Abstract
We are using biochemical and genetic approaches to study Rtf1 and the Spt4–Spt5 complex, which independently have been implicated in transcription elongation by RNA polymerase II. Here, we report a remarkable convergence of these studies. First, we purified Rtf1 and its associated yeast proteins. Combining this approach with genetic analysis, we show that Rtf1 and Leo1, a protein of unknown function, are members of the RNA polymerase II-associated Paf1 complex. Further analysis revealed allele-specific genetic interactions between Paf1 complex members, Spt4–Spt5, and Spt16–Pob3, the yeast counterpart of the human elongation factor FACT. In addition, we independently isolated paf1 and leo1 mutations in an unbiased genetic screen for suppressors of a cold-sensitive spt5 mutation. These genetic interactions are supported by physical interactions between the Paf1 complex, Spt4–Spt5 and Spt16–Pob3. Finally, we found that defects in the Paf1 complex cause sensitivity to 6-azauracil and diminished PUR5 induction, properties frequently associated with impaired transcription elongation. Taken together, these data suggest that the Paf1 complex functions during the elongation phase of transcription in conjunction with Spt4–Spt5 and Spt16–Pob3.
Keywords: Paf1 complex/Rtf1/Spt4–Spt5/Spt16–Pob3/transcription elongation
Introduction
Synthesis of an RNA transcript by RNA polymerase II (RNA pol II) requires the successful completion of at least four steps in the transcription cycle: promoter binding and initiation; promoter clearance; elongation; and termination. Transcription factors that modulate these steps often interact directly with the polymerase, and several RNA pol II-associated regulatory complexes have been characterized. These include the Srb-mediator complex that functions as a transcriptional coactivator (Hampsey and Reinberg, 1999) and the Paf1 complex, which has been proposed to act as an alternative to the Srb-mediator (Shi et al., 1997). While many studies have focused on the regulation of initiation, more recent studies have demonstrated that transcription elongation is also controlled by a multitude of transcription factors, many of which bind to the polymerase. For example, DSIF, the human counterpart of the yeast Spt4–Spt5 complex, binds to RNA pol II and regulates transcription elongation (Wada et al., 1998a,b).
SPT4, SPT5 and the related gene SPT6 were identified originally by a genetic selection for factors that affect transcription in yeast (Winston et al., 1984). Biochemical and genetic experiments using yeast and human cells suggest that Spt4–Spt5/DSIF and Spt6 may have both positive and negative roles in transcription elongation (Hartzog et al., 1998; Wada et al., 1998b; Wu-Baer et al., 1998; Ivanov et al., 2000). Studies involving Drosophila polytene chromosomes have demonstrated that the Drosophila homologs of Spt5 and Spt6 co-localize with the hyperphosphorylated, elongating form of RNA pol II at many actively transcribed genes, indicating a broad involvement of these factors in transcription elongation in vivo (Andrulis et al., 2000; Kaplan et al., 2000).
SPT16/CDC68 was identified by the same genetic selection used to find SPT4, SPT5 and SPT6 (Malone et al., 1991). Along with Pob3, Spt16 is a subunit of the yeast CP or SP complex (Brewster et al., 2001; Formosa et al., 2001). The human counterpart of CP/SP, FACT, was purified as a factor that promotes elongation by RNA pol II through chromatin in vitro (Orphanides et al., 1998, 1999). Genetic and biochemical studies suggest that, similarly to Spt4–Spt5, CP/SP/FACT can both promote and repress transcription (Malone et al., 1991; Rowley et al., 1991; Brewster et al., 1998; Evans et al., 1998; Wada et al., 2000).
The differential effects of these proteins on elongation may relate to the chromatin state of the template. FACT promotes elongation through nucleosomes, and both FACT and the yeast CP/SP complex (in association with Nhp6) can bind nucleosomes (Orphanides et al., 1998, 1999; Formosa et al., 2001). Like mutations in the histone-encoding genes HTA1-HTB1 (Clark-Adams et al., 1988), spt4, spt5, spt6 and spt16 mutations can suppress the loss of the Swi–Snf chromatin remodeling complex (Malone et al., 1991; Swanson and Winston, 1992; Evans et al., 1998). In addition, overexpression of SPT5, SPT6 or SPT16 confers mutant phenotypes similar to those caused by overexpression of HTA1-HTB1 (Clark-Adams and Winston, 1987; Clark-Adams et al., 1988; Malone et al., 1991; Swanson et al., 1991), and Spt6 exhibits nucleosome assembly activity in vitro (Bortvin and Winston, 1996).
Genetic studies in yeast have revealed additional factors with potential roles in transcription elongation. The yeast RTF1 gene exhibits several attributes of a gene involved in this process. For example, mutations in RTF1 cause sensitivity to the base analog 6-azauracil (6AU) and a range of synthetic phenotypes when combined with mutations in known elongation factor genes, including SPT4, SPT5 and DST1, which encodes TFIIS (Costa and Arndt, 2000). Moreover, a genetic screen for mutations that cause lethality in combination with an rtf1Δ mutation identified three genes involved in transcription elongation, CTK1, FCP1 and POB3 (Costa and Arndt, 2000). CTK1 encodes the catalytic subunit of the CTDK-I kinase that phosphorylates the C-terminal domain (CTD) of RNA pol II (Lee and Greenleaf, 1991). The transition from the initiation to the elongation stage of the transcription cycle correlates closely with phosphorylation of the CTD (Prelich, 2002). Four different CTD kinases have been described in yeast (Prelich, 2002). Of these, CTDK-I may be especially important for transcription elongation, since it promotes efficient elongation by RNA pol II in vitro (Lee and Greenleaf, 1997), and ctk1 mutations interact genetically with mutations in genes encoding known elongation factors (Jona et al., 2001; Lindstrom and Hartzog, 2001). Fcp1, the only known CTD phosphatase in yeast, interacts with the general initiation and elongation factor TFIIF (Archambault et al., 1997), and possesses a positive elongation function independent of its phosphatase activity (Cho et al., 1999). Moreover, in a very recent study, Fcp1 and Ctk1 were found to localize to the coding regions of transcribed genes and to affect the state of CTD phosphorylation associated with elongating RNA pol II (Cho et al., 2001).
To elucidate the function of Rtf1 in transcription, we have affinity-purified Rtf1 and its associated proteins. Identification of these proteins revealed that Rtf1 and Leo1, a protein about which little is known, are intimately associated with the Paf1 complex. The Paf1 complex was identified originally through its physical association with RNA pol II (Wade et al., 1996); however, the function of this complex in transcription is not known. Together with our previous studies on RTF1 (Costa and Arndt, 2000), our current findings suggest that the Paf1 complex plays a role in transcription elongation. This hypothesis is strongly supported by our observations that RNA pol II, Spt5 and Spt16–Pob3 specifically co-purify with the Paf1 complex, that defects in the Paf1 complex cause sensitivity to 6AU and impaired induction of PUR5 transcription, and that members of the Paf1 complex exhibit extensive genetic interactions with SPT4, SPT5 and SPT16. Collectively, our findings indicate that the Paf1 complex collaborates with a set of Spt proteins to regulate transcription elongation.
Results
Rtf1 and Leo1 are members of the Paf1 complex
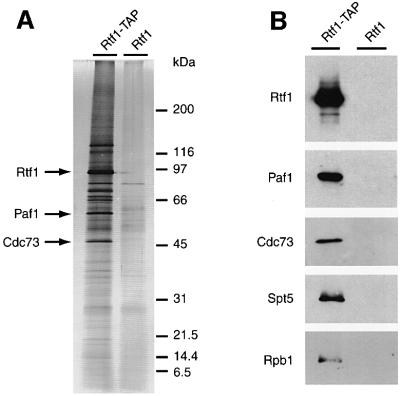
To identify proteins that associate with Rtf1, we used the tandem affinity purification (TAP) method. The TAP fusion cassette encodes the calmodulin-binding peptide, a cleavage recognition site for the TEV protease, and the two IgG-binding units of Staphylococcus aureus protein A (Rigaut et al., 1999). This tripartite tag allows protein complex purification by a two-step affinity chromatography procedure under native conditions (see Materials and methods). Using extracts of a yeast strain carrying a functional RTF1–TAP tag fusion gene, we affinity purified an ∼97 kDa protein corresponding to Rtf1–TAP (Figure 1). When a control strain expressing untagged Rtf1 was processed in parallel, no Rtf1 was observed in the final column eluate. Importantly, silver staining revealed several polypeptides that specifically co-purified with Rtf1–TAP (Figure 1A).
