Abstract
Despite intensive investigation, controversial results have been obtained concerning the precise signaling pathway(s) regulated by K-ras in different cell types. We show that in primary fetal liver erythroid progenitors, erythropoietin activates all three Ras isoforms, but preferentially N- and K-ras. In K-ras-/- fetal liver cells (FLC), erythropoietin- or stem cell factor-dependent Akt activation is greatly reduced, whereas other pathways including Stat5 and p44/p42 MAP kinase are activated normally. We further studied the effects of reduced cytokine-dependent Akt activation in erythroid differentiation. We find that freshly isolated K-ras-/- FLC show an ≈7-fold increase of apoptosis and delayed erythroid differentiation, but only at the stage of erythroid progenitors and very early erythroblasts. When K-ras-/- erythroid progenitors are cultured in vitro, there is a significant delay in erythroid differentiation but little increase in apoptosis. Furthermore, we show that partial pharmacologic inhibition of the phosphatidylinositol 3-kinase/Akt pathway in wild-type erythroid progenitors leads to a delay in erythroid differentiation similar to that observed in K-ras-/- FLC. Taken together, our data identify K-ras as the major regulator for cytokine-dependent Akt activation, which is important for erythroid differentiation in vivo.
Keywords: erythropoiesis
In mammals three different ras gene loci encode four highly homologous 21-KD proteins: H-ras, N-ras, K-ras.4A, and K-ras.4B (1). These proteins share an identical N-terminal effector domain to which all three major Ras effectors [Raf, p110 subunit of phosphatidylinositol 3-kinase (PI3K), and RalGEF] bind (2). Although each of these Ras proteins has the potential to activate all three major pathways (Raf/ERK, PI3K/Akt, and RalGEF/Ral), accumulating data suggest that Ras isoforms vary in their ability to activate different downstream signaling pathways in different cells. Yan et al. (3) reported that, when overexpressed in COS cells, K-ras is a more potent activator of the Raf/Erk pathway than H-ras, whereas H-ras is a more potent activator of the PI3K/Akt pathway than K-ras. Similarly, Hamilton et al. (4) found that Raf shows a higher affinity for N-ras than H-ras. The highly variable C terminus of Ras proteins might account for their different abilities to activate a particular pathway. The C-terminal region of H-ras regulates its interaction with Raf and PI3K (5), and mutations in this region of H-ras change effector pathway utilization (6). Moreover, the highly variable C terminus of Ras is critical for its subcellular localization, which in turn is important for differential pathway activation by Ras (7–9).
In contrast to these observations made in cell lines, several groups reported that when oncogenic K-ras is expressed from its endogenous locus in vivo, there is no elevation of the Raf/ERK pathway activation in unstimulated cells (10–13). These discrepancies might be explained by differences in the expression levels of K-ras, different cell types, and different extracellular stimuli.
Knockout mice deficient in H-ras, N-ras, K-ras.4A, or K-ras (both 4A and 4B) have been generated and potentially can be used to study activation of Ras downstream signaling pathways. K-ras.4A is dispensable for mouse growth and development, and H-ras and N-ras are dispensable both individually and in combination (14–16). In contrast, mice deficient in K-ras die at early embryonic stages [embryonic day (E) 12–E13 in a pure 129/Sv genetic background] with various defects (17, 18). At E12.5, K-ras-/- embryos are anemic; their fetal livers are pale and reduced in cellularity by 2- to 8-fold, although some erythroid cells in K-ras-/- embryos are able to reach the end stage of differentiation and become enucleated red blood cells (17). Defects in both hepatocytes and hematopoietic cells might contribute to the anemic phenotype (17, 19), but in neither hepatocytes nor erythroid progenitors have the potential defects resulting from the absence of K-ras signaling been identified.
Recently, we developed three experimental tools to study erythropoiesis in mouse fetal liver (20). First, we developed a flow cytometry assay that allows quantitative evaluation of erythroid differentiation in vivo and in vitro. Based on the expression of erythroid-specific TER119 and nonerythroid-specific CD71 (transferrin receptor) surface proteins, erythroid cells are divided into five populations (R1–R5), with R1 being the least and R5 being the most differentiated. Second, we developed a single-step procedure to purify large amounts of erythroid progenitors and early erythroblasts from mouse fetal livers with ≈75–85% purity. These cells express high levels of receptors for erythropoietin (Epo) and stem cell factor (SCF); thus, biochemical studies can be conducted by using a pure primary cell population after treatment with Epo or SCF. Third, we established an in vitro culture system that supports normal terminal proliferation and differentiation of erythroid progenitors. During the 2-day culture period the cell number increases 15- to 20-fold, and we monitor erythroid differentiation step-by-step and quantitatively using flow cytometric analysis of CD71 and TER119 double-stained cells. Taken together, these experimental tools provide a valuable and unique resource to study K-ras signaling in a defined primary cell type in response to defined extracellular stimuli.
