Abstract
TNF-α is a potent proinflammatory cytokine that regulates immune and inflammatory responses and programmed cell death. TNF-α stimulation causes nuclear translocation of several NF-κB dimers, including RelA/p50 and RelB/p50. However, contrary to RelA, RelB entering the nucleus in response to TNF-α cannot bind to DNA in mouse embryonic fibroblasts, strongly suggesting that RelB DNA-binding activity is modulated by additional nuclear mechanisms. Here, we demonstrate that TNF-α promotes the association of RelA with RelB in the nucleus and that TNF-α-induced RelA/RelB heterodimers do not bind to κB sites. Remarkably, we show that RelA serine-276, the phosphorylation of which is induced by TNF receptor ligation, is crucial for RelA/RelB complex formation and subsequent inhibition of RelB DNA binding. In the absence of RelA phosphorylation on serine-276, TNF-α stimulation leads to a strong increase in the expression of endogenous NF-κB-responsive genes, such as Bcl-xL, whose transcriptional up-regulation is mainly controlled by RelB. Our findings demonstrate that RelA has a major regulatory role serving to dampen RelB activity in response to TNF-α and define a previously unrecognized mechanism that represents an essential step leading to selective NF-κB target gene expression.
Keywords: NF-κB, phosphorylation, target gene expression
The NF-κB family of transcription factors is essential for the control of immune and inflammatory responses as well as cell proliferation, differentiation, and apoptosis (1–7). Activation of NF-κB antagonizes programmed cell death, in particular, that triggered by TNF-α. It has recently emerged that the antiapoptotic activity of NF-κB is also crucial for tumor development and resistance to the chemotherapeutic drugs used in many common cancer therapies (5, 8–10). In mammals, the NF-κB family consists of five members that form homodimeric and heterodimeric complexes: RelA (p65), RelB, cRel (Rel), NF-κB1 (p50 and its precursor, p105), and NF-κB2 (p52 and its precursor, p100) (11, 12). Under nonstimulated conditions, NF-κB dimers are sequestered in the cytoplasm through interactions with inhibitory proteins of the IκB family (13, 14). After stimulation with a broad range of stimuli such as cytokines, various stress signals, and bacterial or viral products, the IκB molecules are phosphorylated by the IκB kinase (IKK) complex at specific serine residues, leading to their ubiquitination and degradation by the proteasome pathway (15, 16). As a result, the NF-κB dimers are released and become free to translocate into the nucleus to activate the transcription of various target genes.
In resting fibroblasts, the most abundant form of NF-κB is the heterodimer RelA/p50, whereas RelB is expressed at low levels and is sequestered in the cytoplasm due to its association with p100 (17, 18). TNF-α stimulation triggers the activation of RelA/p50 complexes via the activation of the canonical NF-κB pathway, which relies mostly on IKKβ- and IKKγ-dependent (two subunits of the IKK complex) IκBα degradation. Once in the nucleus, RelA/p50 activates the transcription of a large number of genes, some of which include cytokines and antiapoptotic proteins (15, 16). In addition, TNF-α stimulation results in a strong increase of RelB levels in both the cytoplasm and the nucleus (17, 19, 20). Surprisingly, the increased RelB protein level in the nucleus does not lead to an increased DNA-binding activity, suggesting a nuclear control of RelB in these cells. p100 and phosphorylation of RelB on serine 368 seem to contribute to this negative control (19–21), but additional mechanisms clearly account for the nuclear control of RelB DNA-binding activity in these cells.
Analysis of RelB-deficient mice has shown that RelB is essential for the development of medullary epithelium, mature dendritic cell function, and secondary lymphoid tissue organization (22–25), suggesting that RelB exerts a crucial positive effect for these developmental processes that cannot be compensated for by the presence of other NF-κB proteins. RelB-/- mice also spontaneously develop a multiorgan inflammatory syndrome that contributes significantly to their premature mortality (26). Interestingly, analysis of RelB-deficient fibroblasts has revealed that RelA and RelB can exert opposite effects on the regulation of specific subsets of genes. For instance, RelB represses the expression of several NF-κB target genes, such as RANTES and IP-10 (27, 28), whereas the same genes are positively controlled by RelA (29). It has been suggested that RelB reduces RelA activity through modulation of IκBα stability and/or by direct complex formation (28, 30). More recently, the recruitment of both RelA and RelB on specific target promoters was observed in monocyte-derived dendritic cells (31). Importantly, RelB recruitment to some genes has been shown to correlate with transcriptional down-regulation (IL-12p40), whereas in other cases [ELC (EBV-induced molecule 1 ligand chemokine) and MDC (macrophage-derived chemokine)], it increases transcriptional activity over the level achieved by RelA (31). The mechanisms explaining the alternative effects of RelB are unknown, and the possibility that RelA and RelB might regulate each other once in the nucleus is a crucial issue with profound implications for the overall transcriptional activity of a given target. In the study presented here, we investigated how RelA might regulate RelB activity in TNF-α-stimulated fibroblasts and the effects of their cross-regulation on TNF-α-induced NF-κB target gene expression.
Materials and Methods
Reagents and Antibodies. Murine recombinant TNF-α was purchased from Sigma. J. Hiscott (McGill University, Montréal) and N. Rice (National Cancer Institute, Frederick, MD) generously provided anti-p52/p100 and anti-p50/p105 polyclonal antibodies. Anti-RelA phosphoserine-276 was a kind gift from P. Cohen (University of Dundee, Dundee, U.K.). The remainder of the antibodies were purchased from Santa Cruz Biotechnology (RelA, RelB, p105/p50, cRel, and phospholipase C-γ-1), Upstate Biotechnology (Lake Placid, NY) (p100/p52, p105/p50, and Sir2), and Abcam (Cambridge, MA) (anti-RelA phosphoserine-276).