Fig. 1. Rtf1–TAP co-purifies with Paf1, Cdc73, Ctr9, Leo1, Spt5 and Rpb1. Yeast strains expressing untagged and TAP-tagged forms of Rtf1 were processed in parallel as described in Materials and methods. (A) Silver-stained gel of purified proteins derived from ∼150 mg of protein extract. Identification of bands as Rtf1, Paf1 and Cdc73 was based upon immunoblot analyses carried out in parallel. Similar assignments were not made for Ctr9 and Leo1 (predicted masses of ∼125 and 54 kDa, respectively) because antibodies to these proteins were not available and because individual bands were not excised for our mass spectrometry analysis. (B) Immunoblots of purified proteins, visualized with antisera directed against Rtf1, Paf1, Cdc73 and Spt5. The monoclonal antibody 8WG16 was used to probe for Rpb1.
We performed mass spectrometry analysis on Rtf1–TAP preparations to identify the proteins that co-purified with Rtf1 (see Materials and methods). The technique reproducibly identified four Rtf1-associated proteins, Paf1, Cdc73, Ctr9 and Leo1 (Table I). Bands with the expected mobilities for each of these proteins were observed on the silver-stained gel (Figure 1A), and the co-purification of Paf1 and Cdc73 with Rtf1 was confirmed by immunoblot analysis (Figure 1B).
Table I. Mass spectrometric analysis of Rtf1–TAP purified proteins.
| Protein name | No. of peptides identified |
||
|---|---|---|---|
| Experiment 1a | Experiment 2a | Experiment 3 | |
| Rtf1 | 4 | 5 | 13 |
| Paf1 | 1 | 3 | 1 |
| Cdc73 | 2 | 4 | |
| Ctr9 | 2 | 2 | 2 |
| Leo1 | 4 | 4 | 2 |
aExperiments 1 and 2 also identified ribosomal subunits and the translation elongation factors Tef1/2. In experiment 3, these abundant proteins were removed by an ultracentrifugation step (see Materials and methods).
Several of these Rtf1-associated proteins have been studied previously. Notably, Jaehning and co-workers partially purified a novel RNA pol II-associated protein complex that contained Paf1 and Cdc73 (Shi et al., 1996, 1997; Chang et al., 1999), and Koch et al. (1999) identified an ∼670 kDa protein complex that contained Paf1, Cdc73 and Ctr9. In contrast to the other Rtf1-interacting proteins, very little is known about Leo1 (left open reading frame; Magdolen et al., 1994). The predicted Leo1 protein has a mass of 53.7 kDa and contains 43.8% charged amino acids, with short stretches of consecutive aspartate and glutamate residues located in two acidic domains. None of the genes encoding these five proteins is essential, although all have potential metazoan sequence homologs (Koch et al., 1999; data not shown).
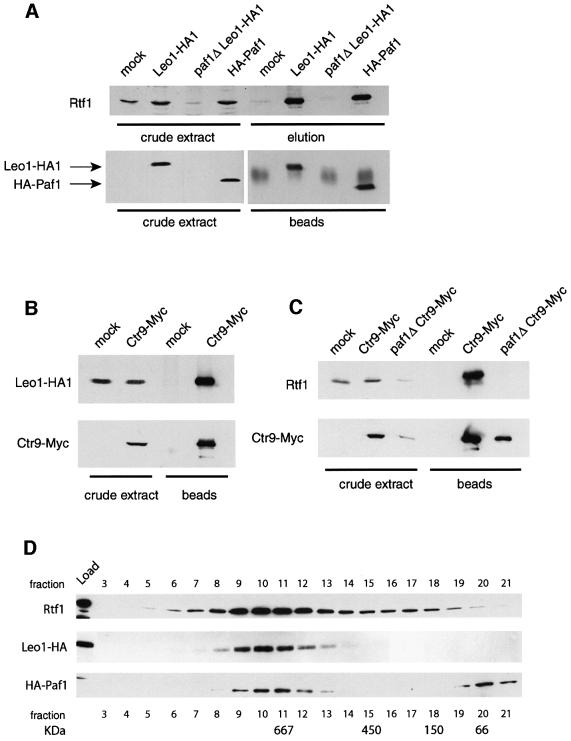
We used functional, epitope-tagged derivatives of several of these proteins and immunoprecipitated them to determine if Rtf1 and Leo1 are members of the Paf1 complex. Proteins that co-precipitated with the bound antigen were eluted from the beads by incubation in 1 M KCl (IP elution), separated by SDS–PAGE and analyzed by immunoblotting. In some cases, the bound antigen and other tightly associated proteins were released from the beads by boiling in loading buffer (beads), and then analyzed as above. As shown in Figure 2A, Rtf1 co-immunoprecipitated with HA1 epitope-tagged derivatives of Leo1 and Paf1, and failed to precipitate from untagged control extracts (mock). In addition, Leo1-HA1 and Rtf1 co-precipitated with a Myc-tagged derivative of Ctr9 (Figure 2B and C). As previously observed, Ctr9 levels were reduced in a paf1Δ mutant (Koch et al., 1999; Figure 2C). We also found that Rtf1 and Leo1 levels are greatly reduced in a paf1Δ mutant (Figure 2A and C). This suggests that these proteins may only be stable when assembled into a complex with Paf1. When we size fractionated yeast extracts on a Superose 6 column, we observed that Rtf1, Leo1 and Paf1 co-eluted in a peak at ∼670 kDa, similar to that observed previously for Paf1, Ctr9 and Cdc73 (Figure 2D; Koch et al., 1999). Thus, we conclude that Rtf1 and Leo1 associate with each other and with Paf1, Ctr9 and Cdc73.
Fig. 2. Rtf1 and Leo1 are members of the Paf1 complex. (A) Rtf1 co-immunoprecipitates with Leo1-HA1 and HA1-Paf1. Anti-HA1 immunoprecipitations were performed from extracts of strains lacking an HA1-tagged protein (mock, GHY1102), containing either HA1-tagged Paf1 (GHY1147) or Leo1 (GHY1141), and from a LEO1-HA1 paf1Δ strain (GHY1146). Proteins that precipitated with the HA1 antibody-bound beads were eluted with 1 M KCl (elution) and separated by SDS–PAGE. Proteins that remained bound to the beads after the 1 M KCl elution were liberated by boiling in sample buffer (beads). The HA1-tagged proteins and Rtf1 were visualized by immunoblotting. (B) Leo1-HA1 co-immunoprecipitates with Ctr9-Myc. Immunoprecipitations were performed on extracts of a Leo1-HA1 (mock, GHY1092) and a Ctr9-Myc Leo1-HA1 strain (GHY1184). (C) Rtf1 co- immunoprecipitates with Ctr9-Myc. Immunoprecipitations were performed on extracts of a strain lacking a myc tag (mock, GHY1002), a Ctr9-Myc strain (GHY1184) and a paf1Δ Ctr9-Myc strain (GHY1222). (D) Paf1, Leo1 and Rtf1 co-fractionate on a gel filtration column. Whole-cell extracts derived from a Leo1-HA1 strain (GHY1140) and a HA1-Paf1 strain (GHY1147) were fractionated on a Superose 6 column, and the indicated fractions from the eluates were analyzed by immunoblotting. Each experiment was performed at least twice, with peak elution of each protein reproducibly occurring in fraction 10.
Because of the previously reported association between RNA pol II and the Paf1 complex (Shi et al., 1996, 1997; Chang et al., 1999), we probed immunoblots of TAP-purified Rtf1 with an antibody that recognizes Rpb1, the largest subunit of RNA pol II, and found that a fraction of the cellular polymerase also co-purified with Rtf1–TAP (Figure 1B). However, examination of the relative intensities of protein bands corresponding to Rpb1 (predicted mass of ∼190 kDa) and members of the Paf1 complex, visualized either by immunoblotting or by silver staining (Figure 1), suggests that RNA pol II is associated with only a subset of the Paf1 complexes in the cell. Alternatively, the interaction between RNA pol II and the Paf1 complex may not be stable under our fractionation conditions.