Using these experimental tools, we studied K-ras signaling in primary erythroid progenitors. Here, we report that in fetal liver erythroid progenitors Epo activates all three Ras isoforms, but preferentially N- and K-ras. Epo and SCF are two cytokines essential for mouse definitive erythropoiesis (21–23). In K-ras-/- fetal liver cells (FLC), both Epo- and SCF-dependent Akt activation are greatly reduced, whereas other pathways including STAT5 and p44/42 MAPK are activated normally. To assess the effects of reduced Akt activation in erythropoiesis in vivo, we analyzed freshly isolated K-ras-/- FLC. K-ras deficiency leads to an ≈7-fold increase of apoptosis in FLC and delayed erythroid differentiation, but only in erythroid progenitors and early erythroblasts. When K-ras-/- erythroid progenitors are cultured in vitro, there is a significant delay in erythroid differentiation but little increase in apoptosis. In summary, our data identify K-ras as the major regulator for cytokine-dependent Akt activation in erythropoiesis in vivo.
Materials and Methods
Mice. Timed-pregnant BALB/c female mice were purchased from The Jackson Laboratory and used in the experiments described in Fig. 1 and Table 2. All of E12–E13 WT (K-ras+/+), heterozygous (K-ras+/-), and knockout (K-ras-/-) K-ras embryos were generated from intercrosses of K-ras+/- mice in a pure 129sv/Jae background (kindly provided by Tyler Jacks, Massachusetts Institute of Technology). Genomic DNA samples were isolated from adult tails or embryonic tissues by using the method of Laird et al. (25), and the genotypes of adult mice and embryos were determined by PCR as described in ref. 17.
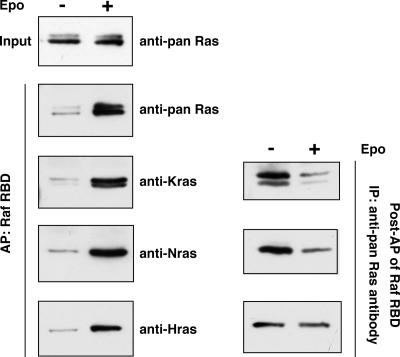
Fig. 1.
Epo-dependent activation of all three Ras isoforms. TER119- cells were purified from WT embryos, deprived of serum and growth factors for 1 h, and stimulated with 5–10 units/ml Epo for 10 min. Levels of Ras-GTP, the active form of Ras, were analyzed by affinity purification (AP) of lysates using a GST fusion with the RBD of Raf immobilized on agarose beads. The top panel in Left illustrates the total input levels of Ras proteins in unstimulated and stimulated cells. The remaining panels in Left show the Epo-dependent activation of different Ras isoforms. Right displays the remaining levels of different Ras isoforms in lysates from unstimulated and stimulated cells after affinity purification of Ras-GTP.
Table 2. Inhibition of Akt activation by a PI3K inhibitor delays erythroid differentiation in vitro.
| Concentration of LY294002, μM
|
Day 1
|
Day 2
|
||
|---|---|---|---|---|
| R1 + R2, % | R3 + R4 + R5, % | R1 + R2, % | R3 + R4 + R5, % | |
| Vehicle control | 23.8 | 68.8 | 6.1 | 85.4 |
| 25 | 38.3 | 51.6 | 10.6 | 82.4 |
| 50 | 59.3 | 24.5 | 29.6 | 58.2 |
| 100 | 74.3 | 2.6 | 73.7 | 9.2 |
TER119– cells were purified from WT Balb/c embryos and cultured in vitro for 2 days in the presence of varying concentrations of LY294002. The cells were stained with FITC-conjugated anti-mouse CD71 and PE-conjugated anti-mouse TER119 on day 1 and day 2. At least three independent experiments were performed, and the results from one representative experiment are presented here. The results are presented as percentages of the total live cells analyzed.
Cells and Chemical Inhibitor. FLC were isolated from E13.5–E15.5 BALB/c embryos or E12–E13 embryos generated from intercrosses of K-ras+/- mice. FLC were mechanically dissociated by pipetting in PBS containing 2% FBS. For FLC isolated from BALB/c embryos, single-cell suspensions were prepared by passing the dissociated cells twice through 70-μm cell strainers. TER119- cells were purified and cultured as previously described (20). For FLC isolated from individual embryos of K-ras+/- intercrosses, single cell suspensions were prepared by passing the dissociated cells through a 25-μm cell strainer. TER119- cells were purified from individual fetal liver by magnetic beads according to the manufacturer's protocol (Stem-Cell Technologies, Vancouver). Purified cells were then cultured for 2 days as described in ref. 20.
LY 294002 (Calbiochem) was dissolved in DMSO according to the manufacturer's instructions. The inhibitor was added to primary cell cultures on day 0 and replaced on day 1 when the culture medium was changed.
Immunostaining and Flow Cytometric Analysis. To obtain erythroid differentiation profiles, freshly isolated FLC and cultured cells were simultaneously stained for CD71 and TER119 as described in ref. 20. To perform apoptosis analysis, freshly isolated FLC and cultured cells were stained first for CD71 and TER119, followed by Annexin V and 7-AAD staining according to the manufacturer's instructions (BD Pharmingen). Flow cytometry was carried out on a FACSCalibur machine (BD Biosciences).
