTs. Total cellular RNA was isolated from senescing leaf tissue of burley lines DH91-1307-46(NC) and DH91-1307-46(Con) by using the TRIzol reagent according to the manufacturer's protocol (Invitrogen). Poly(A)+ RNA was isolated from total RNA by using the MessageMaker system (Invitrogen), and cDNA was subsequently synthesized and directionally cloned into the λZAP II phage vector by using the ZAP-cDNA Synthesis and Gigapack III Gold Cloning Kit (Stratagene). Aliquots of the phage libraries were converted to pBluescript-based plasmid libraries following the mass excision protocol outlined by Stratagene. High-throughput automated DNA sequencers were used to generate EST information for 11,136 randomly chosen cDNAs from the converter library and 11,904 cDNAs randomly selected from the nonconverter library. Single pass sequencing runs were conducted on the presumed 5′ ends of the cDNAs by using the T3 primer. The local alignment search tool blastx (18) was used to compare the predicted protein sequence of each tobacco cDNA with the nonredundant protein database curated by the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Access to all of the sequences in the converter and nonconverter databases is available through the Tobacco Genome Initiative web site maintained at North Carolina State University (www.tobaccogenome.org). Entries corresponding to the converter EST database are prefixed by “RED0”; entries corresponding to the nonconverter database are prefixed by “REDTWO.”
Preparation of DNA Chips. DNA chips were prepared as described by Eisen and Brown (19). Briefly, cDNA inserts were PCR-amplified with M13 forward and reverse sequencing primers (Qiagen, Valencia, CA) and purified by using the Millipore Multiscreen PCR or Montage PCRμ96 systems. Amplified DNAs were spotted onto amino silane-coated slides (Corning GAPS II) by using an Affymetrix (Santa Clara, CA) GMS 417 array printer. DNAs were immobilized to the slide surface by UV crosslinking (≈120 mJ/m2), followed by baking at 75°C for 2 h.
Microarray Hybridization and Analysis. The amino allyl dUTP-based indirect method of dye incorporation described by the Institute of Genome Research (http://pga.tigr.org) was used to generate labeled cDNAs from nonconverter and converter RNAs by using the Cy3 and Cy5 fluorescent dyes (Amersham Pharmacia Biosciences). Prehybridization, cDNA hybridization to the DNA slides, and the posthybridization washes were conducted as outlined by Alba et al. (20). Microarrays were scanned by using scanarray 2.1 (GSI Lumonics, Billerica, MA) or scanarray express (PerkinElmer). Acquired array images were quantified for signal intensity with quantarray analysis software (PerkinElmer) by using the histogram method.
Cloning Full-Length and Additional Members of the CYP82E2 Gene Family. To obtain the complete coding regions of CYP82E2 and CYP82E3 we used a modified 5′-RACE strategy with a pBlue-script II vector-specific forward primer (Bluescript SK; 5′-CGCTCTAGAACTAGTGATC-3′) and a set of gene-specific reverse primers located in the 3′ UTR of the respective genes, with our converter cDNA library serving as template. The partial 3D_C12-15 fragment was recovered from a PCR using the Bluescript SK forward primer and a reverse primer complementary to a sequence within the coding region of CYP82E2 (5′-GTTAGATTTATCGTACTCGAATT-3′). In addition to the 3D_C12-15 fragment, this amplification yielded the 5′ terminal region of CYP82E4v2. Full-length CYP82E4v2 was recovered by using a gene-specific 5′ primer (5′-ATGCTTTCTCCCATAGAAGCC-3′) and a pBluescript-specific reverse primer (5′-TCGAGGTCGACGGTATC-3′). CYP82E4v1 was amplified with a forward primer complementary to the 5′ terminus of the 3D_C12-15 coding region (5′-ATGGTTTTTCCCATAGAAGCC-3′) in conjunction with the pBluescript-specific reverse primer. All PCRs and the subsequent cloning of amplification products into T/A cloning vectors for sequence analysis were carried out as described (21). Nucleic acid and predicted protein sequences of the various members of the CYP82E2 gene family were analyzed and compared by using the blastx (18), clustalw (22), and gap (GCG) algorithms.