Rtf1 and Leo1 are important for Paf1 complex function
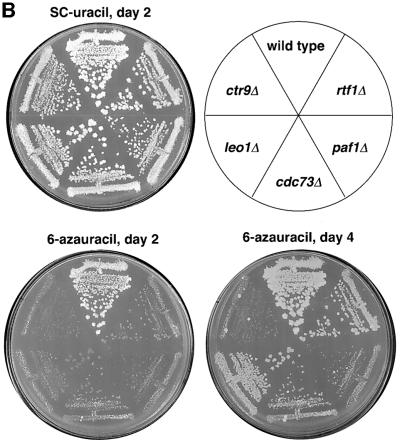
The finding that Rtf1 and Leo1 are members of the Paf1 complex indicates that they may contribute to its function. If so, we would expect leo1 and rtf1 mutant strains to share phenotypes with paf1, cdc73 and ctr9 mutant strains. Deletion mutations of RTF1 or LEO1 only caused mild growth defects, in contrast to the moderate growth defect observed for cdc73Δ cells and the strong growth defect observed for paf1Δ and ctr9Δ strains (Shi et al., 1996, 1997; Koch et al., 1999; data not shown). However, mutations in these five genes do confer several common phenotypes. For example, we previously found that rtf1 null mutations confer an Spt– phenotype (Stolinski et al., 1997). We therefore assayed paf1, cdc73, ctr9 and leo1 null mutations and found that they also confer Spt– phenotypes of varying strengths (Figure 3A). We also observed several other mutant phenotypes characteristic of defects in transcription in these strains. The paf1Δ and ctr9Δ mutations exhibited strong Ino– phenotypes, and leo1Δ and rtf1Δ strains are weak inositol auxotrophs (data not shown). Finally, similarly to an rtf1Δ mutation, paf1, cdc73, ctr9 and leo1 deletion mutations show strong synthetic interactions with fcp1 and ctk1 mutations, including lethality (Table II). Thus, as expected for genes that contribute to a common function, mutation of any of these five genes leads to an overlapping set of mutant phenotypes.
Fig. 3. Mutant phenotypes shared by Paf1 complex members. (A) Strains with the indicated genotypes and containing the his4-912δ and lys2-128δ insertions were grown on YPD media, replica-plated to minimal media supplemented with histidine, lysine, leucine, uracil and tryptophan (complete), and SC-his and SC-lys plates, and incubated for 3 days at 30°C. Strains: WT, FY118; rtf1Δ, KY522; paf1Δ, GHY917; cdc73Δ, GHY1083; leo1Δ, GHY240; ctr9Δ, GHY1157. (B) Strains with the indicated genotypes were grown on YPD media and then replica-plated to SC-ura media lacking or containing 50 µg/ml 6AU. Photographs were taken after 2 and 4 days of growth at 30°C. Strains: wild-type, KY286; rtf1Δ, KY425; paf1Δ, KY686; cdc73Δ, KY689; leo1Δ, KY690; ctr9Δ, KY695.
Table II. Genetic interactions between CTK1, FCP1 and members of the Paf1 complex.
| Genotype | Synthetic phenotypesa |
|---|---|
| rtf1Δ ctk1-217 | dead |
| rtf1Δ fcp1-110 | microcolonies |
| paf1Δ ctk1-217 | dead |
| paf1Δ fcp1-110 | dead or microcolonies |
| cdc73Δ ctk1-217 | slow growth, Ts–, Cs–, strongly sensitive to 0.3 M LiCl and 1.4 M NaCl |
| cdc73Δ fcp1-110 | slow growth, CafS, Ts–, Ino– |
| leo1Δ ctk1-217 | slow growth, Cs–, strongly sensitive to 1.4 M NaCl |
| leo1Δ fcp1-110 | Ino–, strongly sensitive to 1.4 M NaCl |
| ctr9Δ ctk1-217 | very slow growth, Ts– |
| ctr9Δ fcp1-110 | very slow growth, Cs–, HUS, Gal– |
aTs–, temperature sensitivity for growth at 37°C; Cs–, cold sensitivity for growth at 15°C; CafS, inability to grow on media containing 15 mM caffeine; Ino–, inositol auxotrophy; HUS, sensitivity to 100 mM hydroxyurea; Gal–, inability to use galactose as the sole carbon source. The rtf1Δ results were reported previously in Costa and Arndt (2000) and are presented here for completeness.
If Rtf1 and Leo1 are required for the function of the Paf1 complex, then double mutant combinations of rtf1Δ or leo1Δ with deletions of other Paf1 complex genes may not lead to any new mutant phenotypes. Consistent with this hypothesis, we did not observe any dramatic new mutant phenotypes when an rtf1Δ or leo1Δ mutation was combined with deletion mutations in PAF1, CDC73 or CTR9 (data not shown). However, the rtf1Δ leo1Δ double mutant did exhibit a strong Ino– phenotype, and the rtf1Δ cdc73Δ double mutant showed an enhanced Spt– phenotype with the lys2-128δ insertion allele (data not shown). Importantly, the observation that no double mutants exhibited severe synthetic phenotypes is consistent with the hypothesis that Rtf1 and Leo1 function as members of the Paf1 complex and that once one member of the complex is deleted, loss of other complex members causes no additional decrement in function. Similar conclusions were drawn from the analysis of double mutant combinations involving paf1Δ, cdc73Δ and ctr9Δ (Shi et al., 1997; Koch et al., 1999). In summary, the genetic and biochemical data presented above are most consistent with the idea that Rtf1 and Leo1 both associate with and are required for the normal function of the Paf1 complex.
In addition to the phenotypes described above, we also observed that, similarly to RTF1 (Costa and Arndt, 2000), deletions of PAF1, CDC73, CTR9 and LEO1 all cause varying degrees of sensitivity to 6AU, with paf1Δ and ctr9Δ strains exhibiting the most severe phenotypes (Figure 3B). Sensitivity to 6AU is often but not always indicative of a transcription elongation defect (Wind and Reines, 2000). Reines and colleagues have demonstrated that PUR5, which encodes inosine monophosphate dehydrogenase, is transcriptionally induced by exposure of yeast cells to 6AU (Shaw and Reines, 2000). Mutations that cause transcription elongation defects and 6AU sensitivity also prevent the induction of PUR5 transcription in response to 6AU treatment. In contrast, mutations that cause 6AU sensitivity but do not affect transcription elongation do not interfere with PUR5 induction. Thus, a 6AU-sensitive phenotype coupled with the inability to induce PUR5 in response to 6AU is suggestive of a defect in transcription elongation. We therefore isolated RNA from wild-type, rtf1Δ and paf1Δ cells before and after 6AU treatment and performed a northern analysis of PUR5 mRNA levels (Figure 4). This analysis showed that deletion of PAF1 or RTF1 significantly reduces PUR5 induction, and therefore suggests that the Paf1 complex may play a role in transcription elongation.
Fig. 4. rtf1Δ and paf1Δ strains are defective in PUR5 induction. Northern analysis of PUR5 transcription was performed on total RNA samples prepared from wild-type (KY589), rtf1Δ (KY453) and paf1Δ (KY687) cells. The strains were grown to ∼1 × 107 cells/ml in SC-ura media and then divided. 6AU was added to a final concentration of 75 µg/ml to one half of the culture and both cultures were grown at 30°C for the indicated times, in hours, prior to RNA isolation. Each lane contained 10 µg of total RNA. The same filters that were probed for PUR5 mRNA were stripped and probed for SED1 mRNA to serve as a loading control.