Plasmids. pBabe-puro retroviral vectors for WT RelA, RelA-S276A, and RelA-C216S were provided by T. Gilmore (Boston University, Boston). Expression vector for RelA was obtained from M. Benkirane (Institut de Génétique Humaine, Montpellier, France), and p50 and RelB plasmids were supplied by M. Körner (Institut André Lwoff, Villejuif, France).
Cell Culture and Infections. RelA-/- and NF-κB2-/- mouse embryonic fibroblasts (MEFs) were kind gifts from A. Beg (Columbia University, New York) and J. Caamano (University of Birmingham, Birmingham, U.K.), respectively. MEFs were grown in DMEM (Life Technologies, Paisley, Scotland) supplemented with 10% heat-inactivated FBS/2mM l-glutamine/1 mM sodium pyruvate/100 units/ml penicillin/100 μg/ml streptavidin. Production of infectious recombinant retroviruses was performed by transient transfection of Phenix EcoIII packaging cells as described in ref. 32. For infections, 1 × 105 cells in 35-mm dishes were transduced with 1 ml of retroviral supernatant in the presence of 4 μg/ml polybrene. Three days after infection, cells were stably selected on 1 μg/ml puromycin.
EMSA. Nuclear extracts were prepared and analyzed for DNA-binding activity by using the HIV-LTR tandem κB oligonucleotide as a κB probe (33). For supershift assays, nuclear extracts were incubated with specific antibodies for 30 min on ice before incubation with the labeled probe. In vitro translated NF-κB subunits were produced by using a transcription and translation (TNT)-coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions.
Coimmunoprecipitation and Immunoblotting. Coimmunoprecipitation and immunoblotting were performed as reported in ref. 19.
RT-PCR. RT-PCR were performed as described in ref. 34. Linear response ranges were determined for each gene to semiquantify their expression levels. Primer sequences are available upon request.
Short Interfering RNA (siRNA) Transfections. Cells were seeded at a density of 2 × 105 per 60-mm dish and transfected on the following day with 2 μg of siRNA oligonucleotides by using lipofectamine reagent (Invitrogen). After 48 h, the transfected cells were treated with TNF-α (10 ng/ml) for 8 h or left untreated and harvested for Western blotting or RT-PCR analysis. All siRNA duplex oligonucleotides were synthesized by Proligo (Paris). Sequences are available upon request.
Chromatin Immunoprecipitation (ChIP) Assays. The ChIP assays were performed as described in ref. 35. Samples were analyzed by quantitative PCR. Sequences of promoter-specific primers and a detailed experimental protocol are available upon request.
Apoptosis Assays. Apoptosis was monitored by annexin V and propidium iodide double-staining with an annexin V-FITC apoptosis kit (Roche Molecular Biochemicals). Stained cells were analyzed by flow cytometry.
Results
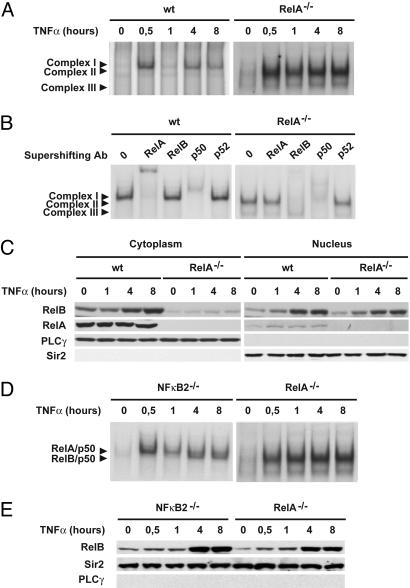
Absence of RelA Leads to Rapid and Sustained RelB Activation in TNF-α-Stimulated Cells. To determine whether interfering with the canonical NF-κB pathway affects RelB activation in response to TNF-α, we first evaluated TNF receptor (TNFR)-mediated activation of NF-κB complexes in MEFs deficient for RelA. As shown in Fig. 1A, TNF-α treatment of WT MEFs resulted in a characteristic biphasic induction of κB DNA binding (complex I). In contrast, after only 30 min of TNF-α treatment, we observed a strong increase of a faster migrating complex that further increased over 8 h in RelA-deficient MEFs (complex II). A constitutive binding of a third complex (complex III) was also observed in these cells. The subunit composition of these NF-κB DNA-binding complexes was next examined by supershift analysis of nuclear extracts from WT and RelA-deficient fibroblasts stimulated for 8 h with TNF-α (Fig. 1B). Antibodies directed against RelA and p50 supershifted complex I almost completely in WT fibroblasts. Not surprisingly, antibodies to RelA did not recognize κB-binding complexes in extracts derived from RelA-/- cells. Instead, complex II was effectively supershifted with anti-RelB and anti-p50 antibodies, and complex III was ablated with antibody directed against p50. Antibodies to p52 (Fig. 1B) and cRel (data not shown) had very little effect on either complex.
Fig. 1.
