Abstract
New plant cells arise at the meristems, where they divide a few times before they leave the cell-cycle program and start to differentiate. Here we show that the E2Fa–DPa transcription factor of Arabidopsis thaliana is a key regulator determining the proliferative status of plant cells. Ectopic expression of E2Fa induced sustained cell proliferation in normally differentiated cotyledon and hypocotyl cells. The phenotype was enhanced strongly by the co-expression of E2Fa with its dimerization partner, DPa. In endoreduplicating cells, E2Fa–DPa also caused extra DNA replication that was correlated with transcriptional induction of S phase genes. Because E2Fa–DPa transgenic plants arrested early in development, we argue that controlled exit of the cell cycle is a prerequisite for normal plant development.
Keywords: Arabidopsis thaliana/cell cycle/cell differentiation/E2F–DP transcription/endoreduplication
Introduction
Cell division and differentiation rely on the coordinated function of cell cycle genes. Typically, differentiation transforms an actively dividing cell into a non-dividing cell with a specialized function. In mammals, the decision of cells to continue or stop dividing depends largely on the activity of the E2F–DP heterodimeric transcription factor. Inhibition of E2F–DP activity arrests dividing cells at G1 (Wu et al., 1996), whereas E2F overexpression forces serum-starved cells to enter S phase (Johnson et al., 1993; Qin et al., 1994; Lukas et al., 1996).
E2F was originally identified as a cellular factor that stimulates the expression of the adenovirus E2 promoter (Kovesdi et al., 1986), but subsequently has been shown to regulate the activity of a wide variety of genes, including cell cycle and regulatory genes [e.g. cyclins, retinoblastoma (Rb), c-myc], genes required for DNA replication and repair (e.g. DNA polymerase α, CDC6, ORC1) and genes encoding structural proteins of chromatin (such as histones; Lavia and Jansen-Dürr, 1999).
In mammals, six E2F genes and two DP genes have been identified (Dyson, 1998). Typically, the E2F and DP proteins contain a highly conserved DNA-binding domain and a dimerization domain. The latter allows the dimerization of E2F and DP, which is a prerequisite for high-affinity, sequence-specific DNA binding (Bandara et al., 1993; Helin et al., 1993; Krek et al., 1993). Most E2F proteins (except E2F6) contain a strong C-terminal activation domain overlapping with a Rb-binding site. Because Rb binding masks the E2F activation domain, Rb-bound E2F–DP complexes work as transcriptional repressors. Not only does Rb shield the E2F transactivation domain, but it also recruits histone deacetylase (Brehm et al., 1998; Luo et al., 1998; Magnaghi-Jaulin et al., 1998). Histone deacetylase is believed to repress promoter activity through the deacetylation of nucleosomes resulting in chromatin condensation. The inhibitory action of Rb is counteracted by the expression of D-type cyclins upon mitogenic stimulation. D-type cyclins bound to cyclin-dependent kinases (CDKs) initiate the phosphorylation of Rb, resulting in the release of transcriptionally active E2F–DP (Weinberg, 1995) and consequential transcription of genes involved in G1–S and S phase transitions.
In plants, post-embryonic development relies on iterative cell division in the meristems. Cells in the meristem remain in an indeterminate state, whereas cells that stop dividing start differentiating. Although many key genes controlling plant cell division have been described, it is still unclear which genes determine cells to divide or differentiate (Stals and Inzé, 2001). Because of their described effects in animals, we postulate that plant E2F proteins are the likely candidates to regulate the proliferative identity of plant cells. E2F genes have been identified recently in tobacco, wheat, carrot and Arabidopsis (Ramínez-Para et al., 1999; Sekine et al., 1999; Albani et al., 2000; Magyar et al., 2000). The encoded E2F proteins display a domain organization similar to their mammalian counterparts and bind Rb. DP proteins have been demonstrated in plants also (Magyar et al., 2000; Ramínez-Para and Gutierrez, 2000). Moreover, plant E2F–DP complexes were shown to bind and activate in vitro the activity of genes containing the consensus E2F–DP target site (Chabouté et al., 2000), suggesting that the general mechanism for G1–S transition and S phase entry is conserved in mammals and plants. Nevertheless, the in planta role of the E2F and DP genes in the regulation of cell division, cell differentiation and plant development is still unclear.
In the Arabidopsis thaliana genome, three E2F (designated E2Fa, E2Fb and E2Fc) and two DP (DPa and DPb) genes could be identified (de Jager et al., 2001; Vandepoele et al., 2002). Here we show that the E2Fa–DPa transcription factor of Arabidopsis is a positive regulator of plant cell division. Increased E2Fa–DPa levels up-regulate the expression levels of S phase-specific genes, resulting in ectopic cell divisions correlated with a delay in cell differentiation. Moreover, we identified E2Fa–DPa as a key regulator of the endocycle.
Results
Expression analysis of the E2Fa and DPa genes
To unravel the function of the E2Fa and DPa genes in the plant cell cycle and development, we analysed their spatial distribution pattern by mRNA in situ hybridization. The low abundance of the E2Fa and DPa transcripts required the use of full-length antisense mRNA sequences as probes. DNA gel-blot analysis showed that the full-length DPa probe did not cross-hybridize with the other DP gene from Arabidopsis. Minor cross-hybridization was seen between the E2Fa probe and the two other Arabidopsis E2F genes (data not shown). Because this cross-hybridization was low, we believe that the observed hybridization pattern is likely to reflect mainly E2Fa expression.