The Paf1 complex interacts with transcription elongation factors Spt5 and Spt16–Pob3
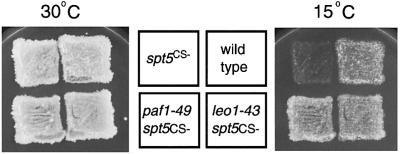
Previously, we sought to identify proteins that interact with the transcription elongation factor Spt5 by selecting for genetic suppressors of a cold-sensitive spt5 mutation (Hartzog et al., 1998). In our initial analysis, we isolated suppressor mutations in RPB1, which encodes the largest subunit of RNA pol II. We have identified two additional suppressors from this selection, PAF1 and LEO1 (Figure 5, see Materials and methods). The paf1 and leo1 suppressor mutations are recessive, and deletions of PAF1 and LEO1 also suppress the spt5Cs– mutation (Table III). We performed genetic crosses to obtain double mutants of spt5Cs– with null alleles of genes encoding other members of the Paf1 complex. As summarized in Table III, a ctr9 mutation was able to suppress spt5Cs–, but rtf1 and cdc73 mutations did not. We extended this analysis by crossing strains carrying either spt5-194, a different allele of spt5, or an spt4 null mutation to strains defective for members of the Paf1 complex (Table III). We observed a variety of severe synthetic growth defects in the double mutant strains, ranging from temperature sensitivity to inviability. Thus, SPT4 and SPT5 exhibit extensive allele-specific genetic interactions with genes encoding the Paf1 complex, consistent with the model suggesting that the Paf1 complex plays a role in elongation.
Fig. 5. Suppression of the spt5Cs– growth defect by paf1 and leo1 mutations. Strains with the indicated genotypes were replica-plated to YPD media and incubated for 2 (30°C) or 4 (15°C) days. Strains: spt5Cs–, GHY92; wild type, FY653; paf1-49 spt5Cs–, GHY144; leo1-43 spt5Cs–, GHY141.
Table III. Genetic interactions between SPT and Paf1 complex genesa.
| spt5Cs– | spt5-194 | spt4Δ | spt16-197 | |
|---|---|---|---|---|
| paf1Δ | Cs+/– | dead | dead | very poor growth |
| cdc73Δ | slow growth | Ts–, slow growth | dead | ND |
| ctr9Δ | Cs+/– | dead | dead | very poor growth |
| leo1Δ | Cs+ | Ts– | dead | no effect |
| rtf1Δ | Cs– | Ts– | very poor growth, Ts–b | no effectb |
aOnly synthetic mutant phenotypes are indicated.
bOriginally described in Costa and Arndt (2000).
Slow growth = small colonies after 3 days at 30°C; very poor growth = small colonies after 5 days at 30°C; Ts– = no growth at 37°C; Cs–, Cs+/–, Cs+ = no, nearly normal and wild-type growth at 15°C. ND = not done.
We next asked if Spt5 physically associates with the Paf1 complex. We probed immunoblots of the Rtf1–TAP fractions and observed that Spt5 had co-purified specifically with Rtf1 (Figure 1B). To confirm the interaction of Spt5 with the Paf1 complex, we performed immunoprecipitation experiments with epitope-tagged forms of Paf1, Leo1 and Ctr9, and assayed for co-precipitation of Spt5. We found that Spt5 co-precipitates with tagged derivatives of Paf1 (Figure 6A), Leo1 (Figure 6A), Ctr9 (Figure 6B) and Cdc73 (data not shown). We also found that Spt5 specifically co-immunoprecipitates with Rtf1, in agreement with the Rtf1–TAP affinity purification results (Figure 6C). In contrast, TBP did not co-precipitate with Rtf1, providing additional evidence that the Rtf1–Spt5 interaction is specific (Figure 6C). Finally, the interaction between Spt5 and the Paf1 complex, detected through co-immunoprecipitation with Ctr9, is abolished in the absence of Paf1 (Figure 6B). Thus, the Paf1 complex interacts with Spt5 both physically and genetically.
Fig. 6. Transcription elongation factors associate with the Paf1 complex. (A) Spt5, Spt16 and Pob3 co-immunoprecipitate with HA1-Paf1 and Leo1-HA1. Anti-HA1 immunoprecipitations were performed from extracts of a strain lacking an HA1 epitope tag (mock, GHY1102), strains containing Leo1-HA1 (GHY1184), HA1-Paf1 (GHY1147) and from a paf1Δ Leo1-HA1 strain (GHY1146). The immunoprecipitates were analyzed by immunoblotting for Spt5, Spt16 and Pob3 as indicated. (B) Spt5 co-immunoprecipitates with Ctr9. Anti-Myc immunoprecipitations were performed on extracts of a strain lacking a Myc tag (mock, GHY1002), a Ctr9-Myc strain (GHY1184) and a paf1Δ Ctr9-Myc strain (GHY1222). Bound proteins were eluted from the anti-Myc beads by a 1 M salt elution (elution) and then by boiling the beads in sample buffer (beads). The immunoprecipitates were analyzed by immunoblotting for Spt5 and Ctr9, with an anti-Myc antibody, as indicated. (C) Whole-cell extracts were prepared from yeast strain FY1639 and used for immunoprecipitations of Rtf1. Western blots were probed with anti-Spt5, anti-Rtf1 and anti-TBP antisera. A 20 µg aliquot of whole-cell extract, 7 µg of unbound protein and three-fifths of the immunoprecipitated sample (beads) were analyzed by immunoblotting with the indicated antibodies. (D) Growth defects in paf1Δ dst1Δ and paf1Δ spt16-197 double mutants. Serial dilutions of cells with the indicated genotypes were spotted onto YPD media and grown for 2 days at 30°C. Strains: wild-type, FY653; paf1Δ, GHY917; dst1Δ, GHY285; spt16-197, GHY1059; paf1Δ dst1Δ, GHY1009; paf1Δ spt16-197, GHY1228.
In a recent experiment to isolate Spt5 and its associated proteins, we immunopurified a FLAG-tagged derivative of Spt5 from yeast extracts and found that Spt5 interacts with several protein complexes (D.Lindstrom and G.Hartzog, in preparation). One of these was the yeast CP/SP complex, composed of Spt16 and Pob3 (Brewster et al., 2001; Formosa et al., 2001). Furthermore, we previously identified a pob3 mutation in a screen for mutations that are synthetically lethal with rtf1Δ (Costa and Arndt, 2000). We therefore probed blots of Leo1-HA1 and HA1-Paf1 immunoprecipitates for Spt16 and Pob3 and found that Spt16–Pob3 co-precipitates with the Paf1 complex (Figure 6A). When we crossed strains containing the spt16-197 mutation to paf1Δ, ctr9Δ and leo1Δ strains, we found that the spt16 leo1 double mutants, like spt16 rtf1 double mutants (Costa and Arndt, 2000), did not exhibit any new mutant phenotypes. However, the spt16 paf1 and spt16 ctr9 double mutants exhibited severe growth defects (Figure 6D and Table III). Thus, like Spt5, the Spt16–Pob3 complex interacts biochemically and genetically with the Paf1 complex. Finally, we also found that deletion of DST1, the gene for the general elongation factor TFIIS, also causes a strong synthetic growth defect when combined with a paf1Δ mutation. Thus, the Paf1 complex shows a broad genetic dependence upon the RNA pol II elongation apparatus.
Discussion
Identification of new members of the Paf1 complex
By purifying TAP-tagged Rtf1, we found that it associates with Leo1, a non-essential highly charged protein, as well as the previously known members of the Paf1 complex, Paf1, Cdc73 and Ctr9. These protein–protein interactions were confirmed by co-immunoprecipitation. Gel filtration chromatography under conditions that included moderate detergent and salt concentrations (Figure 2) demonstrated that these proteins are assembled into either a single complex or several complexes of similar size. As pre viously observed for Ctr9 (Koch et al., 1999), Leo1 and Rtf1 protein levels declined in a paf1Δ mutant (Figure 2), suggesting that these proteins are stable only when assembled into a complex with Paf1. Consistent with the idea that Rtf1 is an integral member of the Paf1 complex, when paf1Δ extracts were fractionated by gel filtration, the residual Rtf1 protein was found to behave as a monomeric protein rather than as a member of a large protein complex (S.L.Squazzo and G.A.Hartzog, unpublished data). Double mutant analysis showed a lack of strong genetic interactions among the genes encoding the members of the Paf1 complex (data not shown; Shi et al., 1997; Koch et al., 1999). This observation is consistent with the idea that these proteins function only in the context of an intact complex. Thus, although we have not purified the Paf1 complex to homogeneity, these data are most consistent with the model that the Paf1 complex is minimally composed of Paf1, Ctr9, Cdc73, Rtf1 and Leo1. This conclusion agrees with two recent studies in which a similar protein complex was identified by purification of TAP-tagged proteins (Gavin et al., 2002; Mueller and Jaehning, 2002).
