Abstract
The mammalian hair cycle involves periodic regeneration of a tiny organ, the hair follicle, through a stem-cell-mediated process. The Hairless (Hr) gene encodes a nuclear receptor corepressor (HR) that is essential for hair follicle regeneration, but its role in this process is unknown. Here, we demonstrate that transgenic expression of HR in progenitor keratinocytes rescues follicle regeneration in Hr-/- mice. We show that expression of Wise, a modulator of Wnt signaling, is repressed by HR in these cells, coincident with the timing of follicle regeneration. This work links HR and Wnt function, providing a model in which HR regulates the precise timing of Wnt signaling required for hair follicle regeneration.
Keywords: corepressor, nuclear receptor, skin
Hair is maintained through a cyclic process that includes periodic regeneration of hair follicles in a stem-cell-dependent manner. The hair cycle consists of three defined stages: growth (anagen), followed by regression (catagen) and rest (telogen) (Fig. 1A) (1). Growth of a new hair requires reentry into anagen, a process involving activation of multipotent epithelial stem cells residing in a specialized part of the follicle outer root sheath (ORS) known as the bulge (Fig. 1B) (2–4). Activating signals emanate from adjacent mesenchymal cells (dermal papilla), directing epithelial stem cells to migrate and differentiate to regenerate the hair bulb (Fig. 1B), the structure from which a new hair will emerge (2–5). Multiple signaling pathways, including Wnts, Sonic hedgehog (Shh), and TGF-β family members have been shown to promote anagen initiation, yet the exact mechanism by which hair follicles regenerate is not clear (5–7).
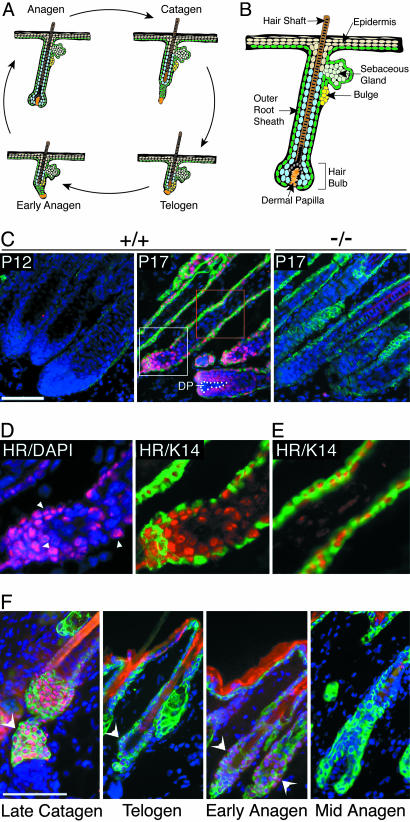
Fig. 1.
HR protein expression in hair follicles during the hair cycle. (A) Schematic representation of the hair cycle depicting changes in follicle structure. (B) Schematic of an anagen-stage hair follicle; relevant structures are indicated. A and B were adapted from ref. 5. (C–F) Immunofluorescent detection of HR protein (red) and keratin 14 (K14) (green). DAPI staining (blue) indicates nuclei. (C) HR protein expression in mouse back skin at P12 and P17. HR is detected in wild type (+/+) hair follicles at P17; the signal is specific because there is no staining in Hr-/- (-/-) follicles (Right). DP is denoted by white dots. Boxed areas are magnified below. (D) Magnification of region in white box shows HR staining in nuclei (HR/DAPI) and in K14-positive ORS and K14-negative hair bulb (HR/K14). Arrowheads denote nuclear staining. (E) Magnification of region in red box shows HR staining in K14-positive ORS. (F) HR expression through the hair cycle. Shown is immunofluorescent staining for HR (red) and K14 (green). Hair cycle stages are indicated. Arrowheads indicate HR signal. (Scale bars, 50 μm.)
Disruption of Hairless (Hr) gene function causes a complex skin phenotype that includes a specific defect in hair follicle regeneration in both humans and mice (8–10). In Hr mutant mice, hair follicle morphogenesis and initial hair growth is normal. However, after the follicles regress (catagen) and the hair is shed, around postnatal day (P)17, telogen stage follicles never reenter anagen, and no new hair is produced, resulting in alopecia (9, 11). Lack of hair regrowth has been attributed to separation of the bulge and dermal papilla (DP) during catagen (3, 12). Alternatively, the defect in anagen initiation may reflect a loss of the relevant epithelial stem cell population or an inability to generate and/or interpret the necessary signal(s).
We have shown that the Hairless protein (HR) is a nuclear receptor corepressor and, therefore, acts by regulating gene expression (13–15). Molecular studies of the epidermal component of the Hr mutant phenotype revealed that HR represses the transcription of genes involved in epidermal differentiation and that misregulation of these genes underlies the formation of abnormal epidermal structures (utricles) (16). Although HR likely regulates gene expression in hair follicle regeneration as well, the role of HR in hair follicles is not known. Here, we address the role of HR in follicle regeneration and provide evidence that HR regulates this process by repressing the expression of a Wnt inhibitor at the proper time in the hair cycle.
