Abstract
Marek's disease virus (MDV) is a highly pathogenic and oncogenic herpesvirus of chickens. MDV encodes a basic leucine zipper (bZIP) protein, Meq (MDV EcoQ). The bZIP domain of Meq shares homology with Jun/Fos, whereas the transactivation/repressor domain is entirely different. Increasing evidence suggests that Meq is the oncoprotein of MDV. Direct evidence that Meq transforms chicken cells and the underlying mechanism, however, remain completely unknown. Taking advantage of the DF-1 chicken embryo fibroblast transformation system, a well established model for studying avian sarcoma and leukemia oncogenes, we probed the transformation properties and pathways of Meq. We found that Meq transforms DF-1, with a cell morphology akin to v-Jun and v-Ski transformed cells, and protects DF-1 from apoptosis, and the transformed cells are tumorigenic in chorioallantoic membrane assay. Significantly, using microarray and RT-PCR analyses, we have identified up-regulated genes such as JTAP-1, JAC, and HB-EGF, which belong to the v-Jun transforming pathway. In addition, c-Jun was found to form stable dimers with Meq and colocalize with it in the transformed cells. RNA interference to Meq and c-Jun down-modulated the expression of these genes and reduced the growth of the transformed DF-1, suggesting that Meq transforms chicken cells by pirating the Jun pathway. These data suggest that avian herpesvirus and retrovirus oncogenes use a similar strategy in transformation and oncogenesis.
Keywords: herpesvirus
Marek's disease virus (MDV) is a highly pathogenic and oncogenic α-herpesvirus of chickens (1). The structural and transcriptional organization of the genome is strikingly similar to herpes simplex virus-1 in the unique (UL and US) regions (2). In stark contrast, the genes in the repeat (RL and RS) regions are unrelated to other herpesviruses and are probably responsible for the lymphotrophic and transforming capabilities of MDV. Several genes, including vIL-8, pp38, and Meq, unique to the onco-serotype of MDV have been identified (2). Among these genes, Meq (MDV EcoQ), a 339-aa protein, is most consistently expressed in all MDV tumor and latently infected cells (3). Meq is characterized by an N-terminal basic leucine zipper (bZIP) domain and is the only herpesviral bZIP protein within the immediate family of the Jun/Fos oncogenes (3). The transforming properties of Meq were first studied on rodent fibroblast cell lines (Rat-2). Overexpression of Meq in these cells led to serum- and anchorage-independent growth, accompanied by morphological changes, a shortened G1 phase, and resistance to apoptosis (4). Strong implication that Meq is an oncogene of MDV came from the recent studies of a Meq-null MDV recombinant mutant virus that replicated well in vitro but was completely nononcogenic in vivo (5). Despite these results, there has never been direct evidence that Meq is able to transform chicken cells and that the partnership with Jun is important in transformation.
In the present study, we wished to explore the transforming pathways of Meq in its natural host. Because of the lack of proper chicken T cell transformation systems, we chose DF-1, a spontaneously immortalized chicken embryo fibroblast (CEF) cell line, developed without deliberate transformation by viruses or chemicals (6). This cell line does not exhibit any transformed phenotypes in vitro or tumorigenicity in vivo, and has been used profitably for studying transformation by a number of avian retroviral oncogenes such as v-ErbB, v-Src, v-Jun, v-Myc, v-Fos, v-Ski and the weakly transforming c-Jun (6). Here, we show that Meq can transform chicken cells, based on morphological changes and serum- and anchorage-independent growth of DF-1. Using microarray and RT-PCR analyses, we interrogated the transcriptional program of Meq and found that growth-related and antiapoptosis genes were up-regulated. Importantly, JTAP-1, JAC, and HB-EGF, known to be transformation-related target genes of v-Jun, were among the up-regulated genes, suggesting Jun as a transforming partner of Meq. Consistent with this notion, c-Jun was found to form dimers and was colocalized with Meq. c-Jun's expression level also increased in Meq-transformed DF-1. RNA interference (RNAi) knockdown (7) of c-Jun or Meq led to reduced cell growth and to lower expression levels of JTAP-1, JAC, and HB-EGF. The data suggest that Meq transforms chicken cells via the Jun pathway, providing the mechanistic insight into transformation by a herpes-virus oncogene.
Methods
Cells and Establishment of Stable DF-1 Cell Clones. DF-1 cells, a gift from D. Foster (University of Minnesota, St. Paul), were cultured in DMEM, 5% FBS, 100 units/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen) at 37°C and 5% CO2. For stable transfection, the full-length coding region of meq was cloned into the pCI-neo expression vector (Promega) between the XhoI and EcoRI sites. Lipofectamine (Invitrogen) was used for transfection, with 0.5 mg (activity)/ml Geneticin (G418, Invitrogen) to select for resistant DF-1 clones. Cell lysates from individual G418-resistant clones were collected (named 5A, 5B, etc.) and tested for Meq expression by Western blotting using anti-Meq Abs (8).
