Graphical abstract
Keywords: Glycyrrhiza, Species classification, Pharmacology, Transcriptional regulation, Synthetic biology
Highlights
-
•
Glycyrrhiza species mainly offer triterpenoids, flavonoids, and polysaccharides with diverse therapeutic effects.
-
•
The review covers Glycyrrhiza species classification, pharmacology, biosynthesis of active compounds, and synthetic biology advancements.
-
•
Research on population genomics, active compounds biosynthesis, and molecular breeding in Glycyrrhiza is key to its future applications.
-
•
Breeding high-yield Glycyrrhiza plants and optimizing biosynthesis of bioactives support resource conservation and drug development.
-
•
Metabolic engineering and green microbial methods boost production efficiency of natural products in licorice.
Abstract
Background
Licorice is extensively and globally utilized as a medicinal herb and is one of the traditional Chinese herbal medicines with valuable pharmacological effects. Its therapeutic components primarily reside within its roots and rhizomes, classifying it as a tonifying herb. As more active ingredients in licorice are unearthed and characterized, licorice germplasm resources are gaining more and more recognition. However, due to the excessive exploitation of wild licorice resources, the degrading germplasm reserves fail to meet the requirements of chemical extraction and clinical application.
Aim of Review
This article presents a comprehensive review of the classification and phylogenetic relationships of species in genus Glycyrrhiza, types of active components and their pharmacological activities, licorice omics, biosynthetic pathways of active compounds in licorice, and metabolic engineering. It aims to offer a unique and comprehensive perspective on Glycyrrhiza, integrating knowledge from diverse fields to offer a comprehensive understanding of this genus. It will serve as a valuable resource and provide a solid foundation for future research and development in the molecular breeding and synthetic biology fields of Glycyrrhiza.
Key Scientific Concepts of Review
Licorice has an abundance of active constituents, primarily triterpenoids, flavonoids, and polysaccharides. Modern pharmacological research unveiled its multifaceted effects encompassing anti-inflammatory, analgesic, anticancer, antiviral, antioxidant, and hepatoprotective activities. Many resources of Glycyrrhiza species remain largely untapped, and multiomic studies of the Glycyrrhiza lineage are expected to facilitate new discoveries in the fields of medicine and human health. Therefore, strategies for breeding high-yield licorice plants and developing effective biosynthesis methods for bioactive compounds will provide valuable insights into resource conservation and drug development. Metabolic engineering and microorganism-based green production provide alternative strategies to improve the production efficiency of natural products.
Introduction
The members of genus Glycyrrhiza, which belongs to subfamily Papilionoideae of family Leguminosae, are perennial herbaceous plants. They have a global distribution, with main production areas in Asia and Europe and a small number found in the tropical and subtropical regions of America and Africa. China is the central zone of licorice resources and is the world’s largest consumer and exporter of licorice [1]. Glycyrrhiza species are widely used in medicine, with G. uralensis, G. glabra, and G. inflata as the source plants for licorice herbal medicine in Chinese, European, and Korean pharmacopoeias [2], [3]. Licorice is one of the traditional Chinese herbal medicines, and its medicinal parts are its roots and rhizomes. It possesses various medicinal properties, such as tonifying the spleen and replenishing qi, clearing heat and detoxification, resolving phlegm and relieving cough, relieving pain, and harmonizing various medicines [3]. In China, licorice is one of the most commonly used bulk medicinal materials, and countries such as the United States and Japan also refer to licorice as “Fairy Herb” or “Divine Herb” [2], [4], [5].
