Abstract
Identification of the genetic basis of common disease may require comprehensive sequence analysis of coding regions and regulatory elements in patients and controls to find genetic effects caused by rare or heterogeneous mutations. In this study, we demonstrate how mismatch repair detection on tag arrays can be applied in a case-control study. Mismatch repair detection allows >1,000 amplicons to be screened for variations in a single laboratory reaction. Variation scanning in 939 amplicons, mostly in coding regions within a linkage peak, was done for 372 patients and 404 controls. In total, >180 Mb of DNA was scanned. Several variants more prevalent in patients than in controls were identified. This study demonstrates an approach to the discovery of susceptibility genes for common disease: large-scale direct sequence comparison between patients and controls. We believe this approach can be scaled up to allow sequence comparison in the whole-genome coding regions among large sets of cases and controls at a reasonable cost in the near future.
Keywords: association studies, autism, high-throughput technology, variation scanning
The study of human genetics is focused on the understanding of the phenotypic consequences of variant nucleotides in the human population. Only a small fraction of nucleotide sites are variant between any two human genomes (π = 1/1,200 nucleotides) (1). Much of this diversity is made up of old mutations in the founder population that have been coinherited by the human populations. Some theoretical work suggested common disease alleles are likely to be common, and others proposed they may be rare and heterogeneous (2-4). Only a few genes contributing to common diseases have already been identified. The alleles involved in these diseases in some instances were common (5), but in several other instances, they were rare (6-9). Comprehensive strategies that can collect the full spectrum of human genetic variation and be scaled to allow researchers to investigate large numbers of genes (or the entire genome) will enable the most powerful approaches to unlocking the power of human genetics. Large-scale technologies that can be used to identify all mutations in large patient and control cohorts are needed to enable this approach. Despite recent dramatic cost reductions, traditional Sanger sequencing can be affordably used only for relatively small studies. At the same time, finding numerous individual mutations in diploid PCR amplicons means that heterozygote detection must be significantly improved beyond the current state of the art to avoid losing the genetic signal amid false-positive signals.
We have developed a technology that allows thousands of individual amplicons to be scanned for all common and rare variations in a fully multiplexed manner. Instead of determining the identity of every base pair, this approach uses a very sensitive bacterial selection assay to find all amplicons that contain mutations relative to all common alleles of this sequence. In this manner, large numbers of nonvariant bases can be rapidly scanned to find the small number of important variant signals.
In this study, we have applied this technique to the study of autism disorder. Autism/autistic disorder [Mendelian Inheritance in Man (MIM) no. 209850], a prototypical pervasive developmental disorder, has a population prevalence of ≈4 in 10,000. Genetic predisposition to autism is complex, with many genes believed to be involved (10). Several genomic regions have been implicated by whole-genome linkage scans, including chromosome 2 that was positive in multiple studies (11-13), even though it was not positive in others (14). To study this peak further, we focused on the 20 Mb that spanned the region within 1 logarithm of odds score point from the maximum of the peak, based on ref. 12. We screened all exons in this region (≈1,200 amplicons) for all mutations in a population consisting of 372 patients and 404 controls.
Materials and Methods
Standard Construction. We have implemented automatic software based on primer 3 (15). Given a set of gene names or chromosome coordinates, the software uses the Ensembl database (www.ensembl.org) to fetch the sequence of the coding regions and design primer pairs. The designed oligos are then used to perform PCR reactions. The PCR products have the AscI site on one side. PCR reactions from genomic DNA are performed and pooled. The pool is then cloned into the mismatch repair detection plasmid vector to generate a “standard” library by AscI on one side and blunt ligation on the other. Different clones in the standard library are shotgun-sequenced by using dideoxy terminator chemistry, and DNA for one clone for each amplicon is pooled to be used in the mismatch repair detection (MRD) reaction.