Immunoprecipitation and Western Blot Analysis. FLC isolated from individual embryos of K-ras+/- intercrosses were starved in Iscove's modified Dulbecco's medium containing 1% BSA or PBS containing 0.5% FBS for 2 h at 37°C. Cells were stimulated with 5–10 units/ml Epo or 25–50 ng/ml SCF for 10 min at 37°C. Stimulated cells were quickly recovered by centrifugation and resuspended in RIPA buffer [1× PBS/1% Igepal CA-630 (Sigma)/0.5% sodium deoxycholate/0.1% SDS] containing complete proteinase inhibitors (Roche), 1 mM sodium orthovana-date, 5 mM sodium fluoride, and 50 nM Calyculin A (Cell Signaling Technology, Beverly, MA). Protein concentration was determined by using the Bio-Rad Protein Assay according to the manufacturer's instructions. For each sample analyzed by gel electrophoresis, 30 μg of lysate protein was loaded. Quantification of Western results was performed by using either molecular analyst (Bio-Rad) or imagej (http://rsb.info.nih.gov/ij) software.
To detect the level of Ras-GTP, 5–10 × 106 TER119- cells were stimulated with Epo as described above. Affinity purification (AP) of Ras-GTP was performed with the Ras Activation Assay kit (Upstate Biotechnology, Lake Placid, NY), using the manufacturer's protocol with the following modifications: 5× MLS buffer was diluted in water, and a double amount of Raf Ras binding domain (RBD) was used. The Ras proteins remaining in solution were further immunoprecipitated by using agarose beads conjugated with anti-pan Ras antibody (Santa Cruz Biotechnology) according to the manufacturer's instructions.
All of the primary antibodies were from Cell Signaling Technology unless otherwise specified. Primary antibodies used for Western blotting were as follows: H-ras (BD Transduction Laboratories), N-ras (Santa Cruz Biotechnology), K-ras (Santa Cruz Biotechnology), pan Ras (Upstate Biotechnology), Phospho-Stat5 (Tyr-694), Stat5b (C-17; Santa Cruz Biotechnology), Phospho-Akt (Ser-473) (587F11), Akt, Phospho-p44/42 MAP kinase (Thr-202/Tyr-204), and p44/42 MAP kinase.
Results
Epo Activates All Three Ras Isoforms in Primary Erythroid Progenitors. All three Ras isoforms are expressed in primary fetal liver erythroid progenitors (Fig. 1). We measured the levels of the active forms of the three Ras isoforms after Epo stimulation. To this end, all Ras proteins that had bound GTP were first purified by affinity purification with the RBD of Raf. Then, different Ras isoforms were detected by isoform-specific antibodies. Our results showed that all three Ras isoforms are activated in response to Epo stimulation (Fig. 1 Left).
To estimate the extent of activation of each of these Ras proteins, we also measured in the same samples the levels of the three Ras isoforms that did not bind to the Raf RBD and are thus presumed to bind GDP (Fig. 1 Right). After Epo stimulation, only a small proportion of H-Ras protein in these cells becomes activated. In contrast, the majority of K- and N-ras proteins bound to the Raf RBD and thus were activated in response to Epo stimulation (see Discussion).
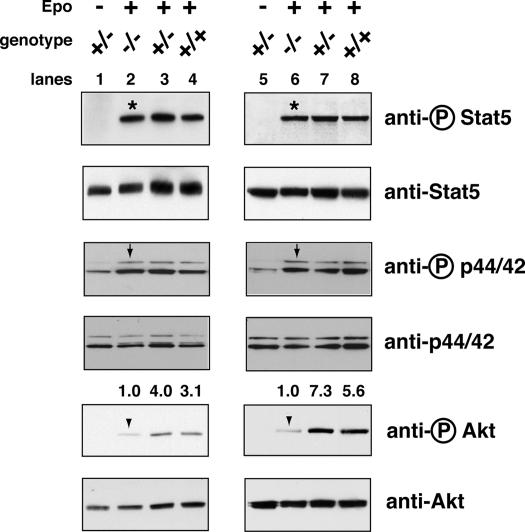
Cytokine-Dependent Akt Activation Is Dramatically Decreased in K-ras-/- Erythroid Progenitors. Because Epo preferentially activates N- and K-ras, we hypothesized that K-ras-/- FLC might show defective cytokine-dependent signaling. Epo and SCF are essential cytokines for definitive erythropoiesis. In primary erythroid progenitors Epo activates three major downstream signaling pathways: Stat5, Akt, and p44/42 ERK (J.Z. and H.F.L., unpublished data). We tested five K-ras-/- embryos isolated from three litters. In all cases, Epo-dependent activation of Stat5 and p44/42 ERK in K-ras-/- FLC was comparable to that in WT or K-ras+/- FLC (Fig. 2). In contrast, Epo-dependent activation of Akt was dramatically decreased in K-ras-/- FLC (3- to 8-fold reduction; Fig. 2).
Fig. 2.
Epo-dependent Akt activation is greatly reduced in K-ras-deficient FLC. Mouse FLC were isolated from individual E12.5 embryos, deprived of serum and growth factors for 2 h, and stimulated with 5–10 units/ml Epo for 10 min. Phosphorylated and total levels of Stat5, p44/42 ERK, and Akt proteins were measured by Western blotting (see Materials and Methods). Activated Akt levels in WT (+/+) and heterozygous (+/-) fetal livers relative to those in knockout (-/-) fetal livers were quantified, relative to the total Akt levels, with molecular analyst software. Results from two representative litters, corresponding to lanes 1–4 and 5–8, are shown here. Arrowheads indicate the phosphorylated Akt, arrows indicate the phosphorylated p44/42 ERK, and asterisks indicate the phosphorylated Stat5.