Soft Agar Colonies. Cells were seeded at a density of 7.5 × 103 per well in six-well plates, in 0.4% Sea Plaque GTG agarose (Cambrex) in 5% FBS-DMEM, on a layer of 0.8% SeaPlaque agarose in DMEM, and incubated for 14 days. Chorioallantoic Membrane (CAM) Assay. The method was derived from Petruzzelli et al. (9), with several modifications. Briefly, specific pathogen-free embryonating chicken eggs (SPAFAS, Wilmington, MA) were opened on day 3 postincubation and placed in a plastic wrap sling in a covered Petri dish, at 37.5°C and 3% CO2. On day 10 postincubation, either vector-DF-1 or Meq-DF-1 cells were grafted onto to the CAM in 50-μl aliquots at a concentration of 4-10 × 107 cells per ml. Each CAM was inoculated in five locations. Two embryos were used in each replicate, and the experiment was repeated three times. The embryos were observed daily. The grafts and adjacent CAMs were harvested on day 5 postincubation, fixed in 10% buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin/eosin. Photomicrographs of representative sections were taken.
RNA Extraction, Microarray, and RT-PCR. Cells were washed and pelleted by centrifugation (500 × g for 3 min), and total RNA was extracted by using the SV Total RNA isolation system (Promega). RNA was sent to the Fred Hutchinson Cancer Institute (Seattle) for hybridization and microarray analysis on a chip containing 3,379 chicken genes and EST sequences. Five hybridizations were performed, three with Meq-DF-1 5G and two with 5F, vs. vector-DF-1. Genes consistently up- or down-regulated were analyzed (10) and confirmed by RT-PCR. RT was performed with SuperScript II reverse transcriptase (Invitrogen), using 2 μg of RNA per sample. Semiquantitative PCR was performed with two template dilutions, using recombinant Taq polymerase (Invitrogen). Reactions were started with 94°C for 3 min, followed by 28 cycles of 94°C for 45 sec, (annealing temperature in degrees) for 45 sec, 72°C for 45 sec, and 72°C for 10 min, and finally kept at 4°C. The primer sequences and annealing temperatures are listed in Table 1, which is published as supporting information on the PNAS web site.
Microarray Data Analysis. Ratios between expression in the MeqDF-1 samples and the vector-DF-1 samples were computed by dividing background-subtracted signals for both Cy5 and Cy3 channels, according to the microarray manufacturer's protocols (11). We used rank consistency scoring (RCoS) (10) to assess differential expression between Meq and vector samples. For each run k, the ratio of expression levels in Meq to levels in the vector reference sample are ranked in descending order. The rank of the ratio at gene g in run k is denoted Rg,k. For every gene g, the rank consistency score Sg is the maximal (i.e., the worst) rank of this gene among all runs, Sg = max1≤k≤5 Rg,k/N, where N is the total number of genes (3,379). To determine the statistical significance of this score, we compute the P value of gene g with score s under the null model of uniform and independent rank vectors. Therefore, P value(s) = s5. Using these computed P values, we can estimate false discovery rates (12).
Dual Luciferase Reporter Assays. Reporter plasmids were constructed by inserting three AP-1 promoter regions upstream of the firefly luciferase coding region (Luc) in the pGL3-Basic vector (Promega). Vector-DF-1 and Meq-DF-1 cells were seeded in 12-well plates at 1 × 105 per well in 1 ml of 10% FBS-DMEM and incubated at 37°C and 5% CO2 for 24 h. For each well, 2.5 μg of plasmid DNA was transfected by using Lipofectamine. All wells were cotransfected with control pRL-SV40 Renilla luciferase plasmids (Promega). Cell lysates were prepared 48 h posttransfection with 1× Passive Lysis Buffer (Promega). The dual luciferase assay was performed according to the manufacturer's protocol, using a Lumat LB 9501 luminometer (Wallac). Three independent experiments were performed.
Serum-Starvation and Analysis of Apoptosis. 1 × 106 vector-DF-1 or Meq-DF-1 cells were grown to confluence in 75-cm2 tissue flasks (Nunc) in 5% FBS for 24 h, when media was replaced to 0.2% FBS for an additional 48 h. Cells were fixed in ice-cold 70% ethanol for cell-cycle analysis, resuspended in 500 μl of PBS, treated with 50 μg of RNase A (MBI Fermentas, Amherst, NY), and stained with 50 μg/ml propidium iodide (Sigma). The cells were analyzed by flow cytometry (FACScan flow cytometer, Becton Dickinson) with lysis ii (Becton Dickinson) software, and the percentage of hypodiploid cells was measured. In situ TUNEL analysis was performed by using the Fluorescein In Situ Cell Death Detection kit (Roche).
siRNA Preparation and Transfections. Twenty-one-nucleotide sequences of annealed, double-stranded siRNAs, with d(TT) in the 3′ overhangs, were synthesized by Qiagen-Xeragon. The targeting sequence of chicken c-Jun was AAGGATTGCCAGGTTGGAAGA, and that for MDV Meq was AAGCAGACGGACTATGTAGAC. The nonsilencing scrambled RNA (scRNA) sequence AATTCTCCGAACGTGTCACGT served as negative control.