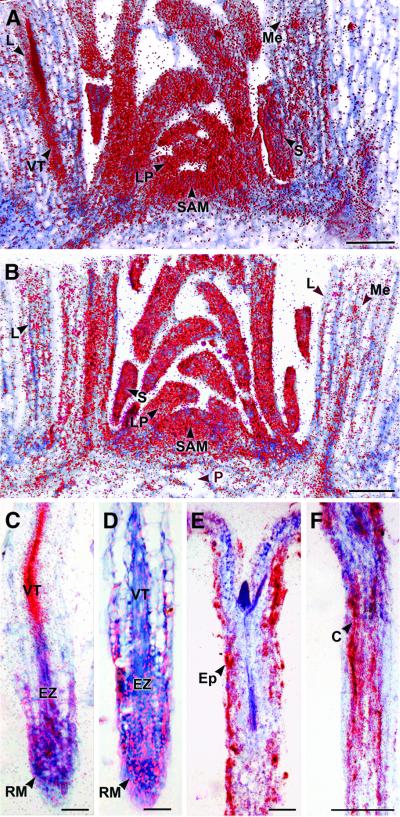
The shoot apex of 2-month-old Arabidopsis plants exhibits various kinds of tissue differing in their state of proliferation and differentiation (Jacqmard et al., 1999). Both E2Fa and DPa were strongly expressed in the actively dividing tissues of the shoot apical meristem, young leaf primordia, the vascular tissues of the maturing leaf primordia and axillary buds (Figure 1A and B). The observed signals were not uniformly distributed, but rather slightly patchy, probably because of their cell-cycle, phase-specific transcription.
Fig. 1. In situ localization of E2Fa and DPa mRNA. Hybridization signals are seen as red dots. (A and B) Expression of E2Fa and DPa in the shoot apex of 2-month-old Arabidopsis plants, respectively. (C and D) E2Fa and DPa expression in the longitudinal section through a root meristem of Arabidopsis, respectively. (E and F) E2Fa expression in a longitudinal section through a hypocotyl of 5-day-old light- and dark-grown seedlings of Arabidopsis, respectively. C, cortex; Ep, epidermis; EZ, elongation zone; L, maturing leaf; LP, leaf primordia; Me, mesophyll; P, pith; RM, root meristem; S, stipule; SAM, shoot apical meristem; VT, vascular tissue. Bars = 100 µm (A, B and F) and 50 µm (C–E).
Previously, using RT–PCR, we have shown that in the roots of 3-week-old plants, the expression of DPa and E2Fa is weak or absent, respectively (Magyar et al., 2000). However, by in situ hybridization analysis, both E2Fa and DPa expression was detected in the apical region and developing vascular tissue where cells divide actively. The hybridization signal became weaker as cells exited the meristematic region (Figure 1C and D). The apparent discrepancy between the RT–PCR and in situ hybridization results can probably be explained by the different type of root material used. The co-expression of E2Fa and DPa in the dividing tissues of the root and shoot suggests that the E2Fa–DPa complex is required for cell cycle progression.
Expression of E2Fa and DPa is not restricted to the regions of mitotic cell division. E2Fa transcripts were also detected in the epidermis and cortex of the hypocotyl of 5-day-old light-grown plants (Figure 1E). At this stage, the cortex is devoid of cell division, but undergoes extensive endoreduplication, which is enhanced in dark-grown plants (Gendreau et al., 1997; Raz and Koornneef, 2001). Likewise, the E2Fa hybridization signal was stronger in the cortical hypocotyl cells of dark-grown plants (Figure 1F). Similarly, DPa signals and, to a lesser extent, E2Fa signals could be observed in the endoreduplicating tissues of the shoot apex (the mesophyll cells of the maturing leaves, the pith cells and the stipules). These results indicate that the E2Fa–DPa complex not only regulates the mitotic cell cycle progression but also plays a role in the endocycle.
Overexpression of E2Fa induces ectopic cell divisions
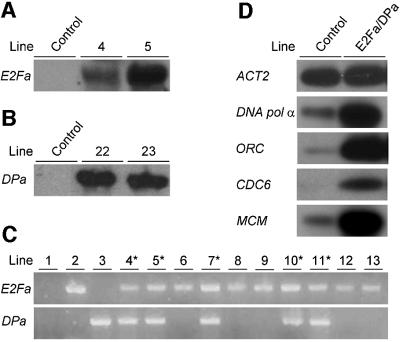
To evaluate the importance of E2Fa and DPa expression in dividing and endoreduplicating cells, transgenic Arabidopsis thaliana plants were generated that contained either the E2Fa or the DPa gene under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter. Out of multiple transgenic lines, two independent CaMV35S-E2Fa (Figure 2A) and two CaMV35S-DPa (Figure 2B) lines were selected, containing only one T-DNA locus. The DPa transgenic plants were morphologically identical to untransformed control plants, whereas E2Fa plants had enlarged cotyledons (Figure 3A and B; Table I). Larger cotyledons can result from larger cells or from an increase in the number of cells. To discriminate between these possibilities, adaxial epidermal cell size was measured. In the strongest E2Fa-overexpressing line, cotyledon cells were less than half the size of control cells. As a consequence of an increase in cotyledon size and a reduction in cell size, transgenic cotyledons contained almost 3-fold the number of cells as the control plants did (Table I). These extra cells could arise from additional cell divisions occurring after seed germination or, alternatively, could be already present in the embryo. To distinguish between these two possibilities, the number of adaxial epidermal cells in cotyledons prior to seed germination were counted. The cotyledons of the weakest E2Fa overexpression line (line 4) had approximately the same number of cells as the control plants, whereas the strongly expressing line (line 5) had a reduced number of cells (Table I). This illustrates that the extra cells found in the cotyledons of 3-week-old E2Fa-overexpressing plants originate from additional cell divisions, which occurred after germination.
Fig. 2. Molecular analysis of E2Fa- and DPa-overexpressing Arabidopsis plants. (A and B) RNA gel blots of independent CaMV35S-E2Fa and CaMV35S-DPa transgenic plants, respectively. (C) Linkage of the observed growth arrest with the presence of both CaMV35S-E2Fa and CaMV35S-DPa transgenes. 1, wild-type plant; 2, cross between a CaMV35S-E2Fa plant and a control plant; 3, cross between a CaMV35S-DPa plant and a control plant; 4–13, individual siblings of a cross between a homozygous CaMV35S-E2Fa plant and a heterozygous CaMV35S-DPa. Lines marked with an asterisk had curled leaves and cotyledons, and were arrested at the seedling stage. Presence of transgenes was tested by PCR. (D) Transcript levels of S phase genes determined by semi-quantitative RT–PCR in control and CaMV35S-E2Fa–DPa plants harvested seven days after sowing.
