Abstract
Although pili have long been recognized in Gram-negative pathogens as important virulence factors involved in adhesion and invasion, very little is known about extended surface organelles in Gram-positive pathogens. Here we report that Group A Streptococcus (GAS), a Gram-positive human-specific pathogen that causes pharyngitis, impetigo, invasive disease, necrotizing fasciitis, and autoimmune sequelae has long, surface-exposed, pilus-like structures composed of members of a family of extracellular matrix-binding proteins. We describe four variant pili and show that each is recognized by a specific serum of the Lancefield T-typing system, which has been used for over five decades to characterize GAS isolates. Furthermore, we show that immunization of mice with a combination of recombinant pilus proteins confers protection against mucosal challenge with virulent GAS bacteria. The data indicate that induction of a protective immune response against these structures may be a useful strategy for development of a vaccine against disease caused by GAS infection.
Keywords: fibronectin-binding, Gram-positive
Many Gram-negative bacterial pathogens use long pili that extend from the bacterial surface in adhesion and invasion of host tissues (reviewed in ref. 1). Immunization designed to target these important functions offer attractive strategies for vaccine development. Although pilus biogenesis and function in Gram-negative pathogens are well understood, little is known about surface organelles in Gram-positive pathogens (2). Pilus-like structures have been identified in Gram-positive Corynebacterium diphtheriae, and their structure and assembly have been elucidated, but little is known of their function (2, 3). More recently, similar structures have been reported in the human pathogen Group B Streptococcus (GBS, Streptococcus agalactiae), and the component proteins were shown to be effective protective antigens against lethal GBS challenge in a mouse model of infection and disease (4, 5).
Here, we report the presence of pili on the surface of Group A Streptococcus (GAS, Streptococcus pyogenes), a Gram-positive bacteria that colonizes the pharynx and skin of humans and is the most common cause of bacterial pharyngitis in children. Although bacterial pharyngitis is usually a self-limiting infection with little long-term pathology, autoimmune sequelae may result in severe cardiac pathology and glomerulonephritis. In addition, GAS causes severe invasive disease, which may result in streptococcal toxic shock syndrome or necrotizing fasciitis (6).
More than 50 years ago, Lancefield and colleagues (7, 8) classified GAS isolates into serotypes on the basis of serum recognition of a variable trypsin-sensitive surface protein, the M protein (7), and a variable trypsin-resistant antigen, the T antigen (8). To date, >100 M serotypes and ≈20 T serotypes have been identified. Epidemiologic studies based on M and T serological typing have been central to our understanding of the biological diversity and disease-causing potential of GAS. Whereas the M protein and its inherent variability have been extensively characterized and despite five decades of study, there is still very little known about the structure and variability of T antigens, although a gene of unknown function has been shown to code for the antigen recognized by T6 sera (9). Here we show that four of the 20 T antigens correspond to trypsin-resistant pili composed of putative adhesion proteins and that recombinant pilus proteins confer protection against lethal GAS challenge in a mouse model of infection and invasive disease.
Materials and Methods
Bacterial Strains, Media, and Growth Conditions. GAS strains used are listed in Table 2, which is published as supporting information on the PNAS web site. Wild-type and mutant strains were grown at 37°C or 30°C in Todd–Hewitt medium supplemented with 0.5% yeast extract (THY) (Difco), or THY agars supplemented with 5% defibrinated sheep blood.
Cell-Wall Fraction Preparation. GAS strains were grown in THY to OD600 = 0.4 at 37°C. Cells were washed once in PBS, suspended in 1 ml of ice-cold protoplasting buffer [40% sucrose/0.1 M KPO4, pH 6.2/10 mM MgCl2/Complete EDTA-free protease inhibitors (Roche)/2 mg/ml lysozime/400 units of mutanolysin (Sigma)] and incubated at 37°C for 3 h. After centrifuging at 13,000 × g for 15 min, the supernatants (cell-wall fractions) were frozen at –20°C.