Does the Paf1 complex contain other members? Using immunoblot analysis, we have demonstrated specific association of the Paf1 complex with Spt5, Pob3, Spt16 and Rpb1. None of these proteins were observed by the mass spectrometry analysis of the Rtf1–TAP fraction, which suggests that they were below the limits of our detection and may be present in amounts substoichiometric to the Paf1 complex. Although we have found previously that all five members of the Paf1 complex co-purify with a FLAG-tagged derivative of Spt5, we were unable to demonstrate a specific Spt5–Paf1 complex interaction in that analysis, as the members of the Paf1 complex were non-specifically retained on anti-FLAG columns (G.A.Hartzog, D.L.Lindstrom, J.Yates and N.Muster, unpublished data). This result was probably due to the presence of a degenerate version of the FLAG epitope in the Rtf1 protein sequence (amino acids 196–202). However, as demonstrated in Figure 6, when using antisera other than anti-FLAG, Spt5 can be co-immunoprecipitated specifically with the Paf1 complex.
Although the Paf1 complex previously was reported to associate with RNA pol II, Hpr1, Ccr4, TFIIB and TFIIF (Shi et al., 1997; Chang et al., 1999), we did not detect these proteins by mass spectrometry. Consistent with the findings of Koch et al. (1999), we found that the Paf1 complex behaved as an ∼670 kDa complex on a gel filtration column. This mass is too small to accommodate all of these proteins in a single complex unless it has an unusual shape that causes it to behave anomalously in a gel filtration column. Furthermore, the elution profiles of Spt5, Pob3 and Spt16 from a gel filtration column overlap with, but are not identical to that observed for the Paf1 complex (S.L.Squazzo, D.L.Lindstrom and G.A.Hartzog, unpublished observations). Thus, we favor the idea that the Paf1 complex transiently or weakly associates with these other proteins. Consistent with these conclusions, Mueller and Jaehning recently found that under low salt conditions, the Paf1 complex fractionates as a 1.7 MDa complex (Mueller and Jaehning, 2002). Elucidation of the potential interactions of the Paf1 complex with Hpr1, Ccr4, TFIIB or TFIIF will require further analysis.
Recently, the five members of the Paf1 complex, along with many other proteins, were affinity purified with the proteasome (Verma et al., 2000). We did not detect proteasome subunits in our Rtf1–TAP preparations by mass spectrometry and we have not attempted to address the issue of Paf1 complex–proteasome interactions directly. However, it is interesting to note that the 19S subunit of the proteasome has been implicated in transcription elongation and interacts physically and genetically with Spt16 (Ferdous et al., 2001).
Evidence for distinct functions of Paf1 complex subunits
It is possible that particular members of the Paf1 complex may carry out distinct or specialized functions. Deletions of the genes encoding members of the Paf1 complex lead to growth defects ranging from mild (RTF1 and LEO1), to moderate (CDC73), to severe (PAF1 and CTR9) on both YPD and 6AU media (Figure 3B; data not shown; Shi et al., 1996, 1997; Koch et al., 1999). Furthermore, muta–tions in these five genes cause variable Spt– phenotypes (Figure 3A) and genetic interactions of differing strength with genes encoding elongation factors (Tables II and III). Alternatively, the strength of these phenotypes and interactions may reflect differential effects on the stability of the remaining members of the Paf1 complex. Thus, further genetic analyses of the structure and function of the Paf1 complex are required to understand how its different subunits contribute to its overall function.
Connections between the Paf1 complex and transcription elongation
Although the Paf1 complex associates with RNA pol II (Figure 1D and Shi et al., 1996, 1997; Wade et al., 1996; Chang et al., 1999), its in vivo function is not known. Analysis of paf1, cdc73 and ctr9 mutants shows pleiotropic effects on the levels of many different mRNAs, and a paf1Δ mutation is lethal when combined with srb5Δ (Shi et al., 1996, 1997; Chang et al., 1999; Koch et al., 1999). Previously, these data were interpreted as indicating a role for the Paf1–RNA pol II complex in initiation, perhaps as an alternative form of the RNA pol II holoenzyme that responds to particular regulatory pathways (Shi et al., 1996, 1997; Wade et al., 1996; Chang et al., 1999). However, these observations could also be explained by an effect of the Paf1 complex on transcription elongation. For instance, biochemical experiments have demonstrated that Srb5 facilitates a step in transcription, possibly CTD phosphorylation, that follows activator-mediated recruitment of the holoenzyme (Lee et al., 1999). Consistent with models for specialization within the Srb-mediator (Lee et al., 1999), we previously found that an rtf1Δ mutation causes extreme sickness in combination with an srb5 mutation but no severe synthetic phenotypes when combined with mutations in five other Srb-mediator genes (Costa and Arndt, 2000).
While we have not investigated the involvement of the Paf1 complex in initiation and our data are not mutually exclusive with this possibility, we have uncovered several lines of evidence that suggest that the Paf1 complex plays a role in transcription elongation. First, defects in the Paf1 complex lead to 6AU sensitivity and reduced PUR5 induction (Figures 3B and 4). Secondly, we previously demonstrated a range of genetic interactions between RTF1 and genes that encode known elongation factors, including Spt4–Spt5 and TFIIS (Costa and Arndt, 2000). In addition, in a screen for mutations that are synthetically lethal with rtf1 mutations, we isolated mutations in the genes for Pob3, Fcp1 and Ctk1 (Costa and Arndt, 2000). Thirdly, we have demonstrated genetic interactions of Paf1 complex genes with SPT4, SPT5 and SPT16 (Table III). Fourthly, we recovered paf1 and leo1 mutations in an unbiased selection for genetic suppressors of a cold-sensitive allele of SPT5 (Figure 5). This spt5Cs– allele is also suppressed by mutations and drug treatments that cause RNA pol II elongation defects (Hartzog et al., 1998). Finally, we have demonstrated biochemical interactions of the Paf1 complex with the Spt4–Spt5 and Spt16–Pob3 transcription elongation factors (Figure 6). In a study complementary to ours, members of the Paf1 complex have also been found to co-purify with Spt16–TAP (Gavin et al., 2002). These data provide strong evidence that the function of the Paf1 complex is tied to that of elongating RNA pol II. Given that both Spt4–Spt5 and Spt16–Pob3 have been implicated in regulating transcription elongation in the context of chromatin, our data raise the intriguing possibility that the Paf1 complex has a functional interaction with chromatin.
RTF1 was identified originally in a genetic selection for suppressors of the spt15-122 mutation, which encodes a form of TBP with altered DNA binding specificity (Stolinski et al., 1997). These results were interpreted previously to indicate a role for Rtf1 in transcription initiation. However, the mechanisms leading to the Spt– phenotype in spt15-122 cells and its suppression in spt15-122 rtf1 cells are not known. Thus, these observations may reflect specific elongation defects in rtf1 cells. In particular, promoter-specific effects on the processivity of RNA pol II or interference with promoter function by elongating polymerases initiating upstream could explain these observations. Furthermore, there are precedents for the idea that TBP may influence events that occur subsequent to initiation; TBP plays a role in loading the polyadenylation factor CPSF onto transcription complexes (Dantonel et al., 1997), and it has been implicated in the function of hnRNP proteins required for mRNA export (Lei et al., 2001).
Other evidence that the Paf1 complex functions during elongation and in the context of chromatin includes the detection of interactions of Rtf1 and Leo1 with Cus1 and Hsh49, components of the U2 snRNP, and of Cdc73 with Msi1, a component of the yeast chromatin assembly complex, in a large-scale two-hybrid analysis of yeast proteins (Ito et al., 2001). Furthermore, we recently have found that spt4 and spt5 mutations lead to defects in pre-mRNA splicing (D.L.Lindstrom and G.A.Hartzog, in preparation). Thus, future studies may reveal more intimate connections of the Paf1 complex with transcript elongation and perhaps even pre-mRNA processing.