Fig. 3. Phenotype of E2Fa- and DPa-overexpressing 12-day-old seedlings. (A) Untransformed control. (B) E2Fa- and (C) E2Fa–DPa-overexpressing plant. All plants were photographed at the same magnification. Bar = 2.5 mm.
Table I. Adaxial epidermal cell size and cell number in cotyledons of E2Fa-overexpressing plants.
| Line | Cotyledona 3 weeks after sowing Size (mm2) | Adaxial epidermal cells 3 weeks after sowing |
Cotyledon prior to germination |
||
|---|---|---|---|---|---|
| Size (µm2) | Estimated number | Size (mm2) | Numberb | ||
| Wild type | 5.3 ± 0.3 | 4612 ± 268 | 1204 ± 104 | 0.099 ± 0.002 | 612 ± 12 |
| E2Fa line 4 | 7.2 ± 0.3 | 2712 ± 272 | 2904 ± 236 | 0.089 ± 0.005 | 610 ± 35 |
| E2Fa line 5 | 7.2 ± 0.3 | 2190 ± 183 | 3389 ± 366 | 0.068 ± 0.003 | 525 ± 32 |
The indicated values are mean ± SE.
aUntransformed and transgenic plants (n = 12) were grown in the same Petri dish to exclude differences in growth conditions. The observed increase in cotyledon size was confirmed in at least three independent experiments. The number of adaxial epidermal cells was determined by the ratio of cotyledon size to adaxial epidermal cell size, which was measured for at least 50 cells in each cotyledon.
bThe number of adaxial cells prior to germination was measured by counting all cells of at least five different cotyledons per line.
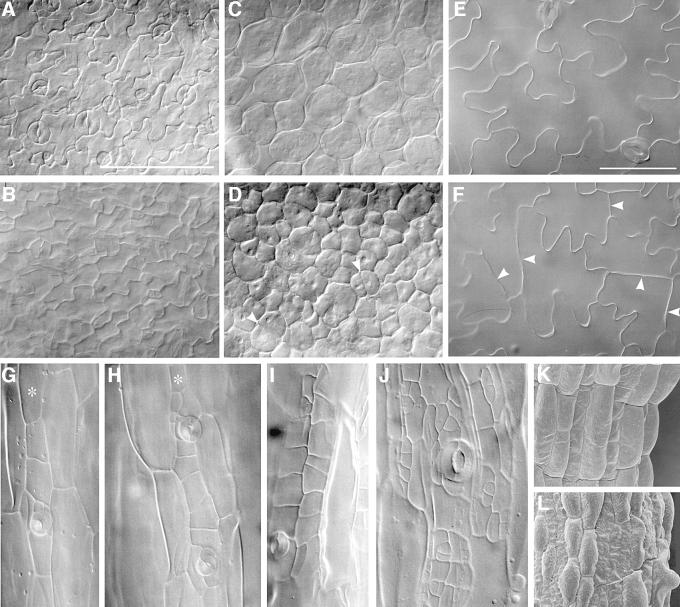
Microscopic analysis revealed that the extra cells formed in the E2Fa-overproducing cotyledons originated from a delay in the onset of cell differentiation. Five days after sowing, the adaxial epidermal cotyledon cells of wild-type plants were differentiated into puzzle-shaped pavement cells and stomata (Figure 4A). In contrast, in 5-day-old E2Fa-overexpressing lines only polygonal cells and few stomata could be observed (Figure 4B). Similarly, instead of typical palisade tissue consisting of round cells with intercellular spaces, CaMV35S-E2Fa palisade cells were still polygonal without intercellular spaces and proliferating (Figure 4C and D). Even in 3-week-old E2Fa cotyledons, epidermal cells were still dividing as indicated by the presumably newly formed cell walls with a straight appearance, whereas in wild-type plants of the same age, only non-dividing puzzle-shaped cells could be observed (Figure 4E and F). In the hypocotyl, cell files consisting of normal epidermal cells alternated with cell files that show extra cell divisions (Figure 4G and H). Cell files that divide ectopically are those in which normally stomata are formed (Berger et al., 1998), indicating here that E2Fa–DPa can sustain cell division only in cells that are competent to divide.
Fig. 4. Microscopic analysis of E2Fa- and E2Fa–DPa-overexpressing Arabidopsis plants. (A and E) Abaxial epidermis of cotyledons of a 5-day-old and 3-week-old control plants, respectively; (B and F) abaxial epidermis of cotyledons of a 5-day-old and 3-week-old E2Fa plant, respectively; (C and D) palisade parenchyma of a 5-day-old control and E2Fa plant, respectively; (G and H) hypocotyl of a 12-day-old control and E2Fa plant, respectively; (I and J) hypocotyl of a 12-day-old and 3-week-old E2Fa–DPa plant, respectively and (K and L) scanning micrographs of (G) and (I). Arrowheads in (D) and (F) point to novel synthesized cell walls; asterisks in (G) and (H) indicate cell file in which stomata are formed. Bars = 100 µm (A–D, K and L, same magnification) and 50 µm (E–J, same magnification).
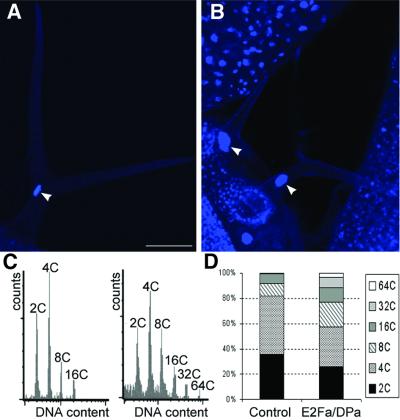
Prolonged cell division in the E2Fa transgenic lines was also evident from flow cytometric analyses performed on 8-day-old cotyledons. Wild-type cotyledons displayed a typical pattern with C values ranging from 2C to 16C. The 8C and 16C peaks were the result of endoreduplication, a common process in plants by which DNA is replicated in the absence of mitosis. When compared with control plants, the amount of nuclei with a 2C and 4C value was significantly higher in the E2Fa transgenic lines, whereas the number of cells with 8C DNA content had decreased by >15 and 10% in lines 4 and 5, respectively (Table II), and the number of cells with 16C value was lower as well. Because cell division and endoreduplication are mutually exclusive, the lower 8C and 16C values again illustrate that E2Fa transgenic plants divide for a longer period than controls. Curiously, in the strongest E2Fa-overexpressing line (line 5), the effects on the 16C ploidy level were less pronounced. This observation can be explained by the positive effect of E2Fa activity on endoreduplication (see below).