DF-1 cells (2 × 105) were seeded in six-well plates in 5% FBS-DMEM. At 24 h, cells were transfected with siRNA by using Oligofectamine (Invitrogen), according to the manufacturer's instructions (7). Cells were lysed for either protein or RNA extraction, 48 h after transfection, or harvested and counted (Coulter Electronics) to analyze the effect of RNAi on growth. Specific silencing was confirmed by three independent experiments.
Western Blots and Coimmunoprecipitation Assays. Cells were pelleted and lysed in 50 μl of EBC buffer (50 mM Tris·HCl, pH 7.5/120 mM NaCl/0.5% Nonidet P-40/50 mM NaF/200 μM Na2VO4/1 mM phenylmethylsulfonyl fluoride) with a protease inhibitor mixture (Sigma) for total-cell lysates, or in NE-PER (Pierce) for nuclear and cytoplasmic extracts. For coimmunoprecipitation, 20 μl of a protein A/protein G on agarose beads (Upstate Biotechnology) was added to the lysate for 2 h at 4°C to reduce nonspecific binding. The cell lysate was reacted with anti-Meq polyclonal Abs (8) (1:100) overnight at 4°C. For Western blotting, samples were boiled for 5 min with 2× SDS sample buffer and subjected to SDS/10% PAGE. Gels were transferred to poly(vinylidene) difluoride membranes (Biotechnology Systems), which were incubated with primary Abs overnight at 4°C. Final dilutions of the Abs were 1:3,000 for the anti-Meq polyclonal Abs (pAbs), 1:500 for anti-GAPDH pAbs (G8140-01, United States Biological, Swampscott, MA), and 1:1,000 for the anti-c-Jun mAb (610326, BD Transduction Laboratories) in TBST/5% skim milk.
Immunofluorescence Assay and BrdUrd Labeling. DF-1 cells were fixed on slides with methanol-acetone (1:1) for 15 min. After blocking for 30 min in PBS-2% BSA, cells were incubated with anti-Meq pAbs (1:500) and anti-c-Jun mAb (1:500) in 2% BSA for 1 h at 37°C. Rhodamine-conjugated anti-rabbit goat IgG F(Ab′)2 (1:1,000; ICN) and FITC-conjugated anti-mouse sheep IgG F(Ab′)2 (1:1,000; ICN) in 2% BSA served as secondary Abs. DNA was stained by using 2.5 μM TO-PRO-3 (Molecular Probes). Imaging was performed by confocal microscope equipped with an argon-krypton laser (LSM 510 MicroSystem, Zeiss). BrdUrd labeling was performed as described in ref. 13.
Results
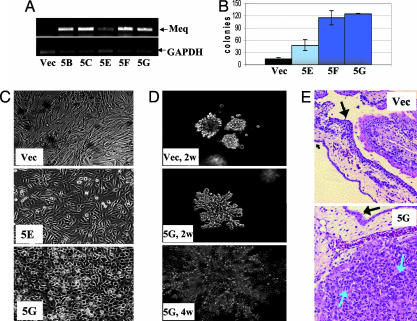
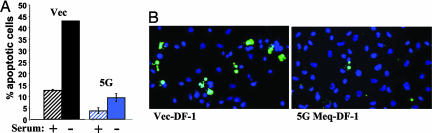
Transformation of DF-1 by Meq. To study the transforming potential of Meq, we transfected CMV-Meq into DF-1, a nontumorigenic, immortalized chicken cell line. Clones expressing different levels of Meq were isolated (Fig. 1A), and their morphologies were examined (Fig. 1B). The vector-transfected control, like the parental DF-1, exhibited a fibroblastic-like morphology. Clone 5G, the high Meq expressor, assumed a round, cuboidal morphology, whereas clone 5E, the low Meq expressor (Fig. 1B) had an intermediate phenotype. When seeded in soft agar medium, the changes were even more dramatic: Meq-DF-1 5G clones grew aggressively and formed numerous large and radiant colonies, eventually spreading into a very large area, whereas the vector-DF-1 showed much more restrained growth potential. Clone 5F behaved similarly to 5G (data not shown). Quantitatively, the number of soft-agar colonies formed by Meq-DF-1 5F and 5G is 3- to 4-fold greater than the vector control (Fig. 1D). Remarkably, the in vitro colony formation potential of these clones is reflected by the tumorigenic assay in CAMs. All grafts were initially visible as a semitransparent focus <1 mm in diameter. Three days after the placement of cells on the CAM, the Meq-DF-1 cells became visible as a small translucent tumor. This tumor increased in size and opacity until 5 days after grafting, when it was harvested. At maximum size, the tumors were ≈2 mm in diameter and white in color. In contrast, the vector-DF-1 cells became increasingly unapparent after 2 days and were not detectable after 4 days. Histologically, the vector-DF-1 inoculated CAMs had no evidence of grafted cells (Fig. 1E). In contrast, the Meq-DF-1 cell grafts were composed of amphophilic fibroblastic cells consistent with the appearance of Meq-transformed cells (blue arrows in Fig. 1E; and see Fig. 8, which is published as supporting information on the PNAS web site). In addition to the changes in growth properties, Meq-transformed DF-1 also became much more resistant to apoptosis, compared with the vector-transfected control. Forty-eight hours of serum starvation induced a significant level of apoptosis in vector-DF-1, whereas the levels of apoptosis in the Meq-transfected cells remained low by both flow cytometry based on propidium iodide analysis (Fig. 2A) and TUNEL staining (Fig. 2B). These data suggest that Meq has the ability to induce morphological changes, anchorage-independent growth, and serum-independent survival of DF-1 cells.