Absence of RelA leads to rapid and sustained RelB activation in TNF-α-stimulated MEFs. (A) Nuclear extracts from WT and RelA-deficient MEFs treated with TNF-α for the indicated periods of time were analyzed by EMSA using a 32P-labeled HIV-LTR tandem κB oligonucleotide as a probe. (B) For supershift, nuclear extracts from WT and RelA-deficient MEFs treated with TNF-α for 8 h were incubated with the indicated antibodies before incubation with the labeled probe. Complex I, RelA/p50; complex II, RelB/p50; complex III, p50/p50. (C) Cytoplasmic and nuclear extracts of WT and RelA-deficient MEFs treated with TNF-α for the indicated periods of time were analyzed by immunoblotting for the indicated proteins. Phospholipase C-γ-1 (PLCγ) and Sir2 were used as quality controls to verify the absence of cytoplasmic contamination in nuclear extracts and nuclear contamination in cytoplasmic extracts, respectively. (D) Nuclear extracts from RelA- and NF-κB2-deficient MEFs treated with TNF-α for the indicated periods of time were analyzed by EMSA as described in A.(E) Nuclear extracts from RelA- and NF-κB2-deficient MEFs treated with TNF-α for the indicated periods of time were analyzed by immunoblotting for the indicated proteins.
To gain further insight into the mechanisms that control the strong and sustained activation of RelB in RelA-deficient fibroblasts, we first compared RelB protein levels and cellular distributions in WT versus RelA-deficient MEFs (Fig. 1C). Although the steady-state level of expression of RelB is low in the cytoplasm of RelA-deficient fibroblasts compared with WT fibroblasts, TNF-α-induced RelB nuclear translocation is clearly not affected by the absence of RelA. Because we have previously reported that p100 negatively controls RelB DNA-binding activity downstream of TNFRs (19), and given that p100 transcription has been reported to be regulated by RelA (36), we also analyzed TNFR-mediated NF-κB DNA-binding activity in NF-κB2-deficient MEFs (i.e., those lacking p100). RelB DNA binding was found to be induced by TNF-α at much lower levels and with a more delayed time course in NF-κB2-deficient fibroblasts compared with RelA-deficient fibroblasts (Fig. 1D), whereas the levels of TNF-α-induced nuclear RelB were similar in RelA- and NF-κB2-deficient cells (Fig. 1E). Taken together, these results strongly suggest that, beyond its effect on p100 expression level, RelA exerts a negative regulatory function on RelB DNA binding downstream of TNFRs.
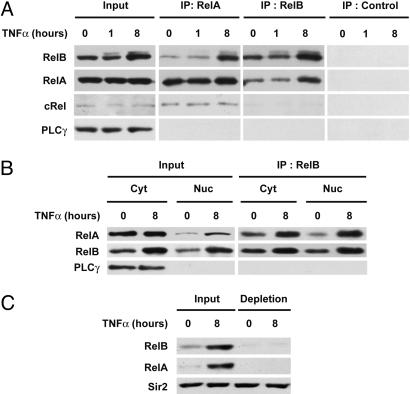
TNF-α Promotes the Formation of RelA/RelB Complexes. Direct binding with other transcription factors such as p53 was previously shown to regulate RelA activity (37, 38). We speculated that dimerization with RelA might impair RelB DNA-binding ability. To test this hypothesis, we first examined whether RelA associates with RelB by coimmunoprecipitation assays using whole-cell extracts from fibroblasts that were either unstimulated or stimulated by TNF-α for 8 h. As a control, similar immunoprecipitation experiments were performed by using nonimmune serum to verify the specificity of the interaction. Reciprocal experiments with RelA- and RelB-specific antibodies showed that endogenous RelB coimmunoprecipitates with RelA (Fig. 2A). Most importantly, TNF-α stimulation resulted in increased association of RelA with RelB. In contrast, no significant interaction between RelB and cRel was detected either in unstimulated or TNF-α-treated MEFs (Fig. 2A). We then examined the cellular distribution of the RelA/RelB complex in TNF-α-stimulated fibroblasts. As shown in Fig. 2B, RelA associates with RelB in both cytoplasmic and nuclear compartments in response to TNF-α stimulation. Importantly, the increase of RelB protein levels parallels the increase of coimmunoprecipitated RelA. Next, we investigated the relative amount of RelB that was complexed with RelA in the nucleus of TNF-α-stimulated fibroblasts. RelA was depleted from nuclear lysates by immunoprecipitation with anti-RelA antibody, and the RelB content was analyzed by immunoblotting. As shown in Fig. 2C, the majority of RelB associates with RelA in the nucleus of TNF-α-treated cells.
Fig. 2.
Increased association of RelA with RelB in the nucleus of TNF-α-treated cells. (A) TNF-α promotes the association of RelA with RelB. Whole-cell extracts from WT MEFs treated with TNF-α for the indicated periods of time were subjected to immunoprecipitation (IP) with anti-RelA, anti-RelB, or nonimmune control antibody and analyzed by immunoblotting for the indicated proteins. (B) TNF-α induces the formation of RelA/RelB complexes in the nucleus of WT MEFs. Cytoplasmic and nuclear extracts from WT MEFs, either untreated or TNF-α-treated (8 h), were subjected to immunoprecipitation with anti-RelB antibody and analyzed for associated RelA. Phospholipase C-γ-1 (PLCγ) was used as a quality control to verify the absence of cytoplasmic contamination in nuclear extracts. (C) The majority of RelB associates with RelA in the nucleus of TNF-α-treated MEFs. Nuclear extracts from WT MEFs, either untreated or TNF-α treated (8 h), were subjected to immunodepletion with anti-RelA antibody and analyzed for RelB content by immunoblotting with anti-RelB antibody. Sir2 was used as a loading control.