Transgenic Plant Analysis. The RNA interference (RNAi)-based gene silencing construct was assembled in a version of the pKYL80 cloning vector (23) that was engineered to contain a 151-bp fragment of the soybean FAD3 gene intron between the XhoI and SacI restriction sites of the polylinker (pKYLX80I). To create a construct in which the FAD3 intron was flanked by a sense and antisense fragment of CYP82E2, a 99-bp region located immediately upstream of the stop codon was cloned between the HindIII-XhoI and SacI-XbaI restriction sites of pKYLX80I in its sense and antisense orientation, respectively. The resulting HindIII-XbaI fragment containing the CYP82E2 sense arm, FAD3 intron, and CYP82E2 antisense arm was subcloned into the pKYLX71 plant binary expression vector (24) between the 35S cauliflower mosaic virus promoter and a rubisco small subunit terminator. Overexpression constructs were created by using the pBI121 plant expression vector as described (21). The pBI121- and pKYLX71-based constructs were transformed into Agrobacterium tumefaciens strain LBA 4404 and introduced into tobacco cultivars Petite Havana and DH98-325-6, respectively, by using established protocols (25).
RNA Blot and Alkaloid Analysis. Total cellular RNAs were isolated from tobacco leaves by using the TRIzol reagent as described by the manufacturer (Invitrogen). RNA immobilization, probe labeling, and signal detection were carried out by using the digoxigenin nucleic acid labeling and detection kits according to the manufacturer's instructions (Roche Applied Science). Alkaloid analysis of the sampled leaf material was performed as described (17).
Expression of the Members of the CYP82E2 P450 Family in Yeast and Metabolism of Nicotine. The subcloning of the plant P450 cDNAs into expression vector pYeDP60, transformation into yeast strain WAT11, and preparation of microsomal extracts were all conducted as outlined (21). Nicotine metabolism assays were conducted in 150-μl reaction volumes containing 50 mM phosphate buffer (pH 7.1), 2.45 μM [pyrrolidine-2-14C] nicotine (Moravek Biochemicals, Brea, CA), 0.75 mM NADPH, and 0.6 mg/ml microsomal protein. After a 45-min incubation at 27°C, the reaction was arrested by adding 50 μl of acetone, followed by centrifugation at 14,000 × g for 2 min. Fifty microliters of the supernatant was spotted onto a 250-μm Whatman K6F silica plate and developed in a chloroform/methanol/ammonia (60:10:1) (vol/vol) solvent system. Radioactive traces were visualized with a Bioscan (Washington, DC) System 400 imaging scanner. The veracity of the [14C]-nornicotine and [14C]-nicotine peaks was determined according to their comigration with nonradiolabeled standards.
Results
Microarray Analysis of Converter Versus Nonconverter Plants. To facilitate transcript profiling of converter versus nonconverter tobacco plants, EST databases were generated by using cDNA libraries derived from mRNAs expressed in senescing leaves of the isogenic doubled haploid lines DH91-1307-46(NC) and DH91-1307-46(Con) (nonconverter and converter, respectively). Each database represented a compilation of >11,000 sequencing runs of randomly selected clones from each library. During the course of this study, two types of DNA chips were synthesized, one corresponding to 4,992 cDNAs selected from the converter cDNA library, and another corresponding to 6,963 cDNAs that represented the complete nonredundant unigene set predicted by clustering analyses of the combined converter and nonconverter EST databases.
Numerous hybridizations were conducted by using RNAs isolated from three closely related converter/nonconverter genotypes that had been subjected to different treatments known to enhance the metabolic conversion of nicotine to nornicotine in converter plants (see Materials and Methods). Collective analysis of multiple, independent microarray hybridizations revealed three highly homologous, yet unique, genes predicted to encode P450 enzymes that consistently showed greater hybridization to fluorescently labeled cDNAs from each of the converter plants in comparison to their nonconverter partners. An ≈2-fold higher signal was observed for each of these P450 genes when hybridized to fluorescently labeled cDNAs derived from converter leaves that are actively metabolizing nicotine to nornicotine than to fluorescently labeled cDNAs from their nonconverter counterparts (Tables 4-6, which are published as supporting information on the PNAS web site). Although the enhancement in hybridization intensity was relatively modest, the results were consistent across numerous independent microarray experiments and were further confirmed with RNA blotting assays (data not shown). A more complete description of the results and analysis of the microarray assays is presented in Supporting Text, which is published as supporting information on the PNAS web site.