Glycyrrhiza plants contain a rich variety of active components, with over 400 secondary metabolites identified [6], [7]. Among them, triterpenoids and flavonoids are two crucial classes of chemical compounds that determine the medicinal effects of licorice. Modern pharmacological research found that licorice has multiple effects, including anti-inflammatory, analgesic, anticancer, antiviral, antioxidant, and hepatoprotective properties [8], [9], [10], [11], [12]. In addition to its medicinal uses, licorice is also employed as an additive and flavoring agent in various industries, such as food, tobacco, cosmetics, and animal husbandry [13], [14]. In addition, licorice is a wild sand-fixing plant under national key protection and management. It protects the ecological environment and prevents land desertification [15]. Owing to its wide-ranging applications and high market demand, licorice has been excessively harvested from the wild, leading to remarkable ecological damage to wild licorice populations and the subsequent scarcity of wild licorice resources [16]. Therefore, cultivated licorice has become the primary source to meet market demand [17]. Owing to the influence of environmental factors and licorice genetic factors, the active ingredients significantly differ between cultivated and wild licorice. In particular, the active ingredients in cultivated licorice are generally fewer and do not meet pharmacopoeia standards. Therefore, cultivated licorice is often diverted to nonmedicinal industries, such as food or confectionery, severely constraining the sustainable development of licorice as a medicinal resource [18]. The importance of key traits and quality in licorice breeding must be emphasized. Therefore, strategies for selecting high-yield licorice plants and developing effective in vivo or in vitro biosynthesis methods for bioactive compounds will provide valuable insights into drug development and resource conservation.
This article provides an overview of the classification, phylogenetic relationships, and pharmacology of Glycyrrhiza species. It discusses licorice genomics and the biosynthetic pathways of active compounds. Finally, it explores the prospects for metabolic engineering and breeding of licorice. Furthermore, it presents future prospects for the development and utilization of Glycyrrhiza resources.
Classification and phylogenetic relationships of Glycyrrhiza species
Genera Glycyrrhiza and Glycyrrhizopsis constitute the Glycyrrhizeae tribe within the licorice family. Licorice holds significant and diverse applications, but its intergeneric and intrageneric taxonomy is controversial. This situation has implications for the medicinal, industrial, and scientific utilization of licorice and the collection and breeding of licorice germplasm resources. For instance, nonmedicinal species have been sold as medicinal licorice in the market, and some scientific studies on pharmacology or chemistry have utilized misidentified licorice materials. Hence, precise and specific delineation is crucial for the proper application of licorice.
Genera Meristotropis and Glycyrrhizopsis were initially considered as subsidiaries of the Glycyrrhiza genus. Whether Meristotropis and Glycyrrhizopsis should be included or excluded from the Glycyrrhiza genus has been debated throughout various taxonomic revisions at the intergeneric level. Some argued that both should be part of the Glycyrrhiza genus, and others proposed that they should remain independent genera. Some suggested that Meristotropis and Glycyrrhiza should be merged into a single Glycyrrhiza genus, with Glycyrrhizopsis as a separate genus [19], [20], [21]. Within the Glycyrrhiza genus, taxonomists have classified licorice species using a series of morphological characteristics, including the number of leaflets per leaf, inflorescence/fruit cluster shape, fruit shape, and fruit appendages. However, a significant number of transitional morphological features have blurred the specific boundaries of licorice species. The Glycyrrhiza genus has undergone five major revisions, resulting in varying species counts ranging from 13 to 36 species [22]. Hence, the debate regarding the current species limitations within the Glycyrrhiza genus is still ongoing.
The latest research involved extracting low-copy nuclear genes from shallow genome sequencing data and constructing phylogenetic trees using chloroplast genomes and nuclear ribosomal DNA. Classification and phylogenetic studies combined this approach with morphological characteristics and specimen image recognition [22], [23]. The results revealed that the Central Asian endemic genus Meristotropis should be merged into the Glycyrrhiza genus, resulting in the inclusion of the following 13 species within this genus: G. acanthocarpa, G. astragalina, G. bucharica, G. echinata, G. foetida, G. glabra, G. gontscharovii, G. lepidota, G. macedonica, G. pallidiflora, G. squamulosa, G. triphylla, and G. yunnanensis. This study proposes a broad concept of G. glabra, which now includes the previously classified G. uralensis, G. glabra, G. inflata, and G. aspera representing the medicinal group containing glycyrrhizic acid. The sister genus, Glycyrrhizopsis, is endemic to the Western Anatolia Plateau in Western Asia and comprises only two species: G. asymmetrica and G. flavescens.