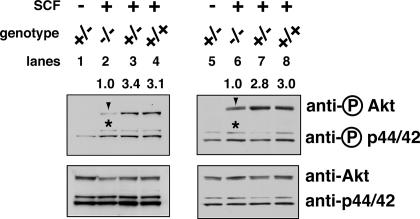
In primary erythroid progenitors, SCF activates two major signaling pathways: Akt and p44/42 ERK, but not Stat5. We tested six K-ras-/- embryos isolated from four litters. In all cases, SCF-dependent activation of p44/42 ERK in K-ras-/- FLC was comparable to that of WT or K-ras+/- cells (Fig. 3). In contrast, SCF-dependent activation of Akt was decreased in five of six K-ras-/- embryos tested (≈3-fold reduction; Fig. 3).
Fig. 3.
SCF-dependent Akt activation is greatly reduced in K-ras-deficient FLC. Mouse FLC were isolated and starved as described in Fig. 2, then stimulated with 25–50 ng/ml SCF for 10 min. Phosphorylated and total levels of Akt and p44/42 ERK proteins were measured and quantified as described in Fig. 2. Results from two representative litters, corresponding to lanes 1–4 and 5–8, are shown here. Arrowheads indicate phosphorylated Akt, and asterisks indicate the phosphorylated p44/42 ERK.
Loss of K-ras Leads to Increased Apoptosis and Delayed Erythroid Differentiation in Mouse Fetal Liver. To access the effects of K-ras deficiency in erythropoiesis in vivo, we analyzed FLC freshly isolated from E12.5 K-ras-deficient embryos and their WT littermates. As reported before (17), FLC from K-ras deficient embryos showed increased apoptosis; both TER119- and TER119+ populations displayed a 7-fold increase in apoptotic rate, as measured by Annexin V binding (data not shown).
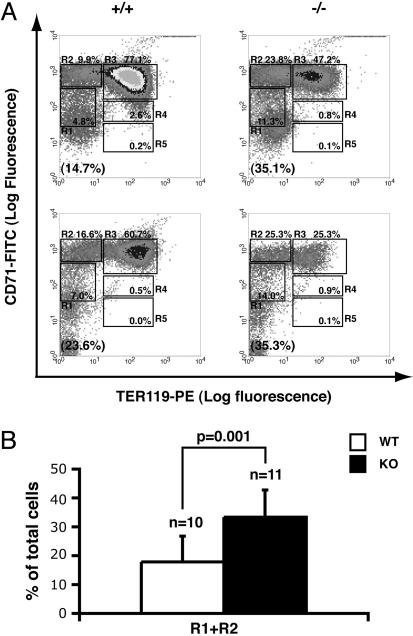
We then examined erythroid differentiation in E12.5 K-ras-deficient and WT littermate embryos using the flow cytometry analysis we developed to monitor definitive erythropoiesis in mouse fetal livers (20). On E12.5, the erythroid progenitor-enriched TER119- population (R1+R2) consisted of 17.8% of total FLC in WT embryos (Fig. 4B). In contrast, the percentage of TER119- cells in K-ras-/- fetal livers was significantly increased to 33.3% (P = 0.001; Fig. 1B). Because the percentage of TER119- cells decreases after erythroid differentiation in vivo, the increased percentage of TER119- cells indicated that erythroid differentiation is delayed in K-ras-deficient embryos. This finding confirms an earlier report that erythroid differentiation is delayed in K-ras-deficient FLC (26). Because some TER119+ cells were present and reticulocytes with normal morphologies were formed in K-ras-/- embryos (Fig. 4 and data not shown) (17), our data indicate that K-ras deficiency only delays but does not block erythroid development from TER119- erythroid progenitors.
Fig. 4.
Erythroid differentiation is delayed in K-ras-deficient fetal livers. (A) Mouse FLC were freshly isolated from E12.5 embryos and double-labeled with FITC-conjugated anti-CD71 mAb and a PE-conjugated anti-TER119 mAb. Dead cells (propidium iodide positive) and debris (low forward scatter) were excluded from the analysis. (Left) Flow cytometry density plots of all viable FLC from two representative WT (+/+) embryos in two different litters. (Right) Density plots of FLC from two K-ras-/- embryos (-/-) in the same litters. Regions R1–R5 are defined as indicated. The fraction of cells in each region, R1–R5, is indicated as a percentage of all viable cells and shown on each plot. The percentage of TER119- cells (R1+R2) is labeled in the brackets at the bottom of each density plot. (B) The percentages of R1+R2 cells in 10 WT (open bar) and 11 K-ras knockout (KO, filled bar) embryos were quantified. The Student t test was performed to evaluate the statistical significance. The data presented here are the averages + SD.
K-ras Deficiency in Erythroid Progenitors per Se Causes Delayed Erythroid Differentiation but Not Increased Apoptosis. Two possible mechanisms might account for the increased apoptosis and delayed erythroid differentiation in K-ras-/- embryos. First, K-ras could act in a cell-autonomous fashion in erythroid progenitors, and loss of K-ras function in these cells per se would cause the increase in apoptosis and delay in differentiation. Alternatively K-ras could act in a cell-nonautonomous fashion on erythroid progenitors in that loss of K-ras function in stromal or other nonerythroid cells leads both to increased apoptosis and delayed differentiation of erythroid progenitors. Indeed, defects in both hematopoietic cells and fetal liver hepatocytes were reported previously in K-ras-deficient embryos (17, 19).