Based on the first member of this gene family observed in our EST database, we collectively refer to these P450s as the CYP82E2 gene family. For cDNAs of this family for which full-length DNA sequence information has been obtained, the standardized “CYP” system of P450 nomenclature is used (26); for those members for which only partial sequence information is known, our laboratory-based codes are used (e.g., 131A_A02, an EST represented in our nonconverter database).
Sequence Analysis of the CYP82E2 Gene Family. CYP82E2 and CYP82E3 were the first cDNAs of the CYP82E2 gene family that were identified by the microarray experiments as candidates for involvement in the conversion process. Because neither of the original cDNAs corresponding to these genes was predicted to contain full-length ORFs, a PCR strategy was used in which vector-specific and gene-specific primer pairs were used to amplify the missing 5′ coding regions of these cDNAs. In addition to yielding the desired full-length coding sequences of CYP82E2 and CYP82E3, three additional closely related members of this gene family were revealed as amplification products. A partial-length cDNA, designated 3D_C12-15, was generated by using a PCR primer internal to the CYP82E2 coding region and a vector-specific primer. In an attempt to generate a full-length version of 3D_C12-15, a primer corresponding to the first seven codons of the reading frame was used in an additional anchored PCR. Instead of yielding a full-length 3D_C12-15 clone, however, an additional member of the gene family, designated CYP82E4v1, was amplified. Similarly, PCR amplifications designed to recover additional sequence information for CYP82E2 resulted in the amplification of CYP82E4v2 (see Materials and Methods). The predicted amino acid sequence of CYP82E4v2 differs from CYP82E4v1 only at the second and third codon positions. Because the 5′ primer used to amplify CYP82E4v1 was defined according to the 3D_C12-15 sequence, it is possible that CYP82E4v1 and CYP82E4v2 originated from a common gene (discussed below).
Nucleotide and predicted amino acid sequence alignments of the members of the CYP82E2 gene family are shown in Figs. 5 and 6, which are published as supporting information on the PNAS web site. Pairwise GAP alignments of the CYP82E2 family show that all members share >90% identity at both the nucleotide and predicted amino acid sequence levels (Tables 7 and 8, which are published as supporting information on the PNAS web site). blastx analysis of members of the CYP82E2 family against the nonredundant GenBank database revealed greatest sequence homology to CYP82E1, a tobacco P450 of unknown function that is up-regulated in response to fungal elicitors (27). CYP82E2 gene family members share <70% amino acid sequence with CYP82E1 and <50% amino acid identity toward any other plant P450 gene currently deposited in the GenBank database (data not shown). Although the CYP82E2 P450 family is currently comprised of the six sequences shown in Fig. 5, it is possible that even more members of this P450 family may reside within the tobacco genome. Consistent with this possibility are the results of Southern blotting assays that reveal very complex patterns when members of the CYP82E2 family are used as hybridization probes (data not shown).
Transgenic Plant Analysis of the CYP82E2 Gene Family. To determine whether members of the CYP82E2 P450 family are involved in the metabolic conversion of nicotine to nornicotine, transgenic plants were generated by using constructs designed to either enhance or inhibit gene expression. To test the effects of down-regulating gene activity, an RNAi strategy was used. Because each member of the CYP82E2 family we have characterized shares >90% DNA sequence identity, RNAi-based constructs synthesized against one member would be expected to silence the entire gene family. An RNAi construct was generated against a 99-bp sequence near the 3′ end of the CYP82E2 coding region (Fig. 5) and transformed into the strong converter burley line DH98-325-6.
Ten independently transformed individuals were selected to assess the effects of the CYP82E2/RNAi construct on the metabolic conversion of nicotine to nornicotine. Leaves from each of the CYP82E2/RNAi individuals, and two plants transformed with the control vector alone, were treated with ethephon and cured for 7 days. Alkaloid analysis of these materials is shown in Table 1. Typical of line DH98-325-6, ethephon treatment and curing resulted in substantial nornicotine production in the two control plants (48.9% and 87.8% conversion of nicotine to nornicotine). In dramatic contrast, 7 of the 10 independent transgenic plants possessing the CYP82E2/RNAi construct displayed minimal nicotine to nornicotine conversion, with conversion percentages ranging from 2.8% to 7.0%. The other three CYP82E2/RNAi lines displayed alkaloid contents similar to the vector-only control plants. Concentrations of the minor alkaloids anabasine and anatabine were not significantly influenced by the presence of the CYP82E2/RNAi transgene (data not shown).
