Abstract
Nitrification, the microbial oxidation of ammonia to nitrite and nitrate, occurs in a wide variety of environments and plays a central role in the global nitrogen cycle. Catalyzed by the enzyme ammonia monooxygenase, the ability to oxidize ammonia was previously thought to be restricted to a few groups within the β- and γ-Proteobacteria. However, recent metagenomic studies have revealed the existence of unique ammonia monooxygenase α-subunit (amoA) genes derived from uncultivated, nonextremophilic Crenarchaeota. Here, we report molecular evidence for the widespread presence of ammonia-oxidizing archaea (AOA) in marine water columns and sediments. Using PCR primers designed to specifically target archaeal amoA, we find AOA to be pervasive in areas of the ocean that are critical for the global nitrogen cycle, including the base of the euphotic zone, suboxic water columns, and estuarine and coastal sediments. Diverse and distinct AOA communities are associated with each of these habitats, with little overlap between water columns and sediments. Within marine sediments, most AOA sequences are unique to individual sampling locations, whereas a small number of sequences are evidently cosmopolitan in distribution. Considering the abundance of nonextremophilic archaea in the ocean, our results suggest that AOA may play a significant, but previously unrecognized, role in the global nitrogen cycle.
Keywords: Crenarchaeota, nitrification, ammonia monooxygenase
Nitrogen (N) is an essential nutrient in the ocean and limits biological productivity in most marine ecosystems. At the global scale, biological N fixation is the largest source of N to the ocean, whereas anaerobic microbial processes are responsible for N losses (1, 2). However, biological N fixation and anaerobic N losses are ultimately connected by nitrification, the microbially mediated, two-step conversion of ammonium ( ) to nitrate (
) to nitrate ( ) via nitrite (
) via nitrite ( ). As much as 4 × 1011 kg of N cycles through the ocean each year (1), and nearly all of this N must be nitrified at least once (3). Although nitrification is important throughout the ocean, it plays a critical role in the coastal ocean by linking the decomposition of nitrogenous organic matter to N loss via denitrification (microbial conversion of
). As much as 4 × 1011 kg of N cycles through the ocean each year (1), and nearly all of this N must be nitrified at least once (3). Although nitrification is important throughout the ocean, it plays a critical role in the coastal ocean by linking the decomposition of nitrogenous organic matter to N loss via denitrification (microbial conversion of 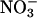 to N2 gas). By removing a large percentage of anthropogenic N pollution from estuaries and continental shelf regions before it can reach the open ocean (4), coupled nitrification/denitrification effectively isolates the marine N cycle from the heavily altered terrestrial N cycle (1). In the open ocean, 30-50% of all N loss occurs in pelagic oxygen minimum zones (OMZs), where massive N losses have recently been attributed to anaerobic ammonium oxidation, or “anammox” (5). During annamox, oxidation of
to N2 gas). By removing a large percentage of anthropogenic N pollution from estuaries and continental shelf regions before it can reach the open ocean (4), coupled nitrification/denitrification effectively isolates the marine N cycle from the heavily altered terrestrial N cycle (1). In the open ocean, 30-50% of all N loss occurs in pelagic oxygen minimum zones (OMZs), where massive N losses have recently been attributed to anaerobic ammonium oxidation, or “anammox” (5). During annamox, oxidation of  occurs at the expense of
occurs at the expense of  produced by either heterotrophic
produced by either heterotrophic  reduction or aerobic ammonia oxidation, the first step of nitrification. Nitrification is therefore a particularly significant process in OMZs and at oxic/anoxic interfaces in coastal sediments, where the complex interplay between nitrification, denitrification, and anammox drives rapid N transformations and large N losses to the atmosphere.
reduction or aerobic ammonia oxidation, the first step of nitrification. Nitrification is therefore a particularly significant process in OMZs and at oxic/anoxic interfaces in coastal sediments, where the complex interplay between nitrification, denitrification, and anammox drives rapid N transformations and large N losses to the atmosphere.
Only a few phylogenetically restricted groups of microorganisms are known to perform either of the two steps of nitrification (conversion of  to
to  and
and  to
to  ), all of which are members of the domain Bacteria. Because they catalyze the first and rate-limiting step of nitrification, ammonia-oxidizing bacteria (AOB) have received considerable attention in a wide variety of habitats, including soils, freshwater, marine water columns, estuaries, and sediments (6). Despite their critical biogeochemical role in both pelagic and benthic oceanic environments, AOB often comprise only 0.1% of bacterial assemblages (7). In contrast, microorganisms from the domain Archaea are ubiquitous and often abundant in the ocean (8-10), but their biogeochemical role remains unclear. Archaea represent the third domain of life, evolutionarily distinct from the Eukarya and Bacteria, and were previously thought to inhabit mostly extreme environments (11). Nonextremophilic archaea (Crenarchaeota and Euryarchaeota) are now recognized to be widespread in the ocean (12, 13), but our current understanding of their physiology and biogeochemical function remains largely speculative. Although some planktonic archaea may be heterotrophic because of their uptake of amino acids (14, 15), there is evidence that some marine archaea may be chemoautotrophs (16), capable of light-independent carbon (C) fixation (15, 17). Further insight into potential metabolic capabilities of planktonic archaea comes from recent work by Venter et al. (18), who investigated microbial genomic diversity in the Sargasso Sea by shotgun DNA sequencing. Based on their discovery of a unique ammonia monooxygenase gene on an archaeal-associated scaffold, Venter et al. suggest that some archaea may be capable of performing chemoautotrophic nitrification.