Recombinant Proteins and Antisera. Genes coding for surface-expressed proteins were cloned by PCR from genomic DNA into Escherichia coli plasmid vectors to produce ether 6Xhistidine or GST as described by Maione et al. (5). Recombinant fusion proteins were purified by affinity chromatography as in Montigiani et al. (10). Mouse antisera specific for the recombinant proteins were produced by immunizing groups of four CD1 mice with the purified recombinant proteins.
Primers used to clone and sequence major pilus genes are listed in Table 3, which is published as supporting information on the PNAS web site.
Immunoblotting. Cell-wall preparations were separated by 3–8% gradient gels (NuPAGE Tris-acetate gels, Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad) for immunoblot analysis with mouse polyclonal antisera (1:500) and ECL enhanced chemiluminescence detection (SuperSignal West Pico chemiluminescent substrate, Pierce). The secondary antibody (ECL, horseradish-peroxidase-linked anti-mouse IgG, GE Healthcare) was used at a 1:5,000 dilution.
In the experiment with recombinant proteins, commercially available standard anti-T-typing sera (11) were obtained from Sevapharma (Prague) and used at a 1:500 dilution and the secondary antibody (horseradish peroxidase-linked anti-rabbit IgG, Bio-Rad) was used at a 1:10,000 dilution.
Flow Cytometry. Bacteria were grown in THY to OD600 = 0.4, washed twice with PBS, suspended in newborn calf serum (Sigma), incubated for 20 min at room temperature, and dispensed in a 96-well plate (20 μl per well). Eighty microliters of preimmune or immune mouse sera diluted in PBS/0.1% BSA was added to the bacterial suspension to a final dilution of 1:200. Incubation was performed on ice for 30 min. The bacteria were washed, incubated on ice for 30 min in 10 μl of goat anti-mouse IgG, and conjugated with F(ab′)2 fragment-specific R-phycoerythrin (Jackson Immunoresearch Laboratories) in PBS/0.1% BSA/20% newborn calf serum to a final dilution of 1:100. The stained bacteria were analyzed with a FACSCalibur cytometer (Becton Dickinson) and cellquest software (Becton Dickinson). In experiments in which trypsin treatment was required and before antibody reaction, bacteria were suspended in 20 μl of 40% PBS plus 2, 20, or 200 μg of trypsin. After 30 min of incubation at 37°C, 80 μl of PBS and 2 μl of PMSF were added to the samples (to a 2 mM final concentration), which were then incubated at room temperature for 10 min and washed with PBS.
Construction of In-Frame Deletion and Complementation Mutants. Mutations were constructed by using splicing-by-overlap-extension PCR as described in ref. 12. Primers used for the construction and screening of deletion alleles are listed in Table 4, which is published as supporting information on the PNAS web site. Restriction sites that were used in cloning are shown in boldface type in Table 4. The PCR deletion construct was ligated to the temperature-sensitive allelic exchange vector pJRS233 (13), and transformation and allelic exchanges were performed as described in refs. 14–16. Transformants were selected on THY plates with 1 μg/ml erythromycin (Sigma) at 30°C. Drug-sensitive colonies were screened and deletions were confirmed by PCR assay.
The complementation vector (pAM401::Spy128) was constructed with the appropriate primers (see Table 4) to amplify the fragment that includes the Spy128 gene, the predicted promoter and the ρ-independent terminator.
Mouse Immunization and Challenge. Five-week-old female CD1 mice (10 mice per group) were immunized with a mixture of 15 μg (each) of recombinant proteins (Cpa + M1_128 + M1_130) administered i.p. at 0, 21, and 35 days with complete Freund adjuvant the first time and with incomplete Freund adjuvant the two following times. Blood samples were collected before the first and after the third immunizations. Immunized mice were challenged intranasally with 106 colony-forming units of ISS-3348 strain. The colony-forming unit titer of the infecting dose was verified by plating on THY/blood plates. The final survival rate for the mice was calculated after 10 days. Statistical analysis was performed using Fischer's exact test.
Bioinformatics. Computer programs included in the gcg wisconsin package 10.0 were used to analyze and compare the sequences from different GAS strains.