Table 1. Alkaloid analysis of DH 98-325-6 plants independently transformed with the CYP82E2/RNAi construct (and vector control).
| Sample | % Nicotine* | % Nornicotine* | % Conversion† |
|---|---|---|---|
| CYP82E2/RNAi | |||
| 1 | 3.149 | 0.100 | 2.8 |
| 2 | 2.569 | 0.193 | 7.0 |
| 3 | 2.175 | 0.064 | 2.9 |
| 4 | 3.517 | 0.125 | 3.4 |
| 5 | 1.085 | 0.868 | 44.4 |
| 6 | 0.025 | 2.260 | 98.9 |
| 7 | 0.027 | 1.867 | 98.6 |
| 8 | 2.268 | 0.128 | 5.3 |
| 9 | 2.197 | 0.133 | 5.7 |
| 10 | 2.434 | 0.112 | 4.4 |
| Vector control | |||
| 3 | 1.811 | 1.1735 | 48.9 |
| 11 | 0.290 | 2.090 | 87.8 |
Leaves were treated with ethephon and cured for 7 days.
Percentage of leaf dry weight.
[% nornicotine/(% nicotine + % nornicotine)] × 100.
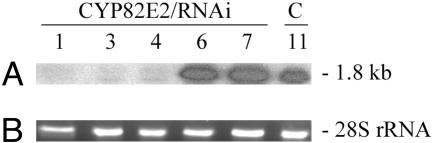
To confirm that gene silencing of the CYP82E2 gene family was indeed correlated with the low nornicotine phenotype, an RNA blot analysis was conducted by using RNAs isolated from three transgenic plants possessing CYP82E2/RNAi constructs and displaying low nornicotine phenotypes, two individuals transformed with the CYP82E2/RNAi construct yet still showing high levels of nornicotine and one vector-only control plant. In this experiment, the CYP82E4v1 cDNA was used as the probe under standard hybridization and wash conditions where cross-hybridization to the entire CYP82E2 gene family would be expected. As shown in Fig. 2, a strong hybridization signal was detected in each plant showing a high nornicotine phenotype, and minimal hybridization was detected in the plants transformed with the CYP82E2/RNAi construct that displayed low nornicotine phenotypes. We thus conclude that silencing of the CYP82E2 gene family inhibits the metabolic conversion of nicotine to nornicotine in tobacco.
Fig. 2.
RNA blot analysis of transgenic plants possessing the CYP82E2/RNAi construct. (A) Hybridization of the CYP82E4v1 probe to RNAs isolated from ethephon-treated, cured leaves of transgenic plants displaying low nornicotine phenotypes (CYP82E2/RNAi-1, -3, and -4) and high nornicotine phenotypes (CYP82E2/RNAi-6 and -7) and a vector control plant (C-11). Estimated size of the hybridizing band is indicated in kb. (B) Ethidium bromide staining of the portion of the gel used in A that contains the 28S ribosomal RNA to show RNA loading equivalence.
To assess the effects of overexpression of gene activity, cDNAs from the three members of the CYP82E2 gene family for which we first obtained full-length sequence information (CYP82E2, CYP82E3, and CYP82E4v1) were cloned in their sense orientations downstream of the 35S cauliflower mosaic virus promoter. These constructs were subsequently introduced into the N. tabacum cultivar Petite Havana, an experimental tobacco known for its comparatively short generation time. Although the Petite Havana plants in our possession were strong converters, alkaloid assays were conducted with green noncured leaves, a tissue type where the 35S cauliflower mosaic virus promoter is very active, yet endogenous conversion is not manifest.
Alkaloid analysis of the Petite Havana transgenic plants is shown in Table 2. Four independently transformed plants expressing the CYP82E2 and CYP82E4v1 constructs were tested along with seven individuals expressing CYP82E3 and three plants transformed with the pBI121 control vector. As expected, the green, noncured leaves of the three vector-only control plants accumulated minimal amounts of nornicotine. Likewise, all plants transformed with the CYP82E2 and CYP82E3 constructs showed minimal metabolic conversion of nicotine to nornicotine. In contrast, all four plants independently transformed with CYP82E2v1 contained nornicotine as the predominant alkaloid in the green, untreated leaf; nicotine to nornicotine conversion percentages ranged from 94.6 to 98.6.