Materials and Methods
Mouse Lines. Hr-/- mice and TOP-Gal mice have been described in refs. 16 and 17. K14-rHr transgenic mice were made by cloning the coding sequence of the rHr cDNA downstream of a human K14 promoter (18, 19) and were generated by the Transgenic Core Facility of the Johns Hopkins Medical Institutions. Animal care was in accordance with institutional guidelines.
Histology, Immunostaining, and in Situ Hybridization. Histological analysis was as described in ref. 16. For immunofluorescence staining for HR, frozen sections (8 or 20 μm) were fixed in 10% neutral-buffered formalin, blocked and permeabilized, and incubated with HR-specific antisera (1:250) (20). Anti-rabbit Cy3-coupled antibody (1:2,000, Jackson Immunoresearch) was used to detect HR antiserum. For subsequent detection of K14, sections were blocked in 1 μg/ml rabbit IgG and incubated with FITC-coupled rabbit anti-K14 antibody (1:500; Covance). Cell nuclei were visualized with DAPI (4′,6-diamidino-2-phenylindole) (Molecular Probes).
For in situ hybridization, frozen sections (20 μm) were hybridized with digoxygenin-labeled cRNA probes, as described in ref. 15. Plasmids used to make probes were as follows: Shh, pBS-shh (16); Wise, IMAGE clone 5066413 (American Type Culture Collection); Axin2, IMAGE clone 6827741; Scd1, IMAGE clone MGC-6427. For immunohistochemistry to detect HR protein, endogenous peroxidase activity was blocked, and sections were incubated with HR antisera (1:250). HR antibody was detected with biotinylated goat anti-rabbit antibody followed by VECTASTAIN Elite ABC kit (Vector Laboratories) and DAB (3,3′-diaminobenzidine) (Sigma).
For alkaline phosphatase staining to identify DP, frozen sections (20 μm) were fixed in neutral-buffered formalin, stained with 4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate and counterstained with Nuclear Fast red (Vector Laboratories). β-gal was detected in frozen sections (20 μm), as described in refs. 17 and 21. Subsequent immunostaining for HR protein was as described above.
Photomicrographs were prepared by digital picture capture and processed with Adobe photoshop, as described in refs. 13 and 16. Hair follicles were staged by examination of follicle morphology as described in ref. 22.
RNA and Protein Analysis. Protein preparation and Western analysis were as described in ref. 16. Total RNA was prepared from back skin from one male and one female for each genotype with Trizol (Sigma). Northern analysis and quantification was as described in refs. 16 and 23. Probes were prepared from cDNAs for Wise and β-tubulin [kindly provided by A. Lanahan (Dartmouth University, Hanover, NH)]. Wise (24) was also identified as sclerostin-domain-containing gene 1 (Unigene: 25), sclerostin-like (26), ectodermal bone morphogenetic protein inhibitor (Ectodin) (27) and USAG-1 (28). Wise was not cited in previous microarray results because misexpression was not significant at P12 (<2-fold) (16).
Cell Culture and Transfection Assays. To isolate the promoter of the Wise gene, the Vista Genome Browser (http://pipeline.lbl.gov/cgi-bin/gateway2?bg=mm4&selector=vista) was used to identify conserved regions between mouse and human genomic sequences. PCR of mouse genomic DNA with gene-specific oligonucleotides was used to isolate a fragment (-1430 to +65), which was cloned into pGL2-Basic (Promega); reporter gene activity was increased >100-fold compared with the promoter-less vector.
Expression plasmids for rHR and silencing mediator of retinoid and thyroid hormone receptors (SMRT) (kindly provided by R. Evans, Salk Institute) were described in refs. 13 and 29. TR-interacting domains (TR-ID) (13) are mutated in mtHR; TR-ID1 overlaps the site of vitamin D receptor binding (15). Expression plasmids for Wnt1 and 3a were kindly provided by J. Nathans (Johns Hopkins School of Medicine). The Wnt 10b expression plasmid was made by using a partial Wnt10b cDNA [kindly provided by G. Shackleford (University of Southern California, Los Angeles)] and RT-PCR; the coding sequence was cloned into pCMX.
The GC cell line [kindly provided by P. Mellon (University of California, San Diego)], which supports HR corepressor activity, was grown in DMEM with 10% horse serum and 5% FBS. Super TOP-Flash (STF) cells (kindly provided by Dr. J. Nathans) are 293 cells that stably express a reporter gene with seven lymphoid enhancer factor/T cell-specific-factor (LEF/TCF)-binding sites driving luciferase expression (30). STF cells were grown in DMEM: F12 with 10% FBS.
For GC transfections, cells were plated in 24-well plates and transfected with 150 ng of reporter plasmid, 50–75 ng of expression plasmid and 150 ng of RSV-βgal by using Lipofectamine 2000 (Invitrogen). Cells were harvested in passive lysis buffer (Promega) and assayed for β-gal and luciferase activity. Luciferase activity was normalized to β-gal activity to correct for transfection efficiency. Data are the average of four experiments done in duplicate. For STF transfections, cells were transfected with 75 ng of each expression plasmid and 25 ng of CMV-βgal by using Lipofectamine 2000. Cells were processed as for GC; data are the average of three experiments done in duplicate.