Results and Discussion
Identification of Pilus-Like Proteins in GAS Genomes. We have recently described the presence of covalently polymerized pilus-like structures in the related bacterium GBS (4) and shown that recombinant forms of the major pilin proteins confer protection in a mouse model of GBS disease (5). To date, these covalently linked pili have been identified in C. diphtheriae (2) and GBS (4). In both organisms the pili are formed by the sortase enzyme-catalyzed covalent linkage of surface proteins containing the cell-wall-sorting signal LPXTG and conserved pilin motifs involved in the linkage reaction (2, 3). These data prompted us to search available GAS genomes for genes coding for similar pili to test the hypothesis that pilin components may represent protective antigens also against GAS disease. Because we failed to identify pilus genes in GAS by gene or protein similarity with the pilin genes of GBS, our strategy involved a search in the five complete genomes (GenBank accession nos. AE004092, AE014074, AE009949, BA000034, and CP000003) and all GAS sequences in the available databases for closely linked genes coding for surface proteins containing LPXTG motifs in close proximity to genes coding for variant sortase enzymes.
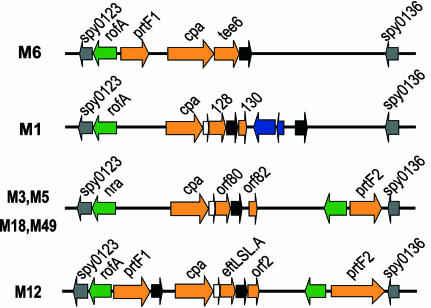
Genes with these characteristics were found in all genomes in an ≈11-kilobase, highly variable pathogenicity island known as the fibronectin-binding, collagen-binding T antigen (FCT) region (17) because it contains members of a family of genes coding for extracellular matrix (ECM)-binding proteins, some of which have been shown to be involved in adhesion and invasion (reviewed in ref. 18). Interestingly, the FCT region in one M6 strain (MGAS10394) contains the gene coding for the Lancefield T6 antigen (9). Four classes of FCT region have been characterized by the types and order of the genes contained, and the region is flanked in all strains by two highly conserved genes, Spy0123 and Spy0136. The FCT region of strains of types M3, M5, M18, and M49 has a similar organization, whereas those of M6, M1, and M12 differ (shown schematically in Fig. 1). Between three and five genes in each of the FCT regions (see Fig. 1) code for proteins containing motifs similar to the LPXTG motif found in cell-wall-anchored proteins and all belong to the family of genes coding for ECM-binding adhesins.
Fig. 1.
Schematic representation of the FCT region from seven GAS strains. Genes coding for LPXTG-containing proteins are represented with orange arrows, whereas transcriptional regulators (rofA, nra, and msmRL) are in green and conserved flanking genes (Spy0123 and Spy0136) are in gray. At least one sortase is present in every FCT region (black arrows), and a signal peptidase, lepA, is present in three of four regions (white arrows). In the FCT region of the M1 strains, two transposable elements are also present (blue arrows).
The T6 Antigen Corresponds to a Pilus. To determine whether the FCT islands do code for pilus-like structures, we have raised antisera against recombinant products of these genes and used them to explore the expression of the proteins in the GAS strains from which they derive. In an immunoblot of reducing SDS/PAGE for cell wall extracts of strain M6_ISS3650, antiserum specific for the T6 protein recognized a band corresponding to the predicted molecular mass of the product and a ladder of high-molecular-mass bands ranging in mobility from ≈70 kDa to beyond the resolution of the 3–8% gradient gels used (Fig. 2A). This pattern of high-molecular-mass products is similar to that observed in immunoblots of the covalently linked protein components of pili identified in C. diphtheriae (2, 3) and more recently in Streptococcus agalactiae (4). In fact, immunogold EM of strain M6_ISS3650 with antisera specific for the product of tee6 revealed abundant surface staining and long pilus-like structures extending to >500 nm from the bacterial surface (Fig. 3A). Hence, the T6 protein, one of the antigens recognized in the original Lancefield serotyping system, forms long polymerized pilus structures.