Fig. 1.
The effect of Meq on DF-1 cells. (A) RT-PCR detection of Meq in stably transfected DF-1 clones, compared with vector cells. Clone 5E showed a lower level of Meq expression. (B) DF-1 cell morphology in culture. Clone 5G cells that express high levels of Meq have a rounded appearance, whereas clone 5E maintain an intermediate appearance, closer to the fibroblast shape of the vector cells. (C) Soft agar colonies were grown for 2 (Top and Middle) and 4 (Bottom) weeks. The Meq-expressing clone acquires a large and radiant appearance, whereas the vector colonies remain small and less invasive. (D) Soft agar colonies were counted at 14 days. Colonies of clones 5F and 5G were more numerous than those of clone 5E. Only few colonies of the vector-DF-1 cells were seen. (E) CAMs grafted with DF-1 cells. The location of the vector-DF-1 cell graft and the surrounding membranes are shown (Upper). Vector-DF-1 cells were not evident microscopically. (Magnification: ×200.) In the CAM grafted with Meq-DF-1 cells (Lower), a 2-mm tumor was visible macroscopically, and fibroblastic cells, phenotypically consistent with Meq-DF-1 cells, are evident microscopically. The black arrows indicate the CAM, and the blue arrows show individual Meq-DF-1 cells. (Magnification: ×400.)
Fig. 2.
Meq protects DF-1 cells from apoptosis. (A) Untreated (hatched bars) and serum-starved (filled bars) vector-DF-1 (black) and Meq-DF-1 5G (blue) cells were analyzed by flow cytometry for percentage of hypodiploid cells. (B) In situ TUNEL analysis of cells depicts apoptotic cells (stained green) in vector-DF-1 (Left) and Meq-DF-1 (Right) serum-starved cultures.
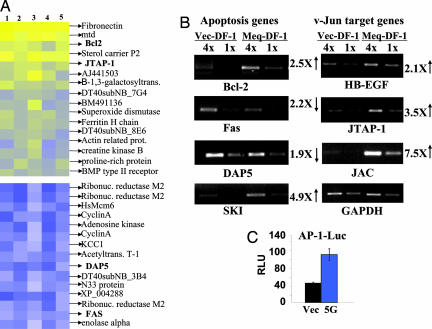
Host Genes Regulated by Meq. Meq is a transcriptional factor, which carries a bZIP domain with homology to the Jun/Fos family. To gain a better understanding of the transformation mechanism, we wished to identify host genes regulated by Meq, using the recently developed chicken cDNA microarray (11). This array carries close to 3,400 confirmed chicken EST sequences, derived from a mixture of cDNA libraries. We compared the expression profiles of Meq-DF-1 clones against vector-DF-1. The result is derived from five independent microarrays, of five independent cultures (three of clone 5G and two of 5F against the vector-DF-1). No statistically significant difference in the expression profiles measured for clones 5G and 5F was observed. We used rank consistency scoring (RCoS) to assess Meq-induced differential expression. Only genes that showed consistent changes with statistical significance (P < 0.001) were selected for further pursuit. By these criteria, of the 3,379 ESTs, 95 were overexpressed and 115 were underexpressed [both at false discovery rate (FDR) = 0.04]. Here, we highlight those which are considered to be relevant to transformation (Fig. 3A). Based on these results, we searched for other related genes, which together with the microarray data were analyzed and confirmed by a semiquantitative RT-PCR, using two dilutions for each template (Fig. 3B). The fold changes are based on RT-PCR densitometric quantitation. The complete list of genes can be found as Tables 2 and 3, and the FDR curves can be found as Fig. 9, all of which are published as supporting information on the PNAS web site. Among the survival pathways, we found that Bcl-2 and Ski, a protooncogene that interacts with Smads and intercepts the TGF-β pathway (14, 15), are up-regulated, whereas Fas and DAP5 are down-modulated, consistent with the strong antiapoptotic properties of Meq-DF-1. Very little information is available concerning the regulation of these genes in chickens; however, in mammals many of them are regulated by transcriptional factors of the Jun pathway and carry AP-1 sites in the promoters. Perhaps most interesting is the finding that v-Jun's transformation-related target genes (JTAP-1, JAC, and HB-EGF) are up-regulated (16-18), suggesting that Meq may transform chicken cells in part via the v-Jun pathway. A corollary to this hypothesis is that the AP-1 activity should be enhanced in Meq-DF-1 cells vs. vector-DF-1 cells. A reporter gene driven by AP-1 promoter was transfected into these two cell types, and the reporter activities were measured. As shown in Fig. 3C, the AP-1 activity is highly elevated in Meq-DF-1 cells. The up-regulation of Ski is also noteworthy (Fig. 3B); v-Ski is an avian retroviral oncogene known to transform chicken embryo cells into a radiant phenotype (15). Interestingly, a recent report showed that Jun and Ski work in concert to affect the antiapoptosis pathway (19).