Not all species within the Glycyrrhiza genus are considered medicinal plants. Only species with glycyrrhizic acid in their roots have medicinal value, constituting the medicinal group. The ancestors of the Glycyrrhiza genus lack glycyrrhizic acid, and the medicinal groups in Eurasia share a common ancestor. Their descendants have all arisen from two rapid divergence events within the last million years [3], [19]. This phenomenon has also resulted in significant morphological overlap and a high confusion degree among subspecies within this group [22]. As mentioned earlier, the medicinal group is traditionally composed of four widely accepted species: G. uralensis, G. glabra, G. inflata, and G. aspera. However, recent research indicated that the North American species G. lepidota also contains glycyrrhizic acid, albeit in relatively low quantities [19], [24]. According to previous phylogenetic findings, the group containing glycyrrhizic acid is not monophyletic [25]. Although G. aspera contains glycyrrhizic acid, its long and relatively short roots yield a low quantity and it has therefore been considered unsuitable for medicinal or industrial purposes [19]. This circumstance may also explain why the Chinese Pharmacopoeia only includes G. uralensis, G. glabra, and G. inflata as licorice sources [3].
Although G. uralensis, G. glabra, and G. inflata are all medicinal species, their contents of active ingredients significantly differ, resulting in substantial variations in their quality and medicinal efficacy [26], [27], [28]. Accurate identification of these three species is essential to ensure the effectiveness and safety of clinical medication. However, these three original licorice species belong to the same genus, and their medicinal parts are their roots. Their microscopic characteristics are highly similar, making it difficult to effectively distinguish them through traditional trait identification and microscopic methods. Specific variable sites in the ITS/ITS2 and psbA-trnH sequences have been utilized for the identification of these three distinct original licorice species [29], [30]. In the ITS2 sequence, three mutation sites are located at positions 16–18 bp, which can be divided into two genotypes: I2-i (TGC), representing the single haplotype of G. uralensis; and I2-ii (CAA), representing the single haplotype of G. glabra and G. inflata. The ITS sequence at positions 187 and 411–413 bp (equivalent to positions 16–18 bp in the ITS2 sequence) can be classified into two genotypes: I-i (CTGC), representing the single haplotype of G. uralensis; and I-ii (TCAA), representing the single haplotype of G. glabra and G. inflata. The psbA-trnH sequence has three variant positions located at 189, 235, and 288 bp, which can be classified into four single haplotypes: PT-i (ACG), PT-ii (ATG), PT-iii (CTG), and PT-iv (ATA). Among them, PT-i and PT-ii are the haplotypes of G. uralensis, PT-iii is the haplotype of G. uralensis and G. glabra, and PT-iv is the haplotype of G. inflata. The combination of specific variable sites in the ITS/ITS2 and psbA-trnH sequences allows for the identification of these three licorice species.
Primary active constituents of Glycyrrhiza plants
Glycyrrhiza species exhibit a diverse array of chemical components. Among the predominantly reported chemical constituents, those primarily found in the three medicinal species include triterpenoids, flavonoids, polysaccharides, coumarins, alkaloids, volatile oils, and trace elements.
Triterpenoids
Triterpenoids are a major class of compounds in licorice. They are also a type of natural sweetener, consisting of polymers based on the unit of six molecules of isoprene. Triterpenoids are characterized by their high content, strong physiological activity, and high medicinal value. Their basic skeletal structure is typically composed of 3β-hydroxyoleanane-type pentacyclic triterpenoids, which can be considered β-amyrin derivatives [1]. The triterpenoid components in G. uralensis include glycyrrhizic acid, glycyrrhetinic acid, uralsaponin, and licoricesaponin, all of which are derivatives of 3β-oleanane-type compounds [31], [32]. The triterpenoid compounds isolated from G. glabra include glycyrrhizic acid, glycyrrhetinic acid, 29-hydroxyl-glycyrrizin, licoricesaponin, and 22β-acetoxyl-glycyrrizin [33], [34]. G. inflata constitutes over 60 % of wild licorice reserves in China [35] and contains triterpenoids such as glycyrrhizic acid, glycyrrhetinic acid, methyl glycyrrhetinate, 11-deoxyglycyrrhetinic acid, glycyrrhetinate acetylates, and 3-oxo-glycyrrhetinic acid [36], [37], [38], [39], [40].