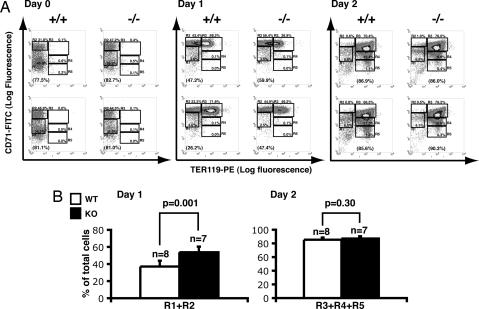
To distinguish between these two possibilities, we purified TER119- cells from individual fetal livers and cultured them in vitro (Fig. 5A). As expected, on day 0 >99% of these purified cells were TER119-, and ≈80% of the purified cells were R1+R2 cells, both from WT and K-ras-/- embryos (Fig. 5A). Strikingly, during culture K-ras-/- erythroid progenitors showed comparable low apoptosis rates as those from WT littermates (Table 1). The dramatic increase in apoptosis we observed in freshly isolated cells from K-ras-/- fetal livers likely represents a requirement for K-ras expression in nonerythroid cells.
Fig. 5.
K-ras-deficient erythroid progenitors show delayed differentiation in culture. (A) TER119- cells were purified from individual E12.5 fetal livers (Day 0). Purified cells were cultured in vitro for 2 days as described in ref. 20. After 1 day (Day 1) or 2 days (Day 2) in culture, erythroid cells were harvested, and the differentiation profiles were analyzed by the flow cytometry analysis described in Fig. 4. The percentage of TER119- cells (R1+R2) is labeled in the brackets at the bottom of each density plot on the Day 0 and Day 1 sets. The percentages of TER119+ cells (R3+R4+R5) are labeled in the brackets at the bottom of each density plot on the Day 2 panels. (B) The percentages of R1+R2 cells from the day 1 culture and that of the R3+R4+R5 cells from the day 2 culture of cells from multiple embryos were quantified. The results of the statistical tests are presented as described in Fig. 4.
Table 1. Cultured K-ras-deficient FLC do not show enhanced apoptosis rate.
| Genotype | Annexin V+;7-AAD–, % in R1 + R2 population | Annexin V+;7-AAD–, % in R3 + R4 population |
|---|---|---|
| K-ras+/+ (n = 2) | 0.37 ± 0.11 | 0.25 ± 0.04 |
| K-ras+/– (n = 6) | 0.69 ± 0.01 | 0.26 ± 0.00 |
| K-ras–/– (n = 3) | 0.60 ± 0.01 | 0.33 ± 0.00 |
TER119– cells were purified from individual embryos and cultured in vitro for 1 day. The cells were stained with FITC-conjugated anti-mouse CD71 and PE-conjugated anti-mouse TER119, followed by incubation with APC-conjugated Annexin V and 7-AAD. The Annexin V+;7-AAD– population (apoptotic cells) were presented as a percentage of the total TER119– (R1 + R2) or TER119+ (R3 + R4) cells analyzed.
However, we did observe a delay in erythroid differentiation of cultured K-ras-/- erythroid progenitors, similar to that seen in freshly isolated K-ras-/- FLC. After one day in culture, ≈58% of erythroid progenitors isolated from WT embryos had up-regulated CD71 expression, induced TER119, and differentiated into R3 cells; ≈37% cells remained TER119-negative (R1+R2) (Fig. 5). In contrast, only ≈42% of K-ras-/- erythroid progenitors differentiated into R3 cells, and ≈54% cells remained in the R1 and R2 fractions. However, after 2 days in culture, ≈87% of K-ras-/- erythroid progenitors had differentiated into TER119+ cells (R3+R4+R5), a fraction comparable to those from WT littermate embryos (≈85%; Fig. 5). Moreover, reticulocytes formed from K-ras-/- erythroid progenitors appeared indistinguishable from those formed from WT progenitors (data not shown). Moreover, partial inhibition of the PI3K/Akt pathway in WT erythroid progenitors by a dominant negative p85 or by addition of LY 294002 leads to a delay in erythroid differentiation similar to that observed in K-ras-/- FLC (Table 2 and data not shown). Taken together, our data indicate that the loss of K-ras function in erythroid progenitors per se causes delayed erythroid differentiation of erythroid progenitors and early erythroblasts, and this phenotype is likely caused by reduced cytokine-dependent Akt activation.
Discussion
Here, we provide several insights into K-ras signaling as well as its role in cytokine signaling. First, and in contrast to previous work that described activation of Akt directly through phosphorylated tyrosine residues on cytokine receptors that activate PI3K, our data identify K-ras as the major regulator for cytokine-dependent Akt activation in vivo. This primary role of K-ras in cytokine signaling has not been documented previously. Second, despite the presence of all three Ras isoforms in primary erythroid progenitors, K-ras deficiency leads to a great reduction of Akt activation, suggesting that Akt is preferentially activated by K-ras in primary erythroid progenitors in vivo. Third, the defect in K-ras signaling leads to delayed differentiation of primary erythroid progenitors.