Fig. 3.
Regulation of host genes by Meq. (A) Color rendition of analyzed expression ratios of five microarray experiments. Columns represent the five runs of vector-DF-1 vs. Meq-DF-1 clones 5F and 5G, as described in the text. Genes with statistically significant rank consistency scores (P < 5 × 10-5) are depicted as overexpressed (extent of yellow; Upper) and underexpressed (extent of blue; Lower). Genes in bold are discussed in the text. (B) Confirmation of gene regulation by RT-PCR. RNA from vector-DF-1 (Left) and Meq-DF-1 (Right) cells was extracted and reverse-transcribed to cDNA, which was added to the PCR reactions at 1:1 (first and third lanes) or 1:4 (second and fourth lanes) concentrations. Fold change of expression was compared densitometrically to the respective GAPDH bands. (C) Luciferase reporter activation driven by AP-1. Vector-DF-1 (black bar) and Meq-DF-1 (blue bar) cells were transfected with plasmids containing three AP-1 promoter binding-sites and collected 2 days later for analysis. RLU, relative luciferase units.
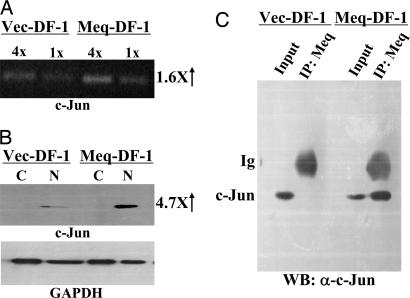
Meq's Association with and Activation of c-Jun in DF-1. To explore the possibility that Meq imparts its transforming potential through the Jun-pathway, we investigated the partnership between Meq and Jun in Meq-DF-1. We first studied the level of expression of c-Jun. The level of Jun in Meq-DF-1 is slightly increased at the transcriptional level (Fig. 4A) but much more so at the protein level. Virtually all of the overexpressed c-Jun molecules are found in the nucleus (lanes N in Fig. 4B). We then examined the interaction of Meq and Jun by coprecipitation (Fig. 4C) and by colocalization (data not shown). In both situations, the two molecules were in the same complex. Previously, we showed that Meq and Jun formed stable heterodimers, co-recruited to and cooperated to transactivate AP-1 containing promoters (20). Taken in that light, our results here suggest that in vivo, Meq stabilizes and activates Jun through dimerization to mediate its transformation potential.
Fig. 4.
Expression and coimmunoprecipitation of Meq and Jun in DF-1 cells. (A) RT-PCR for c-Jun expression was performed as described in Fig. 3B.(B) Western blotting for cytoplasmic (lanes C) and nuclear (lanes N) extracts of vector-DF-1 and Meq-DF-1 cells was performed with anti-Jun (Upper) and anti-GAPDH (Lower) antibodies. (C) Vector-DF-1 (left lanes) and Meq-DF-1 (right lanes) cell extracts were precipitated by antibodies against Meq, followed by Western blotting with c-Jun antibody. Input consisted of 10% of total cell lysate.
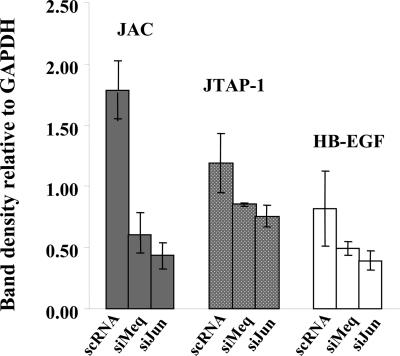
The Involvement of Jun in Meq-Mediated Transformation. To study whether the Jun pathway is relevant to Meq-mediated gene regulation and transformation, we used the RNAi technology (7) to knock down the expression of Meq and Jun, to see their effects on gene expression patterns. As shown in Fig. 5A, our siRNA sequences specifically knock down Meq or Jun (Fig. 5), and the knockdown of either molecule reduced the expression of JTAP-1, JAC, and HB-EGF, based on RT-PCR analysis (Fig. 6). This experiment provides evidence that both Meq and Jun play a role in the activation of the v-Jun target genes in our model.
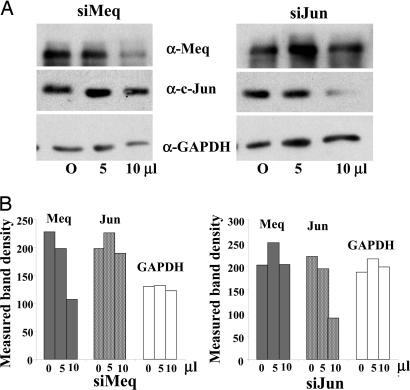
Fig. 5.