MRD Reaction. The MRD reactions are done in a 96-well format, allowing 96 patients to be tested per plate (across the full set of amplicons). The process starts with the amplicon generation process, where we generate the 96-plex PCR amplicons for each of 96 individuals in 12 plates for the 1,200 exons. Each of the 12 plates contains an identical well layout for the 96 individuals. After PCR, gel quality control (QC) steps are run to quality control both sample and PCR pool failures. After the gel QC, the 12 plates are consolidated to generate one plate, which contains all amplicons for one individual in each well. The pool of PCR amplicons in each well of the 96-well plate is desalted in plate format and put through a dam methylation (NEB, Beverly, MA) reaction as well as AscI restriction digest (NEB) to create ends for later ligation. Following these protocols, the PCR amplicon pool plate is mixed with methylated linearized vector DNA carrying the inactive Cre (cre-) gene and the pool of unmethylated standards containing the active Cre (cre+) gene. This mixture is denatured and reannealed to form the hemimethylated heteroduplexes. This plate is then desalted (Edge Biosystems, Gaithersburg, MD). Closed circles are formed by Taq ligase (NEB), and the reaction is then treated with exonuclease III and T7 exonuclease (United States Biochemical) to convert noncircular DNA to single-stranded (ss)DNA. This ssDNA is cleaned up by binding to a single-stranded binding resin, and the mixture is put on a desalting column to eliminate salt and resin (Edge Biosystems).
Next, this heteroduplex DNA is transformed into an electro-competent mutation-sorter strain. After initial growth, the cells are separated into two duplicate plates, and each plate is treated with specific antibiotic: carbenicillin + tetracycline or carbenicillin + streptomycin.
After growth in the selective media, DNA from the two plates is prepared by using the Qiaprep 96 Turbo kit (Qiagen, Valencia, CA). The DNA preps are treated with DraI, whose site is present between the tag and the genomic priming sequence. This releases the tag from the rest of the amplicon. The tag sequences in the two plates are then labeled by using linear amplification with common vector primers with two different modifications. The DNA grown in the tetracycline DNA is labeled with primer carrying the 6-carboxyfluorescein (FAM) fluorescent group, and the DNA grown in the tetracycline DNA is labeled with primer carrying biotin. The two plates are then consolidated into one, and each well of this final plate is hybridized onto one GenFlex tag chip (Affymetrix, Santa Clara, CA). After overnight hybridization per the manufacturer's recommendation, the arrays are stained by using a streptavidin-phycoerythrin conjugate that binds the biotin molecules. The arrays are then scanned, and the measured fluorescence signals in the two channels are used to make the variant, heterozygous, or nonvariant calls.
Cluster Caller/Data Analysis. We have used automatic software to make nonvariant/heterozygous/variant calls starting from the array hybridization signal results based on a clustering algorithm. Data from each array undergo background subtraction and spectral overlap correction. Then a finite mixed model of the data was evaluated (for each marker separately) by a maximum-likelihood approach known as the E/M algorithm (16). This model assumes Gaussian distributions for the three classes of data: nonvariant N/N, heterozygous N/V, and variant V/V. The first step in the analysis is to transform the 2D measurement space of the signals from the variant and nonvariant pools into a 1D space (x) that is derived from the ratio of these two signals. Then, the E/M algorithm is applied in this 1D space, where we expect homozygous N/N samples to form a cluster at (approximately) x = -1, heterozygous N/V samples to form a cluster at (approximately) x = 0, and homozygous 2/2 samples to form a cluster at (approximately) x = 1. The best-fit parameter values of the clusters (weights, means, and sigmas) are derived from the E/M algorithm. Finally, each sample is associated with a single cluster so long as its probability of being in the given cluster is greater than its probability of being in any other cluster by a factor F = 50, which is an input parameter of the fitting process.
DNA Samples. The patient samples were probands from a total of 372 unrelated families (of a set of 411). These include most of the families used in the initial positive linkage (12) and have a high overlap with families used in a subsequent scan that was not positive (14). The families were either recruited by the Seaver Autism Research Center at Mount Sinai School of Medicine (n = 40), corecruited by the Seaver Autism Research Center and the Autism Genetic Resource Exchange (AGRE) (n = 127), or recruited by the AGRE (n = 244). All parents provided written informed consent, and potentially affected individuals were assessed with the Autism Diagnostic Interview-Revised (ADI-R). Individuals meeting ADI-R criteria for autism or borderline autism were defined as affected. The cohort and research diagnosis definitions used in the current study have been described (33).
