Abstract
The K3 protein of a human tumor-inducing herpesvirus, Kaposi’s sarcoma-associated herpesvirus (KSHV), down-regulates major histocompatibility complex (MHC) class I surface expression by increasing the rate of endocytosis. In this report, we demonstrate that the internalization of MHC class I by the K3 protein is the result of multiple, consecutive trafficking pathways that accelerate the endocytosis of class I molecules, redirect them to the trans-Golgi network (TGN), and target MHC class I to the lysosomal compartment. Remarkably, these actions of K3 are functionally and genetically separable; the N-terminal zinc finger motif and the central sorting motif are involved in triggering internalization of MHC class I molecules and redirecting them to the TGN. Subsequently, the C-terminal diacidic cluster region of K3 is engaged in targeting MHC class I molecules to the lysosomal compartment. These results demonstrate a novel trafficking mechanism of MHC class I molecules induced by KSHV K3, which ensures viral escape from host immune effector recognition.
Keywords: endocytosis/intercellular trafficking/ KSHV K3/MHC class I/viral immune evasion
Introduction
Internalization and transport of membrane-associated receptors to specific compartments of the endosomal– lysosomal system are largely dependent upon the sorting signals contained within their cytosolic domains. The most extensively characterized motif contains a critical tyrosine residue within the canonical sequence, YXXφ (where Y is a tyrosine, X is any amino acid and φ is a hydrophobic amino acid). This YXXφ tyrosine-based sorting signal has been shown to bind directly with the medium subunits µ1, µ2 and µ3 of the adaptor protein (AP) complexes AP-1, AP-2 and AP-3, respectively (Fuhrer et al., 1994; Boll et al., 1996; Ohno et al., 1998; Owen and Evans, 1998; Piguet et al., 1998). Interactions between the YXXφ motif and APs result in selective incorporation of the motif-containing proteins into coated vesicles that carry proteins to different destinations within the cell. Another less well-characterized endocytosis motif is the dileucine-based sorting signal (Aiken et al., 1994; Honing et al., 1998). While not as much is known about these signals, acidic cluster dileucine signals have recently been shown to direct sorting from the trans-Golgi network (TGN) to the lysosomes by interacting with a new set of cellular cargo proteins named the Golgi-localized, γ-ear-containing, ARF-binding proteins (GGAs) (Nielsen et al., 2001; Puertollano et al., 2001; Takatsu et al., 2001).
A major immune defense against viral infection is mediated by cytotoxic T lymphocytes (CTLs), which recognize and lyse infected cells upon engagement of the T cell receptor with major histocompatibility complex (MHC) class I molecules presenting viral peptides. Many diverse mechanisms of viral interference with cellular immunity are becoming increasingly apparent (Ploegh, 1998; Alcami and Koszinowski, 2000). Human and simian immunodeficiency virus (HIV and SIV, respectively) nef proteins have been shown to down-regulate MHC class I molecules by accelerating their endocytosis (Aiken et al., 1994; Le Gall et al., 1998). In the presence of HIV-1 nef, MHC class I molecules are internalized from the cell surface by endocytosis and relocalized to the TGN through interactions between nef and the cellular phosphofurin acidic cluster sorting protein-1 (PACS-1) (Piguet et al., 2000).
A human tumor-inducing virus, called Kaposi’s sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8, has been consistently identified in Kaposi’s sarcoma tumors, primary effusion lymphoma and an immunoblast variant of Castleman’s disease (Chang et al., 1994; Cesarman et al., 1995; Renne et al., 1996). KSHV has been shown to contain a battery of genes whose products down-regulate host immune responses at various levels (Schulz et al., 1998; Choi et al., 2001). Two proteins, KSHV K3 and K5, dramatically down-regulate MHC class I molecules (Coscoy and Ganem, 2000; Ishido et al., 2000b; Haque et al., 2001). While K5 additionally down-regulates ICAM and B7-2 from the cell surface, expression of K3 specifically decreases MHC class I surface expression (Ishido et al., 2000a; Coscoy and Ganem, 2001). Biochemical analyses have demon strated that similar to HIV nef, expression of either K3 or K5 decreases cell surface MHC class I levels through an increased rate of class I molecule endocytosis. Interestingly, the proteins exhibit 40% amino acid identity to each other (Russo et al., 1996; Nicholas et al., 1997) and contain C4HC3 zinc finger motifs at the N-terminus with hydrophobic transmembrane regions in the central region, but are of varying size in the C-terminal tail. However, despite this similarity, K3 and K5 differ in their specificity. K3 drastically down-regulates HLA-A, -B, -C and -E, whereas K5 is active in down-regulating, primarily, HLA-A and -B (Ishido et al., 2000b).
In this report, we demonstrate that the putative zinc finger motif in the N-terminal region, the YAAV sorting motif in the central region and diacidic clusters at the C-terminal region of K3 are required for efficient down-regulation of MHC class I surface expression. Surprisingly, diacidic cluster mutants of K3, defective for MHC class I down-regulation, maintain a significant ability to induce internalization of MHC class I molecules. This puzzling observation led to the discovery that the down-regulation of MHC class I molecules by the K3 protein is the result of multiple consecutive trafficking steps. After triggering internalization, K3 targets MHC class I molecules to the TGN. Subsequently, the diacidic clusters of K3 are involved in directing MHC class I molecules to the lysosomal compartment. Thus, KSHV K3 and HIV nef down-regulate MHC class I molecules in similar but distinct ways to escape host immune effector recognition.
Results
Effect of deletion mutations in the C-terminal region of K3 on MHC class I down-regulation
Both K3 and K5 open reading frames contain two C4HC3 zinc finger motifs at the N-terminus in addition to hydrophobic transmembrane domains in the central region, but are of varying size in the C-terminal tail, with extensive sequence variation. Despite this variation, several motifs for endocytosis and signal transduction are conserved in both the K3 and K5 C-terminal regions. These include sequences containing potential tyrosine-based endocytosis motifs (Trowbridge et al., 1993), Y152AAV for K3 and Y156AAN for K5, and PX2–3P potential SH3-binding motifs (Figure 1). In addition, both K3 and K5 contain conserved diacidic cluster regions, which resemble HIV nef (Piguet et al., 1999; Figure 1).
Fig. 1. Conserved motifs between K3 and K5. KSHV K3 and K5 amino acid sequences were aligned based on a computer prediction of their structure. The conserved motifs between K3 and K5 are indicated with colored boxes in the center panel. Tm (transmembrane region), blue boxes; Yxxφ (the potential tyrosine-based sorting motif), red box; NTRV (conserved residues), green box; DE1 (acidic cluster 1) and DE2 (acidic cluster 2), yellow boxes; SH3B designates the proline-rich potential SH3 binding motif. A magnification of the zinc finger-containing sequences is shown at the top, while a magnification of the K3 and K5 central regions is shown at the bottom.