Down-regulation of Meq and Jun gene expression by RNAi. (A) Meq-DF-1 cells were transfected with different levels (5 or 10 μl) of synthetic RNAi molecules specific for Meq (Left) or Jun (Right), or with 10 μl of a nonsilencing sequence (O), as described in the text. Western blots were performed with extracts from the transfected cells, with anti-Meq (Top), anti-Jun (Middle), and anti-GAPDH (Bottom) Abs. (B) Measured band intensities for the protein bands indicate the specific decrease in the expression of Meq (by siMeq; Right) and of c-Jun (by siJun; Left).
Fig. 6.
Effect of siRNA treatments on v-Jun-related genes. RT-PCR was performed on RNA extracted from Meq-DF-1 cells, treated with sequences of the following: scRNA, nonsilencing negative control; siMeq, down-regulation of Meq; siJun, down-regulation of c-Jun. Band intensities for JAC, JTAP-1, and HB-EGF were corrected vs. respective GAPDH band densities.
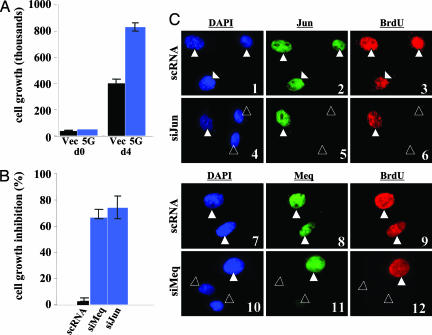
If Meq and Jun both play critical roles in the transformation of DF-1, we surmised that the RNAi knockdown of either gene should diminish the ability of DF-1 to grow. Fig. 7 A and B revealed that this was indeed the case, and in those cells where either Meq or Jun were knocked down, the BrdUrd labeling was lower, compared with those retaining their expressions (Fig. 7C). These data thus demonstrated that the sustained expression of Meq and Jun are vital components involved in the growth of the transformed clones.
Fig. 7.
The effect of siRNA on cell growth. (A) Untreated vector-DF-1 (black bars) and Meq-DF-1 (blue bars) cells were trypsinized and counted for growth comparison at day 0 (left bars) and day 4 (right bars). (B) Meq-DF-1 cells were transfected with scRNA (negative control; black), siMeq (blue), or siJun (blue), trypsinized and counted on day 4. No significant difference was observed between siMeq and siJun, while both significantly inhibited growth compared to scRNA. (C) Immunostaining of siRNA-treated MeqDF-1 cells confirmed the decrease in expression of c-Jun (slide 5, open triangles) and Meq (slide 11, open triangles). The staining of c-Jun and Meq (both green) in untreated cells is shown in slides 2 and 8, respectively. siRNA-knockdown of c-Jun and Meq affected the cell cycle, as indicated by BrdUrd levels (slides 6 and 12; open triangles). DAPI and BrdUrd staining are shown in blue and red, respectively.
Discussion
MDV is among the most oncogenic herpesviruses (1). It induces a rapid onset of T cell lymphomas with an aggressiveness rivaling oncogene-carrying acute retroviruses. As such, it has been postulated that MDV may carry a direct-acting oncogene of its own. MDV is also unique among oncogenic herpesviruses in that it has a genome organization resembling alpha-herpesvirus (1). There are three serotypes of MDV, with type I being the oncogenic serotype. All three serotypes share significant sequence homology throughout the genomes, with the exception of the repeat-long (RL) regions. MDV oncogene(s), if existing, are thus most likely derived from the divergent repeat region and from genes unique to serotype I MDV. Previously, MDV has been shown to be capable of transforming CEF, when lytic infection is suppressed (21). Meq is predominantly expressed in latent or tumor cells (22), and it was demonstrated that blocking the expression of Meq reduces the anchorage-independent growth of an MDV-transformed cell line (23). More recently, MDV mutants devoid of Meq have been generated, showing replication in culture but displaying no oncogenicity in vivo (5). The evidence collectively suggests that Meq is a critical gene involved in MDV oncogenesis. In this report, we provide evidence that Meq is able to transform chicken cells. The cells undergo morphological changes and display anchorage-independent growth and serum-independent survival. In addition, the CAM assays revealed the in vivo tumorigenic properties of Meq. The data further support Meq as a potential transforming protein of MDV.