Table 2. Alkaloid analysis of individual Petite Havana plants transformed with CYP82E2, CYP82E3, and CYP82E4v1 constructs or the pBl121 vector control.
| Sample | % Nicotine* | % Nornicotine* | % Conversion† |
|---|---|---|---|
| Vector control | |||
| 2 | 0.673 | 0.018 | 2.6 |
| 8 | 0.605 | 0.014 | 2.3 |
| 10 | 0.694 | 0.017 | 2.4 |
| CYP82E2 | |||
| 1 | 0.706 | 0.005 | 0.7 |
| 2 | 0.814 | 0.022 | 2.6 |
| 3 | 0.630 | 0.010 | 1.6 |
| 4 | 0.647 | 0.010 | 1.5 |
| CYP82E3 | |||
| 1 | 0.761 | 0.011 | 1.4 |
| 2 | 0.507 | 0.009 | 1.7 |
| 4 | 0.653 | 0.015 | 2.2 |
| 5 | 0.643 | 0.013 | 2.0 |
| 6 | 0.521 | 0.007 | 1.3 |
| 7 | 0.716 | 0.015 | 2.1 |
| 8 | 0.701 | 0.027 | 3.7 |
| CYP82E4v1 | |||
| 1 | 0.005 | 0.347 | 98.6 |
| 2 | 0.006 | 0.255 | 97.4 |
| 3 | 0.017 | 0.300 | 94.6 |
| 4 | 0.010 | 0.384 | 97.5 |
Green leaves were harvested and analyzed without treatment.
Percentage of leaf dry weight.
[% nornicotine/(% nicotine + % nornicotine)] × 100.
Although all Petite Havana plants, other than those expressing construct CYP82E4v1, showed low levels of nornicotine in green leaves, one plant expressing a CYP82E2 construct, CYP82E2 (1), appeared to accumulate even less nornicotine than the control plants. We speculated that the low nornicotine content of CYP82E2(1) may be the result of cosuppression of the CYP82E2 gene family in this specific plant, a phenomenon frequently observed in transgenic plants even when a transgene is expressed in its sense orientation (28). To test this prediction, alkaloid profiles were determined on ethephon-treated, cured leaves of CYP82E2 (1) and two vector-only control plants. As shown in Table 3, ethephon treatment and curing resulted in >97% nicotine to nornicotine conversion in the two control plants, whereas similarly treated CYP82E2 (1) leaves displayed negligible conversion (0.6%). Leaves from five other plants expressing either CYP82E2 or CYP82E3 transgenes also were subjected to ethephon treatment and curing. In each case, a high nornicotine phenotype was observed, similar to the vector-only control plants (data not shown).
Table 3. Alkaloid analysis of CYP82E2 (1) and pBI121 vector control plants.
| Sample | % Nicotine* | % Nornicotine* | % Conversion† |
|---|---|---|---|
| Vector control | |||
| 8 | 0.009 | 0.425 | 97.9 |
| 10 | 0.008 | 0.560 | 98.6 |
| CYP82E2 (1) | 1.185 | 0.007 | 0.6 |
Leaves were treated with ethephon and cured for 7 days.
Percentage of leaf dry weight.
[% nornicotine/(% nicotine + % nornicotine)] × 100.
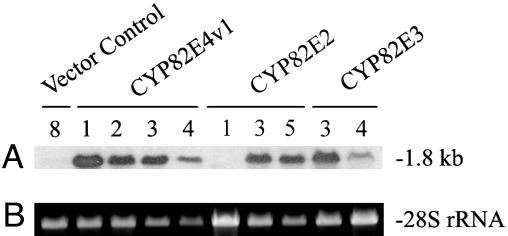
RNA blot assays were conducted on select plants representing each of the Petite Havana transgenic genotypes (Fig. 3). Using the CYP82E4v1 cDNA as a hybridization probe, minimal signal was detected with RNA isolated from green, untreated leaves of the vector-only control plant. In contrast, hybridization was easily detected in RNA samples from all four independent transgenic plants possessing the CYP82E4v1 construct. A strong hybridization signal was similarly observed by using RNA from all other transgenic plants tested that were transformed with either the CYP82E2 or the CYP82E3 constructs, with the exception of the low nornicotine-containing plant CYP82E2 (1). The RNA blotting assays showed that the 35S cauliflower mosaic virus promoter was effective in mediating a high level of transcript accumulation for each of the three members of the CYP82E2 gene family tested in this study. Failure to detect a hybridization signal in plant CYP82E2 (1) is consistent with the interpretation that the CYP82E2 gene family has been silenced by cosuppression in this individual.