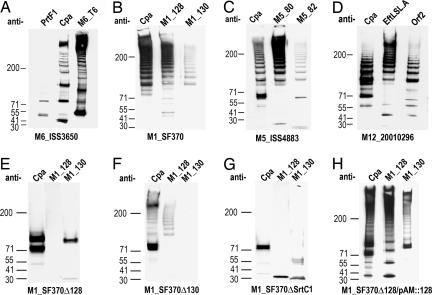
Fig. 2.
FCT proteins form high-molecular-mass structures. (A–D) Immunoblots of cell-wall fractions of GAS strains with antisera specific for LPXTG proteins of strain M6_ISS3650 (A), M1_SF370 (B), M5_ISS4883 (C), and M12_20010296 (D). (E–H) Immunoblots of cell-wall fractions of deletion mutants M1_SF370Δ128 (E), M1_SF370Δ130 (F), M1_SF370ΔSrtC1 (G), and the M1_128 deletion strain complemented with plasmid pAM::128, which contains the M1_128 gene (H) with antisera specific for the pilin components of M1_SF370. The antisera used are given above the lanes, and the strains used are indicated below the lanes.
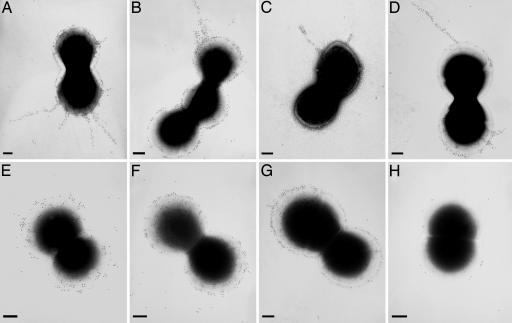
Fig. 3.
Pilus-like structures observed by electron microscopy. Immunogold labeling and transmission electron microscopy of four GAS strains with antisera raised against pilus proteins. (A) Anti-T6 antisera staining of strain M6_ISS3650. (B) Anti-M1_128 staining of strain M1_SF370. (C) Anti-M5_orf80 staining of strain M5_ISS4883. (D) Anti-M12_EftLSL.A staining of strain M12_20010296. (E) Anti-M6_Cpa staining of strain M6_ISS3650. (F) Anti-M1_Cpa staining of strain M1_SF370. (G) Anti-M1_130 staining of strain M1_SF370. (H) Anti-M1_128 staining of strain M1_SF370Δ128, which lacks the M1_128 gene. (Scale bars, 200 nm.)
The FCT region in M6_ISS3650 contains, in addition to the tee6 gene, two other genes (prtF1 and cpa) predicted to code for surface-exposed proteins, characterized by the cell wall attachment motif of the form LPXTG. Antiserum specific for PrtF1 recognized a single molecular species with electrophoretic mobility corresponding to the predicted molecular mass of the protein and one smaller band of unknown origin, whereas antisera specific for Cpa recognized a high-molecular-mass, covalently linked ladder (Fig. 2 A). Antiserum specific for Cpa also revealed abundant surface staining but only occasional gold particles extended from the surface (Fig. 3E).
The FCT Islands of M1, M5, and M12 Strains Also Code for Pili. The finding of genes coding for pili in the FCT region of strain M6_ISS3650 prompted us to examine the predicted surface-exposed proteins in the variant FCT regions of GAS strains of different M type (M1_SF370, M5_ISS4883, and M12_20010296) representing the other three FCT variants. Each gene was cloned and expressed, and antisera raised against each recombinant protein were used to probe mutanolysin extracts of the respective strains. In the FCT region of M1_SF370, there are three predicted surface proteins, and the respective antisera reacted with a ladder of high-molecular-mass material (Fig. 2B). Immunogold staining with antiserum specific for M1_128 revealed pili structures similar to those seen in the M6 strain (Fig. 3B). Antisera raised against Cpa and M1_130 revealed abundant surface staining and occasional gold beads extending from the bacteria (Fig. 3 F and G). Elimination of M1_128 gene by in-frame deletion resulted in the loss of staining in immunoblot (Fig. 2E) and surface and pilus staining in EM (Fig. 3H), which confirms that in this strain the antisera were specific for M1_128 protein. Although sera raised against the pilus proteins are likely to crossreact with variants in different strains, there is only one FCT region in each strain, and these data demonstrate that there is no crossreaction with other proteins in the same strain.