In >90% of the cases, the subjects are Caucasians. The Caucasian control population was obtained from multiple sources: 260 from the Corriell Cell Repositories (Coriell Institute for Medical Research, Camden, NJ (from the 200-Caucasian panel as well as other Caucasian collections), 81 from Caucasian volunteers in Texas, and 63 from volunteers at Stanford University, Stanford, CA. The male-to-female ratio of the cases was 4:1, and that for the control group was close to 1:1.
Genomic Control Analysis. The average χ2 for all of the fragments with variant frequency of >5% (not including the top two variants) was 1.3, and the 95% upper confidence limit was 1.56. This was computed as described (17).
Results
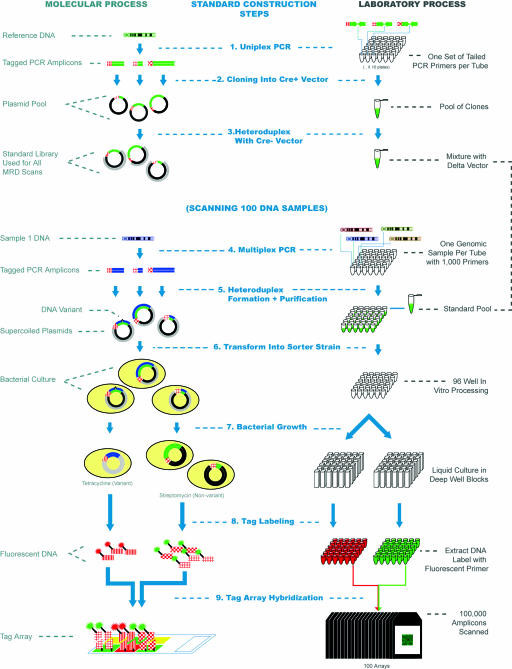
MRD on Tag Arrays. A method for using bacteria to sort heteroduplex molecules into a pool that contains mismatches and another pool that does not has been described (18). This technique, called MRD, forms the basis of the methodology that we utilize as is shown schematically in Fig. 1. A pool of reference plasmids is generated for each amplicon under investigation by cloning PCR amplicons from a haploid source into an MRD vector containing an active Cre gene (cre+). Importantly, PCR primers are used that incorporate one of a set of ≈10,000 21-mer tag sequences (19, 20) at the 5′ end of a standard PCR priming sequence. In this way, the PCR amplicons generated contain a unique tag whose sequence has no homology to the genomic amplicon sequence, which can be used to separate the amplicons later. With the pool of standards in place, tagged PCR amplicons are then generated from each of the patient samples under study. To make the MRD method more fully multiplexed, a PCR technique is used that allows unoptimized 100-plex PCR to be carried out followed by a normalization process that involves hybridization to a limiting capture probe followed by reamplification with common primers (ref. 21; J.Z. and M.F., unpublished results). In this way, 10 multiplexed PCR reactions can be used to study 1,000 exons in each individual sample. Heteroduplexes are formed between the test amplicon pool and the reference pool along with vector DNA containing a crucial 5-bp deletion in the Cre gene (cre- plasmid). In this manner, heteroduplexes are formed in which one strand consists of the reference standard, and the second strand consists of the test amplicon and the cre- vector sequence. In vitro purification steps are then carried out to leave only supercoiled heteroduplexes that can be transformed en masse into the mutation sorter strain. After selective growth in two antibiotic media, the tetracycline pool is enriched for those cells containing amplicons with variations from the reference sequence, whereas the streptomycin pool contains those cells whose plasmids matched the reference DNA. Heterozygous amplicons will appear in both pools. Plasmid DNA from these two pools can then be extracted and labeled with two fluorescent primers. Hybridization to a universal tag array allows the tag content of each of the two pools to be detected by using a two-color chip scanner. Calls of nonvariant, heterozygous, or variant are made automatically by using a clustering algorithm, as described in Materials and Methods.
Fig. 1.