), all of which are members of the domain Bacteria. Because they catalyze the first and rate-limiting step of nitrification, ammonia-oxidizing bacteria (AOB) have received considerable attention in a wide variety of habitats, including soils, freshwater, marine water columns, estuaries, and sediments (6). Despite their critical biogeochemical role in both pelagic and benthic oceanic environments, AOB often comprise only 0.1% of bacterial assemblages (7). In contrast, microorganisms from the domain Archaea are ubiquitous and often abundant in the ocean (8-10), but their biogeochemical role remains unclear. Archaea represent the third domain of life, evolutionarily distinct from the Eukarya and Bacteria, and were previously thought to inhabit mostly extreme environments (11). Nonextremophilic archaea (Crenarchaeota and Euryarchaeota) are now recognized to be widespread in the ocean (12, 13), but our current understanding of their physiology and biogeochemical function remains largely speculative. Although some planktonic archaea may be heterotrophic because of their uptake of amino acids (14, 15), there is evidence that some marine archaea may be chemoautotrophs (16), capable of light-independent carbon (C) fixation (15, 17). Further insight into potential metabolic capabilities of planktonic archaea comes from recent work by Venter et al. (18), who investigated microbial genomic diversity in the Sargasso Sea by shotgun DNA sequencing. Based on their discovery of a unique ammonia monooxygenase gene on an archaeal-associated scaffold, Venter et al. suggest that some archaea may be capable of performing chemoautotrophic nitrification.
amoA encodes the catalytic α-subunit of ammonia monooxygenase, the enzyme responsible for catalyzing the rate-limiting step in bacterial ammonia oxidation. Because amoA is well conserved and required by all AOB, this gene has been used extensively as a molecular marker for cultivation-independent studies of AOB communities. The possibility that some archaea carry an amoA was confirmed by Schleper et al. (19), who found a Sargasso Sea-like amoA homolog on the same 43-kb metagenomic fragment as a 16S rRNA gene from soil Crenarchaeota, suggesting that archaea capable of ammonia oxidation may be present in soils. A definitive link between this novel amoA and archaeal ammonia oxidation was only recently established by cultivation of an ammonia-oxidizing member of the marine group 1 Crenarchaeota, the first cultivated representative from this dominant archaeal lineage (20). How widespread ammonia-oxidizing archaea (AOA) are in the marine environment, and their relative importance in the global N cycle, remain unknown.
In this study, we use unique PCR primers specifically targeting the archaeal amoA to document the distribution and diversity of AOA in water columns and sediments of the ocean.
Materials and Methods
Site Descriptions, Sample Collection, and DNA Extraction. Water samples were collected from three depths (σT = 15.7, 15.8, and 15.9) bracketing the secondary nitrite maximum in the suboxic zone of the Black Sea (April 2003) and two stations (designated C1 and M2) in Monterey Bay (April 2005) by using Niskin bottles mounted on a hydrography wire. Water samples were either filtered directly onto Sterivex capsules (0.2 μM filter, Millipore) with positive pressure (Black Sea) or a peristaltic pump was used (Monterey Bay). After filtration, capsules were capped, packed in polypropylene bags, frozen in liquid nitrogen, and stored at -80°C until DNA extraction. Briefly, DNA was extracted from Sterivex capsules by using 1.6 ml of lysis buffer (20 mM EDTA/400 mM NaCl/750 mM sucrose/50 mM Tris), 200 μl of 10% SDS, and 100 μl of proteinase K (10 mg/ml), incubated at 55°C for 2 h. After the addition of 1 ml of 100% ethanol, lysates were purified by using DNeasy columns (Qiagen, Valencia, CA).
Ten liters of water was collected in November 2003 from the secondary nitrite maximum (200-m depth) within the OMZ off the Pacific coast of Mexico, filtered onto a SpiralCap capsule, frozen, and stored below -70°C. For lysis, 12 ml of Tris-EDTA/1% SDS and 750 μl of proteinase K (10 mg/ml) was added to the capsules, followed by incubation at 55°C for 2 h. The lysate was removed, placed on ice for 90 min, and centrifuged for 40 min at 17,000 × g to remove cell debris. DNA was ethanol-precipitated overnight, pelleted at 32,000 × g for 30 min, resuspended in 300 μl of sterile water, and further purified by using Qiagen DNeasy columns.