Flavonoids
Flavonoids primarily refer to compounds with a basic nucleus of 2-phenylchromone. To date, over 300 flavonoids have been isolated and identified from Glycyrrhiza species worldwide [41]. The flavonoids in licorice can be categorized into flavones, flavonols, isoflavones, chalcones, dihydroflavones, dihydroisoflavones, and isoflavanes [42]. The types and quantities of flavonoids can vary in different species of licorice or in the same species from different geographical locations [43]. The flavonoids found in G. uralensis include liquiritin, liquiritigenin, licoflavonol, isolicoflavonol, glycyrol, licoricone, and glicoricone [44]; those found in G. glabra include liquiritin, liquiritigenin, glabridin, hispaglabridin, and 4ʹ-O-methylglabridin [45]; and those found in G. inflata include liquiritin, liquiritigenin, licochalcone, 3-OH licochalcone, licoagrochalcone, and 3,4-didehydroglabridin [36], [37], [38], [39], [40].
Polysaccharides
Polysaccharides are one of the active components of licorice. They have complex structures and diverse functions, and current research is mainly focused on their relationship with biological activities. The polysaccharides in G. uralensis are primarily composed of rhamnose monohydrate, dextran, arabinose, and galactose [46], [47]. In G. inflata, the predominant polysaccharide components are acidic polysaccharides, including L-arabinose, L-rhamnose monohydrate, D-glucose, D-galactose, and D-galacturonic acid [48]. Researchers have found variations in the polysaccharide content among different licorice varieties. Although the monosaccharide compositions are the same in the polysaccharides of different licorice varieties, there are differences in the proportions of these monosaccharides [49], [50].
Coumarins
Coumarin-type active ingredients are mainly found in G. uralensis. Their typical characteristic is the presence of a phenyl ring substitution on the carbonyl adjacent position, and their structure conforms to the C6-C3-C6 rule. Some scholars classify coumarins as flavonoid components; a minority of compounds in this group have a fused furan ring between the substituted benzene ring and the nucleus, forming coumestan (II)-type structures [1]. Lu et al. [51] investigated the chemical composition of waste residues from G. uralensis and discovered new coumarin-type compounds, naming them “huai coumarins”. Liu [52] first isolated 7,2ʹ,4ʹ-trihydroxy-5-methoxy-3-coumaranone from Glycyrrhiza plants. The chemical name of coumarin A in G. inflata is 4-(p-hydroxybenzene)-6-isoprenyl-7-hydroxycoumarin [53].
Alkaloids
Considerable research has focused on the analysis of flavonoid and triterpenoid components in licorice. Some studies were conducted on alkaloid components in licorice, but their variety of origin is often disregarded. Zhang et al. [54] found that the alkaloids in G. uralensis, G. inflata, G. glabra, and G. pallidiflora belong to the same category, possibly quinoline derivatives. The content of these alkaloids is relatively high and similar among the four licorice species. G. uralensis contains over six alkaloids with a mass fraction of 0.26 %.
Other components
In addition to the mentioned chemical components, licorice contains volatile oil compounds, amino acids, and trace elements. Ma et al. [55] determined that the major volatile components in G. uralensis roots are alkanes and aromatic hydrocarbons. Amino acids play a crucial role in tissue growth, metabolism, maintenance, and repair. G. uralensis is rich in amino acids, containing all eight essential amino acids required by the human body [56]. The trace elements in licorice roots may play a role in its authenticity and quality. Liang et al. [57] discovered a significant positive correlation among the Zn content, Zn/Cu ratio, and glycyrrhizic acid level in G. uralensis roots.