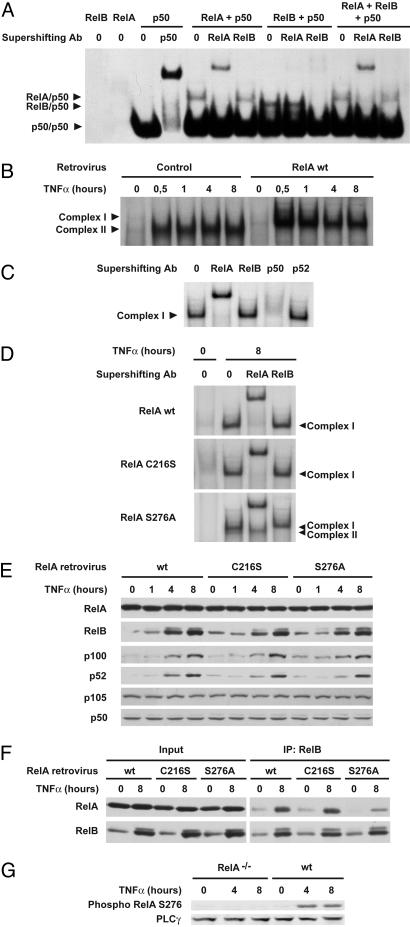
RelA Serine-276 Is Critical for RelA/RelB Complex Formation and Inhibition of RelB DNA Binding. To determine whether the formation of RelA/RelB complexes might be responsible for inhibition of RelB/p50 DNA-binding activity, we analyzed the κB-binding activity of RelB/p50 dimers in the presence or absence of RelA by using in vitro translated NF-κB proteins. In this experiment, we used an excess of p50 to avoid its titration by RelA. As shown in Fig. 3A, RelA/p50, RelB/p50, and p50/p50 dimers alone exhibited binding to the κB probe. In contrast, RelB/p50 binding was nearly abolished in the presence of RelA, although protein expression levels for in vitro translated RelB and p50 were comparable in the absence or presence of RelA, and RelB heterodimerization with p50 was not affected by RelA coexpression (Fig. 6, which is published as supporting information on the PNAS web site). Importantly, no binding of RelA/RelB heterodimers was observed, strongly suggesting that they are unable to bind κB sites in vitro.
Fig. 3.
RelA serine-276 is critical for RelA/RelB complex formation and consequent inhibition of RelB DNA binding. (A) RelA inhibits RelB/p50 DNA binding in vitro. The κB probe was incubated with the indicated in vitro translated NF-κB proteins, and DNA binding was analyzed by EMSA. For supershift, in vitro translated NF-κB proteins were incubated with the indicated antibodies before incubation with the labeled probe. (B) Reexpression of RelA in RelA-deficient MEFs abolishes TNF-α-induced RelB/p50 DNA binding. Nuclear extracts from RelA-deficient MEFs stably transduced with retroviruses encoding either the parental empty vector (control) or WT RelA and treated with TNF-α for the indicated periods of time were analyzed by EMSA. (C) For supershift, nuclear extracts from RelA-deficient MEFs reexpressing WT RelA treated with TNF-α for 8 h were incubated with the indicated antibodies before incubation with the labeled probe. (D) RelA serine-276 is critical for inhibition of TNF-α-induced RelB DNA binding. Nuclear extracts from RelA-deficient MEFs stably transduced with the indicated RelA retroviruses, either untreated or treated with TNF-α for 8 h, were analyzed for NF-κB activity by EMSA. For supershift, nuclear extracts were incubated with the indicated antibodies before incubation with the labeled probe. (E) Protein expression levels of NF-κB family members in RelA-deficient MEFs reexpressing WT RelA or C216S or S276A RelA mutants. Whole-cell extracts from RelA-deficient MEFs infected with the indicated RelA retroviruses and treated with TNF-α for the indicated periods of time were analyzed by immunoblotting for the indicated proteins. (F) RelA serine-276 is critical for association with RelB. Whole-cell extracts from RelA-deficient MEFs infected with the indicated RelA retroviruses, either untreated or treated with TNF-α for 8 h, were subjected to immunoprecipitation (IP) with anti-RelB antibody and analyzed for associated RelA. (G) TNF-α induces RelA serine-276 phosphorylation. Whole-cell extracts from either WT MEFs or RelA-deficient MEFs treated with TNF-α for the indicated periods of time were analyzed by immunoblotting for RelA serine-276 phosphorylation.
To assess whether RelA might inhibit RelB/p50 DNA-binding activity in vivo, we first infected RelA-deficient cells with either an empty retrovirus or a retrovirus carrying WT RelA and analyzed the κB-binding activity. Whereas TNF-α stimulation of empty virus-infected RelA-/- fibroblasts induced RelB DNA binding (complex II) with kinetics that parallel what is seen in noninfected cells, reintroduction of WT RelA induced strong RelA/p50 DNA binding (complex I) and completely abolished RelB-binding activity (Fig. 3 B and C). As was the case in vitro, we were unable to detect any DNA binding of RelA/RelB complexes in vivo. Together, these data indicate that RelA can repress RelB DNA binding both in vitro and in vivo, most likely through its sequestration in RelA/RelB heterodimers.
Because RelB requires p50 or p52 as a dimerization partner to bind DNA, it is possible that sequestration of RelB complex partners p50 and p52 by RelA might account for the lack of RelB DNA binding. To test this possibility, we infected RelA-deficient cells with retrovirus carrying two mutant forms of RelA, S276A and C216S, which cannot form homodimers but can still form heterodimers with p50 and p52 (refs. 39 and 40 and Fig. 7, which is published as supporting information on the PNAS web site) and analyzed their ability to suppress RelB DNA-binding activity in response to TNF-α. Introduction of C216S into RelA-deficient cells induced strong RelA/p50 DNA binding (complex I) and completely abolished RelB-binding activity (complex II) (Fig. 3D), thus acting just like WT RelA. In contrast, whereas expression of RelA S276A induced RelA/p50 DNA binding to a level similar to that seen with WT RelA and C216S, it only weakly inhibited RelB DNA-binding activity. Importantly, C216S- and S276A-infected cell lines expressed RelA at levels similar to that of WT RelA (Fig. 3E). Moreover, protein expression levels for RelB, p100, p105, p52, and p50 were comparable in RelA-deficient MEFs reconstituted with WT, S276A, and C216S RelA (Fig. 3E). Finally, just as it does in vivo, C216S but not S276A RelA mutant inhibited RelB/p50 DNA binding in vitro (Fig. 8, which is published as supporting information on the PNAS web site). These results strongly suggest that RelA can repress RelB DNA-binding activity downstream of TNFRs independently of the sequestration of RelB dimerization partners p50 and p52 and pinpoint RelA serine-276 as a regulator of RelA/RelB complex formation.