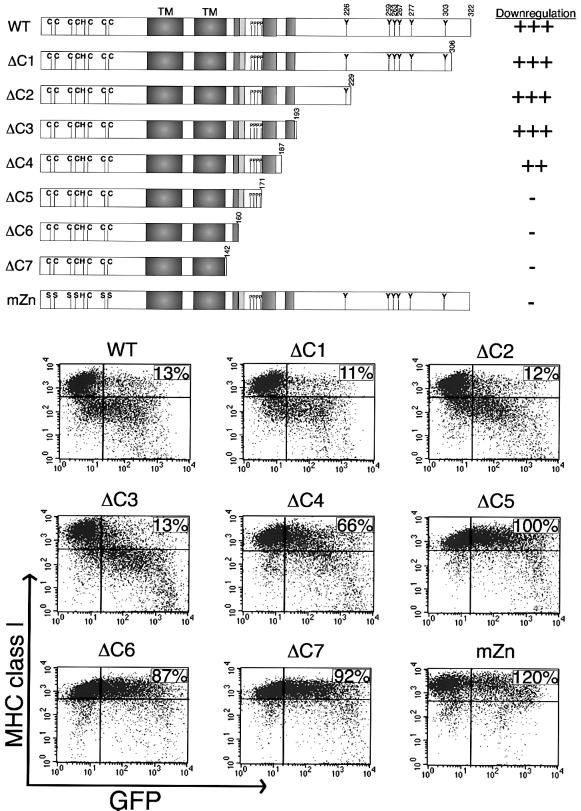
Since MHC class I down-regulation by K3 is more pronounced than that by K5 (Ishido et al., 2000b), K3 was selected for detailed mutational analysis. Besides the conserved Y152 residue in the C-terminal region, K3 also contains Y226, Y259, Y263, Y267, Y277 and Y303 residues that may function as potential tyrosine-based endocytosis motifs (Figure 1). To determine whether these tyrosine-containing sequences were required for MHC class I down-regulation, a series of deletion mutations were introduced into the C-terminal region of K3 (Figure 2). These were ΔC1, ΔC2 and ΔC3 (Figure 2). Additional deletion mutations, ΔC4, ΔC5 and ΔC6, were targeted to determine the roles of the diacidic clusters, PX2–3P and conserved NTRV residues in MHC class I down-regulation (Figure 2). The ΔC7 mutation further removed the YAAV motif located C-terminal of the transmembrane spanning regions.
Fig. 2. Point mutations at N-terminal zinc finger motifs and deletion mutations at the C-terminal region of K3. A schematic of the location of various mutations of the K3 protein is shown at the top. Deletion mutations at the C-terminal region of K3 were generated as follows: ΔC1, a deletion of residues 307–322; ΔC2, a deletion of residues 230–322; ΔC3, a deletion of residues 194–322; ΔC4, a deletion of residues 188–322; ΔC5, a deletion of residues 172–322; ΔC6, a deletion of residues 161–322; ΔC7, a deletion of residues 143–322. The cysteine residues at 9, 12, 24, 26, 50 and 53 in the putative zinc finger motifs at the N-terminal region of K3 were replaced with serine residues to generate the mZn mutant. Along the right side, the ability of wild-type K3 and each mutant to down-regulate MHC class I, as shown underneath the schematic, was given a relative score. ‘+++ and –’ indicate strong and no activity of the K3 mutant in MHC class I down-regulation, respectively. Below the schematic, differential MHC class I down-regulation activity of K3 mutants is shown. To examine the ability of K3 and its mutants to down-regulate MHC class I, BJAB cells were electroporated with pTracer–GFP–K3 (WT), GFP–K3 mZn (mZn) or one of the deletion mutants. Cell surface levels of MHC class I were assessed 48 h post-transfection by staining for MHC class I with its specific antibody (y axis) and gating the GFP-positive cell population (x axis) by flow cytometry. Numbers in the upper right side quadrant represent the percentage mean channel florescence (MCF) of MHC class I staining in the GFP-positive, K3-transfected cells as compared with that in the GFP-positive, vector-transfected cells. The data were reproduced in at least two independent experiments.
BJAB cells were electroporated with pTracer vector alone or vector expressing mutant or wild-type (wt) K3. The effects of expression of K3 and its mutants on the cell surface levels of MHC class I were assessed 48 h post-transfection in the green fluorescent protein (GFP)-positive cell population by flow cytometry. As shown previously, wt K3 expression dramatically down-regulated MHC class I surface expression (Figure 2). Three constructs, the ΔC1, ΔC2 and ΔC3 mutants, were capable of down-regulating MHC class I molecules at levels equivalent to wt K3 (Figure 2). This indicated that sequences containing the Y226, Y259, Y263, Y267, Y277 and Y303 residues were not required for MHC class I down-regulation (Figure 2). In contrast, each of the other deletion mutations dramatically abrogated the ability of K3 to down-regulate MHC class I (Figure 2). All K3 mutants were expressed at equivalent levels to wt K3 in these cells (see Supplementary figure 1 at The EMBO Journal Online). These results indicate that the region containing the putative tyrosine-based sorting YAAV motif, the PX2–3P SH3 binding motif and diacidic clusters of K3 are required for MHC class I down-regulation.
Mutations in sequence-specific motifs of K3 for MHC class I down-regulation
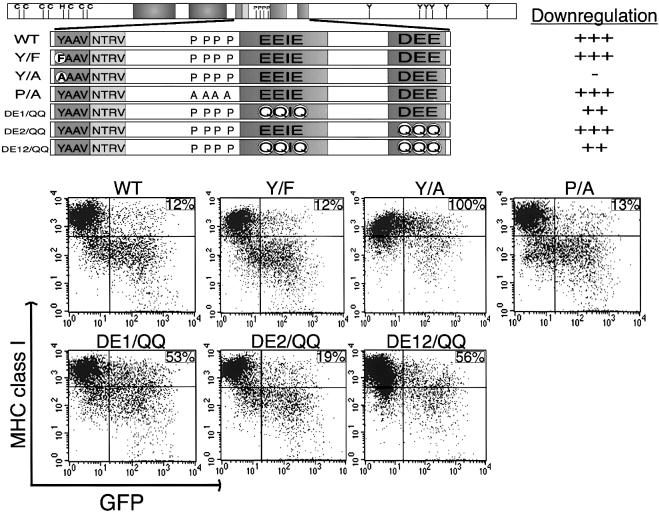
Subsequent point mutational analysis was targeted at the putative zinc finger motifs at the N-terminal region, YAAV and PX2–3P motifs in the central region, and diacidic clusters at the C-terminus of K3. The cysteine residues at positions 9, 12, 24, 26, 50 and 53 in the putative zinc finger motifs were replaced with serine residues to generate the mZn mutant (Figure 2). The tyrosine residue Y152 in the putative tyrosine-based endocytosis motif was replaced with phenylalanine (F) or alanine (A) to generate, respectively, the Y152/F and Y152/A mutants (Figure 3). Additionally, the proline residues P163, P166, P168 and P171 in the putative SH3-binding motif were replaced with alanine to generate the P/A mutant (Figure 3). To define a role for the diacidic clusters of K3 in MHC class I down-regulation, several mutants were produced: residues E176, E177 and E179 in the first acidic cluster were replaced with glutamine (Q) residues to generate the DE1/QQ mutant; and residues D191, E192 and E193 in the second acidic cluster were replaced with glutamine residues to generate the DE2/QQ mutant. These two mutants were combined to generate the DE12/QQ mutant (Figure 3).