In summary, research has primarily focused on the components of G. uralensis and G. glabra, followed by G. inflata. All three licorice species contain triterpenoids, flavonoids, polysaccharides, and alkaloids. However, the chemical composition varies among different varieties. Significant differences in the content of flavonoids and triterpenoids are found among different varieties, but confirming which variety of licorice has a high content is difficult due to the influence of sample variations and experimental design differences. The proportions of monosaccharide compositions in polysaccharides also vary among different varieties. Coumarin-type components are prevalent in G. uralensis. Different licorice varieties have components that are unique to their species. Research has reported 27 biomarkers that can be used to differentiate G. uralensis, G. inflata, and G. glabra [58]. The biomarkers for G. uralensis are licoflavonol, isolicoflavonol, glycyrol, glycycoumarin, glycyuralin E, glyasperin D, licoricone, glicoricone, isoangustone A, 7-O-methylluteone, 6,8-diprenylgenistein, and glicophenone. The biomarkers for G. glabra are glabridin, hispaglabridin B, 4ʹ-O-methylglabridin, 3ʹ-hydroxy-4ʹ-O-methylglabridin, hispaglabridin A, parvisoflavones-A, glycybridin C, erybacin B, and 3-hydroxy-glabrol. The biomarkers for G. inflata are licochalcone A, licochalcone C, 3-OH licochalcone A, licochalcone E, licoagrochalcone C, and 3,4-didehydroglabridin. These biomarkers can provide a basis for the identification and quality control of different licorice varieties.
Pharmacological activities of active components in plants of the Glycyrrhiza genus
Antimicrobial and anti-inflammatory
Modern pharmacological studies suggested that the flavonoids and triterpenoids in licorice may be the main active components responsible for its antimicrobial and anti-inflammatory effects. This type of research could have implications for the development of new drugs targeting drug-resistant bacteria and the study of preservatives with high salt and protease contents as antimicrobial agents in food. However, the safety and efficacy of these compounds still require further investigation. G. uralensis has demonstrated good inhibitory effects against Gram-positive bacteria such as Staphylococcus aureus, Streptococcus, and Enterococcus [59]. Licochalcone A extract from G. inflate could inhibit the growth of Bacillus subtilis in a concentration-dependent manner [60]. It can also effectively improve the clinical symptoms of acetic acid-induced ulcerative colitis in mice by exerting its antioxidant effects through the inhibition of the nuclear factor-kappa B (NF-κB) signaling pathway and the promotion of the nuclear factor erythroid 2-related factor 2 (Nrf2) signaling pathway [61]. Glabridin from G. glabra hinders biofilm formation in methicillin-resistant Staphylococcus aureus by modulating cell surface proteins and reducing the abundance of surface-associated adhesins [62]. It can also effectively and dose-dependently inhibit the production of inflammatory mediators leukotrienes and thromboxanes in HL-60 cells, a type of acute promyelocytic leukemic cells [63]. Glabridin inhibits dendritic cell maturation by reducing pro-inflammatory cytokines, enhancing antigen capture, inhibiting migration, and impairing T cell activation, through blocking NF-κB and MAPK signaling pathways [64]. Glycyrrhizic acid has demonstrated potential against various bacterial and viral pathogens, positioning it as a candidate for alternative therapeutic applications. This compound significantly inhibits the growth of Pseudomonas aeruginosa by altering membrane permeability, efflux activity, and biofilm formation [65]. Meanwhile, glycyrrhizic acid has anti-inflammatory and immunoregulatory effects. It inhibits inflammation and apoptosis induced by Mycoplasma gallisepticum in chickens by suppressing the MAPK signaling pathway, indicating its potential as an antibiotic alternative [66]. Glycyrrhizic acid exerts anti-asthmatic effects by modulating Th1/Th2 cytokines and enhancing regulatory T cells (Tregs) in an ovalbumin-sensitized mouse model [67]. Glycyrrhizic acid exhibits remarkable antimicrobial activities against a diverse array of bacterial. Its mechanisms encompass inhibiting bacterial growth and biofilm formation, and modulating immune responses. These discoveries imply that glycyrrhizic acid might serve as a precious alternative to conventional antibiotics drugs, possessing potential applications in the treatment of infections and inflammatory conditions.