Surface sediments were collected from San Francisco Bay and Bahiáa del Tóbari, Mexico (Gulf of California), using box cores deployed off small vessels in July 2004 and October 2004, respectively. Sediments were collected by hand from three sites in the Elkhorn Slough estuary (from head to mouth: Hudson's Landing, Hummingbird Island, and Vierra Marsh) in October 2004 and from the surf zone at the mouth of the unconfined Huntington Beach aquifer in July 2004. Soil aggregates were collected in June 2004 from the saturated C horizon at a noncontaminated “background” site near Oak Ridge, TN. Cores were collected from each environment by using cut-off 5-cc syringes, frozen on dry ice, and stored at -80°C. DNA was extracted from ≈0.25 g of sediment (0-0.5 cm depth interval) or soil by using the FastDNA SPIN kit for soil (Qbiogene, Carlsbad, CA).
PCR Primer Design, Amplification, and Cloning of Archaeal amoA Fragments. PCR primers were designed based on alignments of archaeal amoA genes and deduced amino acid sequences from the Sargasso Sea (GenBank accession no. AACY01435967) and German soil (GenBank accession no. AJ627422). Gene fragments (635 bp) were amplified by using the PCR primers Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′) with the following protocol: 95°C for 5 min; 30 cycles consisting of 94°C for 45 s, 53°C for 60 s, and 72°C for 60 s; and 72°C for 15 min. Triplicate PCRs were pooled (to minimize PCR bias), gel-purified, and cloned with the TOPO-TA cloning kit (Invitrogen). White colonies were transferred to 96-well plates containing LB broth (with 50 μg/ml kanamycin), grown overnight at 37°C, and PCR-screened directly for the presence of inserts by using T7 and M13R vector primers.
Sequencing and Phylogenetic, Rarefaction, and Statistical Analyses. Sequencing of T7/M13 PCR products was performed by using vector primers on Applied Biosystems 3730xl capillary sequencers. Nucleotide sequences were assembled and edited by using sequencher, version 4.1 (Gene Codes, Ann Arbor, MI). A 606-bp region of 377 archaeal amoA sequences was chosen for phylogenetic analysis. Nucleotide and amino acid alignments (based on 202 residues) were generated by using macclade (http://macclade.org). Neighbor-joining phylogenetic trees were constructed based on alignments of DNA sequences (using Jukes-Cantor corrected distances) and amino acid sequences within paup*4.0b10. Bootstrap analysis was used to estimate the reliability of phylogenetic reconstructions (1,000 replicates). To compare the archaeal amoA-based richness within each clone library, rarefaction analysis and Chao1 nonparametric richness estimations were performed by using dotur (21). Operational taxonomic units (OTUs) were defined as sequence groups in which sequences differed by ≤2% or 5%.
Results and Discussion
Ubiquity of AOA. Nonextremophilic Crenarchaeota have been previously detected in a wide variety of environments and are widespread in the ocean (12, 13); however, their role in the marine C or N cycle has remained a mystery. Our results suggest that archaea capable of ammonia oxidation are ubiquitous in marine water columns and sediments. An extensive PCR survey of archaeal amoA genes using our primers resulted in amplification of 635-bp fragments (corresponding to essentially the entire gene) from multiple sites or depths within eight different geographic locations: the water columns of Monterey Bay (base of the euphotic zone), the Eastern Tropical North Pacific (ETNP; OMZ), and the Black Sea (suboxic zone); coastal and estuarine sediments from San Francisco Bay, Elkhorn Slough, and Huntington Beach, CA, and Bahiáa del Tóbari, Mexico, and for purposes of comparison, a saturated (suboxic) soil sample from Oak Ridge, TN. In total, we produced 15 clone libraries and a database of 377 archaeal amoA sequences from water columns, sediments, and soil (Fig. 1).
Fig. 1.
Phylogenetic relationships among archaeal amoA sequences from water columns, sediments, and soil. Sequences are color-coded according to location; symbols are used to distinguish different sites or depths within a given location. One representative of sequence groups ≥99% identical is shown; additional symbols show the total number of clones represented by a sequence. Database sequences are shown in black. Bootstrap values (>50%) are indicated at branch points. Brackets highlight phylogenetic clusters referred to in the text. HI, Hummingbird Island; HL, Hudson's Landing; VM, Vierra Marsh.
Primers were designed based on the novel archaeal amoA sequences from German soil and the Sargasso Sea water column (18, 19). These two genes shared 76% DNA identity, but 85% identity and 91% similarity at the amino acid level. Because these archaeal amoA genes are distant homologs of known bacterial amoA genes from β- and γ-Proteobacteria (19), it is not possible to recover them from environmental DNA samples with bacterial amoA (or pmoA) primers. Although designed based on a limited number of available sequences, our primers recovered 377 archaeal amoA sequences ranging from 69% to 96% identity to the German soil sequence and from 69% to 99% identity to the Sargasso Sea sequence. Based on this extensive database, it may be possible to design new, more degenerate primers that capture even greater archaeal amoA diversity.