Schematic of the MRD on tag arrays process. A molecular representation of the various steps in the process is shown on the left, whereas the corresponding laboratory process is shown on the right (1). The process starts with the generation of a set of plasmid standards for each of the sequences to be scanned. PCR amplicons are generated for each target by using tailed primers that allow tag sequences to be incorporated. Colors are used to distinguish DNA from different genomic sources (DNA from the reference genome is green). The red textured line segments represent different tag sequences, whereas individual PCR amplicons are shown as being of different lengths, to distinguish them (2). The PCR amplicons are cloned into the cre+ vector (shown as black), pooled, and mixed (3) with digested cre- vector (shown in gray) to create a pool of standards in a single tube that are used for all individual MRD scans (4). PCR amplicons are then generated from each DNA sample under investigation by using the same priming sequences as for the standards. In this process, >1,000 amplicons are generated for each unique DNA sample in each well of a 96-well plate (5). The PCR products are denatured and then mixed with the standard pool. Heteroduplex molecules are then formed between the unknown DNA sequences and the standard DNA. The nicks are closed by using a ligase enzyme, and the supercoiled plasmids are purified in a 96-well format. Each well then contains the heteroduplex plasmids from 1,000 amplicons for a single individual (6). Material from each well is then used to transform the mutation sorter strain (7). The resulting culture is split into two deep 96-well plates. Streptomycin is added to one and tetracycline to the second, and the bacteria are grown as liquid cultures overnight to complete the separation of variant and nonvariant plasmids (8). Plasmid DNA is then extracted from the cultures in 96-well format, and run-off reactions with a labeled primer are used to add a fluorescent label. The primers used to amplify DNA from the tetracycline cultures have a 6-carboxyfluorescein (FAM) group (arbitrarily shown as red), and those used to amplify the streptomycin cultures have a biotin group that ultimately binds to streptavidin-phycoerythrin (9). Finally, the two labeled DNAs from each sample are pooled and hybridized to the universal tag microarray. A total of 96 arrays are used to scan the 96 DNA samples under investigation. The fluorescence intensities of each tag feature allow the variation status of each of the 1,000 amplicons to be determined. Steps (4)-(9) are then repeated for each set of 96 DNA samples in the experiment.
MRD Standard Construction. MRD assays were developed for the exons of all known genes lying within logarithm of odds 1 of the autism chromosome 2 linkage peak (165-185 Mb in Ensembl build 28) (www.ensembl.org). This yielded a total of 1,522 segments that spanned exons from the 140 genes in the region, 20 candidate genes from other sections in the genome, and one segment of the linkage peak (containing 10 homeobox genes) that was scanned in its entirety. The amplicon sizes ranged from 150 to 500 bp. Ninety percent of the primers designed (1,333/1,485) yielded an amplification product, and 95% of these produced a clone (1,268/1,335), as summarized in Table 1.
Table 1. MRD performance metrics.
| Metric | Value |
|---|---|
| Standard construction success rate | 1,268/1,485 = 85% |
| Assay success rate for successful standards | 939/1,268 = 74% |
| Total number of amplicons screened in study | ≈730,000 |
| Total number of base pairs screened in study | >180 Mb |
| Call rate for converted targets | 94% |
| Repeatability | 99.6% |
| Accuracy (concordance with Sanger sequencing) | 99.2% |
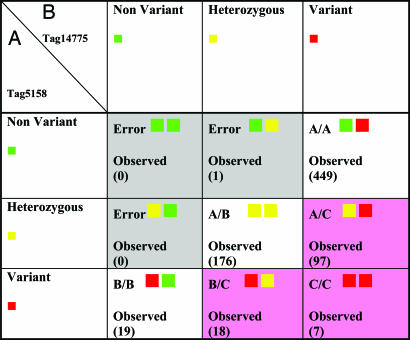
As mentioned above, one feature of the MRD assay on tag arrays is that it can be used to distinguish known alleles from other unknown alleles of the same amplicon, thus combining the power of genotyping common alleles with the power to detect previously unknown mutations. This capability was tested in this experiment. A representative group of amplicons known to have common minor alleles were selected. Two primer sets were prepared with identical priming sequences but containing distinct tag sequences. Two standards were constructed: tag 1 was used to construct standards from the major allele, and tag 2 was used to construct standards from the minor allele. Both standards were then added to the probe pool. Table 2 shows data from this experiment that demonstrate the ability of the MRD assay on tag arrays by using these two standards to reconstruct all combinations of rare and common alleles.