Fig. 3. Mutations in sequence-specific motifs of K3 and MHC class I down-regulation. The schematic at the top shows the location of various mutations of the K3 protein. Six mutations, Y152/F (Y/F), Y152/A (Y/A), P/A, DE1/QQ, DE2/QQ and DE12/QQ were introduced into K3. The location of these mutants is described in greater detail in the text. Along the right side of the schematic, the ability of wild-type K3 and each mutant to down-regulate MHC class I, as shown in the below the schematic, was given a relative score. ‘+++, ++, + and –’ indicate strong, weak, very weak and no activity of the K3 mutant in MHC class I down-regulation, respectively. This shows differential MHC class I down-regulation activity of various K3 mutants. To examine the activity of K3 and its mutants in MHC class I down-regulation, BJAB cells were electroporated with the pTracer–GFP–K3 (WT) or one of the various point mutants. Cell surface levels of MHC class I were assessed 48 h post-transfection by staining for MHC class I with a PE-conjugated antibody (y axis) and gating the GFP-positive cell population (x axis) by flow cytometry. Numbers in the upper right side quadrant represent the MCF of MHC class I staining in the GFP positive population, calculated as described in the legend to Figure 2. The data were reproduced in at least two independent experiments.
BJAB cells were electroporated with pTracer vectors containing GFP alone or GFP plus each of the various constructs. Effects of expression of K3 and its mutants on the cell surface levels of MHC class I were assessed 48 h post-transfection in the GFP-positive cell population. Mutation of the putative zinc binding motifs completely abolished K3-mediated down-regulation of MHC class I (Figure 2). Similarly, mutation of Y152 to A in the putative tyrosine-based endocytosis motif of K3 abrogated its ability to down-regulate MHC class I, whereas the Y152/F mutation did not (Figure 3). This suggests that the YAAV sequence in the cytoplasmic region of K3 is a potential tyrosine-based sorting motif, and that sorting activity through this sequence is likely to be independent of tyrosine phosphorylation. Unlike the SH3-binding motif of HIV nef (Swigut et al., 2000), the P/A mutation in the putative SH3-binding motif of K3 had no effect on MHC class I down-regulation ability (Figure 3). Finally, the DE2/QQ mutation of the second acidic cluster of K3 had only a small effect on its ability to reduce cell surface expression of MHC class I, whereas the DE1/QQ mutation in the first acidic region of K3 impaired MHC class I down-regulation activity to a greater extent (Figure 3). In addition, the DE12/QQ construct, containing mutations of both acidic clusters of K3, possessed a significantly reduced MHC class I down-regulation activity (Figure 3). All K3 mutants were expressed at an equivalent level to wt K3 in these cells (see Supplementary figure 1). Thus, mutational analysis demonstrates that the putative zinc finger motifs at the N-terminal region, the tyrosine-based sorting motif in the central region and the diacidic clusters at the C-terminal region of K3 are necessary for efficient down-regulation of MHC class I surface expression. However, unlike the SH3-binding motif of HIV nef, which is required for MHC class I down-regulation (Swigut et al., 2000), the potential proline-rich SH3-binding motif of K3 is apparently not involved in down-regulation of MHC class I molecules.
Conserved NTRV residues of K3 are required for MHC class I down-regulation
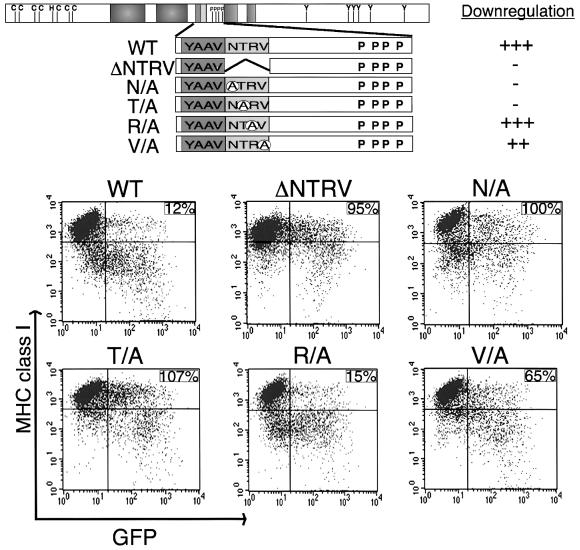
Despite dramatic sequence variation at the C-terminal region, the NTRV residues downstream of the YAAV motif are completely conserved between K3 and K5 (Figure 1), indicating that this sequence may play a role in MHC class I down-regulation. To test this hypothesis, the NTRV residues were deleted from the cytoplasmic region of K3 and this mutant, called ΔNTRV, was examined for its MHC class I down-regulation activity (Figure 4). Cytometry showed that the ΔNTRV mutant did not down-regulate surface expression of MHC class I molecules, in contrast to the strong down-regulation by wt K3 under the same conditions (Figure 4). To define further a role for the NTRV sequences in MHC class I down-regulation, each amino acid was replaced with alanine to generate the N156/A, T157/A, R158/A and V159/A mutants (Figure 4). Effects of mutant expression on the cell surface levels of MHC class I molecules were assessed 48 h post-transfection in the GFP-positive cell population by flow cytometry. The N156/A and T157/A mutations completely abrogated the ability of K3 to down-regulate MHC class I, whereas the V159/A mutation reduced class I down-regulation activity to a lesser extent (Figure 4). In contrast, the R158/A mutation had little effect on K3 MHC down-regulation activity under the same conditions (Figure 4). These results demonstrated that the conserved NTRV sequence is apparently engaged in efficient down-regulation of MHC class I molecules.
Fig. 4. NTRV sequence at the cytoplasmic region of K3 is required for MHC class I down-regulation. A schematic of the location of various mutations of the K3 protein is shown at the top. Five mutations, K3 ΔNTRV (ΔNTRV), K3 N156/A (N/A), K3 T157/A (T/A), K3 R158/A (R/A) and GFP K3 V159/A (V/A), were introduced into K3. Each mutant is described in greater detail in the text. Along the right side, the ability of wild-type K3 and each mutant to down-regulate MHC class I, as shown below the schematic, was given a relative score. ‘+++, ++, + and –’ indicate strong, weak, very weak and no activity of the K3 mutant in MHC class I down-regulation, respectively. The differential MHC class I down-regulation activity of several K3 mutants is shown below the schematic. To examine activity of K3 and its mutants in MHC class I down-regulation, BJAB cells were electroporated with pTracer–GFP–K3 (WT) or one of the K3 mutant vectors. Cell surface level of MHC class I was assessed 48 h post-transfection by staining for MHC class I (y axis) and gating the GFP-positive cell population (x axis) by flow cytometry. Numbers in the upper right side quadrant represent the MCF of MHC class I staining in the GFP-positive population, calculated as described in the legend to Figure 2. The data were reproduced in at least two independent experiments.