Antiviral
Since the 1980 s when Japanese researchers first reported the anti-HIV effects of glycyrrhizic acid, significant research has been conducted on the antiviral properties of licorice. The studies involved various active compounds, such as flavonoids, triterpenoids, coumarins, and polysaccharides, and explored licorice’s potential to combat various viruses, including HIV, atypical pneumonia viruses, hepatitis viruses, human respiratory syncytial virus, and influenza viruses. Adianti et al. [68] discovered that methanol extracts from G. uralensis roots and its chloroform fraction exhibited antiviral activity against hepatitis C virus. Wang et al. [69] found that the antiviral activity of G. uralensis is related to the glycyrrhizic acid content of its extracts. Glycyrrhizic acid also dose-dependently blocks the replication of enterovirus 71 and coxsackievirus A16. Soufy et al. [70] isolated glycyrrhizic acid from G. glabra roots through in vitro and in vivo experiments; when used alone or in combination with duck hepatitis vaccine, glycyrrhizic acid exhibited strong immune stimulation and antiviral effects against duck hepatitis virus infection. Glycyrrhizic acid exerts a direct antiviral effect on HBV by inhibiting the secretion of hepatitis B surface antigen (HBsAg) and improving liver function in patients with chronic hepatitis B, ultimately enhancing immune status [71]. Additionally, glycyrrhizic acid and its derivatives exhibit antiviral activity against a wide range of viruses, including herpes virus [72], SARS-CoV-2 [73], COVID-19 [74], [75], and Epstein-Barr virus (EBV) [76]. Glycyrrhizic acid demonstrates significant antiviral properties against a variety of viruses through mechanisms such as inhibition of viral entry and replication. The development of glycyrrhizic acid derivatives further enhances its antiviral efficacy, making it a promising candidate for alternative antiviral therapies.
Antioxidant
Licorice has antioxidant activity, which may be related to the content, concentration, extraction temperature, and extraction methods of its triterpenoids, polysaccharides and flavonoids [77]. Researchers evaluated the antioxidant activity of licorice using methods such as measuring the hydrogen peroxide system and scavenging 1,1-diphenyl-2-picrylhydrazyl (DPPH) radicals and superoxide anion radicals. Xu et al. found that glycyrrhizic acid can activate the expression of myocardial nuclear factor E2-related factor 2/heme oxygenase-1 (Nrf2/HO-1) and inhibit the nuclear factor kappa B (NF-κB) signaling pathway, thereby improving myocardial ischemic injury and preventing myocardial infarction [78]. Chen et al. [79] obtained a polysaccharide component, glycyrrhiza polysaccharide II, from G. uralensis roots; it showed excellent concentration-dependent hydroxyl radical scavenging activity and significant DPPH radical scavenging ability. G. glabra polysaccharides (GGP) treatment can enhance immune activities and reduce oxidative stress in high-fat mice [80]. Glabridin is an isoflavone obtained from G. glabra root, and in vitro and in vivo experiments have shown that it possesses excellent antioxidant properties [81]. Ojha et al. [82] found that G. glabra extract exhibited protective effects against ischemia–reperfusion injury induced by left anterior descending coronary artery ligation in rats by alleviating oxidative stress and favorably regulating the cardiac function. Cong et al. [83] extracted and found that the polysaccharides in G. inflate have certain scavenging effects on DPPH radicals, hydroxyl radicals, and superoxide anion radicals. Pan et al. [84] identified a polysaccharide called GIBP from G. inflate that has antioxidant and antiaging effects. In vitro experiments confirmed its strong scavenging activity against hydroxyl radicals. Liquiritin can reduce the content of acetaldehyde in the human body, increase the levels of superoxide dismutase and glutathione peroxidase, alleviate aging, and enhance antioxidant capacity [85]. Glycyrrhiza, through its difference extracts, exhibits significant antioxidant properties by enhancing antioxidant enzyme activities, reducing oxidative stress markers, and providing hepatoprotective effects.