The Role of K-ras in Cytokine Signaling. Our identification of K-ras as the major activator of Akt in erythroid progenitors reveals an unexpected role of K-ras in cytokine signaling in vivo. Previously, we and other groups identified K-ras-independent mechanisms by which Epo addition leads to Akt activation (reviewed in ref. 27). For example, (phospho)Y479 of the Epo receptor (EpoR) is essential for binding to the p85 subunit of PI3K and activating the PI3K/Akt pathway (28–30). In addition, EpoR-associated IRS-2 and Src tyrosine kinase provide an alternative mechanism for activation of PI3K in response to Epo (31, 32). However, these results were obtained by using hematopoietic cell lines overexpressing various EpoR mutants. Such studies cannot provide an accurate metric of the contribution of these pathways to Epo-dependent Akt activation in vivo. In contrast, the present results were obtained by using primary cells expressing EpoR at its physiological level, indicating that K-ras is the major regulator of Epo-dependent Akt activation in vivo.
Ras proteins can be activated by Epo through two mechanisms. First, Grb2, possibly in concert with Shc, binds to the canonical Grb2 binding site surrounding (phospho)Y464 of the EpoR and couples to mSos (33–35). This leads to Ras activation. Second, Ras proteins are activated through the phosphorylated Shc/Grb2/mSos pathway (36). In either case, activated Ras in turn activates the downstream Akt pathway. Because Epo might activate Akt through H- and N-ras as well as other Ras-independent mechanisms, it is not surprising that in K-ras-/- erythroid progenitors some Epo-dependent Akt activation, although greatly reduced, still occurs.
We identified K-ras as an important regulator for Akt activation in response not only to Epo but also to SCF. The receptors for Epo and SCF belong to different cytokine receptor groups. EpoR, a class I cytokine receptor, does not contain a tyrosine kinase activity in its cytoplasmic domain. Activation of all known signaling pathways downstream of the EpoR requires activation of the tightly associated protein tyrosine kinase Jak2 (27). In contrast, c-Kit (SCF receptor) is a class III transmembrane tyrosine kinase receptor (37). Activation of its downstream signaling pathways depends on transphosphorylation of the kinase that forms an integral part of the cytosolic domain of these receptors. Similar to EpoR, phosphorylated c-Kit activates Akt through direct binding and activation of PI3K (37). However, we found that K-ras is the major activator of Akt in response to SCF in vivo. It is likely that K-ras is also important for Akt and presumably PI3 kinase activation downstream of other class I and III protein tyrosine kinase receptors.
Activation of Akt Through K-ras. Activation of Akt involves several steps. Akt contains a pleckstrin homology (PH) domain that tightly binds phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] and PI 3,4-bisphosphate [PI(3,4)P2], the products of PI3K (38). Maximum activation of Akt requires, besides binding to PI(3,4,5)P3 and PI(3,4)P2 via the PH domain, activation loop phosphorylation at Thr-308 by PDK1 and also phosphorylation within the C terminus at Ser-473 by PDK2 (39–41). PDK1 is a Ser/Thr kinase ubiquitously expressed in human tissues (41, 42). It consists of an N-terminal kinase domain and a C-terminal PH domain. Activation of PDK1 requires binding to PI(3,4,5)P3 or PI(3,4)P2. PDK2 was recently identified as the Rictor-mTOR complex (43), and its kinase activity also depends to some extent on PI3K activation.
Given this incremental and complex mechanism of Akt activation, the exact mechanism by which K-ras activates Akt is not fully understood in any system. Because the p110 subunit of PI3K directly binds to a Ras effector domain that is identical in all Ras proteins, one possible scenario is that K-ras preferentially binds to p110 and thus stimulates Akt activation mainly through activating PI3K activity. Alternatively, K-ras could induce Akt activation mainly through regulating the activity of PDK2, the Rictor-mTOR complex.
Although normal K-ras is necessary for maximum Akt activation in erythroid progenitors, it remains unclear whether oncogenic (constitutively active) K-ras is sufficient to elevate Akt activation in the absence of cytokine stimulation. It was reported that, when overexpressed in cell lines, K- and N-ras activate the Raf/ERK pathway more potently than do H-ras (3, 4). Thus, it is interesting and important to understand which pathway(s) is abnormally activated by oncogenic K-ras when it is expressed in erythroid progenitors at its physiological level, as opposed to its overexpression.
Reduced Cytokine-Dependent Akt Activation Leads to Delayed Erythroid Differentiation in K-ras-/- Erythroid Progenitors. Consistent with a previous report (26), we found that K-ras-/- erythroid progenitors show delayed erythroid differentiation (Fig. 4). Furthermore, we showed that this phenotype is caused by the intrinsic loss of K-ras in erythroid progenitors and not in the associated stromal cell population (Fig. 5). Partial inhibition of the PI3K/Akt pathway in WT erythroid progenitors by a dominant negative p85 or by addition of LY 294002 leads to a delay in erythroid differentiation similar to that observed in K-ras-/- FLC (Table 2 and data not shown). These results strongly suggest that the delayed erythroid differentiation in K-ras-/- erythroid progenitors resulted from reduced cytokine-dependent Akt activation.