Results and Discussion
To determine where and when HR acts in hair follicles, we localized nuclear HR protein throughout the hair cycle using HR-specific antisera. We find that follicles actively growing hair in anagen (P12) do not contain detectable HR (Fig. 1C). HR protein is detected as follicles enter catagen (P17) (Fig. 1C), which coincides with the onset of phenotypic alterations in hair follicles of Hr mutant mice (12, 16). Within the follicles, HR protein is found in the nuclei of keratin 14 (K14)-positive cells in the ORS, which includes the bulge region (Fig. 1 C–E). HR is also detected in K14-negative hair bulb cells but not in the DP (Fig. 1 C and D). HR expression is maintained in the ORS through late catagen into telogen and the early part of the next anagen (Fig. 1F). Once the hair bulb has reformed in mid-anagen, HR protein is again undetectable (Fig. 1F). Thus, HR protein expression is spatially and temporally regulated during the hair cycle, consistent with function of HR in both follicle regression and anagen reinitiation.
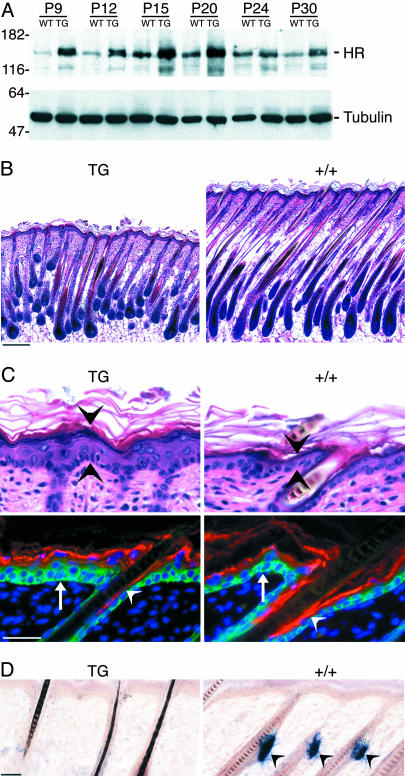
Because HR protein is localized to K14-positive ORS cells during the rest and reinitiation phases of the hair cycle, we predicted that expression of HR in this subset of progenitor keratinocytes would be sufficient to initiate the first postnatal anagen and rescue hair growth in Hr-/- mice. Therefore, we attempted to rescue the hair phenotype of Hr-/- mice by creating mice that express HR only in progenitor keratinocytes. First, we generated transgenic mice that constitutively express HR in progenitor keratinocytes by using the K14 promoter (K14-rHr). In K14-rHr skin, HR protein expression is high early in postnatal development (P9, P12) and remains above wild type levels as development proceeds (Fig. 2A). Analysis of the K14-rHr phenotype showed shorter hair (Fig. 2B), associated with decreased proliferation of matrix cells (data not shown). Thicker epidermis (Fig. 2C) is attributed to an expansion of the undifferentiated compartment (shown by K14 staining) and a decrease in the relative number of terminally differentiated keratinocytes (shown by filaggrin staining), suggesting that epidermal differentiation is delayed. Sebaceous differentiation is similarly delayed, because sebaceous glands are not detected at P9 (Fig. 2D) but are visible in some follicles at P12 (data not shown). Together, these results are consistent with our model that HR normally promotes differentiation toward hair cell fate and suppresses or delays differentiation into epidermis and sebaceous glands (16).
Fig. 2.
Transgenic expression of HR in progenitor keratinocytes. (A) Western analysis showing HR protein expression in wild type (WT) and K14-rHr (TG) mouse skin at the indicated postnatal ages. Detection of β-tubulin (Lower) shows protein loading. Molecular mass markers are indicated in kDa. (B) Hematoxylin–eosin staining of sections from TG and wild type (+/+) back skin at P9. (Scale bar, 100 μm). (C) (Upper) Hematoxylin–eosin staining of TG and wild type (+/+) epidermis. (Bottom) Immunofluorescent detection of K14 (green, arrows) and filaggrin (red, arrowheads). DAPI (blue) staining indicates nuclei. (D) In situ hybridization for Scd1, a sebaceous gland marker, in TG and wild type (+/+) back skin at P9. Signal is detected only in wild type (arrowheads). (Scale bar, 20 μm in C and D.)