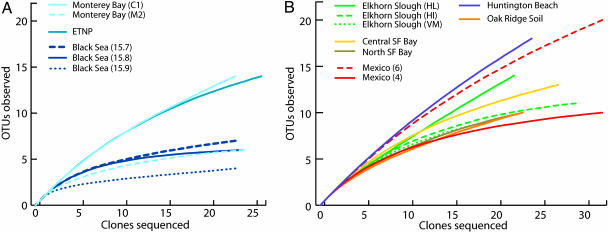
AOA Diversity. Overall, we recovered a total of 138 unique OTUs (based on a 2% cutoff) from 377 individual archaeal amoA sequences. Both observed and extrapolated AOA richness varied widely within water column and sediment libraries (Fig. 2), with Chao1 richness estimates ranging from 5 to 37 OTUs for water column libraries and 11 to 48 OTUs for sediment libraries (see Table 1, which is published as supporting information on the PNAS web site). These richness levels are comparable to, or higher than, those observed for bacterial amoA clones obtained from water columns and sediments (22, 23). Based on phylogenetic analysis of PCR clone libraries, individual sample locations often had unique archaeal amoA sequences associated with them (Fig. 1); however, clades of these unique AOA sequences clustered according to habitat. For example, all sequences from North San Francisco Bay (NB-1), our only freshwater site, fell exclusively into one coherent cluster. Similarly, all amoA sequences recovered from Tennessee soil fell into one large, phylogenetically distinct cluster (see soil/sediment cluster in Fig. 1). This cluster also included the amoA sequence from soil metagenomic clone 54d9, which is derived from a member of the most widely distributed Crenarchaeota lineage (group 1.1b) known in soils (19, 24). Interestingly, 141 of 142 water column sequences from the Black Sea, Monterey Bay, and the ETNP, despite widely different geographical locations and biogeochemical characteristics, fell into three distinct clusters (A, B, and C; Fig. 1). The largest of these clusters (cluster A) contains >70% of the water column sequences, including the Sargasso Sea sequence. It is tempting to speculate that these water column sequences correspond to strictly “planktonic” AOA; however, several sequences from Elkhorn Slough sediments also fell into these clades and were >99% identical to Black Sea and Monterey Bay water column sequences (Fig. 1). A number of sedimentary sequences also fell into the soil/sediment cluster. Based on Fig. 1, it is evident that while all but one of the water column sequences fell into the water column clusters, and all soil sequences fell into the large soil/sediment cluster, sequences within these clusters are not found exclusively in those environments. This finding in part reflects the wide range of archaeal amoA sequences recovered from sediments, with sequences falling in numerous clusters throughout the phylogenetic tree (Fig. 1).
Fig. 2.
Rarefaction curves indicating archaeal amoA richness within clone libraries derived from water columns (A) and sediments and soil (B). OTUs were defined as groups of sequences differing by ≤2% at the DNA level. Curves are color-coded as in Fig. 1. HI, Hummingbird Island; HL, Hudson's Landing; VM, Vierra Marsh.
AOA Diversity in the Water Column: The Black Sea, Monterey Bay, and ETNP. Based on the identification of an archaeal amoA gene in a Sargasso Sea surface water genomic library, we anticipated that similar genes would be present in other marine water column environments where nitrification plays a significant role. The Black Sea is the world's largest chemically stratified anoxic marine basin and encompasses a pronounced suboxic (O2 <5 μM) zone in which nitrogen cycling (e.g., denitrifcation, anammox) is extremely important (25, 26). Archaeal amoA was amplified from multiple depths within the suboxic zone of the Black Sea. Interestingly, this gene was present at densities (σT) similar to those where anaerobic ammonia oxidation has been reported (26). The majority (>88%) of the archaeal amoA sequences obtained from the Black Sea suboxic zone fell into cluster A and showed 90-92% nucleotide identity to the Sargasso Sea gene sequence. Within this cluster, there were four predominant sequence types differing by ≈1-2% at the DNA level (visible in an expanded tree, Fig. 3, which is published as supporting information on the PNAS web site), primarily at the third position. The remaining Black Sea sequences fell into clusters B and C. Overall, the Black Sea clone libraries were among the least diverse in this study (Fig. 2), and we appear to have recovered most or all of the OTUs predicted by the Chao1 richness estimator (Table 1).