In summary, using primary fetal liver erythroid progenitors, we showed that in K-ras-deficient erythroid progenitors cytokine-dependent Akt activation but not other signaling pathways is greatly reduced. This signaling defect resulted in delayed erythroid differentiation in a cell-autonomous fashion. Our data thus identified K-ras as the major regulator of cytokine-dependent Akt activation in vivo. This unexpected finding greatly advances our understanding of both cytokine signaling and Ras downstream signal transduction.
Acknowledgments
We thank Dr. Tyler Jacks for generously providing K-ras heterozygous mice; Drs. Qiang Chang and Wei Tong for helpful discussion and critical comments on the manuscript; Yangang Liu for excellent technical help; and RoxAnne Shaffer, Stacey Sullivan, and Tony Chavarria for help with mice. This work was supported by National Institutes of Health Grant P01 HL 32262 (to H.F.L.) and a Fellow Grant from the Leukemia and Lymphoma Society (to J.Z.).
Abbreviations: En, embryonic day n; Epo, erythropoietin; EpoR, Epo receptor; FLC, fetal liver cells; PI3K, phosphatidylinositol 3-kinase; RBD, Ras binding domain; SCF, stem cell factor.
References
- 1.Barbacid, M. (1987) Annu. Rev. Biochem. 56, 779-827. [DOI] [PubMed] [Google Scholar]
- 2.Polakis, P. & McCormick, F. (1993) J. Biol. Chem. 268, 9157-9160. [PubMed] [Google Scholar]
- 3.Yan, J., Roy, S., Apolloni, A., Lane, A. & Hancock, J. F. (1998) J. Biol. Chem. 273, 24052-24056. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton, M. & Wolfman, A. (1998) Oncogene 16, 1417-1428. [DOI] [PubMed] [Google Scholar]
- 5.Jaumot, M., Yan, J., Clyde-Smith, J., Sluimer, J. & Hancock, J. F. (2002) J. Biol. Chem. 277, 272-278. [DOI] [PubMed] [Google Scholar]
- 6.Booden, M. A., Sakaguchi, D. S. & Buss, J. E. (2000) J. Biol. Chem. 275, 23559-23568. [DOI] [PubMed] [Google Scholar]
- 7.Choy, E., Chiu, V. K., Silletti, J., Feoktistov, M., Morimoto, T., Michaelson, D., Ivanov, I. E. & Philips, M. R. (1999) Cell 98, 69-80. [DOI] [PubMed] [Google Scholar]
- 8.Apolloni, A., Prior, I. A., Lindsay, M., Parton, R. G. & Hancock, J. F. (2000) Mol. Cell. Biol. 20, 2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, V. K., Bivona, T., Hach, A., Sajous, J. B., Silletti, J., Wiener, H., Johnson, R. L., II, Cox, A. D. & Philips, M. R. (2002) Nat. Cell Biol. 4, 343-350. [DOI] [PubMed] [Google Scholar]
- 10.Tuveson, D. A., Shaw, A. T., Willis, N. A., Silver, D. P., Jackson, E. L., Chang, S., Mercer, K. L., Grochow, R., Hock, H., Crowley, D., et al. (2004) Cancer Cell 5, 375-387. [DOI] [PubMed] [Google Scholar]
- 11.Guerra, C., Mijimolle, N., Dhawahir, A., Dubus, P., Barradas, M., Serrano, M., Campuzano, V. & Barbacid, M. (2003) Cancer Cell 4, 111-120. [DOI] [PubMed] [Google Scholar]
- 12.Chan, I. T., Kutok, J. L., Williams, I. R., Cohen, S., Kelly, L., Shigematsu, H., Johnson, L., Akashi, K., Tuveson, D. A., Jacks, T. & Gilliland, D. G. (2004) J. Clin. Invest. 113, 528-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun, B. S., Tuveson, D. A., Kong, N., Le, D. T., Kogan, S. C., Rozmus, J., Le Beau, M. M., Jacks, T. E. & Shannon, K. M. (2004) Proc. Natl. Acad. Sci. USA 101, 597-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plowman, S. J., Williamson, D. J., O'Sullivan, M. J., Doig, J., Ritchie, A. M., Harrison, D. J., Melton, D. W., Arends, M. J., Hooper, M. L. & Patek, C. E. (2003) Mol. Cell. Biol. 23, 9245-9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteban, L. M., Vicario-Abejon, C., Fernandez-Salguero, P., Fernandez-Medarde, A., Swaminathan, N., Yienger, K., Lopez, E., Malumbres, M., McKay, R., Ward, J. M., et al. (2001) Mol. Cell. Biol. 21, 1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umanoff, H., Edelmann, W., Pellicer, A. & Kucherlapati, R. (1995) Proc. Natl. Acad. Sci. USA 92, 1709-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, L., Greenbaum, D., Cichowski, K., Mercer, K., Murphy, E., Schmitt, E., Bronson, R. T., Umanoff, H., Edelmann, W., Kucherlapati, R. & Jacks, T. (1997) Genes Dev. 11, 2468-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koera, K., Nakamura, K., Nakao, K., Miyoshi, J., Toyoshima, K., Hatta, T., Otani, H., Aiba, A. & Katsuki, M. (1997) Oncogene 15, 1151-1159. [DOI] [PubMed] [Google Scholar]