The variant FCT regions of strains M5_ISS4883 and M12_20010296 are more complex. The M5 strain contains genes for four predicted surface-exposed proteins, whereas the M12 strain contains five (see Fig. 1). Antisera against three of the four products of the FCT region of M5_ISS4883 (Cpa, M5_orf80, and M5_orf82) and three of the five products of the FCT region of M12_20010296 (Cpa, EftLSL.A, and Orf2) stained high-molecular-mass ladders (Fig. 2 C and D). Long pili were also visible in both of these strains when antisera against M5_orf80 or EftLSL.A were used (Fig. 3 C and D).
Pilus Assembly Requires One LPXTG Protein and Sortases. To better understand the structure of the pili, we conducted a genetic analysis of the region in the transformable strain M1_SF370. Elimination of the M1_128 gene in M1_SF370 resulted in the loss of M1_128 staining and in the loss of the high-molecular-mass ladder stained by antisera against the products of Cpa and M1_130 (Fig. 2E), indicating that the M1_128 protein is necessary for polymerization of these proteins. This result was confirmed by transformation of the deletion strain with a plasmid containing the gene for M1_128, which resulted in reexpression of the high-molecular-mass ladders recognized by Cpa- and M1_130-specific sera (Fig. 2H). Elimination of the M1_130 gene eliminated staining with anti-130 antiserum but did not eliminate polymerization of M1_128, although the amount of high-molecular-mass material appeared diminished (Fig. 2F). Therefore, the M1_130 protein, although not being essential for polymerization of M1_128, may stabilize the structure or promote more extensive polymerization. Pilus polymerization in C. diphtheriae depends on particular sortase enzyme whose gene resides at the same genetic locus as the pilus components (2, 3). Likewise in GAS, elimination of the srtC1 gene from the FCT of M1_SF370 abolished polymerization of all three proteins and assembly of pili (Fig. 2G). Hence, the pili in GAS resemble those previously described for both C. diphtheria (2, 3) and S. agalactiae (4) in that each pilus is formed by a backbone component that abundantly stains the pili in EM and is essential for the incorporation of the other components. It is not clear, however, why the accessory proteins in GAS do not stain the full length of the pili. One explanation is that these components are preferentially incorporated into shorter forms of the polymeric structures.
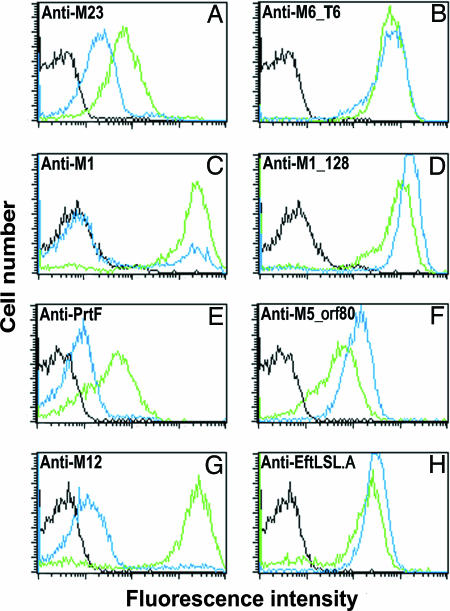
The Major Pili Proteins Are Lancefield T Antigens. The pilus backbone proteins in the four strains (T6, M1_128, M5_orf80, and EftLSL.A) share between 23% and 65% amino acid identity in any pairwise comparison. We hypothesized that each pilus may represent a different Lancefield T antigen. In support of this hypothesis, when bacteria harboring each of the FCT types were treated with trypsin, surface expression of the pilus proteins was barely affected, as assayed by indirect immunofluorescence and flow cytometry (Fig. 4). In contrast, staining with sera specific for the respective M proteins or a surface protein not associated with pili was substantially reduced by trypsin treatment.
Fig. 4.