The majority of sequences from the upper water column of Monterey Bay fell into cluster A along with the closely related Black Sea sequences (Fig. 1). Although archaeal amoA was successfully amplified from all depths analyzed between 30 and 80 m, for brevity we report here only sequence data derived from 30 m at a coastal site (C1) and 40 m at an offshore site (M2). The coastal station (C1) was used in part to determine the extent of overlap between planktonic AOA communities in Monterey Bay and benthic communities in Elkhorn Slough. (Because Elkhorn Slough is a small estuary that is tidally flushed by Monterey Bay seawater, Monterey Bay represents a potential source and sink for nutrients and microorganisms to the slough.) Near the offshore site (M2), nitrification rates (27) and AOB diversity (22) have recently been examined. Although there was some compositional overlap between the coastal (C1) and offshore (M2) archaeal amoA clone libraries, the coastal library was more diverse (Fig. 2). One subcluster within cluster A is comprised of seven C1 sequences (and one ETNP sequence) that share 93-99% identity with the Sargasso Sea sequence at the DNA level. Translation of these sequences revealed that all are identical to the Sargasso sequence at the amino acid level. These results indicate that the “microdiversity” within these coastal Monterey Bay sequences corresponds entirely to synonymous changes, with no effect on archaeal ammonia monooxygenase function. Additionally, most of the microdiverse (1-2% divergent) clusters of Black Sea and Monterey Bay sequences in cluster A are identical at the amino acid level. Given the differences between the suboxic zone of the Black Sea and the oxygenated euphotic zone of Monterey Bay, the compositional overlap between the archaeal amoA sequences from these two environments is surprising and suggests that planktonic AOA can tolerate a wide range of O2 levels.
This conclusion is consistent with our results from the ETNP, one of the largest pelagic OMZs in the ocean, where archaeal amoA was amplified successfully from a 200-m depth under low oxygen conditions (O2 = 3.1 μM) at the secondary nitrite maximum ( ). The ETNP and coastal Monterey Bay sequences were the most diverse of the six water column samples in the study (Fig. 2), with sequences from the ETNP falling into water column clusters A and B and one sediment cluster (see below). In contrast to the Black Sea and Monterey Bay libraries, the majority (21 of 26) of the ETNP sequences fell into cluster B, which included only nine sequences from the other water column libraries. Overall, we appear to have identified several key phylogenetic groups/clusters of planktonic AOA, based on the six archaeal amoA clone libraries generated from these three geographically and biogeochemically distinct water column environments.
). The ETNP and coastal Monterey Bay sequences were the most diverse of the six water column samples in the study (Fig. 2), with sequences from the ETNP falling into water column clusters A and B and one sediment cluster (see below). In contrast to the Black Sea and Monterey Bay libraries, the majority (21 of 26) of the ETNP sequences fell into cluster B, which included only nine sequences from the other water column libraries. Overall, we appear to have identified several key phylogenetic groups/clusters of planktonic AOA, based on the six archaeal amoA clone libraries generated from these three geographically and biogeochemically distinct water column environments.
Significance of AOA in the Water Column. Previous studies have shown marine crenarchaeal abundance in the water column to increase after blooms in surface phytoplankton and to be negatively correlated with chlorophyll concentrations (28). Archaea also dominate the microbial community in regions of the ocean where nitrification is known to be important (9) and, in the Santa Barbara Channel, crenarchaeal abundance and  concentrations were found to be significantly correlated over time (29). Many of the spatial and temporal patterns observed for marine Crenarchaeota as a group are therefore similar to patterns expected for nitrifying microorganisms. We suggest here that a significant portion of the archaeal community may be oxidizing remineralized
concentrations were found to be significantly correlated over time (29). Many of the spatial and temporal patterns observed for marine Crenarchaeota as a group are therefore similar to patterns expected for nitrifying microorganisms. We suggest here that a significant portion of the archaeal community may be oxidizing remineralized  (i.e., performing the first step of chemoautotrophic nitrification), and that some of these broader archaeal patterns in the water column may be driven in part by AOA.
(i.e., performing the first step of chemoautotrophic nitrification), and that some of these broader archaeal patterns in the water column may be driven in part by AOA.
In the Black Sea, for example, our AOA data are consistent with earlier work focused on marine Archaea as a group. Roughly 10% of the archaeal 16S rRNA clones previously reported from the chemocline (i.e., suboxic zone) of the Black Sea were most closely related to marine group 1 Crenarchaeota (30). Diversity was extremely low and only two 16S rRNA sequences (sharing 97% identity) were identified. In the present study, we recovered distinct phylogenetic clusters from the suboxic zone, but found archaeal amoA diversity to be quite low compared with other water column environments.