Table 2. An example of data revealing both common and rare alleles.
This is an example of one amplicon where a common allele is known to be present. To distinguish this common allele from other alleles, two standards carrying both alleles of the known variant were constructed. The standard carrying allele (A) of this example amplicon is attached to tag number 5158. The standard carrying allele (B) of the same amplicon is attached to tag number 14775. Assessment of the variation status for this amplicon in a sample is done through comparison of the sample's two alleles with the two standards carrying the two tags. The array information from each of these tags (corresponding to the two standards with the two alleles) can be assigned to one of three states: nonvariant (green square), heterozygous (yellow square), or variant (red square). There are thus nine possible states created by using these two tags. The number of observations of each of these genotypes within the population scanned is shown within each respective genotype box. The gray shaded area indicates allele combinations that are impossible to create (e.g., nonvariant to both known alleles). Indeed, only a single call is misassigned to these genotypes. The white areas indicate genotypes in which the individuals carried only common alleles (A/A, A/B, and B/B). The pink areas indicate genotypes in which the individuals contained an allele distinct from the two alleles represented in the two standards: allele C. In this case, allele C occurs at moderate frequency. Random resequencing of individuals confirmed both the genotypes called and identified allele C as a single SNP of moderate frequency.
MRD Assay. The MRD procedure was performed to scan the 1,268 amplicons in 372 patients with autism and 404 controls. The initial PCR was done in 96-plex, and the later steps were done in 1,268-plex. The results are summarized in Table 1. Of the 1,268 amplicons, 939 yielded successful assays (74%). More than half of the failures were due to PCR failure. The amplicons in the homeobox region had a high PCR failure rate. Excluding this segment, amplicon size was not an important determinant for yielding a successful assay. The average size for amplicons yielding successful assays was 275 bp, and those that failed for any reason in the process had an average size of 278 bp. The call rate for the successful assays was 94%, that is, 6% of the calls were ambiguous and were not called. Repeat concordance measured from 85 repeated samples with ≈80,000 data points was 99.6%. Concordance with dideoxy sequencing (surrogate for accuracy) estimated by sequencing >1,031 traces is 99.2%. In total, ≈730,000 amplicons were scanned for variations in this study. Given an average size of 255 bp per amplicon (not including the primers), the sequence scanned was ≈240 kb per individual and >180 Mb in all individuals.
Variations Detected. The 240 kb of DNA scanned comprise ≈140 kb of exonic DNA and 100 kb of nonexonic DNA. The amplicons tested can be divided into those that cover homeobox genes and those that do not. The homeobox amplicons have an average size of 390 bp (not counting the priming sequences), cover ≈55 kb, and are >80% nonexonic. The nonhomeobox amplicons have an average size of 230 bp (not including the priming sequences), cover ≈184 kb, and are 71% exonic (i.e., 71% of the bases analyzed are within exons).
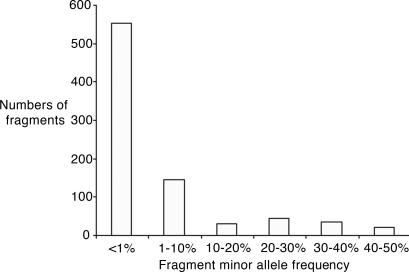
Variants were detected in 832 amplicons. These range from private polymorphisms (a single sample with the second allele) to common variants with minor allele frequencies as high as 50%. It should be noted that some of the amplicons might contain more than one variant complicating the calculation of nucleotide diversity. This calculation is further complicated, because SNP locations are not randomly distributed (22). Taking the conservative assumption that each amplicon contains only one variant, the observed heterozygosity per base pair, π, is 2.7 × 10-4, and the mutation parameter, φ, is 4.4 × 10-4. The diversity measures π and φ for the homeobox amplicons are 3.4 × 10-4 and 4.5 × 10-4, respectively, and for nonhomeobox amplicons π and φ are 2.5 × 10-4 and 4.2 × 10-4. As expected, the homeobox amplicons that are mostly intronic sequences are more variable than the nonhomeobox sequences. This underestimates the difference, because the homeobox amplicons are significantly larger and therefore more likely to have more than one variable site in them. These values are somewhat lower than previously published (23-26). This is in part due to our conservative assumption of one SNP per amplicon and may also be due to the fact that most reports address intronic sequences and/or diverse ethnic populations. Indeed, in a large-scale published study (25), the value for φ in coding variants in Caucasians (4.5 × 10-4) is close to our observations. Fig. 2 shows the distribution of the minor alleles and demonstrates that the majority of the variants are rare. Given the complication mentioned above of the possibility of more than one SNP in an amplicon, it is difficult to compare the observed distribution with the expected distribution, because multiple alleles on the same amplicon appear as a common allele, resulting in underestimation of the rarer alleles. Nevertheless, it is clear that the proportion of all of the variants that are rare is greater than expected, consistent with previous results (27).