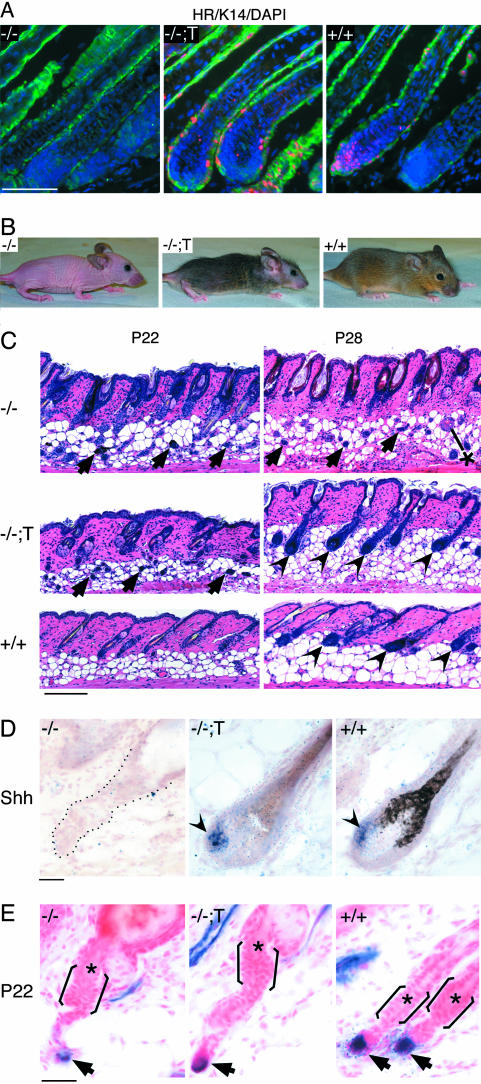
To generate mice that express HR only in K14-positive cells (“transgenic rescue”), we crossed the K14-rHr transgenic mice with Hr-/- mice (Fig. 3 A and B). At P22, both Hr-/- and transgenic rescue mice exhibit hair loss, consistent with histological analysis that shows prominent utricles and clusters of cells in the subdermal fat layer (Fig. 3C). Despite the prior defect, the transgenic rescue mice subsequently regenerate hair and eventually display thick fur (Fig. 3B). By P28, the transgenic rescue hair follicles resemble wild type follicles (Fig. 3C) and express Shh mRNA, a marker of early anagen (6) (Fig. 3D, arrowheads), in contrast to Hr-/- mouse skin, which remains grossly abnormal (Fig. 3C) and does not express Shh (Fig. 3D) (16). Thus, HR expression in K14-expressing cells rescues the ability of Hr-/- hair follicles to regenerate via the normal pathway. Failure to rescue hair loss may be due to a function of HR in K14-negative cells (Fig. 1D). The location of the DP is similar in Hr-/- and transgenic rescue skin (Fig. 3E), indicating that separation of the DP and bulge is not responsible for lack of hair regrowth and supporting the idea that a diffusible signal between DP and bulge reinitiates hair growth (3).
Fig. 3.
Hr expressed by the K14 promoter rescues hair regrowth in Hr-/- skin. (A) Immunofluorescent detection of HR protein (red) and K14 (green) in Hr-/- (-/-), transgenic rescue (-/-;T), and wild type (+/+) back skin. DAPI (blue) staining indicates nuclei. (Scale bar, 50 μm.) (B) Mice of the indicated genotypes at 7 weeks. (C) Hematoxylin–eosin staining of back skin sections from the indicated ages in Hr-/- (-/-), transgenic rescue (-/-;T), and wild type (+/+) mice. Arrows indicate cells in the dermis of Hr-/- and transgenic rescue mice at P22. Arrowheads indicate reformed hair bulbs in transgenic rescue and wild type mice at P28. *, cyst in Hr-/- at P28. (Scale bar, 100 μm.) (D) In situ hybridization detecting Shh mRNA in P28 mouse back skin. Shh expression (arrowheads) is detected in transgenic rescue (-/-;T) and wild type (+/+) hair follicles. Black dots outline follicle remnant in Hr-/- mice (-/-). (Scale bar, 20 μm.) (E) Alkaline phosphatase staining localizing the DP (arrows) in Hr-/- (-/-), transgenic rescue (-/-;T), and wild type (+/+) back skin at P22. Brackets approximate position of the bulge; asterisks indicate club hair. Sections are counterstained with Nuclear Fast red. (Scale bar, 20 μm.)