In addition, the presence of AOA in the suboxic water columns of the Black Sea and the ETNP is consistent with a peak in the concentration of crenarchaeol (the distinct membrane lipid biomarker for the Crenarchaeota) (31, 32) previously reported in the OMZ of the northwestern Arabian Sea (33). Because of the abundance of crenarchaeotal membrane lipids at oxygen levels <1 μM, Sinninghe Damste et al. (33) proposed that these organisms are likely facultative anaerobes, possibly capable of denitrification. Our data suggest that these Crenarchaeota may be AOA. Many bacteria capable of oxidizing ammonia, although they require oxygen to do so, are microaerophilic or facultatively anaerobic and known to produce nitric oxide and nitrous oxide (N2O) by “nitrifier denitrification” under low oxygen conditions (34). AOB have been shown to carry a copper-containing nitrite reductase gene (nirK) that may be involved in this N2O production pathway (35), and some AOA also carry a nirK gene (19). In pelagic OMZs, both nitrification and denitrification represent major sources of N2O to the atmosphere (36); however, the extent to which AOA are actively oxidizing ammonia and/or producing N2O under low-oxygen conditions is unknown.
AOA Diversity in Sediments: Elkhorn Slough, Bahiáa del Tóbari, San Francisco Bay, and Huntington Beach. Sediment DNA extracts from four geographically distinct locations were PCR-screened to determine the distribution and diversity of archaeal amoA sequences in coastal and estuarine sediments. Unique AOA communities were associated with each geographic location, and within these locations, different clusters were often specifically associated with individual sample sites. In Elkhorn Slough, different levels of richness (Fig. 2) and surprisingly little compositional overlap were found among clone libraries generated from three sites that span the estuarine gradient from head to mouth of the slough. For each slough site, at least one distinct clade of sequences was site-specific, with no overlap with sequences from other sites (Fig. 1). Similar shifts in AOA community composition and richness were observed in Bahiáa del Tóbari, a small estuary located along the west coast of Mexico in the Gulf of California. Multiple sequences from the interior of Tóbari (site 4) fell into the soil/sediment cluster, yet no sequences from the mouth of the bay (site 6) fell into this cluster (Fig. 1); in addition, the archaeal amoA library from the interior site was among the least diverse of the sediment libraries analyzed in this study, whereas the mouth site was among the most diverse (Fig. 2 and Table 1).
The factors that influence AOA diversity and community structure in Elkhorn Slough and Bahiáa del Tóbari are currently unknown. Although salinity has been shown to be a key factor influencing the distribution and diversity of AOB in estuarine systems, based on bacterial 16S rRNA (37) and amoA genes (23), differences between the sites within Elkhorn Slough and Bahiáa del Tóbari are probably not salinity-driven; the slough experiences daily tidal flushing by Monterey Bay seawater, and Tóbari receives only episodic inputs of fresh water via agricultural runoff (38). However, differences in AOA community composition in San Francisco Bay, the largest estuary on the west coast of the United States, are likely associated with salinity. As alluded to earlier, sequences from the 30.5-practical salinity units (psu) Central San Francisco Bay (CB-20) site were distributed throughout a number of different regions of the tree, whereas all of the low-salinity (0.5 psu) North San Francisco Bay (NB-1) sequences fell exclusively into one distinct phylogenetic cluster (Fig. 1). This clustering suggests that the North San Francisco Bay archaeal amoA sequences may represent a unique, low-salinity AOA type. Many of the Central San Francisco Bay (CB-20) sequences fell into a well supported cluster with clones from two well flushed Elkhorn Slough sites (Hummingbird Island and Vierra Marsh). These sequences share 93-100% identity at the DNA level and may represent a ubiquitous AOA group in coastal estuarine sediments of this geographic region.
As the primary substrate required for nitrification, NH4+ might also be expected to influence AOA community structure. Coastal permeable sediments from Huntington Beach are characterized by consistently elevated NH4+ concentrations caused by hydrologic connection with groundwater (39), which is highly enriched in NH4+ (groundwater [NH4+] > 150 μM). The archaeal amoA library from this site was the most diverse in this study (Fig. 2), yet many of the Huntington Beach sequences fell into a distinct cluster with a single sequence from the most NH4+-rich site sampled in Elkhorn Slough (Hudson's Landing).
The four sedimentary environments included in this study were selected in part because of highly variable physical and biogeochemical conditions found within and between them. Despite these pronounced differences, one broad phylogenetic cluster (located directly above cluster A) included sequences from all four of the marine sedimentary environments we sampled (Fig. 1). This cluster also included the only sequence from the ETNP that did not fall into any of the water column clusters. Given the differences between these five environments, this cluster may represent a cosmopolitan group of marine AOA. Preliminary characterization of ammonia-oxidizing enrichment cultures further supports this conclusion, as several archaeal amoA sequences recovered from both Elkhorn Slough sediment enrichments and Monterey Bay seawater enrichments fall into this cluster (K.J.R. and C.A.F., unpublished data).