Alteration of MHC class I endocytosis rate by K3 and its mutants
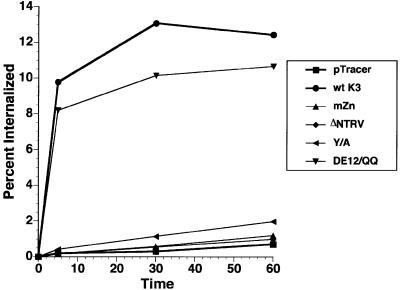
Detailed mutational analysis indicated that the zinc finger motif at the N-terminal region, the YAAV and NTRV sequences in the central region and the diacidic clusters at the C-terminal region of K3 are required for efficient down-regulation of MHC class I molecules. To delineate further their roles in MHC class I down-regulation, we examined effects of K3 and its mutants on the endocytosis rate of MHC class I molecules using a flow cytometry-based endocytosis assay. BJAB cells were electroporated with pTracer containing GFP alone, GFP with wt K3 or GFP with each K3 mutant. Cells were labeled 48 h post-transfection with a PE-conjugated MHC class I antibody and analyzed for MHC class I internalization in the GFP-positive population by flow cytometry. Over the time of the analysis, the internalization of MHC class I molecules in control cells transfected with pTracer-GFP was negligible (Figure 5). In contrast, wt K3 expression dramatically increased the endocytosis rate of MHC class I molecules (Figure 5). The K3 mZn, ΔNTRV and Y152/A mutants, shown to be incapable of down-regulating surface expression of MHC class I molecules, did not increase the endocytosis rate correspondingly (Figure 5). Surprisingly, the K3 DE12/QQ mutant, which exhibited a significant defect in down-regulation of MHC class I surface expression (Figure 3), increased the endocytosis rate of MHC class I molecules as efficiently as wt K3 (Figure 5). These results demonstrate that the zinc finger motifs in the N-terminal region, and the YAAV sorting motif and NTRV sequences in the central region of K3 are involved in an increased MHC class I molecule internalization rate. However, the diacidic clusters at the C-terminal region of K3, while necessary for efficient down-regulation of MHC class I surface expression, are likely engaged in processes other than the internalization of MHC class I molecules.
Fig. 5. Alteration of MHC class I endocytosis rate by K3 and its mutants. BJAB cells were electroporated with the pTracer–GFP (Tracer), GFP–K3 (wt K3), GFP–K3 mZn (mZn), GFP–K3 ΔNTRV (ΔNTRV), GFP–K3 Y152/A (Y/A) and GFP–K3 DE12/QQ (DE12/QQ) constructs. The internalization rates of MHC class I molecules were measured using a FACS-based endocytosis assay as described in the Materials and methods section. The results are representative of two independent experiments.
K3 and K5 targets MHC class I molecules to the TGN
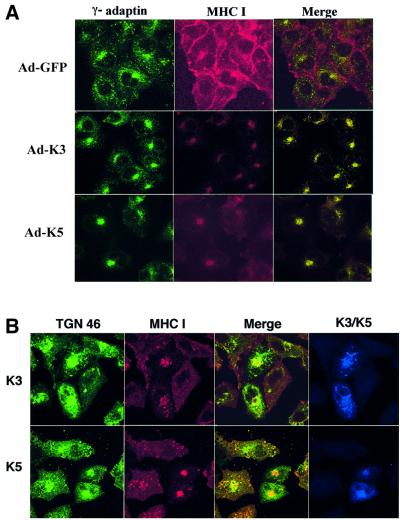
To investigate effects of K3 on the intracellular localization of MHC class I molecules, we used confocal immunofluorescence analysis. Since internalization of MHC class I molecules and their movement through the endocytic pathway is rapid in lymphocytes, we initially used A7 human melanoma cells, in which constitutive internalization of MHC class I molecules is somewhat slow and its relatively large cytoplasmic content facilitates the identification of various sorting compartments. A7 cells were infected with the same titer of adenovirus– GFP (Ad–GFP), adenovirus–GFP/K3 (Ad–K3) and adenovirus–GFP/K5 (Ad–K5) virus. The A7 cells were 100% GFP-positive 48 h post-infection, at which time cells were stained with a PE-labeled anti-MHC class I antibody, incubated at 37°C for 4 h, fixed with paraformaldehyde and subjected to confocal analysis. Extensive surface staining of MHC class I molecules was detected in Ad–GFP-infected A7 cells (Figure 6A). In striking contrast, MHC class I molecules were rapidly internalized and accumulated in the perinuclear region of Ad–K3-infected A7 cells and Ad–K5-infected A7 cells (Figure 6A). Since HIV nef redirects MHC class I molecules to the TGN (Le Gall et al., 1998), recombinant adenovirus-infected A7 cells were further stained with an antibody specific for the γ-subunit of AP-1 adaptor complexes, a protein commonly used to identify the TGN (green) (Traub et al., 1993). Merged images in multiple optical sections showed that MHC class I molecules were extensively colocalized with the γ-subunit of AP-1 adaptor complexes in Ad–K3- and Ad–K5-infected A7 cells, but not in Ad–GFP-infected A7 cells. In contrast, MHC class I molecules were not colocalized with the α-subunit of AP-2 adaptor complexes under the same conditions (data not shown). These results indicate intracellular accumulation of MHC class I molecules in a γ-adaptin-positive compartment, which is likely to be the TGN (Figure 6A).
Fig. 6. K3 and K5 targets MHC class I molecules to the trans-Golgi network. (A) TGN accumulation of MHC class I molecules by recombinant adenovirus-mediated K3 and K5 expression. Human A7 melanoma cells were infected with recombinant adenoviruses, Ad–GFP, Ad–K3 or Ad–K5. Forty-eight hours post-infection, cells were stained with an anti-MHC class I antibody (red) at 4°C and incubated at 37°C for 4 h. Cells were then fixed with paraformaldehyde, permeabilized and reacted with an anti-γ-adaptin antibody (green). Immuno fluorescence was examined using a Leica confocal immunofluorescence microscope, and a single representative optical section is presented. The yellow color in the merged image indicates overlapping staining of MHC class I and γ-adaptin. The pseudocolor assignment of green to γ-adaptin was done to facilitate data analysis. (B) TGN accumulation of MHC class I molecules by transient expression of K3 and K5. Human A7 melanoma cells were transfected with pEF–K3–His or pEF–K5–His vector. Forty-eight hours post-transfection, cells were stained with an anti-MHC class I antibody (red) at 4°C, incubated at 37°C for 4 h, fixed, permeabilized and then stained with a mouse anti-TGN46 antibody (green) and a rabbit polyclonal anti-His antibody (blue) specific for K3 and K5. Immunofluorescence was examined using a Leica confocal immunofluorescence microscope. The anti-class I and anti-TGN images were merged and any regions of yellow color in the final optical section indicates overlapping staining of MHC class I and TGN46. (C) Mutational analysis of K3 for TGN accumulation of MHC class I molecules. Human A7 melanoma cells were transfected with pEF–K3 mZn, pEF–K3 ΔC1, pEF–K3 ΔC2 or pEF–K3 ΔC7. At 48 h post-transfection, cells were stained with an anti-MHC class I antibody (red) at 4°C and incubated at 37°C for 4 h. Cells were then fixed with paraformaldehyde, permeabilized and stained with a rabbit polyclonal anti-His antibody specific for the K3 mutants (green). Immunofluorescence was examined using a Leica confocal immunofluorescence microscope and a single, representative optical section was selected for presentation. (D) The acidic cluster region of K3 is not engaged in the process of TGN accumulation of MHC class I molecules. Live BJAB–K3 and BJAB–K3 DE12/QQ cells were labeled with an anti-MHC class I antibody (red) at 4°C, incubated at 37°C for 10 min and fixed with paraformaldehyde. These cells were then reacted with an antibody specific for the γ subunit of AP-1 complexes (green). The yellow color in the merged optical section indicates overlapping staining of MHC class I and γ-adaptin.