Next, we directly compared the association of C216S and S276A RelA mutants with RelB. RelA C216S coimmunoprecipitates with RelB at levels similar to those seen with WT RelA (Fig. 3F). In contrast, RelA S276A only weakly associates with RelB. Thus, RelA serine-276 is required for complex formation with RelB, just as it is for the inhibition of RelB/p50 DNA binding (Fig. 3D). Because phosphorylation of RelA on serine-276 is increased by TNF-α stimulation in MEFs (refs. 41 and 42 and Fig. 3G), these results suggest that RelA serine-276 phosphorylation might be required for RelA/RelB complex formation and subsequent repression of RelB activity downstream of TNFRs.
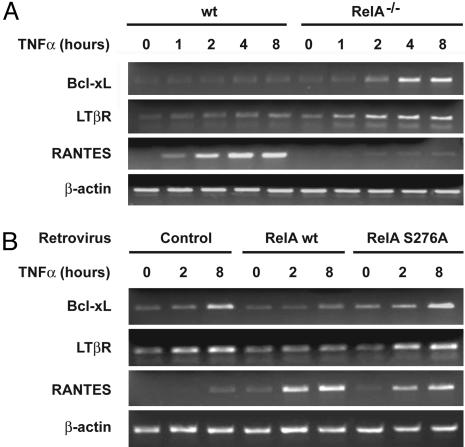
RelA Exerts a Serine-276-Dependent Repression of Endogenous NF-κB Target Gene Expression. It was important to determine whether these inhibitory effects could also be seen on endogenous NF-κB target gene expression. Therefore, using semiquantitative RT-PCR, we first compared the expression of 20 known NF-κB target genes, including antiapoptotic genes, inflammatory chemokines and cytokines, and their specific receptors, in WT and RelA-deficient MEFs. Fig. 4A presents the results obtained for three genes that represent diverse classes of NF-κB targets and are differently induced in response to TNF-α. These genes encode the antiapoptotic protein Bcl-xL, the receptor for lymphotoxin-β (a cytokine that induces the NF-κB alternative pathway), and the chemokine RANTES. As expected, TNF-α strongly induced the gene encoding RANTES in WT fibroblasts, whereas no such activation was found in the absence of RelA. In contrast, whereas the expression of Bcl-xL and the lymphotoxin-β receptor (LTβR) was only slightly increased by 8 h of TNF-α stimulation in WT fibroblasts, loss of RelA resulted in a much stronger time-dependent increase of Bcl-xL and LTβR mRNA levels (Fig. 4A). Analyses using two different WT and RelA-deficient cell lines demonstrated reproducibility (data not shown). We also analyzed the mRNA expression levels of these three genes in RelA-/- cells reconstituted with WT RelA and the S276A mutant. Reintroduction of WT RelA but not S276A mutant reversed the strong TNF-α-induced expression of Bcl-xL and LTβR, indicating that RelA serine-276 is required to restrain Bcl-xL and LTβR expression downstream of TNFRs (Fig. 4B). In contrast, reintroduction of either WT RelA or S276A RelA mutant restored a strong induction of RANTES expression in response to TNF-α (Fig. 4B). Taken together, these results indicate that RelA not only acts as an activator of its target genes but also exerts a serine-276-dependent selective repressive function serving to dampen NF-κB activity.
Fig. 4.
RelA serine-276 is required for restraining Bcl-xL and LTβR gene expression downstream of TNFRs. (A) Bcl-xL and LTβR expression is increased in the absence of RelA. Semiquantitative RT-PCR analysis was performed with specific primer pairs for the indicated genes by using total RNAs prepared from either WT or RelA-deficient MEFs treated with TNF-α for the indicated periods of time. All of the RT-PCRs were performed in the linear range for each transcript and normalized with β-actin as a reference control. (B) RelA serine-276 is required to restrain Bcl-xL and LTβR gene expression downstream of TNFRs. RelA-deficient MEFs stably transduced with the indicated retroviruses were treated with TNF-α for the indicated periods of time, and semiquantitative RT-PCR analysis was performed as described in A.