Significance of AOA in Marine Sediments. Although phylogenetically diverse crenarchaeal 16S rRNA phylotypes have been previously identified in sediments of freshwater lakes (40, 41), estuaries (42), continental margins (43), and the deep sea (44), the biogeochemical role of Crenarchaeota in these sedimentary environments is unknown. Clearly, the ubiquity and diversity of AOA in coastal and estuarine sediments suggests that they may be pivotal in benthic N cycling. In coastal and estuarine ecosystems, nitrification is known to link the mineralization of organic nitrogen to the loss of fixed nitrogen by sedimentary denitrification. A common characteristic of all of the sediment samples analyzed in this study, as well as the suboxic water column and soil samples, is that denitrification functional genes (i.e., nitrite reductase) have also been successfully amplified from each of them. These results suggest that AOA may be intimately associated with denitrifiers and, like AOB, play an important role in coupled nitrification-denitrification. Interestingly, bacterial amoA could not be amplified from some of the samples analyzed in this study (e.g., Tóbari samples), which may indicate that archaeal amoA is more widespread than bacterial amoA, and that AOA are actually more widespread than AOB in sedimentary environments. Future studies focused on quantifying the relative abundance and expression of bacterial versus archaeal amoA genes under nitrifying conditions will be imperative for understanding the relative contributions of what are apparently two functionally equivalent, but evolutionarily distinct, groups of microorganisms.
Supplementary Material
Acknowledgments
We thank Amal Jayakumar for collecting the ETNP water sample and geochemical data; Scott Wankel for collecting sediment samples during the San Francisco Bay RMP sediment cruise; and Brendan Bohannan, John Spear, and Gregory O'Mullan for helpful comments on the manuscript. This work was supported in part by National Science Foundation Grant MCB-0433804 (to C.A.F.), a National Science Foundation Graduate Research Fellowship (to A.E.S.), and a National Science Foundation Dissertation Improvement Grant (to J.M.B.). B.B.O. was supported by National Science Foundation Microbial Observatory Grant MCB-0132101 and the National Aeronautics and Space Administration Astrobiology Institute.
Abbreviations: amoA, ammonia monooxygenase α-subunit; AOA, ammonia-oxidizing archaea; AOB, ammonia-oxidizing bacteria; ETNP, Eastern Tropical North Pacific; OMZ, oxygen minimum zone; OTU, operational taxonomic unit.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ14825-DQ14848 and DQ148573-DQ148905).
References
- 1.Galloway, J. N., Dentener, F. J., Capone, D. G., Boyer, E. W., Howarth, R. W., Seitzinger, S., Asner, G. P., Cleveland, C. C., Green, P. A., Holland, E. A., et al. (2004) Biogeochemistry 70, 153-226. [Google Scholar]
- 2.Gruber, N. & Sarmiento, J. L. (1997) Glob. Biogeochem. Cycles 11, 235-266. [Google Scholar]
- 3.Karl, D. M. (2002) Trends Microbiol. 10, 410-418. [DOI] [PubMed] [Google Scholar]
- 4.Seitzinger, S. (1988) Limnol. Oceanogr. 33, 702-724. [Google Scholar]
- 5.Kuypers, M. M. M., Lavik, G., Woebken, D., Schmid, M., Fuchs, B. M., Amann, R., Jørgensen, B. B. & Jetten, M. S. M. (2005) Proc. Natl. Acad. Sci. USA 102, 6478-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowalchuk, G. A. & Stephen, J. R. (2001) Annu. Rev. Microbiol. 55, 485-529. [DOI] [PubMed] [Google Scholar]
- 7.Ward, B. B. (2000) in Microbial Ecology of the Oceans, ed. Kirchman, D. (Wiley, New York), pp. 427-454.
- 8.Stein, J. L. & Simon, M. I. (1996) Proc. Natl. Acad. Sci. USA 93, 6228-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karner, M. B., DeLong, E. F. & Karl, D. M. (2001) Nature 409, 507-510. [DOI] [PubMed] [Google Scholar]
- 10.DeLong, E. F., Wu, K. Y., Prézelin, B. B. & Jovine, R. V. M. (1994) Nature 371, 695-697. [DOI] [PubMed] [Google Scholar]
- 11.Woese, C. R. (1987) Microbiol. Rev. 51, 221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., McCallum, K. & Davis, A. A. (1992) Nature 356, 148-149. [DOI] [PubMed] [Google Scholar]
- 13.DeLong, E. F. (1992) Proc. Natl. Acad. Sci. USA 89, 5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouverney, C. C. & Fuhrman, J. A. (2000) Appl. Environ. Microbiol. 66, 4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herndl, G. J., Reinthaler, T., Teira, E., van Aken, H., Veth, C., Pernthaler, A. & Pernthaler, J. (2005) Appl. Environ. Microbiol. 71, 2303-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson, A., McNichol, A. P., Benitez-Nelson, B. C., Hayes, J. M. & Eglinton, T. I. (2001) Geochim. Cosmochim. Acta 65, 3123-3137. [Google Scholar]