- 19.Matsui, T., Kinoshita, T., Morikawa, Y., Tohya, K., Katsuki, M., Ito, Y., Kamiya, A. & Miyajima, A. (2002) EMBO J. 21, 1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, J., Socolovsky, M., Gross, A. W. & Lodish, H. F. (2003) Blood 102, 3938-3946. [DOI] [PubMed] [Google Scholar]
- 21.Wu, H., Liu, X., Jaenisch, R. & Lodish, H. F. (1995) Cell 83, 59-67. [DOI] [PubMed] [Google Scholar]
- 22.Qiu, F. H., Ray, P., Brown, K., Barker, P. E., Jhanwar, S., Ruddle, F. H. & Besmer, P. (1988) EMBO J. 7, 1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nocka, K., Majumder, S., Chabot, B., Ray, P., Cervone, M., Bernstein, A. & Besmer, P. (1989) Genes Dev. 3, 816-826. [DOI] [PubMed] [Google Scholar]
- 24.Kim, S. C., Hahn, J. S., Min, Y. H., Yoo, N. C., Ko, Y. W. & Lee, W. J. (1999) Blood 93, 3893-3899. [PubMed] [Google Scholar]
- 25.Laird, P. W., Zijderveld, A., Linders, K., Rudnicki, M. A., Jaenisch, R. & Berns, A. (1991) Nucleic Acids Res. 19, 4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalaf, W. F., White, H., Wenning, M. J., Orazi, A., Kapur, R. & Ingram, D. A. (2005) Blood 105, 3538-3541. [DOI] [PubMed] [Google Scholar]
- 27.Ghaffari, S., Huang, L., Zhang, J. & Lodish, H. (2003) in Erythropoietins and Erythropoiesis: Molecular, Cellular, Preclinical, and Clinical Biology, eds. Molineux, G., Foote, M. & Elliott, S. (Birkhauser, Basel), pp. 65-85.
- 28.Damen, J. E., Mui, A. L., Puil, L., Pawson, T. & Krystal, G. (1993) Blood 81, 3204-3210. [PubMed] [Google Scholar]
- 29.Damen, J. E., Cutler, R. L., Jiao, H., Yi, T. & Krystal, G. (1995) J. Biol. Chem. 270, 23402-23408. [DOI] [PubMed] [Google Scholar]
- 30.Klingmuller, U., Wu, H., Hsiao, J. G., Toker, A., Duckworth, B. C., Cantley, L. C. & Lodish, H. F. (1997) Proc. Natl. Acad. Sci. USA 94, 3016-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdier, F., Chretien, S., Billat, C., Gisselbrecht, S., Lacombe, C. & Mayeux, P. (1997) J. Biol. Chem. 272, 26173-26178. [DOI] [PubMed] [Google Scholar]
- 32.Kubota, Y., Tanaka, T., Kitanaka, A., Ohnishi, H., Okutani, Y., Waki, M., Ishida, T. & Kamano, H. (2001) EMBO J. 20, 5666-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravichandran, K. S., Lorenz, U., Shoelson, S. E. & Burakoff, S. J. (1995) Mol. Cell. Biol. 15, 593-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gram, H., Schmitz, R., Zuber, J. F. & Baumann, G. (1997) Eur. J. Biochem. 246, 633-637. [DOI] [PubMed] [Google Scholar]
- 35.Wu, H., Klingmuller, U., Acurio, A., Hsiao, J. G. & Lodish, H. F. (1997) Proc. Natl. Acad. Sci. USA 94, 1806-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miura, Y., Miura, O., Ihle, J. N. & Aoki, N. (1994) J. Biol. Chem. 269, 29962-29969. [PubMed] [Google Scholar]
- 37.Taylor, M. L. & Metcalfe, D. D. (2000) Hematol. Oncol. Clin. North Am. 14, 517-535. [DOI] [PubMed] [Google Scholar]
- 38.Walker, E. H., Pacold, M. E., Perisic, O., Stephens, L., Hawkins, P. T., Wymann, M. P. & Williams, R. L. (2000) Mol. Cell 6, 909-919. [DOI] [PubMed] [Google Scholar]
- 39.Alessi, D. R., Andjelkovic, M., Caudwell, B., Cron, P., Morrice, N., Cohen, P. & Hemmings, B. A. (1996) EMBO J. 15, 6541-6551. [PMC free article] [PubMed] [Google Scholar]
- 40.Stokoe, D., Stephens, L. R., Copeland, T., Gaffney, P. R., Reese, C. B., Painter, G. F., Holmes, A. B., McCormick, F. & Hawkins, P. T. (1997) Science 277, 567-570. [DOI] [PubMed] [Google Scholar]
- 41.Stephens, L., Anderson, K., Stokoe, D., Erdjument-Bromage, H., Painter, G. F., Holmes, A. B., Gaffney, P. R., Reese, C. B., McCormick, F., Tempst, P., et al. (1998) Science 279, 710-714. [DOI] [PubMed] [Google Scholar]
- 42.Alessi, D. R., James, S. R., Downes, C. P., Holmes, A. B., Gaffney, P. R., Reese, C. B. & Cohen, P. (1997) Curr. Biol. 7, 261-269. [DOI] [PubMed] [Google Scholar]
- 43.Sarbassov, D. D., Guertin, D. A., Ali, S. M. & Sabatini, D. M. (2005) Science 307, 1098-1101. [DOI] [PubMed] [Google Scholar]