Table II. Ploidy levels in wild-type and E2Fa-overexpressing cotyledons eight days after sowing.
| C value | Control (%) | E2Fa line 4 (%) | E2Fa line 5 (%) |
|---|---|---|---|
| 2C | 25.1 ± 1.1 | 32.5 ± 2.5 | 30.8 ± 1.7 |
| 4C | 28.6 ± 0.6 | 39.0 ± 1.1 | 30.2 ± 0.2 |
| 8C | 35.4 ± 0.2 | 20.3 ± 0.7 | 24.6 ± 0.9 |
| 16C | 4.8 ± 0.9 | 1.8 ± 0.2 | 4.2 ± 0.6 |
Data represent average ± SD.
E2Fa and DPa synergistically induce ectopic cell divisions
Mammalian E2F and DP proteins activate E2F-dependent transcription in a synergistic manner. To analyze whether E2F and DP proteins in plants cooperate also, plants homozygous for the CaMV35S-E2Fa gene were crossed with heterozygous CaMV35S-DPa lines. Half of the offspring developed normally, whereas 50% of the plants displayed cotyledons and leaves curled along their proximal–distal axis (Figure 3C). PCR analysis on individual plants confirmed that plants with the curled leaf phenotype contained both the CaMV35S-E2Fa and CaMV35S-DPa constructs, whereas the phenotypically normal siblings contained the CaMV35S-E2Fa gene only (Figure 2C).
Microscopic analysis showed that the phenotype seen in the E2Fa-overexpressing lines was strongly enhanced in the CaMV35S-E2Fa–DPa plants. In the hypocotyl, many more cell divisions were observed, resulting in islands of small irregular cells (Figure 4I). This phenotype became more pronounced in older hypocotyls (Figure 4J). Scanning electron microscopy showed that the typical epidermal differentiation pattern found in wild-type hypocotyls was totally disrupted, displaying a mixture of small isodiametric cells and elongated bulging cells (Figure 4K and L).
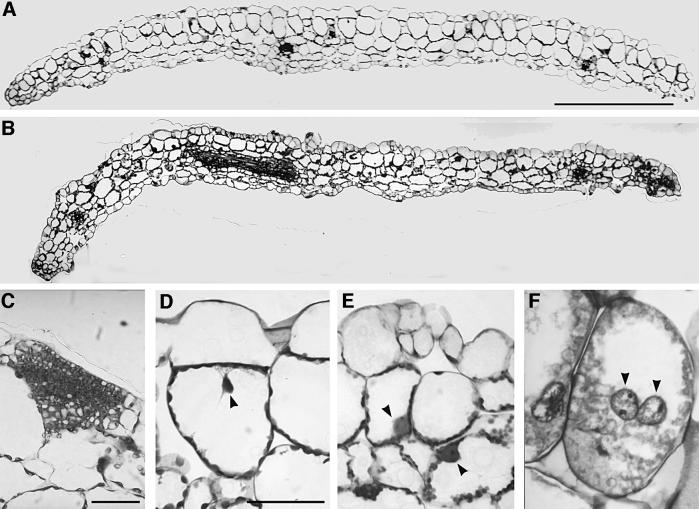
A cross-section through a mature cotyledon illustrated that the epidermal layers of the E2Fa–DPa-overproducing plants contained many more cells, most of which were smaller. Cells of the inner tissues were smaller or equal in size when compared with wild-type cells (Figure 5A and B). Occasionally, a part of the cotyledon was converted to a meristem-like structure consisting of a group of non-vacuolated cells with a dense cytoplasm (Figure 5C), demonstrating that E2Fa–DPa activity is a potent trigger of cell division.
Fig. 5. Microscopic analysis of a mature cotyledon of control and E2Fa–DPa-overexpressing plants. (A and B) Transverse section through the central part of a cotyledon of a control and E2Fa–DPa plant, respectively. (C) Group of small non-vacuolated cells with dense cytoplasm located at the epidermis of a cotyledon of an E2Fa–DPa-overexpressing plant. (D and E) Detail of cotelydon palisade parenchyma cells of wild-type and E2Fa–DPa plants, respectively. (F) E2Fa–DPa cotyledon palisade parenchyma cell containing two giant nuclei. Arrowheads point to nuclei (D–F). Scale bars: 500 µm (A and B, same magnification) and 50 µm (C and D; D–F, same magnification).
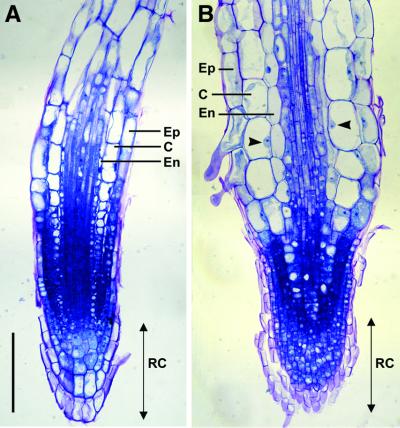
Roots of E2Fa–DPa transgenic plants were shorter than those of wild-type plants (data not shown). Microscopic analysis of the root tip showed that the root cap of the transgenic plants was approximately the same size as that seen in control plants, but it contained many more cells that were significantly smaller (Figure 6). Remarkably, the mature root had an ∼1.5-fold greater diameter than that of wild-type plants. This increase in thickness was not the result of extra cell layers being formed, but rather of radial expression of cortex and endodermis tissues.
Fig. 6. Microscopic analysis of root tissue. Median, longitudinal section through a 3-week-old control (A) and E2Fa–DPa plant (B). Arrowheads point to nuclei. C, cortex; En, endodermis; Ep, epidermis; RC, root cap. Scale bar = 100 µm (A and B, same magnification).