- 17.Wuchter, C., Schouten, S., Boschker, H. T. S. & Sinnighe Damsté, J. S. (2003) FEMS Microbiol. Lett. 219, 203-207. [DOI] [PubMed] [Google Scholar]
- 18.Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., Eisen, J. A., Wu, D., Paulsen, I., Nelson, K. E., Nelson, W., et al. (2004) Science 304, 66-74. [DOI] [PubMed] [Google Scholar]
- 19.Schleper, C., Jurgens, G. & Jonuscheit, M. (2005) Nat. Rev. Microbiol. 3, 479-488. [DOI] [PubMed] [Google Scholar]
- 20.Könneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B. & Stahl, D. A. (2005) Nature 437, 543-546. [DOI] [PubMed] [Google Scholar]
- 21.Schloss, P. D. & Handelsman, J. (2005) Appl. Environ. Microbiol. 71, 1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Mullan, G. D. & Ward, B. B. (2005) Appl. Environ. Microbiol. 71, 697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis, C. A., O'Mullan, G. D. & Ward, B. B. (2003) Geobiology 1, 129-140. [Google Scholar]
- 24.Ochsenreiter, T., Selezi, D., Quaiser, A., Bonch-Osmolovskaya, L. & Schleper, C. (2003) Environ. Microbiol. 5, 787-797. [DOI] [PubMed] [Google Scholar]
- 25.Codispoti, L. A., Friederich, G. E., Murray, J. W. & Sakamoto, C. M. (1991) Deep-Sea Res. 38, S691-S710. [Google Scholar]
- 26.Kuypers, M. M. M., Sliekers, A. O., Lavik, G., Schmid, M., Jørgensen, B. B., Kuenen, J. G., Sinninghe Damste, J. S., Strous, M. & Jetten, M. S. M. (2003) Nature 422, 608-611. [DOI] [PubMed] [Google Scholar]
- 27.Ward, B. B. (2005) Mar. Ecol. Prog. Ser. 292, 97-109. [Google Scholar]
- 28.Murray, A. E., Preston, C. M., Massana, R., Taylor, L. T., Blakis, A., Wu, K. & DeLong, E. F. (1998) Appl. Environ. Microbiol. 64, 2585-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray, A. E., Blakis, A., Massana, R., Strawzewski, S., Passow, U., Alldredge, A. & DeLong, E. F. (1999) Aquat. Microb. Ecol. 20, 129-145. [Google Scholar]
- 30.Vetriani, C., Tran, H. & Kerkhof, L. (2003) Appl. Environ. Microbiol. 69, 6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLong, E. F., King, L. L., Massana, R., Cittone, H., Murray, A., Schleper, C. & Wakeham, S. G. (1998) Appl. Environ. Microbiol. 64, 1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schouten, S., Hopmans, E. C., Pancost, R. D. & Damste, J. S. S. (2000) Proc. Natl. Acad. Sci. USA 97, 14421-14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinninghe Damste, J. S., Rijpstra, W. I. C., Hopmans, E. C., Prahl, F. G., Wakeham, S. G. & Schouten, S. (2002) Appl. Environ. Microbiol. 68, 2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goreau, T. J., Kaplan, W. A., Wofsy, S. C., McElroy, M. B., Valios, F. W. & Watson, S. W. (1980) Appl. Environ. Microbiol. 40, 526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casciotti, K. L. & Ward, B. B. (2001) Appl. Environ. Microbiol. 67, 2213-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naqvi, S. W. A., Yoshinari, T., Jayakumar, D. A., Altabet, M. A., Narvekar, P. V., Devol, A. H., Brandes, J. A. & Codispoti, L. A. (1998) Nature 394, 462-464. [Google Scholar]
- 37.deBie, M. J., Speksnijder, A. G., Kowalchuk, G. A., Schuurman, T., Zwart, G., Stephen, J. R., Diekmann, O. E. & Laanbroek, H. J. (2001) Aquat. Microb. Ecol. 23, 225-236. [Google Scholar]
- 38.Beman, J. M., Arrigo, K. R. & Matson, P. A. (2005) Nature 434, 211-214. [DOI] [PubMed] [Google Scholar]
- 39.Boehm, A. B., Shellenbarger, G. G. & Paytan, A. (2004) Environ. Sci. Technol. 38, 3558-3566. [DOI] [PubMed] [Google Scholar]
- 40.MacGregor, B., Moser, D., Alm, E., Nealson, K. & Stahl, D. (1997) Appl. Environ. Microbiol. 63, 1178-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schleper, C., Holben, W. & Klenk, H. P. (1997) Appl. Environ. Microbiol. 63, 321-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abreu, C., Jurgens, G., De Marco, P., Saano, A. & Bordalo, A. A. (2001) J. Appl. Microbiol. 90, 713-718. [DOI] [PubMed] [Google Scholar]
- 43.Vetriani, C., Reysenbach, A. L. & Dore, J. E. (1998) FEMS Microbiol. Lett. 161, 83-88. [DOI] [PubMed] [Google Scholar]
- 44.Vetriani, C., Jannasch, H., MacGregor, B., Stahl, D. & Reysenbach, A. (1999) Appl. Environ. Microbiol. 65, 4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.