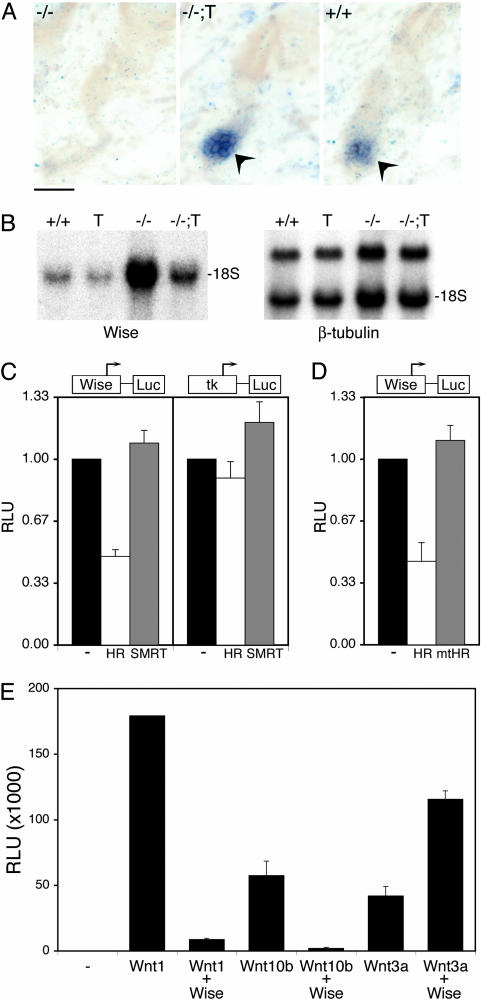
The demonstration that HR function in progenitor keratinocytes is sufficient for hair follicle regeneration led us to investigate whether HR regulates Wnt signaling, because activation of this pathway in these cells has been directly implicated in hair follicle regeneration (31–35). We examined expression of a Wnt-responsive gene, Axin2 (36–38), and found that Axin2 expression is not detected in Hr-/- hair follicles at anagen initiation (Fig. 4A). Remarkably, Axin2 expression is restored in transgenic rescue skin (Fig. 4A). Because HR is a corepressor, we hypothesized that HR may promote Wnt activation by repressing the expression of Wnt inhibitor(s). From a previous microarray screen for HR-regulated genes, we noted that there is, indeed, a potential Wnt inhibitor that is slightly up-regulated in Hr-/- skin at P12, before hair loss (16). This gene encodes Wise (Wnt modulator in surface ectoderm), a protein that has been shown to modulate Wnt signaling (24). To play a role in hair follicle regeneration, at the time of anagen reinitation, Wise expression should (i) increase in Hr-/- skin, (ii) decrease in transgenic rescue skin (relative to Hr-/-), and (iii) repress Wnt signaling in hair progenitor keratinocytes. Northern analysis confirmed that Wise is up-regulated (5.1-fold) in Hr-/- skin and reduced 2-fold (relative to Hr-/-) in transgenic rescue skin (Fig. 4B). Notably, expression is also reduced (1.7-fold) in K14-rHr mice, consistent with overexpression of a repressor and suggesting that HR represses Wise expression.
Fig. 4.
HR regulates expression of a modulator of Wnt signaling (Wise). (A) In situ hybridization for Axin2, a Wnt-responsive gene, in back skin sections from P22 mice of the indicated genotypes: -/- (Hr-/-), -/-;T (transgenic rescue), and +/+ (wild type). Arrowheads indicate Axin2 expression. (Scale bar, 20 μm.) (B) Northern analysis for Wise mRNA expression using RNA from back skin of P24 mice of the indicated genotypes: +/+ (wild type); T (K14-rHr); -/-, (Hr-/-); and -/-;T (transgenic rescue). (Right) β-tubulin hybridization of an identical blot; Wise expression was normalized to β-tubulin expression. 18S, position of 18S RNA. (C) Reporter genes for the Wise promoter (Wise-Luc) and control (tk-Luc) were transfected into cells with the indicated expression vectors. (-), empty expression vector. Results were normalized to vector control for each promoter. (D) The Wise reporter gene was transfected with expression vectors for HR or a HR derivative (mtHR) that has mutated receptor-interaction domains. Equal expression of HR and mtHR was verified by Western analysis (data not shown). (E) Cells stably expressing a Wnt-responsive reporter gene (Super TOP-Flash) were transfected with the indicated expression vectors. For B–D, relative light units (RLU) is luciferase activity relative to β-gal activity (internal control). Results are the average of at least three experiments done in duplicate.
To determine whether the Wise gene is repressed by HR, genomic regions containing the promoter for Wise were cloned upstream of a luciferase reporter gene, and activity was measured in the absence and presence of HR. Expression of HR significantly reduced activity of the Wise reporter gene (Fig. 4C). Repression is specific, because HR did not affect the activity of a minimal thymidine kinase promoter (Fig. 4C). Additionally, expression of another nuclear receptor corepressor, silencing mediator of retinoid and thyroid hormone receptors (SMRT) (39) had no effect on activity (Fig. 4C). The Wise promoter region contains putative binding sites for nuclear receptors that interact with HR, including thyroid hormone receptors (TRs), vitamin D receptor (VDR), and retinoid orphan receptors (13–15). Consistent with repression by nuclear receptors, a HR derivative with mutations in TR- and VDR-binding domains no longer represses the Wise promoter (Fig. 4D).
Considering the hypothesis that repression of Wise by HR regulates hair follicle regeneration requires demonstration that Wise functions as a Wnt inhibitor in this context. Using a cell line that stably expresses a Wnt-responsive reporter gene (Super-TOP-Flash) (30), we find that expression of Wise dramatically inhibits Wnt1-induced reporter gene activity (20-fold) (Fig. 4E). Although the Wnt family member that directs hair follicle regeneration is not known, Wnt 10b is expressed at the time (anagen initiation) and place (follicle epithelial cells) to play a role in hair follicle regeneration (40). We find that Wnt 10b-induced reporter gene activity is reduced 28-fold by expression of Wise (Fig. 4E). Notably, the cells used for this assay express Frizzled-1 and Frizzled-7 (41), Wnt receptors that are found in hair follicles (42), suggesting that Wnt 10b activation in STF cells recapitulates signaling in vivo. Consistent with Wise function as a context-dependent regulator (24), Wise potentiated activation by Wnt3a, a Wnt not expressed at anagen initiation (40). These results show that Wise can inhibit signaling by the Wnt (10b) most likely to play a role in anagen initiation.