E2Fa–DPa plants display enhanced endoreduplication levels
Because the E2F–DP transcription factor is required for endocycle progression in Drosophila melanogaster (Royzman et al., 1997), the effect of E2Fa–DPa overexpression on endoreduplication was analyzed. Microscopic analysis showed that nuclei of some palisade cells of transgenic E2Fa–DPa plants contained conspicuously large nuclei (Figure 5D and E). In addition, in the cotyledons, cells were observed with more than one nucleus (Figure 5F). The nuclear size of mature trichomes had increased dramatically (Figure 7A and B) as well, and in root cells, enlarged nuclei could be seen (Figure 6). Extensive endoreduplication in the CaMV35S-E2Pa–DPa plants was confirmed by flow cytometric analysis. Two-week-old transgenic seedlings showed two additional endocycles when compared with control plants, resulting in DNA values as high as 64C (Figure 7C and D).
Fig. 7. DNA ploidy level in control and CaMV35S-E2Fa–DPa transgenic plants. (A and B) Trichome of control and E2Fa–DPa transgenic plant, respectively. Arrowheads point to the nucleus. (C) Ploidy distribution of control (left) and E2Fa–DPa transgenic seedlings (right) harvested 12 days after germination. (D) Quantification of the results shown in (C). Bar = 50 µm (A and B, same magnification).
E2Fa–DPa overexpression up-regulates the activity of S phase-specific genes
Previously, the E2F consensus binding sequence has been shown to be conserved in plants and mammals (Chabouté et al., 2000). These sequences can be found in the promoters of Arabidopsis genes of which the mammalian counterparts are regulated by E2F–DP and include DNA pol α, ORC, MCM and CDC6. The transcript level of these genes was compared between control and E2Fa–DPa transgenic plants by semi-quantitative RT–PCR analysis. Whereas the expression level of the control gene (actin 2) was not influenced by E2Fa–DPa overexpression, all S phase genes were dramatically up-regulated (Figure 2D). These data demonstrate that E2Fa–DPa overexpression resulted in the increased expression of S phase genes and suggests that the observed phenotypes of the transgenic lines result from mis-expression of these genes.
Discussion
In plants, the mechanisms linking cell proliferation, differentiation and endoreduplication are still unclear. Here we showed that the E2Fa–DPa transcription factor plays a crucial role in all three different processes. Overexpression of E2Fa resulted in plants with more cells than the control plants due to extra post-embryonic cell divisions. The extra cells in the E2Fa plants may be the result of a shortening of the cell cycle or a prolonging of the proliferative phase. We showed that the latter is at least in part responsible, as cell differentiation was clearly delayed in the transgenic lines. A negative effect on cell differentiation was also seen in mammals where overexpression of E2F1 inhibited differentiation of cells into myoblasts and megakaryocytes (Wang et al., 1995; Guy et al., 1996). E2Fa overexpression sustained cell division in tissues otherwise already devoid of proliferation, as seen in the epidermis of 3-week-old epidermal pavement cells. This suggests that the E2Fa transcription factor may be one of the most important rate-limiting factors for cells to divide.
The phenotype observed in the E2Fa-overexpressing plants was strongly enhanced by the co-expression of E2Fa and DPa, clearly demonstrating that both proteins interact as a functional complex. The expression levels of S phase-specific genes were dramatically increased in the E2Fa–DPa transgenics, indicating that the observed phenotypes are due to the uncontrolled expression of cell-cycle genes. Under normal circumstances, expression of E2F–DP-dependent genes is inhibited by the complex formation of E2F–DP with Rb. Therefore, we believe that the most probable mechanism by which the S phase genes are activated in the E2Fa–DPa transgenic lines is inactivation of Rb by out-titration, thereby escaping the Rb-mediated transcriptional repression. According to this hypothesis, overexpression of Rb would counteract the phenotypes seen in the E2Fa–DPa transgenics.
E2Fa–DPa also seems to be a crucial regulator of the endocycle because transgenic plants display huge nuclei and increased ploidy levels. In the E2Fa–DPa transgenic plants, values were observed up to 64C, whereas a maximum of 16C was detected in control plants of the same age. In Arabidopsis, the zone of active endoreduplication in the shoot apex is in direct continuity with the region where mitotic cell divisions occur (Jacqmard et al., 1999), suggesting that the endocycle is achieved by a modification of the mitotic cell cycle. However, the molecular mechanism regulating endoreduplication is still unclear. In Drosophila, E2F–DP induces transcription of cyclin E, leading to the formation of the S phase-specific cyclin E–CDK2 complex. This complex is a central regulator for endoreduplication (Edgar and Orr-Weaver, 2001). Although no clear homologue of cyclin E had been identified in the Arabidopsis genome, the activation of an S phase-specific CDK complex in the endosperm of maize upon the onset of endoreduplication has been reported (Grafi and Larkins, 1995). These results, combined with the data presented here, suggest that plants regulate their endocycle in a similar way to that of Drosophila.
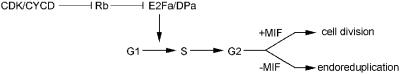
We postulate that the decision of E2Fa–DPa-overexpressing cells to undergo ectopic cell divisions or to endoreduplicate depends on the competence of cells to divide (Figure 8). Ectopic E2Fa–DPa stimulates cell-cycle progression by triggering S phase entry. Cells with a mitosis-inducing factor (MIF) proceed into mitosis, whereas cells lacking this factor are stimulated by the sustained E2Fa–DPa activity to re-enter S phase, leading to increased ploidy levels. This hypothesis is supported by the phenotype observed in the hypocotyl where extra cell divisions occur mainly in the cell files that still have the potential to divide, giving rise to stomata. Currently, the nature of the MIF remains unknown, but most probably depends on the presence of M phase-specific CDK activity. In yeast and Drosophila, down-regulation of the M phase-associated kinase activity is sufficient to drive cells into the endoreduplication cycle (Hayles et al., 1994; Sauer et al., 1995). A similar system might be operational in plants. This is supported by the identification of the fizzy-related ccs52 gene, which induces endoreduplication through activation of the anaphase-promoting complex, resulting in the destruction of mitotic cyclins (Cebolla et al., 1999).