Fig. 2.
Frequency of variations discovered in the study. The frequency distribution of amplicon variation is shown, considering each amplicon as a two-allele marker (variant or nonvariant). The number of amplicons with variations at different frequencies is plotted. As can be seen, most amplicons carry variations at lower than 1% frequency. It is this wealth of data that cannot easily be assessed by using a common SNP scoring approach.
Associations with Autism. Analysis was carried out for all amplicons that had the variant allele observed at least five times in cases and five times in controls. The P value was computed by using a standard χ2 test of independence, looking for significant difference in allele frequencies between cases and controls. Amplicons with P value <0.05 were sequenced in variant individuals to identify the nature of the variant. Tables 3 and 4, which are published as supporting information on the PNAS web site, show the most common variants in these amplicons. The two most significant amplicons result from nonsynonymous changes in the same exon in the CMYA3 gene. Although sequencing of this exon confirmed the MRD results in this population, the effect was not replicated when one of the SNPs was genotyped in an independent population (Elena Maestrini, personal communication).
We have implemented a genomic control correction for stratification, assuming the markers in the study (except the top two associations) were not associated with disease, and computed the upper 95th percentile bound of average χ2 as described (17). The most significant result then generates a P value of 0.07 after Bonferroni correction, potentially explaining its nonreproducibility. We note that both of the corrections are overly conservative, because the fragments are not independent, and some of them may indeed be associated with disease
Discussion
The MRD on tag arrays technology has the potential to enable a new approach to the study of complex human genetics. The ability to scan large numbers of genes for all known and previously unknown variation allows the technology to combine the best of both genotyping and resequencing technologies while being able to operate at high throughput and with unprecedented accuracy. This technology may allow direct detection of causative alleles irrespective of whether they are common, rare, or heterogeneous.
For the direct approach to be effective, the causative variations need to be in the amplicons scanned for variations. Several factors need to be considered to maximize this probability. The first factor is the number of amplicons scanned. The most direct way to increase the odds of scanning the correct amplicons is to increase the throughput and lower the cost of the scanning, as we have demonstrated in this work. One factor that limited the power of this study, however, was the conversion rate, the ratio of amplicons attempted to those that were successfully assayed. Most of this failure was due to low signal-to-noise ratios that lead to poor separation between variants and nonvariants and diffuse data clustering. We addressed both of these issues upon completion of the study. First, tightening of the clustering was achieved through increasing the number of transformants by 20-fold. Second, the assay backgrounds have been significantly reduced through improvements in the vector (J.Z., unpublished results). These improvements improve accuracy as well as conversion.
A second factor in achieving comprehensiveness in this type of study is the validity of the linkage peak(s) or the proper choice of a set of functional candidate genes (28). If a regional approach is taken, the enumeration of all genes in the region is also important. For example, we noted that the newer releases of the Ensembl database (build 34) describe another gene [containing an Isl-1 Mec-3 (LIM) domain present in a set of genes that are frequently involved in developmental regulation (29)] overlapping the gene with our lowest P value. Comparative techniques and experimental approaches are rapidly improving the prediction of genes in public databases (for example, ref. 30). Complicating the issue of comprehensiveness further is the increasing evidence that some of the causative variants may have a regulatory rather than a coding function. Assessing regions that are conserved across species is a powerful way to address this issue. Regions conserved between human and mouse are readily available in public resources (for example, http://genome.ucsc.edu and www.ensembl.org), and prediction of these regions will improve as more species are sequenced (31).