Fig. 3.
RNA blot analysis of transgenic plants possessing sense orientation constructs of members of the CYP82E2 gene family. (A) Hybridization of the CYP82E4 probe to RNAs isolated from untreated leaves of independent transgenic lines expressing CYP82E2, CYP82E3, CYP82E4v1, and a vector control plant. Estimated size of the hybridizing band is indicated in kb. (B) Ethidium bromide staining of the portion of the gel used in A that contains the 28S ribosomal RNA to show RNA loading equivalence.
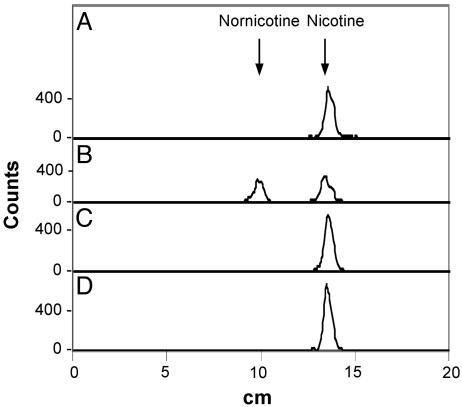
Expression of CYP82E4 cDNAs in Yeast. The transgenic plant results described above could be explained by either of two possibilities: (i) CYP82E4v1 encodes the nicotine demethylase activity that is directly responsible for the metabolic conversion of nicotine to nornicotine; or (ii) CYP82E4v1 produces a signaling molecule or biochemical cofactor that ultimately results in the activation of a separate nicotine demethylase enzyme. To test the former possibility, the CYP82E4v1 cDNA was cloned into the pYeDP60 yeast expression vector and transformed into yeast strain WAT11, a cell line designed to enhance the expression of heterologous plant P450s through the coexpression of an Arabidopsis P450 reductase gene (29). Microsomal membrane preparations from yeast cells expressing CYP82E4v1 or the vectoronly control were incubated in the presence of [14C]-nicotine. As shown in Fig. 4, microsomes from yeast cells expressing the CYP82E4v1 gene actively converted [14C]-nicotine to [14C]-nornicotine; no metabolism was observed by using microsomes isolated from the vector control yeast. The closely related CYP82E4v2 P450 similarly catalyzed the metabolism of [14C]-nicotine to [14C]-nornicotine when expressed in yeast (data not shown). In contrast, no nornicotine synthesis was detected by using microsomal membrane preparations from yeast expressing the closely related CYP82E2 and CYP82E3 cDNAs (Fig. 4).
Fig. 4.
Thin-layer chromatograms of products obtained after yeast microsomal membrane preparations were incubated in the presence of [14C]-nicotine. Assays were conducted by using microsomes isolated from yeast expressing either the vector control (A), CYP82E4v1 (B), CYP82E2 (C), or CYP82E3 (D).
Discussion
The results of this study clearly implicate the CYP82E2 gene family, and the CYP82E4 gene(s) in particular, as playing a major role in the metabolic conversion of nicotine to nornicotine in tobacco. The inhibition of transcript accumulation of this family using an RNAi-based construct effectively suppressed the high nornicotine phenotype of the strong converter burley genotype DH98-325-6. Likewise, cosuppression of this gene family greatly inhibited nornicotine production in the strong converter Petite Havana background. The contrast in alkaloid phenotypes between the cosuppressed CYP82E2 (1) plant and vector-only control Petite Havana plants was most dramatic in leaves that had been ethephon-treated and cured (0.6% conversion versus >97% conversion; Table 3). However, it is noteworthy that the nornicotine content of the CYP82E2 (1) plant also appeared to be reduced even in green, untreated leaves where a high nornicotine phenotype is typically not manifest in converter or nonconverter tobacco plants (0.7% versus 1.3-3.7%).