Potential TGN localization of MHC class I molecules was tested further by a transient expression assay. A7 cells were transfected with either pEF–K3–His or pEF–K5–His expression vectors and, 48 h later, cells were stained with a PE-labeled anti-MHC class I antibody (red) at 4°C, incubated at 37°C for 4 h, fixed with paraformaldehyde and stained with a mouse antibody specific for another common marker of the TGN, the TGN residential protein TGN46 (green), as well as a polyclonal anti-His antibody specific for K3 and K5 (blue) (Figure 6B). This further confirmed that expression of K3 and K5 induced a drastic accumulation of MHC class I molecules in the TGN (Figure 6B).
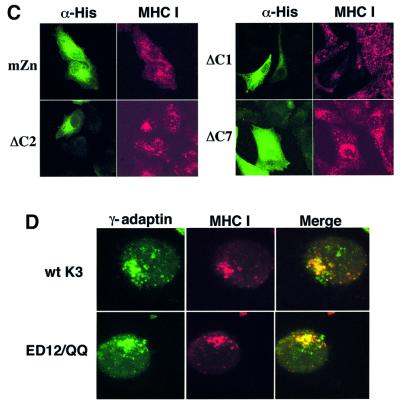
To delineate the roles of the individual motifs of K3 for TGN localization of MHC class I molecules, K3 mutants were subjected to the transient confocal assay as described above. The K3 mutants mZn, Y152/A, ΔNTRV and ΔC7, which were not capable of down-regulating MHC class I surface expression, were also not capable of inducing the accumulation MHC class I molecules in the TGN (Figure 6C; data not shown). In contrast, K3 mutants ΔC1, ΔC2 and ΔC3, which down-regulated MHC class I surface expression, induced dramatic accumulation of MHC class I molecules in the TGN (Figure 6C; data not shown). This suggests that the K3 activity of down-regulation of MHC class I surface expression may be directly linked to that of the TGN accumulation of class I molecules.
K3 containing mutations of the diacidic clusters still induces TGN localization of MHC class I molecules
Since the acidic cluster region of HIV nef interacts with PACS-1, and this interaction is necessary for relocalization of MHC class I molecules to the TGN (Piguet et al., 2000), we transfected BJAB cells with the pEF–K3 or pEF–K3 DE12/QQ expression vectors. Live BJAB–K3 and BJAB–K3 DE12/QQ cells were labeled with a PE-conjugated anti-MHC class I antibody (red) at 4°C, incubated at 37°C for 10 min and fixed with paraformaldehyde. These cells were then reacted with an anti-γ-subunit of AP-1 complexes antibody (green). As seen in K3-expressing A7 cells, MHC class I molecules were primarily localized in the perinuclear region of BJAB-K3 cells and extensively colocalized in multiple optical sections with the γ-subunit of AP-1 adaptor complexes (Figure 6D). Surprisingly, the K3 DE12/QQ mutant also induced an accumulation of MHC class I molecules in the TGN, as efficiently as wt K3 (Figure 6D). When the K3 DE12/QQ mutant was transiently expressed in A7 cells, essentially the same results were obtained (data not shown). Despite the similarities with HIV nef, we were not able to detect a specific interaction between K3 and cellular PACS-1 in repeated experiments (data not shown). This indicates that unlike HIV nef, the diacidic cluster region of K3 is not involved in the process of TGN accumulation of MHC class I molecules.
The K3 diacidic clusters are engaged in directing MHC class I molecules to the lysosome
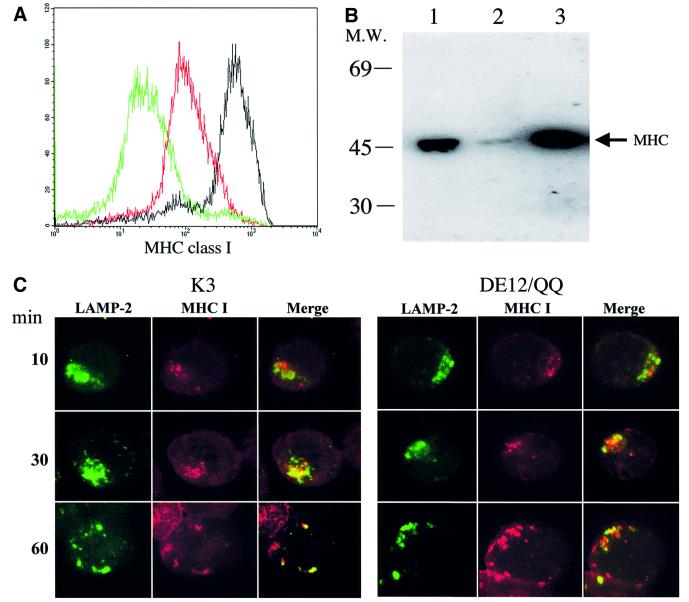
Previously, internalized MHC class I molecules in K3- and K5-expressing cells have been shown to be delivered to lysosomal vesicles, where they undergo degradation (Coscoy and Ganem, 2000). To investigate the fate of internalized class I molecules, cell lines containing the empty pEF expression vector or pEF containing wt K3 or the DE12/QQ mutant were established. Surprisingly, while the levels of MHC class I surface expression on BJAB–K3 DE12/QQ cells were considerably lower than those on BJAB–EF cells, the total amount of MHC class I molecules in BJAB–K3 DE12/QQ cells was equal to that in BJAB–EF cells (Figure 7A and B). In contrast, both the total amount and surface expression level of MHC class I molecules were dramatically lowered in BJAB–K3 cells (Figure 7A and B). This indicates that the total amount of MHC class I molecules correlated with the level of MHC class I surface expression in BJAB–K3 cells, whereas this correlation was not true in BJAB–K3 DE12/QQ cells (Figure 7A and B).
Fig. 7. The diacidic cluster region of K3 is engaged in directing MHC class I molecules to the lysosome for degradation. (A) MHC class I surface expression on BJAB–K3 and BJAB–K3 DE12/QQ cells. G418-resistant BJAB–EF, BJAB–K3 and BJAB–K3 DE12/QQ cells were stained with a PE-conjugated W6/32 pan-class I antibody and analyzed by flow cytometry. Two hundred thousand events were collected on a FACScan flow cytometer. The level of MHC class I expression on BJAB–K3 DE12/QQ is indicated by the red histogram, on the BJAB–K3 cell line by the green histogram and the BJAB–EF cell line by the black histogram. (B) Total amount of intracellular MHC class I heavy chain. The same amount of proteins from BJAB–EF, BJAB–K3 and BJAB–K3 DE12/QQ cells were separated by SDS–PAGE and reacted with a rabbit anti-MHC class I heavy chain antibody. The arrow indicates the location of the MHC class I heavy chain. Lane 1, BJAB–EF; lane 2, BJAB–K3; lane 3, BJAB–K3 DE12/QQ. (C) The acidic clusters of K3 are engaged in directing MHC class I molecules to the lysosome. Live BJAB–K3 and BJAB–K3 DE12/QQ cells were labeled with an anti-MHC class I antibody clone W6/32 (red) at 4°C, incubated at 37°C for 10, 30 and 60 min and fixed with paraformaldehyde. These cells were then stained with an anti-LAMP2 antibody (green). The yellow color in the merged, optical section indicates overlapping staining of MHC class I and LAMP2.