Meq has a structure resembling the retroviral oncogenes Jun, Fos, and Maf (22). The sequence homology of the bZIP domain places Meq in the immediate family of Jun/Fos. Meq is the only herpesvirus bZIP protein that belongs to this family, and its biochemical properties confirm that it functions like a Jun/Fos family protein: Meq dimerizes with Jun and forms a heterodimer that is more stable than each homodimer. Meq/Jun bind AP-1 sequences and transactivate promoters carrying AP-1 motifs. Recently, we showed by chromatin immunoprecipitation experiments that Meq and Jun are co-recruited to the AP-1 site present in the chicken IL-2 promoter in MDV-transformed cells (20). Also of interest to us is the wealth of information concerning v-Jun transformation of CEF. v-Jun of avian retrovirus ASV17, like its cellular homologue c-Jun, is a major component of the AP-1 transcriptional factor complex (24). v-Jun (in retroviral vectors) transforms CEF, by itself (25), and rat embryo fibroblasts, in cooperation with Ras (26). Although overexpressed c-Jun can similarly transform CEF, v-Jun is much more potent, presumably because of the activating mutations found in v-Jun (25). The mutations include a deletion in the N-terminal, JNK-docking domain (27), which is also the site for ubiquitination, and a point mutation of a serine phosphorylation site that affects the recognition by a ubiquitin ligase (28), contributing to the higher stability of v-Jun. It was postulated that Jun mediates transformation by transcriptionally activating transformation-associated genes (16, 29) by pairing with other members of the Jun/Fos family. In an effort to identify the genes responsible for enhanced transformation, chicken genes activated specifically by v-Jun, but not by c-Jun, were identified (16), among which, JTAP-1, JAC, and HB-EGF are significant, because their expression correlates with that of v-Jun. Furthermore, when HB-EGF and JAC were individually placed in retrovirus vectors, they were capable of transforming CEF (17, 18, 30). Perhaps not coincidentally, we found that Meq up-regulates all three v-Jun activated genes and exhibits a strong transforming potential on DF-1, because Meq has some structural features more akin to v-Jun than c-Jun. For instance, one of the activating mutations of v-Jun is the change of serine 243 of c-Jun to phenylalanine in v-Jun, and Meq carries a phenylalanine in this position, too. Given that the protein level of c-Jun is higher in the Meq-transformed cells (Fig. 4B), we suggest that Meq/Jun dimerization stabilizes the c-Jun protein and/or changes its conformation to the extent that it begins to acquire some of the transactivating and transforming properties of v-Jun. This suggestion is consistent with the report that dimerization can significantly influence the stability of Jun/Fos family members (31). Indeed, the soft agar colonies of Meq-DF-1 have a shape resembling colonies of v-Jun-transformed CEFs (32), and a comparison of our microarray data with that of v-jun-transformed CEF (33) on the same type of chip revealed similar expression profiles including the modulation of JTAP-1, Bcl2, and DAP5 discussed in this article.
The siRNA results indicate that Meq and c-Jun are important for the up-regulation of JAC, HB-EGF, and JTAP-1 in our cells (Fig. 6), strengthening the notion that these two oncoproteins play a combined role in transformation of the cells, although we do not rule out the possibility of other bZIP proteins participating in this process.
In addition to the Jun pathway, we found that Meq activates c-Ski, a cellular homologue of the avian retrovirus oncogene v-Ski. Ski is known to intercept the TGF-β pathway by interacting with Smads (19). Lymphoma cells often release TGF-β as means to suppress surrounding immune cells, whereas Ski's activation in these cells is a way to escape immunity-induced growth arrest and apoptosis. In addition to Ski, Meq up-regulated antiapoptosis genes such as Bcl-2 and down-modulated proapoptosis genes such as Fas and DAP5, the latter a protein involved in IFN-γ-induced cell death (34). T cells limit their extended activation by self-destruction, via two separate and independent pathways, Bcl-2 down-regulation on the one hand and Fas/FasL up-regulation on the other (35). Our results suggest that Meq can control both of these pathways, allowing the cells to be continuously activated, an important step for transformation. All of the properties shown here in chicken cells echoed previous findings in Rat-2 cells (4), reinforcing the notion that Meq is a strong antiapoptotic protein. Thus, although for logistic reasons the present studies were carried out in CEF, the information gained provides a useful framework for understanding T cell transformation by MDV.
Supplementary Material
Acknowledgments
We thank Drs. M. Parcells, S. Reddy, and E. Stavnezer for stimulating discussions and sharing of unpublished results. We benefited significantly from Dr. J. Delrow's chicken cDNA microarray service and insightful assistance. This work was supported by National Institutes of Health Grants CA46613, CA91574, and USDA 2001-02390 (to H.J.-K. and L.L.). A.M.L. was supported by Vaadia-BARD (United States-Israel Binational Agricultural Research and Development Fund) Postdoctoral Award FI-323-2001.
Abbreviations: CAM, chorioallantoic membrane; CEF, chicken embryo fibroblast; MDV, Marek's disease virus; RNAi, RNA interference; scRNA, scrambled RNA.
References
- 1.Witter, R. L. & Schat, K. A. (2003) Marek's Disease in Diseases of Poultry (Iowa State Univ. Press, Ames).