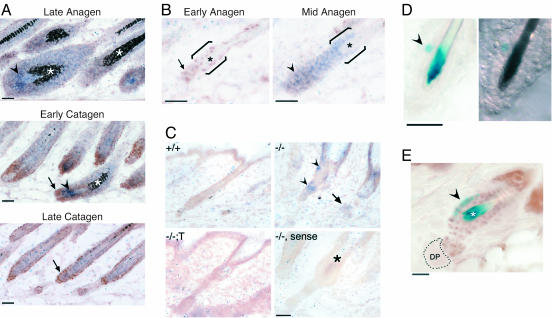
If repression of Wise expression by HR regulates hair follicle regeneration, the appearance of nuclear HR in hair follicles should coincide with both decreased Wise expression and increased Wnt-dependent gene expression. To address this hypothesis, we first combined immunohistochemistry for HR protein with in situ hybridization for Wise (Fig. 5 A and B). In late anagen, when HR protein is undetectable, Wise mRNA is expressed throughout the hair bulb (Fig. 5A). As follicles proceed into early catagen, nuclear HR protein is detected at the base of the bulb, and Wise mRNA expression becomes limited to the upper part of the bulb. In late catagen, Wise expression has almost disappeared and remains absent from cells expressing HR as follicles enter the next growth phase (Fig. 5B, early anagen). By mid-anagen, HR protein is no longer detected, whereas Wise mRNA expression is restored in the bulb and putative bulge (Fig. 5B). The inverse timing of HR protein and Wise mRNA expression in the same cell population suggests that HR is suppressing Wise expression in vivo, allowing hair cycle progression.
Fig. 5.
HR expression is inversely correlated with Wise mRNA expression and directly correlated with Wnt activation in the bulge. (A) In situ hybridization for Wise mRNA (blue, arrowheads) and immunohistochemistry for HR protein expression (brown, arrows) in P17 mouse back skin; follicles are at the indicated stages. Asterisks indicate pigment. (Scale bar, 50 μm.) (B) Expression of Wise mRNA (blue) and HR protein (brown) in P22 back skin; follicles are at the indicated stages. Brackets indicate bulge region; asterisks indicate club hair. (Scale bar, 50 μm.) (C) In situ hybridization for Wise mRNA in P24 mouse back skin. Expression is detected in the follicle remnant (arrowhead) and dermal cells (arrow) in Hr-/- skin (-/-). Sense control probe with adjacent Hr-/- section (-/-, sense); asterisk indicates club hair. (Scale bar, 20 μm.) (D) Detection of β-gal reporter gene expression (blue, arrowhead) in TOP-Gal mouse back skin. Bright-field (Left) and differential interference contrast (Right) microscopy images. (Scale bar, 20 μm.) (E) Detection of reporter gene expression (blue, arrowhead) and immunohistochemical staining for HR (brown). HR protein colocalizes with Wnt/β-catenin activation (blue) in the bulge. (Scale bar, 10 μm.)
Consistent with HR repressing Wise expression, Wise mRNA is present in Hr-/- skin at telogen, a stage at which Wise expression is undetectable in both wild type and transgenic rescue skin (Fig. 5C). Wise mRNA is localized to the lower portion of the follicle remnant (Fig. 5C, arrowhead) and in dermal cells (Fig. 5C, arrow). The follicle remnant and dermal cells express K14 and correspond to the location of new hair formation, thereby linking abnormally persistent expression of Wise with cells unable to reform a hair follicle (16, 43).
To test the prediction that nuclear HR protein is required for Wnt-dependent gene expression in hair follicles, we analyzed HR expression in the skin of TOP-Gal transgenic mice, which express a β-gal reporter gene in response to Wnt/β-catenin activation (17). Similar to previous reports, we detect β-gal expression in cells adjacent to the club hair in the putative bulge region at the telogen–anagen transition (Fig. 5D) (17, 32). Strikingly, we find that β-gal activity coincides with nuclear HR protein in the putative bulge region (Fig. 5E). The significance of HR-expressing cells that lack β-gal activity is not clear but may reflect a role in modulating other signals. Subsequent to this stage, both HR protein and β-gal activity are undetectable (data not shown). Thus, Wnt/β-catenin activation is correlated with the presence of nuclear HR protein at anagen initiation and inversely correlated with Wise mRNA expression.
Based on these data, we propose a model in which HR promotes Wnt activation and, thus, hair growth initiation by repressing expression of a soluble Wnt inhibitor at the proper time in the hair cycle. This work links Hr gene function and Wnt signaling and provides a molecular mechanism for regulating the precise temporal and spatial localization of Wnt signaling required for hair-cycle reinitiation (33, 34, 44). Presumably, HR function is not needed during initial hair morphogenesis, because undifferentiated ectoderm is not subject to the influence of Wnt inhibitors, or an alternative factor may regulate Wnt signaling in this context.
Although it has been proposed that lack of follicle regeneration in Hr mutant mice is due to separation of the DP and bulge, we find that the position of the DP is similar in Hr-/- and transgenic rescue skin (Fig. 3 C and E). Instead, the data described here support the hypothesis that loss of Hr function results in the inability to generate or interpret a diffusible signal. Thus, our model includes a molecular mechanism to account for the failure of hair follicles to regenerate in Hr mutant skin: inhibition of Wnt signaling caused by the persistent expression of Wise and, potentially, other Wnt inhibitors.