Based on these observations, we hypothesized that the K3 DE12/QQ mutant might have a defect in targeting MHC class I molecules to lysosomes for degradation. To test this hypothesis, live BJAB cells transfected with either the K3 or K3 DE12/QQ vector were labeled with a PE-conjugated anti-MHC class I antibody (red) at 4°C, incubated at 37°C for 10, 30 or 60 min and fixed with paraformaldehyde. These cells were then stained with an antibody specific for the lysosomal protein LAMP2 (green). Only a minimal level of colocalization between MHC class I molecules and LAMP2 was detected in BJAB–K3 cells after a 10 min incubation at 37°C (Figure 7C). However, an increased amount of MHC class I molecules were colocalized with LAMP2 in BJAB–K3 cells after 30 and 60 min of incubation, with essentially all of the class I molecules colocalizing with LAMP2 (Figure 7C). In contrast, only minimal levels of colocalization between MHC class I molecules and LAMP2 were detected throughout incubation in BJAB–K3 DE12/QQ cells (Figure 7C).
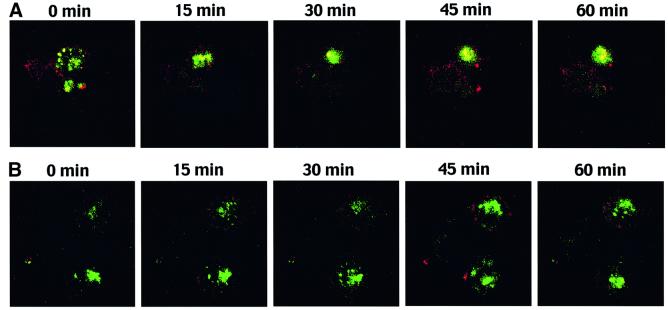
To investigate further the role of the diacidic cluster region of K3 in lysosomal targeting of MHC class I molecules, cell lines stably expressing wt K3 or the DE12/QQ mutant were viably labeled with the Lysotracker cell permeant dye (green) that accumulates in the lysosomal compartment. These cells were then stained with a PE-conjugated anti-MHC class I antibody at 4°C, loaded into an observation chamber and monitored for movement of MHC class I molecules to the lysosome over a 1 h period by confocal microscopy. Time-lapse sequences showed that colocalization between MHC class I molecules and lysosomes in BJAB–K3 cells was evident after ∼20–30 min of incubation and significantly increased during subsequent incubation (see Figure 8A and Supplementary figure 2A). In contrast, few or no MHC class I molecules were delivered to the lysosomal compartment in BJAB–K3 DE12/QQ cells under the same conditions (see Figure 8B and Supplementary figure 2B). Using NIH image, MHC class I colocalization with the lysosomal compartment was calculated in images taken after 30 min of incubation or longer and was found to be 10.5- to 14-fold higher in the BJAB–K3 cells as compared with the BJAB–K3 DE12/QQ cells. These results demonstrate that the K3 DE12/QQ mutant has a defect in delivering MHC class I molecules to the lysosomal compartment, whereas wt K3 does this in an efficient manner.
Fig. 8. Time-lapse confocal analysis of lysosomal localization of MHC class I molecules in K3-expressing cells. (A) MHC class I accumulates in the lysosomes of live BJAB–K3 cells. Live BJAB–K3 cells were viably labeled with a fluorescent, cell-permeable Lysotracker dye (green), which accumulates in the lysosome. Excess dye was washed away and the cells were then labeled with an anti-MHC class I antibody (red) at 4°C. Cells were maintained at 4°C through two washing steps and then loaded into a POC chamber for visualization in the Leica confocal microscope. A representative field of view was rapidly selected and images consisting of eight slices along the z-axis were captured every 3 min throughout the 60 min, room temperature experiment. Merged images of slice 5 at five time points, 0, 15, 30, 45 and 60 min, are presented. The yellow color in the images indicates overlapping staining of the Lysotracker dye and MHC class I. (B) MHC class I does not accumulate in the lysosomes of live BJAB–K3 DE12/QQ cells. Live BJAB–K3 DE12/QQ cells were stained with the Lysotracker dye (green) and anti-MHC class antibody (red), and visualized as described above. Merged images of optical slice 5 at five time points, 0, 15, 30, 45 and 60 min, are presented. The yellow color in the images indicates overlapping staining of the Lysotracker dye and MHC class I.
Discussion
We and others have previously reported that the KSHV K3 and K5 zinc finger proteins drastically down-regulate MHC class I molecules (Coscoy and Ganem, 2000; Ishido et al., 2000b; Haque et al., 2001). In this report, we further demonstrate that the down-regulation of MHC class I molecules by K3 protein is the result of multiple, consecutive trafficking pathways. After triggering their internalization, K3 targeted MHC class I molecules to a γ-subunit of the AP-1 adaptor complex and TGN46-positive compartment, which is likely to be the TGN. Subsequently, the diacidic clusters of K3 were involved in directing MHC class I molecules to the lysosomal compartment. These actions of K3 are functionally and genetically separable; the N-terminal zinc finger motif and the central YAAV sorting motif and conserved NTRV residues are involved in triggering the internalization of MHC class I molecules and redirecting them to the TGN. Subsequently, the C-terminal diacidic cluster region of K3 is engaged in targeting MHC class I molecules to the lysosomal compartment. These results demonstrate a novel trafficking pathway for MHC class I molecules induced by KSHV K3, conferring comprehensive protection of virally infected cells from host immune effector recognition.
The fact that these actions of K3 are functionally and genetically separable indicates that they are most likely accomplished through interactions with multiple cellular partners. While no molecular mechanisms are as yet defined for K3, several of the motifs identified in this study as being important for MHC class I down-regulation and targeting to the TGN and lysosomal compartments resemble those shown previously to act as intracellular targeting sequences (Aiken et al., 1994; Fuhrer et al., 1994; Honing et al., 1998; Owen and Evans, 1998). The zinc finger motifs in the N-terminal region of K3 and K5 have homology with sequences found in the PHD/LAP family of proteins (Saha et al., 1995). Many of these proteins, an example of which is c-cbl (Fang et al., 2001), have been shown to act as ubiquitin ligases, binding to target proteins and mediating the addition of ubiqutin. This results in the down-regulation of the target molecule from the cell surface, and transport to degradatory compartments within the cell (Bonifacino and Weissman, 1998).
The Yxxφ motif is a well-characterized intracellular targeting motif (Fuhrer et al., 1994; Owen and Evans, 1998). This tyrosine-based sorting signal directly interacts with the adaptor complexes (Boll et al., 1996; Ohno et al., 1998; Owen and Evans, 1998; Piguet et al., 1998). Interactions between the YXXφ motif and APs result in selective incorporation of the motif-containing proteins into coated vesicles that carry proteins to different destinations within the cell. Our results suggest that Y152AAV of K3 is likely to function as a tyrosine-based sorting signal. Mutation of the initial tyrosine to phenylalanine to create the sequence FAAV, a naturally occurring variant of the tyrosine-based sorting signal found in a number of proteins including the asialoglycoprotein receptor (Fuhrer et al., 1994), resulted in no change in the ability of K3 to down-regulate MHC class I. In contrast, mutation of this initial tyrosine to alanine abrogated MHC class I down-regulation activity. This suggests that this motif acts as a potential sorting signal, playing a role in the endocytosis of MHC class I molecules by interacting with the adaptor complexes.