- 2.Lee, L. F., Wu, P., Sui, D., Ren, D., Kamil, J., Kung, H. J. & Witter, R. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6091-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones, D., Lee, L., Liu, J. L., Kung, H. J. & Tillotson, J. K. (1992) Proc. Natl. Acad. Sci. USA 89, 4042-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu, J. L., Ye, Y., Lee, L. F. & Kung, H. J. (1998) J. Virol. 72, 388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lupiani, B., Lee, L. F., Cui, X., Gimeno, I., Anderson, A., Morgan, R. W., Silva, R. F., Witter, R. L., Kung, H. J. & Reddy, S. M. (2004) Proc. Natl. Acad. Sci. USA 101, 11815-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Himly, M., Foster, D. N., Bottoli, I., Iacovoni, J. S. & Vogt, P. K. (1998) Virology 248, 295-304. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 8.Liu, J. L., Lee, L. F., Ye, Y., Qian, Z. & Kung, H. J. (1997) J. Virol. 71, 3188-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petruzzelli, G. J., Snyderman, C. H., Johnson, J. T. & Myers, E. N. (1993) Ann. Otol. Rhinol. Laryngol. 102, 215-221. [DOI] [PubMed] [Google Scholar]
- 10.Chen, M. M., Ashley, E. A., Deng, D. X., Tsalenko, A., Deng, A., Tabibiazar, R., Ben-Dor, A., Fenster, B., Yang, E., King, J. Y., et al. (2003) Circulation 108, 1432-1439. [DOI] [PubMed] [Google Scholar]
- 11.Neiman, P. E., Ruddell, A., Jasoni, C., Loring, G., Thomas, S. J., Brandvold, K. A., Lee, R., Burnside, J. & Delrow, J. (2001) Proc. Natl. Acad. Sci. USA 98, 6378-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjamini, Y. & Liu, W. (1999) J. Stat. Planning Interference 82, 163-170. [Google Scholar]
- 13.Beall, E. L., Manak, J. R., Zhou, S., Bell, M., Lipsick, J. S. & Botchan, M. R. (2002) Nature 420, 833-837. [DOI] [PubMed] [Google Scholar]
- 14.Stavnezer, E., Brodeur, D. & Brennan, L. A. (1989) Mol. Cell. Biol. 9, 4038-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stavnezer, E., Gerhard, D. S., Binari, R. C. & Balazs, I. (1981) J. Virol. 39, 920-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu, S. L., Waha, A. & Vogt, P. K. (2000) Oncogene 19, 3537-3545. [DOI] [PubMed] [Google Scholar]
- 17.Hartl, M., Reiter, F., Bader, A. G., Castellazzi, M. & Bister, K. (2001) Proc. Natl. Acad. Sci. USA 98, 13601-13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harding, P. A., Davis-Fleischer, K. M., Crissman-Combs, M. A., Miller, M. T., Brigstock, D. R. & Besner, G. E. (1999) Growth Factors 17, 49-61. [DOI] [PubMed] [Google Scholar]
- 19.Pessah, M., Marais, J., Prunier, C., Ferrand, N., Lallemand, F., Mauviel, A. & Atfi, A. (2002) J. Biol. Chem. 277, 29094-29100. [DOI] [PubMed] [Google Scholar]
- 20.Levy, A. M., Izumiya, Y., Brunovskis, P., Xia, L., Parcells, M. S., Reddy, S. M., Lee, L., Chen, H. W. & Kung, H. J. (2003) J. Virol. 77, 12841-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buranathai, C., Rodriguez, J. & Grose, C. (1997) Virology 239, 20-35. [DOI] [PubMed] [Google Scholar]
- 22.Kung, H. J., Xia, L., Brunovskis, P., Li, D., Liu, J. L. & Lee, L. F. (2001) Curr. Top. Microbiol. Immunol. 255, 245-260. [DOI] [PubMed] [Google Scholar]
- 23.Xie, Q., Anderson, A. S. & Morgan, R. W. (1996) J. Virol. 70, 1125-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogt, P. K. (2001) Oncogene 20, 2365-2377. [DOI] [PubMed] [Google Scholar]
- 25.Bos, T., Monteclar, F., Mitsunobu, F., Ball, A., Chang, C., Nishimura, T. & Vogt, P. (1990) Genes Dev. 4, 1977-1687. [DOI] [PubMed] [Google Scholar]
- 26.Lloyd, A., Yancheva, N. & Wasylyk, B. (1991) Nature 352, 635-638. [DOI] [PubMed] [Google Scholar]
- 27.Sprowles, A. & Wisdom, R. (2003) Oncogene 22, 498-506. [DOI] [PubMed] [Google Scholar]
- 28.Wei, W., Jin, J., Schlisio, S., Harper, J. W. & Kaelin, W. G., Jr. (2005) Cancer Cell 8, 25-33. [DOI] [PubMed] [Google Scholar]
- 29.Bader, A. G., Schneider, M. L., Bister, K. & Hartl, M. (2001) Oncogene 20, 7524-7535. [DOI] [PubMed] [Google Scholar]
- 30.Hadman, M., Gabos, L., Loo, M., Sehgal, A. & Bos, T. J. (1996) Oncogene 12, 135-142. [PubMed] [Google Scholar]
- 31.Papavassiliou, A. G., Treier, M., Chavrier, C. & Bohmann, D. (1992) Science 258, 1941-1944. [DOI] [PubMed] [Google Scholar]
- 32.Nishizawa, M., Fu, S. L., Kataoka, K. & Vogt, P. K. (2003) Oncogene 22, 7931-7941. [DOI] [PubMed] [Google Scholar]
- 33.Black, E. J., Clair, T., Delrow, J., Neiman, P. & Gillespie, D. A. (2004) Oncogene 23, 2357-2366. [DOI] [PubMed] [Google Scholar]
- 34.Marash, L. & Kimchi, A. (2005) Cell Death Differ. 12, 554-562. [DOI] [PubMed] [Google Scholar]
- 35.Van Parijs, L., Biuckians, A. & Abbas, A. K. (1998) J. Immunol. 160, 2065-2071. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.