The ability of genetic linkage and association to identify relevant genes without the need for a priori knowledge of the function of these genes has often been very illuminating (32). Therefore, in the longer term, we believe our approach needs to be extended to cover all of the genes in the genome. By increasing the multiplexing to 10,000-plex,∥ all of the human exons in the genome can be assessed in 20-30 reactions, and all of the human exons and conserved regions can be scanned in 60 reactions. Such large-scale experiments can be performed at costs of ≈$10,000 per patient without any fundamental improvement to the technology. We believe that scanning all exons and conserved regions for variants in a large number of patients and controls will become the definitive tool in the elucidation of the genetic basis of disease.
Supplementary Material
Acknowledgments
We gratefully acknowledge the resources provided by the AGRE Consortium and the participating AGRE families. This work was supported by National Institutes of Health Small Business Funding Opportunities GrantR41HG02640 (to T.D.W.) and by the Seaver Autism Research Center (J.D.B.) as well as National Institutes of Health through a Studies to Advance Autism Research and Treatment (STAART) Network Center Grant MH066673 (to J.D.B.). The AGRE is a program of Cure Autism Now and is supported, in part, by Grant MH64547 from the National Institute of Mental Health (to Daniel H. Geschwind).
Abbreviations: MRD, mismatch repair detection; AGRE, Autism Genetic Resource Exchange.
Footnotes
We have a proof of principle showing that a 3,000-plex reaction is feasible (M.F. and R.W.D., unpublished work)
References
- 1.Kruglyak, L. & Nickerson, D. A. (2001) Nat. Genet. 27, 234-236. [DOI] [PubMed] [Google Scholar]
- 2.Reich, D. E. & Lander, E. S. (2001) Trends Genet. 17, 502-510. [DOI] [PubMed] [Google Scholar]
- 3.Pritchard, J. K. (2001) Am. J. Hum. Genet. 69, 124-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pritchard, J. K. & Cox, N. J. (2002) Hum. Mol. Genet. 11, 2417-2423. [DOI] [PubMed] [Google Scholar]
- 5.Saunders, A. M., Strittmatter, W. J., Schmechel, D., George-Hyslop, P. H., Pericak-Vance, M. A., Joo, S. H., Rosi, B. L., Gusella, J. F., Crapper-MacLachlan, D. R., Alberts, M. J., et al. (1993) Neurology 43, 1467-1472. [DOI] [PubMed] [Google Scholar]
- 6.Hugot, J. P., Chamaillard, M., Zouali, H., Lesage, S., Cezard, J. P., Belaiche, J., Almer, S., Tysk, C., O'Morain, C. A., Gassull, M., et al. (2001) Nature 411, 599-603. [DOI] [PubMed] [Google Scholar]
- 7.CHEK2 Breast Cancer Case-Control Consortium (2004) Am. J. Hum. Genet. 74, 1175-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaisse, C., Clement, K., Durand, E., Hercberg, S., Guy-Grand, B. & Froguel, P. (2000) J. Clin. Invest. 106, 253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, J. C., Kiss, R. S., Pertsemlidis, A., Marcel, Y. L., McPherson, R. & Hobbs, H. H. (2004) Science 305, 869-872. [DOI] [PubMed] [Google Scholar]