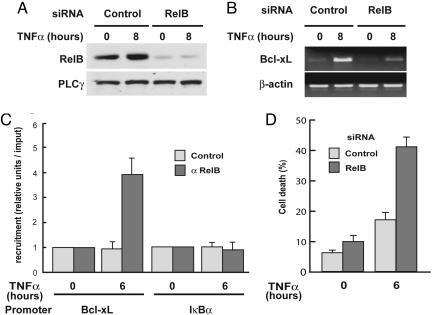
RelB Up-Regulates Bcl-xL Transcription and Protects Cells from TNF-α-Induced Apoptosis in RelA-Deficient MEFs. To establish whether the increased expression of Bcl-xL was RelB-dependent, we used RNA interference (RNAi) to down-regulate RelB levels in RelA-deficient fibroblasts. As shown in Fig. 5A, a siRNA directed against RelB efficiently repressed RelB protein levels; this knockdown persisted even after 8 h of TNF-α stimulation. Importantly, the TNF-α-induced Bcl-xL mRNA expression level was strongly reduced by RelB knockdown in RelA-deficient MEFs (Fig. 5B), whereas RelB knockdown had no effect on Bcl-xL mRNA levels in WT MEFs (Fig. 9, which is published as supporting information on the PNAS web site). Further in vivo evidence for a specific role of RelB on Bcl-xL transcription in RelA-deficient MEFs was obtained by ChIP analysis. As shown in Fig. 5C, TNF-α stimulation induced efficient recruitment of RelB to the Bcl-xL promoter. As a control, RelB was not seen to bind to the IκB promoter. Together, these data indicated that the RelA repressive effects on Bcl-xL expression are exerted through regulation of RelB activity. Because RelA serine-276 is crucial for RelA/RelB complex formation and blockade of RelB DNA binding, our data suggest that heterodimerization of RelA with RelB represses RelB-dependent activation of endogenous NF-κB-responsive genes such as the antiapoptotic gene Bcl-xL. Finally, because Bcl-xL is an antiapoptotic protein, we explored the role played by RelB in mechanisms controlling TNFR-induced programmed cell death and NF-κB-dependent survival. As shown in Fig. 5D, knockdown of RelB expression in RelA-deficient MEFs increased TNF-α-induced apoptosis by >2-fold (18% versus 42%), whereas RelB siRNA did not significantly affect TNF-α-induced apoptosis in WT fibroblasts (data not shown).
Fig. 5.
RelB knockdown by RNAi blocks Bcl-xL expression and promotes TNF-α-induced apoptosis in RelA-deficient MEFs. (A) RelB protein levels are efficiently knocked down by RNAi. RelA-deficient MEFs were transfected with either a siRNA oligonucleotide targeting RelB or a scrambled control. Forty-eight hours after transfection, cells were treated with TNF-α for 8 h or left untreated and then harvested and analyzed for RelB protein expression by immunoblotting. (B) RelB knockdown by RNAi prevents TNF-α induction of Bcl-xL expression in RelA-deficient MEFs. RelA-deficient MEFs were transfected with either a siRNA oligonucleotide targeting RelB or a scrambled control. Forty-eight hours after transfection, cells were treated with TNF-α for 8 h or left untreated and then harvested and analyzed for Bcl-xL and β-actin expression by using semiquantitative RT-PCR. (C) RelB is bound to the Bcl-xL promoter after TNF-α stimulation in RelA-deficient MEFs. RelA-deficient MEFs were treated with TNF-α for 6 h or left untreated, and recruitment of RelB to the Bcl-xL and IκBα promoters was examined by ChIP experiments followed by quantitative PCR analysis. The results are means ± SE of three independent experiments normalized to inputs that reflect relative amounts of sonicated DNA fragments present before immunoprecipitation. (D) RelB knockdown by RNAi promotes TNF-α-induced apoptosis in RelA-deficient MEFs. RelA-deficient MEFs were transfected with either a siRNA oligonucleotide targeting RelB or a scrambled control. Forty-eight hours after transfection, cells were treated with TNF-α (20 ng/ml)for 6 h or left untreated, collected, and incubated with annexin V-FITC for 20 min and analyzed by flow cytometry. Shown are means ± SE of three independent experiments.
Taken together, our results show that RelA negatively modulates RelB DNA-binding activity, most likely through direct complex formation that interferes with the induction of NF-κB target genes encoding proteins such as Bcl-xL.
Discussion
In the study presented here, we have explored the TNFR-mediated signaling events that control RelB activity in fibroblasts. We have found that RelA represses RelB activity by affecting its DNA binding and that inhibition occurs through its sequestration in RelA/RelB heterodimers. It has been previously reported that LPS-stimulated RelB-deficient fibroblasts exhibited a persistent induction of several chemokines that correlates with increased RelA DNA binding (28) and that complex formation with RelB impaired RelA DNA-binding capacity in lymphoid cells (30). Hence, RelA heterodimerization with RelB seems to reciprocally repress both RelA and RelB activity. The inhibitory function of RelB was observed in fibroblasts and lymphoid cells but not in macrophages. Therefore, it is likely that the repressive effects of RelA on RelB downstream of TNFRs are also cell-type specific.
The association of RelA with RelB is markedly increased in response to TNF-α stimulation, and serine-276, located in the Rel homology domain of RelA, seems to be a critical phosphorylation site for TNF-α-induced RelA/RelB complex formation, blockade of RelB DNA binding, and limitation of NF-κB activity. First, using specific anti-RelA phosphoserine-276, we observe, in accordance with previous reports, that in murine fibroblasts this site is phosphorylated in response to TNF-α (42). Second, RelA S276A mutant exhibits a markedly reduced ability to interact with RelB and to repress its DNA binding after TNF-α treatment in vivo. Third, serine-276 is essential for the ability of RelA to limit the expression of endogenous RelB-responsive genes. Two kinases have been shown to phosphorylate RelA on serine-276: the mitogen- and stress-activated kinase (MSK) 1, in response to TNF-α treatment (42), and the activated catalytic subunit of protein kinase A (PKAc), in response to LPS (43, 44). Whether these two kinases are indeed involved in the control of TNF-α-regulated RelB DNA binding is uncertain. PKAc-dependent RelA phosphorylation on serine-276 in response to TNF-α has not been described, and phosphorylation of RelA serine-276 is only partially reduced in TNF-α-treated MSK1-/-MSK2-/- fibroblasts (42). Furthermore, treatment of cells with H89, a potent inhibitor of MSK1, and several other kinases, including PKA, did not abolish RelA phosphorylation on serine-276 in TNF-α-stimulated fibroblasts, suggesting the involvement of multiple kinases in serine-276 phosphorylation (42). Finally, no RelB DNA binding was observed in TNF-α-stimulated MSK1-/-MSK2-/- fibroblasts (unpublished data). Thus, it is likely that other kinases also contribute to specific RelA serine-276 phosphorylation and subsequent increase of sequestration of RelB in RelA/RelB complexes.