Through its restricted expression and corepressor function, HR controls the timing and location of hair follicle regeneration. Although the connection between HR and Wnt signaling is strong, hair follicle regeneration is a complex process requiring coordination of multiple signaling pathways, including Shh and BMPs (45). Molecular mechanisms underlying potential crosstalk between signaling pathways during hair follicle regeneration are not known. Notably, Wise has also been shown to function as a modulator of BMP signaling (26–28). It is likely that HR regulates the expression of multiple genes important for follicle regeneration, which may include modulators of other signaling pathways.
Acknowledgments
We thank J. Zarach for preliminary data and resources, the Coulombe laboratory for support, C.-M. Fan (Carnegie Institution of Washington) for TOP-Gal mice, Q. Xu and J. Nathans (Johns Hopkins School of Medicine) for reagents and advice, and G. Seydoux and K. Smith for critical reading of the manuscript. This work was supported by National Institutes of Health Grants AR44232 (to P.A.C.) and NS41313 (to C.C.T.) and National Research Service Award NS44744 (to G.M.J.B.).
Abbreviations: DP, dermal papilla; ORS, outer root sheath; Pn, postnatal day n; Shh, sonic hedgehog.
References
- 1.Hardy, M. H. (1992) Trends Genet. 8, 55-61. [DOI] [PubMed] [Google Scholar]
- 2.Oshima, H., Rochat, A., Kedzia, C., Kobayashi, K. & Barrandon, Y. (2001) Cell 104, 233-245. [DOI] [PubMed] [Google Scholar]
- 3.Cotsarelis, G., Sun, T. T. & Lavker, R. M. (1990) Cell 61, 1329-1337. [DOI] [PubMed] [Google Scholar]
- 4.Taylor, G., Lehrer, M. S., Jensen, P. J., Sun, T. T. & Lavker, R. M. (2000) Cell 102, 451-461. [DOI] [PubMed] [Google Scholar]
- 5.Alonso, L. & Fuchs, E. (2003) Genes Dev. 17, 1189-1200. [DOI] [PubMed] [Google Scholar]
- 6.Stenn, K. S. & Paus, R. (2001) Physiol. Rev. 81, 449-494. [DOI] [PubMed] [Google Scholar]
- 7.Millar, S. E. (2002) J. Invest. Dermatol. 118, 216-225. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad, W., Faiyaz ul Haque, M., Brancolini, V., Tsou, H. C., ul Haque, S., Lam, H., Aita, V. M., Owen, J., deBlaquiere, M., Frank, J., et al. (1998) Science 279, 720-724. [DOI] [PubMed] [Google Scholar]
- 9.Panteleyev, A. A., Paus, R., Ahmad, W., Sundberg, J. P. & Christiano, A. M. (1998) Exp. Dermatol. 7, 249-267. [DOI] [PubMed] [Google Scholar]
- 10.Cichon, S., Anker, M., Vogt, I. R., Rohleder, H., Putzstuck, M., Hillmer, A., Farooq, S. A., Al-Dhafri, K. S., Ahmad, M., Haque, S., et al. (1998) Hum. Mol. Genet. 7, 1671-1679. [DOI] [PubMed] [Google Scholar]
- 11.Mann, S. J. (1971) Anat. Rec. 170, 485-499. [DOI] [PubMed] [Google Scholar]
- 12.Panteleyev, A. A., Botchkareva, N. V., Sundberg, J. P., Christiano, A. M. & Paus, R. (1999) Am. J. Pathol. 155, 159-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potter, G. B., Beaudoin, G. M., III, DeRenzo, C. L., Zarach, J. M., Chen, S. H. & Thompson, C. C. (2001) Genes Dev. 15, 2687-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moraitis, A. N., Giguere, V. & Thompson, C. C. (2002) Mol. Cell. Biol. 22, 6831-6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh, J. C., Sisk, J. M., Jurutka, P. W., Haussler, C. A., Slater, S. A., Haussler, M. R. & Thompson, C. C. (2003) J. Biol. Chem. 278, 38665-38674. [DOI] [PubMed] [Google Scholar]
- 16.Zarach, J. M., Beaudoin, G. M. J., III, Coulombe, P. A. & Thompson, C. C. (2004) Development (Cambridge, U.K.) 131, 4189-4200. [DOI] [PubMed] [Google Scholar]
- 17.DasGupta, R. & Fuchs, E. (1999) Development (Cambridge, U.K.) 126, 4557-4568. [DOI] [PubMed] [Google Scholar]
- 18.Saitou, M., Sugai, S., Tanaka, T., Shimouchi, K., Fuchs, E., Narumiya, S. & Kakizuka, A. (1995) Nature 374, 159-162. [DOI] [PubMed] [Google Scholar]