- 10.Risch, N., Spiker, D., Lotspeich, L., Nouri, N., Hinds, D., Hallmayer, J., Kalaydjieva, L., McCague, P., Dimiceli, S., Pitts, T., et al. (1999) Am. J. Hum. Genet. 65, 493-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Molecular Genetic Study of Autism Consortium (IMGSAC) (2001) Am. J. Hum. Genet. 69, 570-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buxbaum, J. D., Silverman, J. M., Smith, C. J., Kilifarski, M., Reichert, J., Hollander, E., Lawlor, B. A., Fitzgerald, M., Greenberg, D. A. & Davis, K. L. (2001) Am. J. Hum. Genet. 68, 1514-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao, Y., Cuccaro, M. L., Hauser, E. R., Raiford, K. L., Menold, M. M., Wolpert, C. M., Ravan, S. A., Elston, L., Decena, K., Donnelly, S. L., et al. (2003) Am. J. Hum. Genet. 72, 539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yonan, A. L., Alarcon, M., Cheng, R., Magnusson, P. K., Spence, S. J., Palmer, A. A., Grunn, A., Juo, S. H., Terwilliger, J. D., Liu, J., et al. (2003) Am. J. Hum. Genet. 73, 886-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rozen, S. & Skaletsky, H. (2000) Methods Mol. Biol. 132, 365-386. [DOI] [PubMed] [Google Scholar]
- 16.Aitkin, M. & Rubin, D. B. (1985) J. R. Stat. Soc. 47, 67-75. [Google Scholar]
- 17.Reich, D. E. & Goldstein, D. B. (2001) Genet. Epidemiol. 20, 4-16. [DOI] [PubMed] [Google Scholar]
- 18.Faham, M., Baharloo, S., Tomitaka, S., DeYoung, J. & Freimer, N. B. (2001) Hum. Mol. Genet. 10, 1657-1664. [DOI] [PubMed] [Google Scholar]
- 19.Hardenbol, P., Yu, F., Belmont, J., Mackenzie, J., Bruckner, C., Brundage, T., Boudreau, A., Chow, S., Eberle, J., Erbilgin, A., et al. (2005) Genome Res. 15, 269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 21.Faham, M. & Zheng, J. (2003) U.S. Patent Application 0096291 A1.
- 22.Reich, D. E., Schaffner, S. F., Daly, M. J., McVean, G., Mullikin, J. C., Higgins, J. M., Richter, D. J., Lander, E. S. & Altshuler, D. (2002) Nat. Genet. 32, 135-142. [DOI] [PubMed] [Google Scholar]
- 23.Carlson, C. S., Eberle, M. A., Rieder, M. J., Yi, Q., Kruglyak, L. & Nickerson, D. A. (2004) Am. J. Hum. Genet. 74, 106-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C. R., Lim, E. P., Kalyanaraman, N., et al. (1999) Nat. Genet. 22, 231-238. [DOI] [PubMed] [Google Scholar]
- 25.Stephens, J. C., Schneider, J. A., Tanguay, D. A., Choi, J., Acharya, T., Stanley, S. E., Jiang, R., Messer, C. J., Chew, A., Han, J. H., et al. (2001) Science 293, 489-493. [DOI] [PubMed] [Google Scholar]
- 26.Halushka, M. K., Fan, J. B., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R. & Chakravarti, A. (1999) Nat. Genet. 22, 239-247. [DOI] [PubMed] [Google Scholar]
- 27.Leabman, M. K., Huang, C. C., DeYoung, J., Carlson, E. J., Taylor, T. R., de la Cruz, M., Johns, S. J., Stryke, D., Kawamoto, M., Urban, T. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 5896-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altmuller, J., Palmer, L. J., Fischer, G., Scherb, H. & Wjst, M. (2001) Am. J. Hum. Genet. 69, 936-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freyd, G., Kim, S. K. & Horvitz, H. R. (1990) Nature 344, 876-879. [DOI] [PubMed] [Google Scholar]
- 30.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
- 31.Margulies, E. H., Blanchette, M., Haussler, D. & Green, E. D. (2003) Genome Res. 13, 2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miki, Y., Swensen, J., Shattuck-Eidens, D., Futreal, P. A., Harshman, K., Tavtigian, S., Liu, Q., Cochran, C., Bennett, L. M., Ding, W., et al. (1994) Science 266, 66-71. [DOI] [PubMed] [Google Scholar]
- 33.Geschwind, D. H., Sowinski, J., Lord, C., Iversen, P., Shestack, J., Jones, P., Ducat, L., Spence, S. J. & AGRE Steering Committee (2001) Am. J. Hum. Genet. 69, 463-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.