Materials and methods
Media and genetic methods
Strain construction and other genetic manipulations were carried out by standard methods (Rose et al., 1990). Yeast media, including rich (YPD), minimal (SD), synthetic complete (SC) and 5-fluoro-orotic acid (5FOA) were made as described previously (Rose et al., 1990). 6AU was added to SC-Ura media as noted in the figure legends. All FY, GHY and KY Saccharomyces cerevisiae strains (Table IV) are isogenic to S288C and are GAL2+. All deletion mutations described here remove the entire open reading frame of the gene in question and are therefore true nulls.
Table IV. Saccharomyces cerevisiae strains.
| Strain | Genotype |
|---|---|
| KY286 | MATa leu2Δ1 |
| KY425 | MATa rtf1Δ100::URA3 his4-912δ lys2-128δ ura3-52 |
| KY453 | MATα rtf1Δ100::URA3 his3Δ200 lys2-173R2 leu2Δ 1 ura3-52 trp1Δ63 |
| KY522 | MATa his4-912δ lys2-128δ leu2Δ1 ura3-52 trp1Δ63 rtf1Δ101::LEU2 |
| KY589 | MATα leu2Δ1 trp1Δ63 |
| KY686 | MATa paf1Δ::URA3 ura3(Δ0 or –52) lys2-128δ |
| KY687 | MATα paf1Δ::URA3 ura3(Δ0 or –52) leu2(Δ0 or 1) lys2-173R2 |
| KY689 | MATα cdc73Δ::KANMX4 lys2Δ0 leu2Δ0 |
| KY690 | MATa leo1Δ::URA3 ura3-52 lys2-128δ leu2Δ1 |
| KY695 | MATα ctr9Δ::KANMX4 lys2-128δ leu2Δ1 |
| KY770 | MATa rtf1Δ101::LEU2 his4-912δ lys2-128δ leu2Δ1 ura3-52 trp1Δ63 [pKA61] |
| KY771 | MATa rtf1Δ101::LEU2 his4-912δ lys2-128δ leu2Δ1 ura3-52 trp1Δ63 [pPC59] |
| GHY92 | MATα his4-912δ lys2-128δ leu2Δ1 ura3-52 spt5-242a |
| GHY126 | MATa his4-912δ lys2-128δ leu2Δ1 ura3-52 paf1-49 |
| GHY127 | MATa his4-912δ lys2-128δ leu2Δ1 ura3-52 spt5-242a leo1-43 |
| GHY141 | MATa his4-912δ lys2-128δ leu2Δ1 ura3-52 spt5-242a leo1-43 |
| GHY144 | MATα his4-912δ lys2-128δ leu2Δ1 ura3-52 spt5-242a paf1-49 |
| GHY240 | MATa his4-912δ lys2-128δ ura3-52 leu2Δ1 leo1Δ::URA3 |
| GHY285 | MATα his4-912δ lys2-128δ ura3-52 leu2Δ 1 dst1Δ::HISG-URA3 |
| GHY622 | MATα hisΔ200 lys2-128δ ura3Δ0 |
| GHY917 | MATα his4-912δ lys2-128δ leu2(Δ0 or 1) ura3(Δ0 or 52) paf1Δ::URA3 |
| GHY1002 | MATα his3Δ200 lys2-128δ ura3(Δ0 or 52) 3×HA-PAF1 SPT5-FLAG |
| GHY1009 | MATa his4-912δ lys2-128δ leu2 (Δ0 or 1) ura3(Δ0 or 52) paf1Δ::LEU2 dst1Δ::HISG-URA3 |
| GHY1059 | MATa his3Δ200 lys2-128δ leu2Δ1 spt16-197 |
| GHY1083 | MATa his4-912δ lys2-128δ leu2(Δ0 or 1) ura3(Δ0 or –52) cdc73Δ::KANMX4 |
| GHY1092 | MATa his3Δ200 lys2-128δ leu2Δ1 trp1Δ63 ura3-52 LEO1-3×HA::HIS3 |
| GHY1102 | MATα his4-912δ lys2-128δ leu2Δ1 ura3-52 trp1Δ63 SPT5-FLAG SPT24-13×MYC::TRP1 |
| GHY1140 | MATa his3Δ200a lys2-128δ leu2Δ1 ura3-52 trp1Δ63 LEO1-3×HA-HIS3 SPT24-13×MYC::KANMX4 |
| GHY1141 | MATa his4-912δ lys2-128δ leu2Δ1 trp1Δ63 LEO1-3×HA::HIS3 SPT24-13×MYC::KANMX4 SPT5-FLAG |
| GHY1146 | MATa his4-912δ lys2-128δ leu2(Δ0 or1) ura3Δ0 trp1Δ63 paf1Δ::LEU2 SPT5-FLAG SPT24-13×MYC::KANMX4 LEO1-3×HA::HIS3 |
| GHY1147 | MATa his4-912δ lys2-128δ ura3Δ0 SPT5-FLAG 3×HA-PAF1 SPT24-13×MYC::KANMX4 |
| GHY1157 | MATα his4-912δ lys2-128δ leu2Δ1 ctr9Δ::KANMX4 |
| GHY1184 | MATα his4-912δ lys2-128δ leu2(Δ0 or 1) ura3Δ0 CTR9-6×MYC::LEU2 LEO1-3×HA::HIS3 SPT5-FLAG |
| GHY1222 | MATa his3Δ200b his4-912δ lys2-128δ leu2(Δ0 or 1) ura3(Δ0 or 52) trp1Δ63 paf1Δ::HIS3 3×HA-CDC73::URA3 CTR9-6×MYC::LEU2 SPT5-FLAG |
| GHY1228 | MATα his3Δ200 lys2-128δ leu2(Δ0 or 1) ura3(Δ0 or 52) spt16-197 paf1Δ::URA3 |
| FY118 | MATa his4-912δ lys2-128δ leu2Δ1 trp1Δ63 ura3-52 |
| FY602 | MATa his3Δ200 lys2-128δ leu2Δ1 trp1Δ63 ura3-52 |
| FY653 | MATα his4-912δ lys2-128δ leu2Δ1 ura3-52 |
| FY1639 | MATa HA1-SPT4 HA1-SPT6 SPT5-MYC his4-912δ lys2-128δ leu2Δ1 ura3-52 trp1Δ63 |
aReferred to as spt5Cs– in the text.
bStrain may carry this mutation.
Cloning suppressors of spt5Cs–
The isolation of genetic suppressors of the cold-sensitive growth defect caused by the spt5Cs– mutation, spt5-242, has been described previously (Hartzog et al., 1998). Recessive mutations were assigned to five complementation groups and these assignments were confirmed by linkage analysis. Like the rpb1 suppressors of spt5Cs– (Hartzog et al., 1998), suppressor mutations in two of the other complementation groups caused mild Spt– phenotypes in an SPT5+ background. The genes corresponding to these two complementation groups were cloned by transforming strains GHY126 and GHY127 with plasmid DNA from yeast genomic DNA libraries (Rose et al., 1987; Yoshihisa and Anraku, 1989) and screening for Spt+ transformants. Candidate plasmids were isolated from transformants and retransformed to yeast strains to confirm that they could complement the original suppression phenotype of the mutants. Subcloning showed that PAF1 and LEO1 alone were able to complement their respective suppressor mutations. PAF1, originally cloned from a high-copy plasmid library, was also subcloned to a centromeric vector. This CEN PAF1 plasmid also complemented the suppressor mutation in strain GHY126. Linkage analysis confirmed that PAF1 and LEO1 were allelic to the original suppressor mutations.
Epitope tagging
Plasmid pPC59, a CEN URA3-marked plasmid carrying the Kluyveromyces lactis TRP1 gene and expressing Rtf1 tagged at its C-terminus with the TAP tag, was constructed as follows. Using the Expand High Fidelity PCR system (Roche), a PCR fragment was generated by amplification of plasmid pBS1479 (Rigaut et al., 1999). This PCR product was used to transform KY770, which contains pKA61 (CEN URA3 RTF1; Stolinski et al., 1997), to Trp+ prototrophy, generating pPC59 (strain KY771) by homologous recombination. Rtf1–TAP expressed from pPC59 is fully functional based on complementation of the Spt– phenotype of an rtf1Δ mutant.