Pili are trypsin-resistant structures. Flow cytometry of GAS bacteria treated or not with trypsin and stained with sera specific for the major pilus component or a trypsin-sensitive control protein of the respective strains. The histograms obtained by using immune sera on trypsin-treated bacteria are shown in blue, and those obtained by staining untreated bacteria are shown in green. Preimmune staining is shown in black. Shown are strain M6_ISS3650 stained with sera specific for M6 protein (A) or anti-M6_T6 (B), strain M1_SF370 stained with anti-M1 protein (C) or anti-M1_128 (D), strain M5_ISS4883 stained with anti-PrtF (E) or anti-M5_orf80 (F), and strain M12_20010296 with anti-M12 protein (G) or anti-EftLSL.A (H)
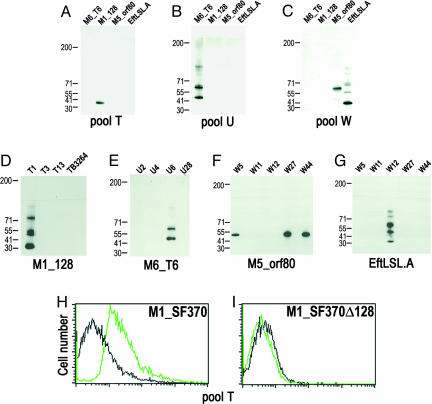
Confirmation that the pili structures do, in fact, represent Lancefield T antigens was obtained by using commercially available T-serotyping sera. First, five polyvalent serum pools (designated T, U, W, X, and Y) were tested for their capacity to recognize recombinant proteins representing the major pilus components of each FCT region. Pool U, which contains the T6 serum, recognized the T6 protein specifically (Fig. 5B), whereas pool T recognized M1_128 specifically (Fig. 5A). In contrast, pool W recognized both M5_orf80 and EftLSL.A (Fig. 5C). Using monovalent sera representative of the components of each polyvalent pool, we confirmed the specificity of the T6 antigen (Fig. 5E) and identified M1_128 as antigen T1 (Fig. 5D), EftLSL.A as antigen T12 (Fig. 5G), and M5_orf80 as a common antigen recognized by the related sera T5, T27, and T44 (Fig. 5F). Furthermore, pool T stained whole bacteria of the M1_SF370 strain (Fig. 5H) but failed to stain the isogenic mutant lacking the M1_128 gene (Fig. 5I). Although Cpa and M1_130 failed to polymerize in the M1_128 knockout strain, they were still detectable on the surface of the bacteria albeit at a reduced level (data not shown). Taken together, these data demonstrate that, in fact, the major pilus component of each strain is a specific Lancefield T antigen.
Fig. 5.
Pilin proteins are Lancefield T antigens. (A–C) Immunoblots of recombinant pilin components with polyvalent Lancefield T-typing sera. The recombinant proteins are shown above the blot, and the sera pool used is shown below the blot. (D–G) Immunoblots of pilin proteins with monovalent T-typing sera. The recombinant proteins are shown below the blot, and the sera used are shown above the blot. (H and I) Flow cytometry analysis of strain M1_SF370 (H) and the deletion strain M1_SF370Δ128 (I) with T-typing antisera pool T.
There are ≈20 known Lancefield T serotypes (19). Here we have demonstrated that the antigens recognized in four of these serotypes are the major components of trypsin-resistant pili whose genes reside in the variable FCT region of the genome. This result raises the question of whether there are sufficient pilus variants to explain all T serotypes. The backbone component of the pili from the four FCT types, although clearly all belonging to the family of ECM binding proteins, show a limited degree of similarity in amino acid sequence (23–65% identity), and these proteins account for four different T serotypes. Analysis of the sequence of the pilus gene in eight GAS strains that share the M3 type of FCT organization revealed three major variants sharing only ≈60% sequence identity. Likewise, two major pilus variants were also identified from the sequence of the pilus gene from four independent strains sharing the M12 type FCT organization. Hence, there are at least seven different variants of the pilus protein, and it is likely that more will be identified as more GAS genomes are sequenced. We cannot, however, exclude that some of the antigens recognized by T-typing sera are not pilus components.