Although our study clearly shows that the control of RelB activity downstream of TNFRs involves the interplay between RelA and RelB in the nucleus, additional factors might also contribute to the control of RelB-dependent gene activation downstream of TNFRs. First, RelB/p50 DNA binding was induced in TNF-α-stimulated NF-κB2-deficient cells, albeit at much lower levels and with delayed kinetics compared with RelA-deficient cells (Fig. 1D), suggesting that p100 accounts for the control of RelB/p50 DNA binding downstream of TNFRs. Second, because selective recruitment of RelB/p52 dimers to Blc and Elc promoters depends on a different κB site whose consensus sequence is distinct from the classical κB site (45), κB site specificity might exist for RelB/p50 dimers. Finally, other mechanisms, such as chromatin remodeling and RelB posttranslational modifications, most likely also influence RelB recruitment to specific promoters. Nonetheless, our results clearly show that an important mechanism of RelB DNA-binding regulation downstream of TNFRs is based on trapping of RelB in RelA/RelB complexes that cannot bind consensus κB-binding sites.
Our findings are of great functional importance because they constitute a significant advance in understanding the mechanisms that control RelB DNA-binding activity once in the nucleus. Together, our data indicate that RelA blocks RelB DNA-binding activity by sequestering it in RelA/RelB heterodimers, and they establish a previously unrecognized mechanism of cross-regulation between RelA and RelB that serves to limit the expression of a subset of RelB-responsive genes.
Supplementary Material
Acknowledgments
We thank A. Beg, F. Weih (Fritz Lipmann Institute, Jena, Germany), M. Karin (University of California at San Diego, La Jolla), J. Caamano, M. Pasparakis (European Molecular Biology Laboratory, Monterotondo, Monterotondo, Italy), S. Ghosh (Yale University, New Haven, CT), T. Gilmore, J. Ye (Louisiana State University, Baton Rouge), M. Benkirane, M. Körner, N. Rice, P. Cohen, G. Haegeman (Ghent University, Ghent, Belgium), and J. Hiscott for valuable cell lines and reagents; Françoise Picard and Claire Francastel for assistance in apoptosis analysis; G. Natoli for advice regarding ChIP experiments; A. Israël for advice and helpful discussion; L. L. Pritchard for critically reading the manuscript; and F. Dreyfus for support. This work was supported by grants from Groupement des Entreprises Françaises dans la Lutte Contre le Cancer, Association pour la Recherche sur le Cancer, Fondation pour la Recherche Médicale, Ministère de la Recherche/Cancéropôle IDF, and Johnson & Johnson (to V.B.) and a fellowship from the Ministère de la Recherche et des Technologies (to E.J.).
Abbreviations: TNFR, TNF receptor; MEF, mouse embryonic fibroblast; ChIP, chromatin immunoprecipitation; siRNA, short interfering RNA; LTβR, lymphotoxin-β receptor; RNAi, RNA interference; MSK, mitogen- and stress-activated kinase.
References
- 1.Barnes, P. J. & Karin, M. (1997) N. Engl. J. Med. 336, 1066-1071. [DOI] [PubMed] [Google Scholar]
- 2.Barkett, M. & Gilmore, T. D. (1999) Oncogene 18, 6910-6924. [DOI] [PubMed] [Google Scholar]
- 3.Silverman, N. & Maniatis, T. (2001) Genes Dev. 15, 2321-2342. [DOI] [PubMed] [Google Scholar]
- 4.Karin, M. & Lin, A. (2002) Nat. Immunol. 3, 221-227. [DOI] [PubMed] [Google Scholar]
- 5.Karin, M., Cao, Y., Greten, F. R. & Li, Z. W. (2002) Nat. Rev. Cancer 2, 301-310. [DOI] [PubMed] [Google Scholar]
- 6.Hatada, E. N., Krappmann, D. & Scheidereit, C. (2000) Curr. Opin. Immunol. 12, 52-58. [DOI] [PubMed] [Google Scholar]
- 7.Weih, F. & Caamano, J. (2003) Immunol. Rev. 195, 91-105. [DOI] [PubMed] [Google Scholar]
- 8.Shishodia, S. & Aggarwal, B. B. (2004) Biochem. Pharmacol. 68, 1071-1080. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin, A. S. (2001) J. Clin. Invest. 107, 241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma, I. M. (2004) Ann. Rheum. Dis. 63, Suppl. 2, ii57-ii61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh, S., May, M. J. & Kopp, E. B. (1998) Annu. Rev. Immunol. 16, 225-260. [DOI] [PubMed] [Google Scholar]
- 12.Siebenlist, U., Franzoso, G. & Brown, K. (1994) Annu. Rev. Cell Biol. 10, 405-455. [DOI] [PubMed] [Google Scholar]
- 13.Baldwin, A. S. (1996) Annu. Rev. Immunol. 14, 649-683. [DOI] [PubMed] [Google Scholar]