Ctr9 was tagged with six copies of the Myc epitope at its C-terminus and Cdc73 was tagged with three copies of the HA1 epitope at its N-terminus as previously described (Koch et al., 1999). In contrast to the other genes expressing epitope-tagged proteins described here, HA1-CDC73 was integrated at URA3 rather than its normal chromosomal locus. A strain expressing Paf1, tagged at its N-terminus with 3×HA1, was created by PCR amplification of pMPY-3×HA (Schneider et al., 1996), and transformation of the PCR product into strain GHY622. Ura+ transformants were purified and replated to 5FOA media to select for recombinants that had lost the URA3 gene and retained the 3×HA1 tag at the 5′ end of PAF1. A strain expressing Leo1 tagged at its C-terminus with 3×HA1 was constructed by PCR amplifying plasmid pFA6a-3HA-HIS3MX6 (Longtine et al., 1998), transforming the product into strain FY602 and selecting His+ transformants.
Co-immunoprecipitation experiments and gel filtration
Anti-Rtf1 immunoprecipitations were performed using 2 mg of protein extract, prepared by bead beating, essentially as described before (Hartzog et al., 1998) except that the immunoprecipitates were washed in a buffer containing 0.2 M ammonium acetate and 0.1 M sodium acetate rather than 0.5 M ammonium acetate and 0.1 M sodium acetate. All other immunoprecipitations were carried out as previously described, using extracts prepared by grinding frozen cell pellets of log-phase cells with a pestle and mortar (Lindstrom and Hartzog, 2001). In some experiments, after immunoprecipitation, antigen-associated proteins were eluted from the beads in 50 mM HEPES-KOH pH 7.6, 1 M potassium chloride, 1 mM magnesium chloride, 1 mM EGTA and 10% glycerol. The eluted proteins were concentrated by trichloroacetic acid (TCA) precipitation, separated by SDS–PAGE and immunoblotted. In other experiments, proteins that remained associated with the beads were released by boiling the beads in SDS–PAGE loading buffer.
Gel filtration chromatography was performed on a Superose 6 column (Pharmacia) equilibrated in lysis buffer (30 mM HEPES-KOH pH 7.4, 200 mM potassium acetate, 1 mM magnesium acetate, 1 mM EGTA, 0.05% Tween-20, 10% glycerol). For whole-cell extracts, ∼5 mg of cleared lysate was injected onto the column and 0.5 ml fractions were collected. Samples (250 µl) of each fraction were precipitated with TCA, separated by SDS–PAGE and transferred to nitrocellulose for immunoblotting.
Tandem affinity purification (TAP) tag experiments
Strains KY770 (RTF1) and KY771 (RTF1–TAP) were grown in SC-Ura media to ∼3.5 × 107 cells/ml. Cells were harvested and washed in lysis buffer containing 100 mM potassium acetate, 2 mM magnesium acetate, 20 mM HEPES-KOH pH 7.4, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol (DTT), 0.1% NP-40 plus protease inhibitors. The cell pellet was frozen in liquid nitrogen. Cells were resuspended in an equal volume of lysis buffer and lysed by bead beating. The lysate was centrifuged at 5000 r.p.m. for 5 min at 4°C in an SA600 rotor. The supernatant was transferred to a clean tube and centrifuged at 10 000 r.p.m. for 10 min at 4°C. In experiment 3, the supernatant derived from this step was clarified by centrifugation at 150 000 g for 1 h at 4°C to remove large protein aggregates and complexes including ribosomes.
Extracts were adjusted to 50 mM sodium acetate and 10 mM Tris-acetate pH 7.5, and ∼40–50 mg of protein extract (derived from ∼3 l of cells) were mixed for 2 h at 4°C with 400 µl of IgG–Sepharose beads (Amersham Pharmacia). To process extract from 9 l of cells, three columns were run in parallel. The subsequent steps in the TAP purification were performed essentially as described by Rigaut et al. (1999) except that all buffers contained acetate instead of chloride ions. The final Rtf1–TAP fractions were eluted from the calmodulin affinity resin with 2.4 ml of IPP150 calmodulin elution buffer (Rigaut et al., 1999) containing 0.01% NP-40. The TAP complexes were precipitated with TCA and resuspended in 100 mM ammonium bicarbonate, 5% acetonitrile. Purified proteins were reduced and alkylated with DTT and iodoacetamide followed by digestion with modified sequencing-grade trypsin (Promega). The peptide mixture was loaded onto a 75 µm ID RP-HPLC column (Poros R2, Perceptive Biosystems) equilibrated in 0.5% acetic acid. Peptides were eluted using a linear gradient of 0–40% acetonitrile over 60 min followed by 40–60% over 10 min at a flow rate of 0.3 µl/min. Eluting peptides were analyzed by electrospray ionization tandem mass spectrometry using an ion trap mass spectrometer (LCQ Deca, ThermoFinnigan; Gatlin et al., 1998). All tandem spectra were searched against the S.cerevisiae open reading frame database (SGD, Stanford University) using the SEQUEST algorithm (Eng et al., 1994). Data processing of the SEQUEST output files into a list of proteins present in the TAP complex has been described (Link et al., 1999).
Antibodies and immunoblots
Immunoblots were performed by standard methods (Harlow and Lane, 1988). The anti- Spt5, -TBP and -Rpb1 (8WG16) antibodies have been described previously (Thompson et al., 1989; Buratowski and Zhou, 1992; Hartzog et al., 1998). The rabbit anti-HA1 antibody was made as described (Mortenson et al., 2002). The anti-Myc antibody, 9E10, was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-Paf1 and anti-Cdc73 antibodies were gifts of Judith Jaehning (Shi et al., 1996, 1997). The anti-Pob3 and anti-Spt16 antibodies were gifts of Tim Formosa (Wittmeyer et al., 1999). The anti-Rtf1 antibody was made by immunizing rabbits with a GST fusion protein containing the first 261 amino acids of Rtf1 and was prepared by Cocalico Biological, Inc. (Reamstown, PA). This antibody was used at dilutions of 1:4000 and 1:3000 for immunoblots and immunoprecipitations, respectively.
Northern analysis
Cells were grown as described in the legend to Figure 4. Cells were harvested, RNA was isolated and northern analysis was performed as described previously (Swanson et al., 1991). Hybridization probes were prepared by random-prime labeling of PCR-generated DNA fragments containing PUR5 and SED1. Oligonucleotides used for the PCR were as described by Shaw and Reines (2000).
Acknowledgments
Acknowledgements
We thank Bertrand Séraphin and Christian Koch for gifts of plasmids, Tim Formosa, Judith Jaehning and Steve Buratowski for gifts of antibodies, and Cherie Mueller and Judith Jaehning for communicating results prior to publication. We are grateful to Greg Prelich and members of the Hartzog and Arndt labs for valuable comments on the manuscript. This work was supported by a Mellon Foundation predoctoral fellowship to P.J.C. and grants from the National Institutes of Health to K.M.A. (GM52593 and AI01816) and G.A.H. (GM60479). A.J.L. was supported by the Vanderbilt-Ingram Cancer Support Grant (5P30CA68485-05), HHMI, and an Ingram Family gift.
Note added in proof
It has recently come to our attention that the HA1-tagged CDC73 gene described in the text (strain GHY1222) was inadvertently integrated at the normal CDC73 locus rather that at the URA3 gene. This HA1-tagged allele of CDC73 is functional, and this finding does not affect the results or conclusions of the experiments reported here.