Recombinant Pilin Proteins Are Protective Antigens. Pili are known to be essential virulence factors and protective antigens in Gram-negative bacteria, where they are often involved in adhesion and invasion of eukaryotic cells (19). From the data presented herein, we infer that the pili in GAS serve a similar function. Each of the pilus components is a member of a family of ECM-binding proteins, which have been shown to be involved in adhesion and invasion (17). Furthermore, mutants in the RofA type regulator, which controls the expression of these proteins, has been shown to reduce cell adhesion and virulence in three independent GAS strains (17).
The fact that GAS pili are likely to be involved in these processes, together with the observation that pilus-like structures in S. agalactiae (5) confer protection in a mouse model of maternal immunization (4), encouraged us to test GAS pilus proteins for their capacity to induce protective immunity. To perform such a test, a combination of the three recombinant pilus-forming proteins of M1_SF370 were used to immunize adult CD1 mice, which were subsequently challenged intranasally with an empirically determined dose of a mouse-adapted M1 strain (ISS-3348) calculated to kill ≈90% of mock-immunized mice. The three protein combination conferred >70% of protection in the immunized mice (Table 1). The level of protection achieved by the pilus proteins was similar to that conferred by immunization with M protein, the most promising vaccine candidate to date. Hence, as with S. agalactiae, the components of the pili are protective antigens and may be valuable candidates for inclusion in a vaccine against GAS disease. It is important to note that, although M-protein-based vaccines are in clinical evaluation in man, there are >100 M types, and there is little crossprotection between types. There are, however, only ≈20 T types that have been described; thus, it may be possible to achieve broader strain coverage with fewer antigens with these proteins.
Table 1. Protection conferred by pilus forming proteins of M1_SF370.
| Antigen | Mice tested | Survival, % |
|---|---|---|
| PBS | 40 | 25 |
| M1 protein | 40 | 82.5 |
| Cpa + M1_128 + M1_130 | 38 | 73.6* |
PBS was considered a negative control; the M1 protein was used as positive control. *, P < 0.00002 compared with PBS control.
Concluding Remarks. Here we show that products of genes located in of each of the four variant FCT regions of the GAS genome form long covalently linked pilus structures similar to those described in C. diphtheriae (2, 3) and GBS (5). As in C. diphtheriae and GBS, pili appear to be formed by a sortase-mediated covalent polymerization of proteins containing the LPXTG motif and incorporation of accessory proteins to the pilus backbone. The Lancefield T6 antigen forms the backbone of one of these pili and the backbone of each of the other three pili was recognized by three individual Lancefield T-typing sera. Hence, the data suggest that pilus-like structures contribute in large part to the molecular explanation of the T-typing system described by Lancefield and colleagues (7, 8) at the very outset of studies on pathogenic streptococci.
In Gram-negative bacteria, pili are known to be important virulence factors, often involved in adhesion and invasion of target cells (20). The fact that the pili in GAS are composed of proteins of the family of ECM-binding proteins strongly suggests that they may also be involved in these processes. Irrespective of the precise function of the pili, the components are effective protective antigens in GAS and GBS (4), indicating an importance in virulence in both of these bacteria. A similar genomic island has also been found in Streptococcus pneumoniae. Previous reports have suggested that the S. pneumoniae and GAS islands share a common origin and that these loci might be subject to horizontal transfer (17, 21). The S. pneumoniae island contains genes coding for proteins with amino acid sequence similarity to Cpa, RofA, the transposase in the FCT of SF370, and weak but definite similarity with the T6 protein. Hence, pilus structures may represent important virulence factors and vaccine candidates for all three of the major human streptococcal pathogens.
Supplementary Material
Acknowledgments
We thank J. M. Musser (Baylor College of Medicine, Houston) for GAS strains; G. Orefici (Istituto Superiore di Sanità, Rome) for GAS strains and T-typing sera; V. Pinto, V. Nardi Dei, and M. Mariani for purification of recombinant proteins; A. Neumann and A. Covre for assistance with FACS analysis and animal challenge experiments; G. Matteucci and M. Tortoli for animal care and experimentation; and J. M. Musser for suggestions on an early version of the paper. This work was supported in part by National Institutes of Health Grant 5U1060595-02.