Although the RNAi- or cosuppression-mediated inhibition of the entire CYP82E2 gene family was effective in reversing the converter phenotypes of DH98-326-6 and Petite Havana plants, the sense expression studies implicated only one of three tested members in directly mediating the nicotine conversion process. Despite the fact that the predicted amino acid sequence of CYP82E4v1 is 92.8% and 94.8% identical to the CYP82E2 and CYP82E3 protein products, respectively, the few differences that do exist are apparently sufficient to enable the proteins to be functionally distinct. The observation that not all members of this closely related gene family participate in the process of nicotine conversion provides a reasonable explanation for the apparent inconsistency between the dramatic alkaloid phenotypes observed in response to the manipulation of this gene family in transgenic plants and the relatively modest increases in transcript accumulation (≈2-fold) observed between converter versus nonconverter plants revealed in the microarray experiments. Cross-hybridization among CYP82E2 family members not involved in nicotine conversion could result in a modest enhancement in overall hybridization signal if, for example, only one member of this gene family was actively induced in the plant materials tested. Expression assays capable of distinguishing the transcript levels of each individual member of the CYP82E2 gene family in converter versus nonconverter plants would be of particular interest for future studies.
The question of whether CYP82E4v1 and CYP82E4v2 represent unique P450s remains open. The protein encoded by the CYP82E4v1 cDNA differs from CYP82E4v2 only at the two amino acid residues immediately after the start methionine (Fig. 6). The codons corresponding to these amino acids were represented in the PCR primer used to generate the CYP82E4v1 cDNA, which was actually based on the sequence of 3D_C12-15, because our original intention was to amplify a full-length cDNA of 3D_C12-15 (as described in Results). Therefore, it is possible that the original mRNA template from which CYP82E4v1 was amplified was the same as that corresponding to the CYP82E4v2 gene, with the PCR primer sequences introducing the changes observed in the second and third amino acid positions. However, three additional sequence polymorphisms were also observed between these cDNAs that did not alter the predicted protein sequence (Fig. 5), suggesting that they, in fact, may be unique. Regardless of whether CYP82E4v1 and CYP82E4v2 represent the same or unique loci within the tobacco genome, the yeast expression assays demonstrated that both enzymes function as N-nicotine demethylases.
In conclusion, our identification and characterization of the CYP82E2 gene family provide insights into the mechanism by which tobacco plants produce nornicotine. The transgene-mediated inhibition of this gene family proved to be very effective in minimizing nornicotine synthesis even in strong converter tobacco lines. Application of this technology should aid in reducing the amount of nicotine that becomes metabolized to nornicotine during the senescence and curing of the leaf, particularly in tobacco lines where the frequency of genetic conversion is high. Lowering nornicotine levels, in turn, should help minimize the subsequent production and accumulation of NNN in the processed leaf.
Supplementary Material
Acknowledgments
We thank Newton Kalengamaliro and Alec Hayes (Philip Morris USA, Richmond, VA) for valuable discussions and review of this publication; Lowell Bush for GC analysis of tobacco alkaloids; Carol Griffin for excellent technical assistance; Earl Wernsman for supplying all of the tobacco lines used in this study and being a continual source of insight and inspiration; Dennis Pompon and Phillippe Urban (Centre de Génétique Moléculaire du Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) for their kind gifts of the pYeDP60 vector and the WAT11 yeast strain; and Randy Dinkins and Arthur Hunt (University of Kentucky) for providing the pKYLX80I and pKYLX71 vectors. This work was supported under a research funding agreement with Philip Morris USA.
Abbreviations: P450, cytochrome P450; RNAi, RNA interference. NNN, N′-nitrosonornicotine.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. DQ131885 (CYP82E4v2), DQ131886 (CYP82E4v1), DQ131887 (CYP82E2), DQ131888 (CYP82E3), DQ131889 (3D_C12-15), and DQ131890 (131A_A02)].