- 14.Whiteside, S. T. & Israel, A. (1997) Semin. Cancer Biol. 8, 75-82. [DOI] [PubMed] [Google Scholar]
- 15.Karin, M. & Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18, 621-663. [DOI] [PubMed] [Google Scholar]
- 16.Hayden, M. S. & Ghosh, S. (2004) Genes Dev. 18, 2195-2224. [DOI] [PubMed] [Google Scholar]
- 17.Bren, G. D., Solan, N. J., Miyoshi, H., Pennington, K. N., Pobst, L. J. & Paya, C. V. (2001) Oncogene 20, 7722-7733. [DOI] [PubMed] [Google Scholar]
- 18.Dobrzanski, P., Ryseck, R. P. & Bravo, R. (1995) Oncogene 10, 1003-1007. [PubMed] [Google Scholar]
- 19.Derudder, E., Dejardin, E., Pritchard, L. L., Green, D. R., Körner, M. & Baud, V. (2003) J. Biol. Chem. 278, 23278-23284. [DOI] [PubMed] [Google Scholar]
- 20.Yilmaz, Z. B., Weih, D. S., Sivakumar, V. & Weih, F. (2003) EMBO J. 22, 121-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier, H. J., Marienfeld, R., Wirth, T. & Baumann, B. (2003) J. Biol. Chem. 278, 39242-39250. [DOI] [PubMed] [Google Scholar]
- 22.Wu, L., D'Amico, A., Winkel, K. D., Suter, M., Lo, D. & Shortman, K. (1998) Immunity 9, 839-847. [DOI] [PubMed] [Google Scholar]
- 23.Weih, F., Warr, G., Yang, H. & Bravo, R. (1997) J. Immunol. 158, 5211-5218. [PubMed] [Google Scholar]
- 24.Weih, F., Carrasco, D., Durham, S. K., Barton, D. S., Rizzo, C. A., Ryseck, R. P., Lira, S. A. & Bravo, R. (1995) Cell 80, 331-340. [DOI] [PubMed] [Google Scholar]
- 25.Weih, D. S., Yilmaz, Z. B. & Weih, F. (2001) J. Immunol. 167, 1909-1919. [DOI] [PubMed] [Google Scholar]
- 26.Weih, F., Durham, S. K., Barton, D. S., Sha, W. C., Baltimore, D. & Bravo, R. (1996) J. Immunol. 157, 3974-3979. [PubMed] [Google Scholar]
- 27.Xia, Y., Pauza, M. E., Feng, L. & Lo, D. (1997) Am. J. Pathol. 151, 375-387. [PMC free article] [PubMed] [Google Scholar]
- 28.Xia, Y., Chen, S., Wang, Y., Mackman, N., Ku, G., Lo, D. & Feng, L. (1999) Mol. Cell. Biol. 19, 7688-7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann, A., Leung, T. H. & Baltimore, D. (2003) EMBO J. 22, 5530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marienfeld, R., May, M. J., Berberich, I., Serfling, E., Ghosh, S. & Neumann, M. (2003) J. Biol. Chem. 278, 19852-19860. [DOI] [PubMed] [Google Scholar]
- 31.Saccani, S., Pantano, S. & Natoli, G. (2003) Mol. Cell 11, 1563-1574. [DOI] [PubMed] [Google Scholar]
- 32.Tchenio, T., Casella, J. F. & Heidmann, T. (2001) Mol. Cell. Biol. 21, 1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feuillard, J., Gouy, H., Bismuth, G., Lee, L. M., Debre, P. & Korner, M. (1991) Cytokine 3, 257-265. [DOI] [PubMed] [Google Scholar]
- 34.Baud, V., Chissoe, S. L., Viegas-Pequignot, E., Diriong, S., N′Guyen, V. C., Roe, B. A. & Lipinski, M. (1995) Genomics 26, 334-344. [DOI] [PubMed] [Google Scholar]
- 35.Saccani, S. & Natoli, G. (2002) Genes Dev. 16, 2219-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liptay, S., Schmid, R. M., Nabel, E. G. & Nabel, G. J. (1994) Mol. Cell. Biol. 14, 7695-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda, A., Sun, X., Li, Y., Zhang, Y., Eckner, R., Doi, T. S., Takahashi, T., Obata, Y., Yoshioka, K. & Yamamoto, K. (2000) Biochem. Biophys. Res. Commun. 272, 375-379. [DOI] [PubMed] [Google Scholar]
- 38.Jeong, S. J., Radonovich, M., Brady, J. N. & Pise-Masison, C. A. (2004) Blood 104, 1490-1497. [DOI] [PubMed] [Google Scholar]
- 39.Gapuzan, M. E., Yufit, P. V. & Gilmore, T. D. (2002) Oncogene 21, 2484-2492. [DOI] [PubMed] [Google Scholar]
- 40.Ganchi, P. A., Sun, S. C., Greene, W. C. & Ballard, D. W. (1993) Mol. Cell. Biol. 13, 7826-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okazaki, T., Sakon, S., Sasazuki, T., Sakurai, H., Doi, T., Yagita, H., Okumura, K. & Nakano, H. (2003) Biochem. Biophys. Res. Commun. 300, 807-812. [DOI] [PubMed] [Google Scholar]
- 42.Vermeulen, L., De Wilde, G., Van Damme, P., Vanden Berghe, W. & Haegeman, G. (2003) EMBO J. 22, 1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong, H., SuYang, H., Erdjument-Bromage, H., Tempst, P. & Ghosh, S. (1997) Cell 89, 413-424. [DOI] [PubMed] [Google Scholar]
- 44.Zhong, H., Voll, R. E. & Ghosh, S. (1998) Mol. Cell 1, 661-671. [DOI] [PubMed] [Google Scholar]
- 45.Bonizzi, G., Bebien, M., Otero, D. C., Johnson-Vroom, K. E., Cao, Y., Vu, D., Jegga, A. G., Aronow, B. J., Ghosh, G., Rickert, R. C. & Karin, M. (2004) EMBO J. 23, 4202-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.