Fig. 8. Model explaining the different phenotypes seen in plants with sustained E2Fa–DPa activity. MIF, mitosis-inducing factor.
We conclude that the E2Fa–DPa transcription factor is a crucial component regulating cell division, differentiation and endoreduplication in plants. Its activity has to be controlled in a stringent way because ectopic expression of E2Fa–DPa results in uncontrolled cell proliferation and delayed differentation. As a result, plants arrest in growth early during post-embryonic development. This growth inhibitory effect is in sharp contrast to the phenotype found upon overexpression of the cyclins Arath;CYCB1;1 and Arath;CYCD;2, which promote plant growth (Doerner et al., 1996; Cockcroft et al., 2000). A major difference between the cyclin-overproducing lines and the E2Fa–DPa transgenic plants is that the former do not show extra proliferation in otherwise differentiated tissues. Thus, in contrast to CYCD2;1 and CYCB1;1, E2Fa–DPa overrides the signals that regulate cell differentiation. The observed negative effect on growth shows that the correct balance between division and differentiation is vital for plant development. Plants may arrest in growth as a consequence of their delayed differentiation or because the required essential cell signalling between different tissue layers is disturbed. The decision between proliferation and differentiation depends upon the concerted expression of genes determining cell fate. The mis-expression of cell-cycle genes induced by E2Fa–DPa could repress the induction of genes needed for differentiation, implying that cell differentiation requires inactivation of E2Fa–DPa transcriptional activity. The accumulation of active Rb proteins in differentiating leaf tissues suggests that, as in mammals, this inactivation is controlled by Rb (Huntley et al., 1998). Nevertheless, our data indicate that the E2Fa–DPa pathways may be used to direct cell division in plants in a specific manner, allowing yield and architecture to be adjusted.
Materials and methods
Regeneration and molecular analysis of E2Fa- and DPa-overexpressing plants
The E2Fa- and DPa-coding regions were amplified by PCR with the 5′-GGCCATGGCCGGTGTCGTACGATCTTCTCCCGA-3′ and 5′-GGG GATCCTCATCTCGGGGTTGAGT-3′ or 5′-GGCCATGGAGTTGTTTGTCACTCC-3′ and 5′-GGAGATCTTCAGCGAGTATCAATGG-3′ primers, respectively. The obtained E2Fa PCR fragment was cut with NcoI and BamHI and the DPa fragment was digested with NcoI and BglII. Subsequently, the restriction fragments were cloned between the CaMV35S-promoter and the nopaline synthase (NOS) 3′ untranslated region in the NcoI and BamHI sites of PH35S (Hemerly et al., 1995), resulting in the PH35SE2Fa and PH35SDPa vectors. The CaMV35S/E2Fa/NOS cassette was released by EcoRI and XbaI and cloned into the EcoRI and XbaI sites of pBinPLUS (van Engelen et al., 1995), resulting in the pBINE2Fa vector, whereas the CaMV35S/DPa/NOS cassette was released by EcoRI and XbaI, made blunt-ended and inserted into the SmaI site of PGSC1704, resulting in the PGSCDPa vector. Both pBINE2Fa and PGSCDPa were mobilized by the helper plasmid pRK2013 into the Agrobacterium tumefaciens C58C1RifR harboring the plasmid pMP90 (Koncz and Schell, 1986). Arabidopsis thaliana (L.) Heynh. ecotype Columbia was transformed by the floral dip method (Clough and Bent, 1998). Transgenic CaMV35S-E2Fa and CaMV35S-DPa plants were obtained on kanamycin- or hygromycin-containing medium, respectively. For all analyses, plants were grown under a 16 h light/8 h dark photoperiod at 22°C on germination medium (Valvekens et al., 1988). RNA gel-blot analysis was performed on 3-week-old plants as described (De Veylder et al., 2001). Linkage of the phenotype with the presence of transgenes was tested by grinding individual plants in 400 µl of DNA extraction buffer (200 mM Tris–HCl pH 7.5, 250 mM NaCl, 25 mM ethylenediaminetetraacetic acid, 0.5% sodium dodecyl sulfate). Extracts were centrifuged at 18 000 g for 2 min. DNA was precipitated by adding 300 µl of isopropanol to a 300 µl extract and subjected to centrifugation for 10 min at 18 000 g. The pellet was rinsed with 70% ethanol, air dried and resuspended in 100 µl of water. For PCR analysis, 5 µl was used with the above mentioned primers. Because the transgenes do not contain introns, they could be distinguished from the endogenous E2Fa and DPa genes based on their size.
Flow cytometric analysis
Plants were chopped with a razor blade in 500 µl of 45 mM MgCl2, 30 mM sodium citrate, 20 mM 3-(N-morpholino)propanesulfonic acid pH 7 and 0.1% Triton X-100 (Galbraith et al., 1991). The supernatant was filtered over a 30 µm mesh, and 1 µl of 4′,6-diamidino-2-phenylindole (DAPI) from a stock of 1 mg/ml was added. The nuclei were analyzed with the BRYTE HS flow cytometer and the WinBryte software (Bio-Rad, Hercules, CA).
Histology and expression analysis
In situ hybridizations on roots and hypocotyls of Arabidopsis thaliana were performed as described (de Almeida Engler et al., 2001). Longitudinal sections of shoot apical buds of vegetative 2-month-old Arabidopsis plants, grown under conditions described by Corbesier et al. (1996), were hybridized as described by Segers et al. (1996). Full-length E2Fa- and DPa-coding sequences were used to generate 35S-labelled RNA probes. No signal was observed in control hybridizations. Images were recorded with an AxioCam digital camera (Zeiss, Jena, Germany). Overlays of bright- and dark-field micrographs were generated to maximally visualize tissue anatomy and mRNA localization.
Tissues for histological examination were placed overnight in ethanol to remove chlorophyll, subsequently cleared, mounted on a microscope slide and stored in lactic acid for microscopy. Cells were observed with differential interference contrast optics on a DMLB microscope (Leica, Wetzlar, Germany). Leaves and roots were sectioned according to Beeckman and Viane (2000). Cell counting in embryos was performed by incubating mature dry seeds overnight in Hoyer medium (Shimizu and Okada, 2000). The seed coat was carefully removed with an injection needle and the embryos were transferred to a microscope slide in a droplet of Hoyer medium. The cellular pattern of the cotyledon epidermis was analyzed with differential interference contrast optics on a Leica DMLB microscope.