Author contributions: M.M., G.B., A.R.T., G.G., and J.L.T. designed research; M.M., G.B., S.C., F.F., C.Z., A.G.O.M., T.M., and A.T. performed research; G.B., S.C., F.F., C.Z., A.G.O.M., T.M., A.R.T., G.G., and J.L.T. analyzed data; and G.G. and J.L.T. wrote the paper.
Conflict of interest statement: Member Rino Rappuoli is employed by Chiron Corporation.
Abbreviations: GAS, Group A Streptococcus; GBS, Group B Streptococcus; ECM, extracellular matrix; FCT, fibronectin-binding collagen-binding T antigen.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ106872–DQ106882).
References
- 1.Sauer, F. G., Mulvey, M. A., Schilling, J. D., Martinez, J. J. & Hultgren, S. J. (2000) Curr. Opin. Microbiol. 3, 65–72. [DOI] [PubMed] [Google Scholar]
- 2.Ton-That, H. & Schneewind, O. (2004) Trends in Microbiol. 12, 228–234. [DOI] [PubMed] [Google Scholar]
- 3.Ton-That, H., Marraffini, L. A. & Schneewind, O. (2004) Mol. Microbiol. 53, 251–261. [DOI] [PubMed] [Google Scholar]
- 4.Lauer, P., Rinaudo, C. D., Soriani, M., Margarit, I., Maione, D., Rosini, R., Taddei, A. R., Mora, M., Rappuoli, R., Grandi, G., et al. (2005) Science 309, 105-. [DOI] [PubMed] [Google Scholar]
- 5.Maione, D., Margarit, I., Rinaudo, C. D., Masignani, V., Mora, M., Scarselli, M., Tettelin, H., Brettoni, C., Iacobini, E. T., Rosini, R., et al. (2005) Science 309, 148–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham, M. W. (2000) Clin. Microbiol. Rev. 13, 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lancefield, R. C. (1928) J. Exp. Med. 47, 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lancefield, R. C. & Dole, V. P. (1946) J. Exp. Med. 84, 449–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneewind, O., Jones, K. F. & Fischetti, V. A. (1990) J. Bacteriol. 172, 3310–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montigiani, S., Falugi, F., Scarselli, M., Finco, O., Petracca, R., Galli, G., Mariani, M., Manetti, R., Agnusdei, M., Cevenini, R., et al. (2002) Infect. Immun. 70, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, D. R., Kaplan, E. L., Sramek, J., Bicova, R., Havlicek, J., Havlickova, H., Motlova, J. & Kriz, P. (1996) Laboratory Diagnosis of Group A Streptococcal Infections (W.H.O., Geneva).
- 12.Horton, R. M., Cai, Z. L., Ho, S. N. & Pease, L. R. (1990) BioTechniques 8, 528–535.2357375 [Google Scholar]
- 13.Biswas, I., Gruss, A., Ehrlich, S. D. & Maguin, E. (1993) J. Bacteriol. 175, 3628–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caparon, M. G. & Scott, J. R. (1991) Methods Enzymol. 204, 556–586. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Casal, J., Price, J. A., Maguin, E. & Scott, J. R. (1993) Mol. Microbiol. 8, 809–819. [DOI] [PubMed] [Google Scholar]
- 16.Framson, P. E., Nittayajarn, A., Merry, J., Youngman, P. & Rubens, C. E. (1997) Appl. Environ. Microbiol. 63, 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bessen, D. E. & Kalia, A. (2002) Infect. Immun. 70, 1159–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreikemeyer, B., Klenk, M. & Podbielski, A. (2004) Int. J. Med. Microbiol. 294, 177–188. [DOI] [PubMed] [Google Scholar]
- 19.Jones, K. F., Schneewind, O., Koomey, J. M. & Fischetti, V. A. (1991) Mol. Microbiol. 5, 2947–2952. [DOI] [PubMed] [Google Scholar]
- 20.Merz, A. J. & So, M. (2000) Annu. Rev. Cell Dev. Biol. 16, 423–457. [DOI] [PubMed] [Google Scholar]
- 21.Hava, D. L. & Camilli, A. (2002) Mol. Microbiol. 45, 1389–1406. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.