- 19.Vassar, R., Rosenberg, M., Ross, S., Tyner, A. & Fuchs, E. (1989) Proc. Natl. Acad. Sci. USA 86, 1563-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potter, G. B., Zarach, J. M., Sisk, J. M. & Thompson, C. C. (2002) Mol. Endocrinol. 16, 2547-2560. [DOI] [PubMed] [Google Scholar]
- 21.Kobielak, K., Pasolli, H. A., Alonso, L., Polak, L. & Fuchs, E. (2003) J. Cell Biol. 163, 609-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller-Rover, S., Handjiski, B., van der Veen, C., Eichmuller, S., Foitzik, K., McKay, I. A., Stenn, K. S. & Paus, R. (2001) J. Invest. Dermatol. 117, 3-15. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, C. C. & Bottcher, M. C. (1997) Proc. Natl. Acad. Sci. USA 94, 8527-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itasaki, N., Jones, C. M., Mercurio, S., Rowe, A., Domingos, P. M., Smith, J. C. & Krumlauf, R. (2003) Development (Cambridge, U.K.) 130, 4295-4305. [DOI] [PubMed] [Google Scholar]
- 25.Schuler, G. D. (1997) J. Mol. Med. 75, 694-698. [DOI] [PubMed] [Google Scholar]
- 26.Balemans, W. & Van Hul, W. (2002) Dev. Biol. 250, 231-250. [PubMed] [Google Scholar]
- 27.Laurikkala, J., Kassai, Y., Pakkasjarvi, L., Thesleff, I. & Itoh, N. (2003) Dev. Biol. 264, 91-105. [DOI] [PubMed] [Google Scholar]
- 28.Yanagita, M., Oka, M., Watabe, T., Iguchi, H., Niida, A., Takahashi, S., Akiyama, T., Miyazono, K., Yanagisawa, M. & Sakurai, T. (2004) Biochem. Biophys. Res. Commun. 316, 490-500. [DOI] [PubMed] [Google Scholar]
- 29.Chen, J. D., Umesono, K. & Evans, R. M. (1996) Proc. Natl. Acad. Sci. USA 93, 7567-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, Q., Wang, Y., Dabdoub, A., Smallwood, P. M., Williams, J., Woods, C., Kelley, M. W., Jiang, L., Tasman, W., Zhang, K., et al. (2004) Cell 116, 883-895. [DOI] [PubMed] [Google Scholar]
- 31.Huelsken, J., Vogel, R., Erdmann, B., Cotsarelis, G. & Birchmeier, W. (2001) Cell 105, 533-545. [DOI] [PubMed] [Google Scholar]
- 32.Merrill, B. J., Gat, U., DasGupta, R. & Fuchs, E. (2001) Genes Dev. 15, 1688-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Mater, D., Kolligs, F. T., Dlugosz, A. A. & Fearon, E. R. (2003) Genes Dev. 17, 1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo Celso, C., Prowse, D. M. & Watt, F. M. (2004) Development (Cambridge, U.K.) 131, 1787-1799. [DOI] [PubMed] [Google Scholar]
- 35.Niemann, C., Owens, D. M., Hulsken, J., Birchmeier, W. & Watt, F. M. (2002) Development (Cambridge, U.K.) 129, 95-109. [DOI] [PubMed] [Google Scholar]
- 36.Leung, J. Y., Kolligs, F. T., Wu, R., Zhai, Y., Kuick, R., Hanash, S., Cho, K. R. & Fearon, E. R. (2002) J. Biol. Chem. 277, 21657-21665. [DOI] [PubMed] [Google Scholar]
- 37.Jho, E. H., Zhang, T., Domon, C., Joo, C. K., Freund, J. N. & Costantini, F. (2002) Mol. Cell. Biol. 22, 1172-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lustig, B., Jerchow, B., Sachs, M., Weiler, S., Pietsch, T., Karsten, U., van de Wetering, M., Clevers, H., Schlag, P. M., Birchmeier, W., et al. (2002) Mol. Cell. Biol. 22, 1184-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, J. D. & Evans, R. M. (1995) Nature 377, 454-457. [DOI] [PubMed] [Google Scholar]
- 40.Reddy, S., Andl, T., Bagasra, A., Lu, M. M., Epstein, D. J., Morrisey, E. E. & Millar, S. E. (2001) Mech. Dev. 107, 69-82. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Z., Shu, W., Lu, M. M. & Morrisey, E. E. (2005) Mol. Cell. Biol. 25, 5022-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy, S. T., Andl, T., Lu, M. M., Morrisey, E. E. & Millar, S. E. (2004) J. Invest. Dermatol. 123, 275-282. [DOI] [PubMed] [Google Scholar]
- 43.Panteleyev, A. A., van der Veen, C., Rosenbach, T., Muller-Rover, S., Sokolov, V. E. & Paus, R. (1998) J. Invest. Dermatol. 110, 902-907. [DOI] [PubMed] [Google Scholar]
- 44.Gat, U., DasGupta, R., Degenstein, L. & Fuchs, E. (1998) Cell 95, 605-614. [DOI] [PubMed] [Google Scholar]
- 45.Botchkarev, V. A. & Sharov, A. A. (2004) Differentiation (Berlin) 72, 512-526. [DOI] [PubMed] [Google Scholar]