For scanning electron microscopy, seedlings were fixed overnight in 4% paraformaldehyde and 1% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2), followed by a post-fixation step in 2% osmiumtetroxide, and a graded ethanol series. Critical-point drying was carried out in liquid carbon dioxide. These seedlings were mounted on stubs, sputter-coated with gold and examined with a scanning microscope (JEOL Ltd, Tokyo, Japan) under an accelerating voltage of 10 kV. Fluorescent staining of nuclei was performed by fixing the seedlings in a mixture of 9:1 (v/v) ethanol and acetic acid. After the samples had been rinsed, they were stained for 24 h with 0.1 µg/ml DAPI and analyzed by using an inverted confocal microscope LSM510 (Zeiss) fitted with a X20 plan-apochromat objective.
RT-mediated PCR analysis
RNA was isolated from plants 7 days after sowing using the Trizol reagent (Amersham Pharmacia Biotech, Little Chalfont, UK). First-strand cDNA was prepared from 3 µg of total RNA with the Superscript RT II kit (Invitrogen, Gaithersburg, MD) and oligo(dT)18 according to the manufacturer’s instructions. A 0.25 µl aliquot of the total RT reaction volume (20 µl) was used as a template in a semi-quantitative RT-mediated PCR amplification, ensuring that the amount of amplified product remained in linear proportion to the initial template present in the reaction. Ten microliters from the PCR was separated on a 0.8% agarose gel and transferred onto Hybond N+ membranes (Amersham Pharmacia Biotech). The membranes were hybridized at 65°C with fluorescein-labelled probes (Gene Images random prime module; Amersham Pharmacia Biotech). The hybridized bands were detected with the CDP Star detection module (Amersham Pharmacia Biotech). Primers used were 5′-TATGGCTGTCTGGGGTTTC-3′ and 5′-CAACTTGAACGTGTGGTTGG-3′ for DNA pol α (DDBJ/EMBL/GenBank accession No. AB020742), 5′-TCGAGTCGGTTGGAAGAAAG-3′ and 5′-CTCATGAACCATAGCCGTCA-3′ for ORC (AL049730), 5′-GCACCGTCAACTGTTGTTTG-3′ and 5′-CAAGCCTCTCCTGCAGAATC-3′ for CDC6 (AC005496), 5′-AGGCTAATGAGGGAGGGGTA-3′ and 5′-GGAACTGGCCTCATTTGTGT-3′ for MCM5 (AC004483), and GTGCCAATCTACGAGGGTTTC and CAATGGGACTAAAACGAAAA for ACT2 (U41998).
Acknowledgments
Acknowledgements
The authors thank the members of the cell-cycle group for fruitful discussions and useful suggestions, Nathalie Detry for technical assistance, Martine De Cock for help in preparing the manuscript and Karel Spruyt and Ruth De Groodt for illustrations. This work was supported by grants from the Interuniversity Poles of Attraction Programme (Belgian State, Prime Minister’s Office, Federal Office for Scientific, Technical and Cultural Affairs; P4/15), the ‘Fonds pour la Recherche Fondamentale et Collective’ (No. 2.4524.96), the European Union (ECCO QLG2-CT1999-00454) and CropDesign N.V. (0235). G.T.S.B. is indebted to the European Union for a Training and Mobility Research Grant (Marie Curie ERBFMI-CT97-2754), L.D.V. is a postdoctoral fellow of the Fund for Scientific Research (Flanders) and S.O. is a Scientific Research Worker of the National Fund for Scientific Research (FNRS).
References
- Albani D., Mariconti,L., Ricagno,S., Pitto,L., Moroni,C., Helin,K. and Cella,R. (2000) DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. J. Biol. Chem., 275, 19258–19267. [DOI] [PubMed] [Google Scholar]
- Bandara L.R., Buck,V.M., Zamanian,M., Johnston,L.H. and La Thangue,N.B. (1993) Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J., 12, 4317–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T. and Viane,R. (2000) Embedding thin plant specimens for oriented sectioning. Biotech. Histochem., 75, 23–26. [DOI] [PubMed] [Google Scholar]
- Berger F., Linstead,P., Dolan,L. and Haseloff,J. (1998) Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev. Biol., 194, 226–234. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Cebolla A., Vinardell,J.M., Kiss,E., Oláh,B., Roudier,F., Kondorosi,A. and Kondorosi,E. (1999) The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J., 18, 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabouté M.-E., Clément,B., Sekine,M., Philipps,G. and Chaubet-Gigot,N. (2000) Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell, 12, 1987–1999. [PMC free article] [PubMed] [Google Scholar]
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cockcroft C.E., den Boer,B.G.W., Healy,J.M.S. and Murray J.A.H. (2000) Cyclin D control of growth rate in plants. Nature, 405, 575–579. [DOI] [PubMed] [Google Scholar]
- Corbesier L., Gadisseur,I., Silvestre,G., Jacqmard,A. and Bernier,G. (1996) Design in Arabidopsis thaliana of a synchronous system of floral induction by one long day. Plant J., 9, 947–952. [DOI] [PubMed] [Google Scholar]
- de Almeida Engler J., De Groodt,R., Van Montagu,M. and Engler,G. (2001) In situ hybridization to mRNA of Arabidopsis tissue sections. Methods, 23, 325–334. [DOI] [PubMed] [Google Scholar]
- de Jager S.M., Menges,M., Bauer,U.-M. and Murray,J.A.H. (2001) Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol. Biol., 47, 555–568. [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beemster,G.T.S., Beeckman,T. and Inzé,D. (2001) CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J., 25, 617–626. [DOI] [PubMed] [Google Scholar]
- Doerner P., Jørgensen,J.-E., You,R., Steppuhn,J. and Lamb,C. (1996) Control of root growth and development by cyclin expression. Nature, 380, 520–523. [DOI] [PubMed] [Google Scholar]
- Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Edgar B.A. and Orr-Weaver,T.L. (2001) Endoreplication cell cycles: more for less. Cell, 105, 297–306. [DOI] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins,K.R. and Knapp,S. (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol., 96, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E., Traas,J., Desnos,T., Grandjean,O., Caboche,M. and Höfte,H. (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol., 114, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi G. and Larkins,B.A. (1995) Endoreduplication in maize endosperm: involvement of M phase-promoting factor inhibition and induction of S phase-related kinases. Science, 269, 1262–1264. [DOI] [PubMed] [Google Scholar]