References
- Andrulis E.D., Guzman,E., Doring,P., Werner,J. and Lis,J.T. (2000) High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev., 14, 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J., Chambers,R.S., Kobor,M.S., Ho,Y., Cartier,M., Bolotin,D., Andrews,B., Kane,C.M. and Greenblatt,J. (1997) An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 14300–14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A. and Winston,F. (1996) Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science, 272, 1473–1476. [DOI] [PubMed] [Google Scholar]
- Brewster N.K., Johnston,G.C. and Singer,R.A. (1998) Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem., 273, 21972–21979. [DOI] [PubMed] [Google Scholar]
- Brewster N.K., Johnston,G.C. and Singer,R.A. (2001) A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol., 21, 3491–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. and Zhou,H. (1992) A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell, 71, 221–230. [DOI] [PubMed] [Google Scholar]
- Chang M., French-Cornay,D., Fan,H.Y., Klein,H., Denis,C.L. and Jaehning,J.A. (1999) A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol., 19, 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.-J., Kobor,M.S., Kim,M., Greenblatt,J. and Buratowski,S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser2 of the RNA polymerase II C-terminal domain. Genes Dev., 15, 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Kim,T.K., Mancebo,H., Lane,W.S., Flores,O. and Reinberg,D. (1999) A protein phosphatase functions to recycle RNA polymerase II. Genes Dev., 13, 1540–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams C.D. and Winston,F. (1987) The SPT6 gene is essential for growth and is required for δ-mediated transcription in Saccharomyces cerevisiae. Mol. Cell. Biol., 7, 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams C.D., Norris,D., Osley,M.A., Fassler,J.S. and Winston,F. (1988) Changes in histone gene dosage alter transcription in yeast. Genes Dev., 2, 150–159. [DOI] [PubMed] [Google Scholar]
- Costa P.J. and Arndt,K.M. (2000) Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics, 156, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel J.C., Murthy,K.G., Manley,J.L. and Tora,L. (1997) Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature, 389, 399–402. [DOI] [PubMed] [Google Scholar]
- Eng J.K., McCormack,A.L. and Yates,J.R.,3rd (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom., 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Evans D.R., Brewster,N.K., Xu,Q., Rowley,A., Altheim,B.A., Johnston,G.C. and Singer,R.A. (1998) The yeast protein complex containing Cdc68 and Pob3 mediates core-promoter repression through the Cdc68 N-terminal domain. Genetics, 150, 1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous A., Gonzalez,F., Sun,L., Kodadek,T. and Johnston,S.A. (2001) The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell, 7, 981–991. [DOI] [PubMed] [Google Scholar]
- Formosa T., Eriksson,P., Wittmeyer,J., Ginn,J., Yu,Y. and Stillman,D.J. (2001) Spt16–Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J., 20, 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatlin C.L., Kleemann,G.R., Hays,L.G., Link,A.J. and Yates,J.R.,3rd (1998) Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem., 263, 93–101. [DOI] [PubMed] [Google Scholar]
- Gavin A. et al. (2002) Functional organization of the yeast proteome by systematic analyses of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Hampsey M. and Reinberg,D. (1999) RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev., 9, 132–139. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Hartzog G.A., Wada,T., Handa,H. and Winston,F. (1998) Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev., 12, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D., Kwak,Y.T., Guo,J. and Gaynor,R.B. (2000) Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol., 20, 2970–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jona G., Wittschieben,B.O., Svejstrup,J.Q. and Gileadi,O. (2001) Involvement of yeast carboxy-terminal domain kinase I (CTDK-I) in transcription elongation in vivo. Gene, 267, 31–36. [DOI] [PubMed] [Google Scholar]
- Kaplan C.D., Morris,J.R., Wu,C. and Winston,F. (2000) Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D.melanogaster. Genes Dev., 14, 2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Wollmann,P., Dahl,M. and Lottspeich,F. (1999) A role for Ctr9p and Paf1p in the regulation G1 cyclin expression in yeast. Nucleic Acids Res., 27, 2126–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M. and Greenleaf,A.L. (1991) CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr., 1, 149–167. [PMC free article] [PubMed] [Google Scholar]
- Lee J.M. and Greenleaf,A.L. (1997) Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J. Biol. Chem., 272, 10990–10993. [DOI] [PubMed] [Google Scholar]
- Lee Y.C., Park,J.M., Min,S., Han,S.J. and Kim,Y.J. (1999) An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol., 19, 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P., Krebber,H. and Silver,P.A. (2001) Messenger RNAs are recruited for nuclear export during transcription. Genes Dev., 15, 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom D.L. and Hartzog,G.A. (2001) Genetic interactions of Spt4–Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics, 159, 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A.J., Eng,J., Schieltz,D.M., Carmack,E., Mize,G.J., Morris,D.R., Garvik,B.M. and Yates,J.R.,3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nature Biotechnol., 17, 676–682. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.,3rd, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Magdolen V., Lang,P., Mages,G., Hermann,H. and Bandlow,W. (1994) The gene LEO1 on yeast chromosome XV encodes a non-essential, extremely hydrophilic protein. Biochim. Biophys. Acta, 1218, 205–209. [DOI] [PubMed] [Google Scholar]
- Malone E.A., Clark,C.D., Chiang,A. and Winston,F. (1991) Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 5710–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortenson E.M., McDonald,H., Yates,J.R.,3rd and Kellogg,D.R. (2002) Cell cycle-dependent assembly of a Gin4–septin complex. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C.L. and Jaehning,J.A. (2002) Ctr9, Rtf1 and Leo1 are components of the Paf1/RNA polymerase II complex: implications for initiation and elongation of transcription. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., LeRoy,G., Chang,C.H., Luse,D.S. and Reinberg,D. (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell, 92, 105–116. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu,W.H., Lane,W.S., Hampsey,M. and Reinberg,D. (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature, 400, 284–288. [DOI] [PubMed] [Google Scholar]
- Prelich G. (2002) The RNA polymerase II CTD kinases: emerging clues to their function. Eukaryotic Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Seraphin,B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol., 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rose M., Novick,P., Thomas,J., Botstein,D. and Fink,G. (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene, 60, 237–243. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Hieter,P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Rowley A., Singer,R.A. and Johnston,G.C. (1991) CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol., 11, 5718–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B.L., Steiner,B., Seufert,W. and Futcher,A.B. (1996) pMPY-ZAP: a reusable polymerase chain reaction-directed gene disruption cassette for Saccharomyces cerevisiae. Yeast, 12, 129–134. [DOI] [PubMed] [Google Scholar]
- Shaw R.J. and Reines,D. (2000) Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol., 20, 7427–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Finkelstein,A., Wolf,A.J., Wade,P.A., Burton,Z.F. and Jaehning,J.A. (1996) Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol., 16, 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Chang,M., Wolf,A.J., Chang,C.H., Frazer-Abel,A.A., Wade,P.A., Burton,Z.F. and Jaehning,J.A. (1997) Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol., 17, 1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolinski L.A., Eisenmann,D.M. and Arndt,K.M. (1997) Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 4490–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M.S. and Winston,F. (1992) SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics, 132, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M.S., Malone,E.A. and Winston,F. (1991) SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol., 11, 4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson N., Steinberg,T., Aronson,D. and Burgess,R. (1989) Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J. Biol. Chem., 264, 11511–11520. [PubMed] [Google Scholar]
- Verma R., Chen,S., Feldman,R., Schieltz,D., Yates,J., Dohmen,J. and Deshaies,R.J. (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell, 11, 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T. et al. (1998a) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev., 12, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Takagi,T., Yamaguchi,Y., Watanabe,D. and Handa,H. (1998b) Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J., 17, 7395–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T. et al. (2000) FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell, 5, 1067–1072. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Werel,W., Fentzke,R.C., Thompson,N.E., Leykam,J.F., Burgess,R.R., Jaehning,J.A. and Burton,Z.F. (1996) A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr. Purif., 8, 85–90. [DOI] [PubMed] [Google Scholar]
- Wind M. and Reines,D. (2000) Transcription elongation factor SII. Bioessays, 22, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Chaleff,D.T., Valent,B. and Fink,G.R. (1984) Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics, 107, 179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J., Joss,L. and Formosa,T. (1999) Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated and copurifies with DNA polymerase α. Biochemistry, 38, 8961–8971. [DOI] [PubMed] [Google Scholar]
- Wu-Baer F., Lane,W.S. and Gaynor,R.B. (1998) Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J. Mol. Biol., 277, 179–197. [DOI] [PubMed] [Google Scholar]
- Yoshihisa T. and Anraku,Y. (1989) Nucleotide sequence of AMS1, the structural gene of vacuolar α-mannosidase of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 163, 908–915. [DOI] [PubMed] [Google Scholar]