Our results show that the conserved NTRV residues are required for efficient down-regulation of MHC class I molecules. Given the need for coordination between a large number of molecules in protein internalization and targeting, it is reasonable that the NTRV sequence mediates binding of one or more cofactors of the endocytosis machinery. Another possibility is that its proximity to the YAAV sorting signal influences the ability of one of the adaptor proteins to bind this motif. Interestingly, mutation of the threonine residue in the NTRV sequence displayed the most significant effect on the ability of K3 to down-regulate MHC class I molecules. This implied that the threonine residue could be a target for phosphorylation, and that phosphorylation might be necessary for efficient down-regulation of MHC class I molecules. However, mutation of the threonine residue to either positively charged arginine or negatively charged glutamic acid abrogated the ability of K3 to down-regulate MHC class I to a similar extent (R.E.Means and S.Ishido, unpublished results). This suggests that the threonine residue is not likely to be a target for phosphorylation, but instead is playing a sequence-specific role.
The acidic cluster region of HIV nef has been shown to interact with cellular PACS-1 and this interaction is important for the endocytosis and TGN accumulation of MHC class I molecules (Piguet et al., 2000). Unlike HIV nef, the acidic cluster region of KHSV K3 neither interacts with cellular PACS-1 nor is engaged in the TGN accumulation of MHC class I molecules. Instead, the acidic cluster region of K3 is involved in targeting MHC class I molecules to the lysosomal compartment. Time-lapse confocal imaging of live cells containing MHC class I and lysosomes tagged with different fluorescent dyes reveals that MHC class I molecules are efficiently targeted to the lysosomal compartment in wt K3-expressing cells, whereas they are not in K3 DE12/QQ mutant-expressing cells. These results suggest that the acidic cluster region of K3 contains a sorting motif that recruits MHC class I molecules to lysosomes. The N-terminus of the GGA family of cellular cargo proteins contain a VHS domain, a stretch of ∼140 amino acids first identified in the proteins VPS27, Hrs and STAM (Lohi and Lehto, 1998). Recently, the VHS domain of the GGAs has been shown to interact with the acidic cluster dileucine signals of integral membrane proteins and this interaction directs sorting of these proteins from the TGN to the lysosomes (Nielsen et al., 2001; Puertollano et al., 2001; Takatsu et al., 2001). Interestingly, the acidic cluster region of K3 and K5 is followed by 183IL and 186VL, respectively (Figure 1), which significantly mimics the acidic cluster dileucine sorting signal recognized by the GGAs. This suggests that the acidic cluster region of K3 potentially interacts with the VHS domain of GGAs and this interaction may direct sorting of MHC class I molecules to lysososmes where they undergo degradation. This hypothesis is under active investigation.
While additional experiments are required to define the detailed mechanisms of K3-mediated endocytosis of MHC class I molecules, a hypothetical model can be drawn from our results. Most likely through an interaction with one of the adaptor proteins, K3 facilitates endocytosis of MHC class I molecules from the cell surface and induces deposition of MHC class I into the TGN. Then, possibly by acting as a target for lysosomal transport proteins, K3 finally relocalizes MHC class I molecules to the lysosomal compartment. How is MHC class I targeted for down-regulation by K3? Since no specific interaction between K3 and MHC class I has been detected (R.E.Means and S.Ishido, unpublished results), this interaction might be weak or transitory. In the case of HIV-1 nef, which also increases MHC class I endocytosis through interactions with the endocytic machinery, no direct interaction with MHC class I has been yet reported. Alternatively, K3 could be ubiquitylating MHC class I or altering lipid composition and raft formation to make MHC class I a better target of the endocytic machinery. Additional experiments are required to understand the molecular nature of this specificity of K3 for MHC class I molecules. In summary, the results presented here reveal a novel viral mechanism for sequestration of MHC class I molecules from the surface, which leads to escape from host immune effector recognition and immune evasion. Thus, a detailed study of the molecular mechanisms of MHC class I down-regulation by KSHV K3 will not only lead to a better understanding of viral persistence and disease progression, but will also provide a novel means for investigating cellular trafficking pathways.
Materials and methods
Cell culture and transfection
BJAB, 293T and A7 cells were grown in RPMI with 10% fetal calf serum (FCS), Dulbecco’s modified Eagle’s medium (DMEM) with 10% FCS or MEM with 2% FCS, and 10% newborn calf serum, respectively. For transient assays, expression vectors were introduced by electroporation at 250 V and 960 µF in serum-free DMEM medium or by Fugene (Roche) transfection. In order to select permanent cell lines, selection medium containing 2 mg/ml of G418 (Sigma) was added 48 h post-electroporation and cells were maintained for 5 weeks.
Mutant plasmid construction
All K3 mutant constructs used in this paper were generated using oligonucleotide-directed mutagenesis (Du et al., 1995) and completely sequenced to verify the presence of the mutation. Each was subcloned into either the GFP co-expression vector pTracer–EF (Invitrogen) or pEF1 expression vector (Invitrogen).
Flow cytometry analysis and antibodies
Cells, 5 × 105 per sample, were washed with RPMI medium containing 10% FCS and incubated with the indicated conjugated monoclonal antibodies for 30 min at 4°C. After washing, each sample was fixed with 2% paraformaldehyde solution and flow cytometry analysis was performed with a FACS Scan (Becton Dickinson Co.). Details of the antisera used are given in the Supplementary data available at The EMBO Journal Online.
Immunoblots
For protein immunoblots, polypeptides in cell lysates corresponding to 105 cells were resolved by SDS–PAGE and transferred to a nitrocellulose membrane filter. Immunoblot detection was performed with a 1:1000 or 1:3000 dilution of primary antibody using the enhanced chemiluminescence system (ECL) (Amersham). The rabbit anti-MHC class I antibody was kindly provided by Dr H.Ploegh (Harvard Medical School) and the HRP-conjugated anti-rabbit IgG antibody was purchased from Santa Cruz Biotechnology.
Immunofluorescence and confocal microscopy
Slides for cells grown in suspension were prepared by cytospin centrifugation. After air drying, cells were fixed with cold acetone for 15 min and blocked with 10% goat serum in phosphate-buffered saline (PBS) for 30 min. Cells were stained with 1:100 diluted primary antibody in PBS containing 1% gelatin for 30 min. After incubation, cells were washed extensively with PBS, incubated with a 1:1000 dilution of Alexa 488 or 568-conjugated secondary antibody (Vector and Molecular Probes) in PBS containing 1% gelatin for 30 min at room temperature. Finally, cells were washed three times with PBS and mounted in mounting media (Vector). Confocal microscopy was performed as described previously (Ishido et al., 2000a).
Endocytosis assay
Experimental details are given in the Supplementary data available at The EMBO Journal Online. Briefly, cells transfected with the various K3 expressing vectors were stained PE-labeled anti-MHC antibodies at 4°C and then incubated for various periods of time at 37°C. They were then washed in an acidic solution to remove uninternalized antibody, fixed and subjected to flow cytometry. The percentage of edocytosis was calculated as a ratio of the fluorescence intensity value of acid-treated PE-labeled cells to that of non-treated PE-labeled cells.
Time-lapse confocal analysis
A detailed procedure is given in the Supplementary data available at The EMBO Journal Online.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We especially thank Drs G.Thomas and H.Ploegh for providing reagents, and G.Pageau and B.Roy for help in preparation of the manuscript. This work was partly supported by U.S. Public Health Service grants CA82057, CA91819 and RR00168, and ACS grant RPG001102. J.J. is a Leukemia and Lymphoma Society Scholar and R.E.M. is a Cancer Research Institute Fellow.