- Guy C.T., Zhou,W., Kaufman,S. and Robinson,M.O. (1996) E2F-1 blocks terminal differentiation and causes proliferation in transgenic megakaryocytes. Mol. Cell. Biol., 16, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J., Fisher,D., Woollard,A. and Nurse,P. (1994) Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell, 78, 813–822. [DOI] [PubMed] [Google Scholar]
- Helin K., Wu,C.-L., Fattaey,A.R., Lees,J.A., Dynlacht,B.D., Ngwu,C. and Harlow,E. (1993) Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev., 7, 1850–1861. [DOI] [PubMed] [Google Scholar]
- Hemerly A., de Almeida Engler,J., Bergounioux,C., Van Montagu,M., Engler,G., Inzé,D. and Ferreira,P. (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J., 14, 3925–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley R. et al. (1998) The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol. Biol., 37, 155–169. [DOI] [PubMed] [Google Scholar]
- Jacqmard A., De Veylder,L., Segers,G., de Almeida Engler,J., Bernier,G., Van Montagu,M. and Inzé,D. (1999) Expression of CKS1At in Arabidopsis thaliana indicates a role for the protein in both the mitotic and the endoreduplication cycle. Planta, 207, 496–504. [DOI] [PubMed] [Google Scholar]
- Johnson D.G., Schwarz,J.K., Cress,W.D. and Nevins,J.R. (1993) Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature, 365, 349–352. [DOI] [PubMed] [Google Scholar]
- Koncz C. and Schell,J. (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet., 204, 383–396. [Google Scholar]
- Kovesdi I., Reichel,R. and Nevins,J.R. (1986) Identification of a cellular transcription factor involved in E1A trans-activation. Cell, 45, 219–228. [DOI] [PubMed] [Google Scholar]
- Krek W., Livingston,D.M. and Shirodkar,S. (1993) Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science, 262, 1557–1560. [DOI] [PubMed] [Google Scholar]
- Lavia P. and Jansen-Dürr,P. (1999) E2F target genes and cell-cycle checkpoint control. BioEssays, 21, 221–230. [DOI] [PubMed] [Google Scholar]
- Lukas J., Otzen Petersen,B., Holm,K., Bartek,J. and Helin,K. (1996) Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol. Cell. Biol., 16, 1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R.X., Postigo,A.A. and Dean,D.C. (1998) Rb interacts with histone deacetylase to repress transcription. Cell, 92, 463–473. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Groisman,R., Naguibneva,I., Robin,P., Lorain,S., Le Villain,J.P., Troalen,F., Trouche,D. and Harel-Bellan,A. (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature, 391, 601–605. [DOI] [PubMed] [Google Scholar]
- Magyar Z., Atanassova,A., De Veylder,L., Rombauts,S. and Inzé,D. (2000) Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett., 486, 79–87. [DOI] [PubMed] [Google Scholar]
- Qin X.-Q., Livingston,D.M., Kaelin,W.G.,Jr and Adams,P.D. (1994) Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl Acad. Sci. USA, 91, 10918–10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E. and Gutierrez,C. (2000) Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Lett., 486, 73–78. [DOI] [PubMed] [Google Scholar]
- Ramírez-Parra E., Xie,Q., Boniotti,M.B. and Gutierrez,C. (1999) The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G1/S regulators. Nucleic Acids Res., 27, 3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V. and Koornneef,M. (2001) Cell division activity during apical hook development. Plant Physiol., 125, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I., Whittaker,A.J. and Orr-Weaver,T.L. (1997) Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev., 11, 1999–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K., Knoblich,J.A., Richardson,H. and Lehner,C.F. (1995) Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev., 9, 1327–1339. [DOI] [PubMed] [Google Scholar]
- Segers G., Gadisseur,I., Bergounioux,C., de Almeida Engler,J., Jacqmard,A., Van Montagu,M. and Inzé,D. (1996) The Arabidopsis cyclin-dependent kinase gene cdc2bAt is preferentially expressed during S and G2 phases of the cell cycle. Plant J., 10, 601–612. [DOI] [PubMed] [Google Scholar]
- Sekine M., Ito,M., Uemukai,K., Maeda,Y., Nakagami,H. and Shinmyo,A. (1999) Isolation and characterization of the E2F-like gene in plants. FEBS Lett., 460, 117–122. [DOI] [PubMed] [Google Scholar]
- Shimizu K.K. and Okada,K. (2000) Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development, 127, 4511–4518. [DOI] [PubMed] [Google Scholar]
- Stals H. and Inzé,D. (2001) When plant cells decide to divide. Trends Plant Sci., 6, 359–364. [DOI] [PubMed] [Google Scholar]
- Valvekens D., Van Montagu,M. and Van Lijsebettens,M. (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl Acad. Sci. USA, 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Raes,J., De Veylder,L., Rouzé,P., Rombauts,S. and Inzé,D. (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell, 14, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen F.A., Molthoff,J.W., Conner,A.J., Nap,J.-P., Pereira,A. and Stiekema,W.J. (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res., 4, 288–290. [DOI] [PubMed] [Google Scholar]
- Wang J., Helin,K., Jin,P. and Nadal-Ginard,B. (1995) Inhibition of in vitro myogenic differentiation by cellular transcription factor E2F1. Cell Growth Differ., 6, 1299–1306. [PubMed] [Google Scholar]
- Weinberg R.A. (1995) The retinoblastoma protein and cell cycle control. Cell, 81, 323–330. [DOI] [PubMed] [Google Scholar]
- Wu C.-L., Classon,M., Dyson,N. and Harlow,E. (1996) Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol. Cell. Biol., 16, 3698–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]