References
- 1.Hecht, S. S. (1998) Chem. Res. Toxicol. 11, 559-603. [DOI] [PubMed] [Google Scholar]
- 2.Williams, D. L. H. (1999) Acc. Chem. Res. 32, 869-876. [Google Scholar]
- 3.Hecht, S. S. & Hoffmann, D. (1989) Cancer Surv. 8, 273-294. [PubMed] [Google Scholar]
- 4.Hoffmann, D., Brunnemann, K. D., Prokopczyk, B. & Djordjevic, M. V. (1994) J. Toxicol. Environ. Health 41, 1-52. [DOI] [PubMed] [Google Scholar]
- 5.Hecht, S. S. (2003) Nat. Rev. Cancer 3, 733-744. [DOI] [PubMed] [Google Scholar]
- 6.Bush, L. P., Cui, M., Shi, H., Burton, H. R., Fannin, F. F., Lei, L. & Dye, N. (2001) Rec. Adv. Tob. Sci. 27, 23-46. [Google Scholar]
- 7.Wernsman, E. A. & Matzinger, D. F. (1968) Tob. Sci. 12, 226-228. [Google Scholar]
- 8.Griffith, R. B., Valleau, W. D. & Stokes, G. W. (1955) Science 121, 343-344. [DOI] [PubMed] [Google Scholar]
- 9.Burk, L. G. & Jeffrey, R. N. (1958) Tob. Sci. 2, 139-141. [Google Scholar]
- 10.Mann, T. J., Weybrew, J. A., Matzinger, D. F. & Hall, J. L. (1964) Crop Sci. 4, 349-353. [Google Scholar]
- 11.Hao, D. Y. & Yeoman, M. M. (1996) Phytochemistry 42, 325-329. [Google Scholar]
- 12.Hao, D. Y. & Yeoman, M. M. (1996) Phytochemistry 41, 477-482. [Google Scholar]
- 13.Hao, D. Y. & Yeoman, M. M. (1998) J. Plant Physiol. 152, 420-426. [Google Scholar]
- 14.Chelvarajan, R. L., Fannin, F. F. & Bush, L. P. (1993) J. Agri. Food Chem. 41, 858-862. [Google Scholar]
- 15.Dickerson, T. J. & Janda, K. D. (2002) Proc. Natl. Acad. Sci. USA 99, 15084-15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fannin, F. F. & Bush, L. P. (1992) Med. Sci. Res. 380, 33-41. [Google Scholar]
- 17.Shi, H. Z., Kalengamaliro, N. E., Krauss, M. R., Hempfling, W. P. & Gadani, F. (2003) J. Agri. Food Chem. 51, 7679-7683. [DOI] [PubMed] [Google Scholar]
- 18.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen, M. B. & Brown, P. O. (1999) Methods Enzymol. 303, 179-205. [DOI] [PubMed] [Google Scholar]
- 20.Alba, R., Fei, Z. J., Payton, P., Liu, Y., Moore, S. L., Debbie, P., Cohn, J., D'Ascenzo, M., Gordon, J. S., Rose, J. K. C., et al. (2004) Plant J. 39, 697-714. [DOI] [PubMed] [Google Scholar]
- 21.Siminszky, B., Corbin, F. T., Ward, E. R., Fleischmann, T. J. & Dewey, R. E. (1999) Proc. Natl. Acad. Sci. USA 96, 1750-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins, D., Thompson, J., Gibson, T., Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schardl, C. L., Byrd, A. D., Benzion, G., Altschuler, M. A., Hildebrand, D. F. & Hunt, A. G. (1987) Gene 61, 1-11. [DOI] [PubMed] [Google Scholar]
- 24.Maiti, I. B., Murphy, J. F., Shaw, J. G. & Hunt, A. G. (1993) Proc. Natl. Acad. Sci. USA 90, 6110-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G. & Fraley, R. T. (1985) Science 227, 1229-1231. [Google Scholar]
- 26.Nelson, D. R., Koymans, L., Kamataki, T., Stegeman, J. J., Feyereisen, R., Waxman, D. J., Waterman, M. R., Gotoh, O., Coon, M. J., Estabrook, R. W., et al. (1996) Pharmacogenetics 6, 1-42. [DOI] [PubMed] [Google Scholar]
- 27.Takemoto, D., Hayashi, M., Doke, N., Nishimura, M. & Kawakita, K. (1999) Plant Cell Physiol. 40, 1232-1242. [DOI] [PubMed] [Google Scholar]
- 28.Fagard, M. & Vaucheret, H. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 167-194. [DOI] [PubMed] [Google Scholar]
- 29.Pompon, D., Louerat, B., Bronine, A. & Urban, P. (1996) Methods Enzymol. 272, 51-64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.