Note added in proof
A study by L.Coscoy, D.J.Sanchez and D.Ganem entitled ‘A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition’ [J. Cell Biol. (2001) 155, 1265–1273] was published during the revision of this manuscript.
References
- Aiken C., Konner,J., Landau,N.R., Lenburg,M.E. and Trono,D. (1994) Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell, 76, 853–864. [DOI] [PubMed] [Google Scholar]
- Alcami A. and Koszinowski,U.H. (2000) Viral mechanisms of immune evasion. Immunol. Today, 21, 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll W., Ohno,H., Songyang,Z., Rapoport,I., Cantley,L.C., Bonifacino,J.S. and Kirchhausen,T. (1996) Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J., 15, 5789–5795. [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S. and Weissman,A.M. (1998) Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell. Dev. Biol., 14, 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E., Chang,Y., Moore,P.S., Said,J.W. and Knowles,D.M. (1995) Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med., 332, 1186–1191. [DOI] [PubMed] [Google Scholar]
- Chang Y., Cesarman,E., Pessin,M.S., Lee,F., Culpepper,J., Knowles,D.M. and Moore,P.S. (1994) Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science, 266, 1865–1869. [DOI] [PubMed] [Google Scholar]
- Choi J., Means,R.E., Damania,B. and Jung,J.U. (2001) Molecular piracy of Kaposi’s sarcoma associated herpesvirus. Cytokine Growth Factor Rev., 12, 245–257. [DOI] [PubMed] [Google Scholar]
- Coscoy L. and Ganem,D. (2000) Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl Acad. Sci. USA, 97, 8051–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L. and Ganem,D. (2001) A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J. Clin. Invest., 107, 1599–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Regier,D.A. and Desrosiers,R.C. (1995) Improved recombinant PCR mutagenesis procedure that uses alkaline-denatured plasmid template. Biotechniques, 18, 376–378. [PubMed] [Google Scholar]
- Fang D., Wang,H.Y., Fang,N., Altman,Y., Elly,C. and Liu,Y.C. (2001) Cbl-b, a RING-type E3 ubiquitin ligase, targets phosphatidylinositol 3-kinase for ubiquitination in T cells. J. Biol. Chem., 276, 4872–4878. [DOI] [PubMed] [Google Scholar]
- Fuhrer C., Geffen,I., Huggel,K. and Spiess,M. (1994) The two subunits of the asialoglycoprotein receptor contain different sorting information. J. Biol. Chem., 269, 3277–3282. [PubMed] [Google Scholar]
- Haque M., Ueda,K., Nakano,K., Hirata,Y., Parravicini,C., Corbellino,M. and Yamanishi,K. (2001) Major histocompatibility complex class I molecules are down-regulated at the cell surface by the K5 protein encoded by Kaposi’s sarcoma-associated herpesvirus/human herpesvirus-8. J. Gen. Virol., 82, 1175–1180. [DOI] [PubMed] [Google Scholar]
- Honing S., Sandoval,I.V. and von Figura,K. (1998) A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J., 17, 1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishido S., Choi,J.K., Lee,B.S., Wang,C., DeMaria,M., Johnson,R.P., Cohen,G.B. and Jung,J.U. (2000a) Inhibition of natural killer cell-mediated cytotoxicity by Kaposi’s sarcoma-associated herpesvirus K5 protein. Immunity, 13, 365–374. [DOI] [PubMed] [Google Scholar]
- Ishido S., Wang,C., Lee,B.S., Cohen,G.B. and Jung,J.U. (2000b) Downregulation of major histocompatibility complex class I molecules by Kaposi’s sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol., 74, 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGall S., Erdtmann,L., Benichou,S., Berlioz-Torrent,C., Liu,L., Benarous,R., Heard,J.M. and Schwartz,O. (1998) Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity, 8, 483–495. [DOI] [PubMed] [Google Scholar]
- Lohi O. and Lehto,V.P. (1998) VHS domain marks a group of proteins involved in endocytosis and vesicular trafficking. FEBS Lett., 440, 255–257. [DOI] [PubMed] [Google Scholar]
- Nicholas J., Ruvolo,V., Zong,J., Ciufo,D., Guo,H.G., Reitz,M.S. and Hayward,G.S. (1997) A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol., 71, 1963–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M.S., Madsen,P., Christensen,E.I., Nykjaer,A., Gliemann,J., Kasper,D., Pohlmann,R. and Petersen,C.M. (2001) The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J., 20, 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H., Aguilar,R.C., Yeh,D., Taura,D., Saito,T. and Bonifacino,J.S. (1998) The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J. Biol. Chem., 273, 25915–25921. [DOI] [PubMed] [Google Scholar]
- Owen D.J. and Evans,P.R. (1998) A structural explanation for the recognition of tyrosine-based endocytotic signals. Science, 282, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguet V., Chen,Y.L., Mangasarian,A., Foti,M., Carpentier,J.L. and Trono,D. (1998) Mechanism of Nef-induced CD4 endocytosis: Nef connects CD4 with the mu chain of adaptor complexes. EMBO J., 17, 2472–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguet V., Gu,F., Foti,M., Demaurex,N., Gruenberg,J., Carpentier,J.L. and Trono,D. (1999) Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of β-COP in endosomes. Cell, 97, 63–73. [DOI] [PubMed] [Google Scholar]
- Piguet V., Wan,L., Borel,C., Mangasarian,A., Demaurex,N., Thomas,G. and Trono,D. (2000) HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nature Cell Biol., 2, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploegh H.L. (1998) Viral strategies of immune evasion. Science, 280, 248–253. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Aguilar,R.C., Gorshkova,I., Crouch,R.J. and Bonifacino,J.S. (2001) Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science, 292, 1712–1716. [DOI] [PubMed] [Google Scholar]
- Renne R., Zhong,W., Herndier,B., McGrath,M., Abbey,N., Kedes,D. and Ganem,D. (1996) Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nature Med., 2, 342–346. [DOI] [PubMed] [Google Scholar]
- Russo J.J. et al. (1996) Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl Acad. Sci. USA, 93, 14862–14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha V., Chaplin,T., Gregorini,A., Ayton,P. and Young,B.D. (1995) The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10 and MLLT6 proteins. Proc. Natl Acad. Sci. USA, 92, 9737–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T.F., Chang,Y. and Moore,P.S. (1998) Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8). In McCance,D.J. (ed.), Human Tumor Viruses. American Society for Microbiology, Washington, DC, pp. 87–133.
- Swigut T., Iafrate,A.J., Muench,J., Kirchhoff,F. and Skowronski,J. (2000) Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility complex antigen expression. J. Virol., 74, 5691–5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H., Katoh,Y., Shiba,Y. and Nakayama,K. (2001) Golgi-localizing γ-adaptin ear homology domain, ADP-ribosylation factor-binding (GGA) proteins interact with acidic di-leucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J. Biol. Chem., 276, 28541–28545. [DOI] [PubMed] [Google Scholar]
- Traub L.M., Ostrom,J.A. and Kornfeld,S. (1993) Biochemical dissection of AP-1 recruitment onto Golgi membranes. J. Cell Biol., 123, 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge I.S., Collawn,J.F. and Hopkins,C.R. (1993) Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell. Biol., 9, 129–161. [DOI] [PubMed] [Google Scholar]