Antineoplastic
The flavonoid components of licorice have demonstrated strong anti-tumor activity against various human cancer cells, including cervical cancer, breast cancer, prostate cancer, and liver cancer cells. They effectively inhibit proliferation and induce apoptosis in these cancer cells. Wei et al. [86] discovered that by downregulating the expression levels of cell cycle proteins D1, cell cycle proteins A, and cell cycle proteins cyclin-dependent kinase 4, liquiritin promotes the expression of p53 and p21 genes and inhibit the proliferation and migration of gastric cancer cells, thus inducing apoptosis and autophagy in cancer cells. Ma et al. [87] discovered that licorice flavonoids have a significant inhibitory and apoptotic effect on cervical cancer HeLa cells, breast cancer Bcap-37 cells, gastric cancer MGC-803 cells, and liver cancer Bel-7404 cells. Park et al. [88] evaluated the hexane–ethanol extract of G. uralensis and found that it significantly inhibits the metastasis and invasion capabilities of malignant prostate cancer cells. Various flavonoids in G. inflate can inhibit cell growth and induce apoptosis. The crude and refined extracts of G. inflate can effectively inhibit the proliferation of cervical cancer cells SiHa and HeLa, thus promoting their apoptosis [89]. Licochalcone A from G. inflate can downregulate the expression of specificity protein 1 and regulate downstream proteins to induce apoptosis in human oral squamous cell carcinoma cells HSC4 and HN22, thereby inhibiting the growth of oral squamous cell carcinoma cells in a concentration- and time-dependent manner [90]. Glycyrrhizic acid and glycyrrhiza polysaccharides also have antitumor effects. Glycyrrhetinic acid exhibit anti-cancer effects by modulating enzymes involved in inflammation and oxidative stress, and downregulating pro-inflammatory mediators, which can suppress tumor cell growth. Cai et al. [91] studied the effects of glycyrrhizic acid on liver cancer cells using proteomics and chemical biology and found that glycyrrhizic acid can inhibit the activity and induce apoptosis in liver cancer stem cells. Glycyrrhetinic acid enhances triple-negative breast cancer treatment by sensitizing cells to etoposide, promoting apoptosis, while glycyrrhizic acid offers protection [92]. Meanwhile, Glycyrrhetinic acid is also therapeutically effective against cervical cancer cells [93]. Additionally, the antitumor mechanism of glycyrrhiza polysaccharides was investigated using a colon cancer CT-26 cell-bearing mouse model based on the gut microbiota [94].
Lowering blood sugar, regulating blood lipids, and antiatherosclerosis
Flavonoids in licorice have demonstrated good hypoglycemic, blood lipid-regulating, and antiatherosclerotic effects and exhibit more effective hypoglycemic activity than positive control drugs. Gou et al. [95] isolated seven flavonoids from G. uralensis roots, all of which exhibited more effective α-glucosidase inhibitory activity than the positive control drug, acarbose. Kent et al. [96] found that glabridin, a compound in G. glabra, inhibits cytochrome P4503A4. This action is believed to contribute to its protective effect against low-density lipoprotein oxidation and its subsequent antiatherosclerotic properties. Carmelli et al. [97] administered glabridin from G. glabra and found a 20 % reduction in plasma low-density lipoprotein oxidation in 22 healthy volunteers within 6 months.
Other pharmacological activities
In addition to the mentioned pharmacological effects, licorice also possesses pharmacological activities such as cardioprotective activity [98], antidepressive activity [99], [100], and neuroprotective activity [101]. Rashedinia et al. [102] found that glycyrrhizic acid can protect cells by promoting mitochondrial biogenesis, maintaining mitochondrial function, and enhancing energy metabolism. Han et al. [103] discovered that glycyrrhizic acid can also exert antiallergic effects by regulating immune cells or immunoglobulins associated with allergies. Zhang et al. [104] reported that liquiritin can inhibit the expression of collagen and alpha-smooth muscle actin, thereby playing a cardioprotective role by suppressing the development of myocardial fibrosis induced by high fructose. Wang et al. [105] demonstrated the antidepressant activity of liquiritin and isoliquiritoside through forced swimming and tail suspension tests. Glycyrrhiza polysaccharides can promote the proliferation of human peripheral blood γδ T cells and dose-dependently increase the secretion of interferon-γ and TNF-α [106]. Ayeka et al. [107] demonstrated the immunomodulatory effects of glycyrrhiza polysaccharides by measuring immune organ quality and indices, immune cell numbers, and serum cytokine levels after administration. Other studies reported the hepatoprotective and neuroprotective effects of G. uralensis [108]; neuroprotective effects, tyrosinase inhibition, and estrogen-like effects of G. glabra [109], [110]; and antiangiogenic effects of G. inflata [111] (Fig. 1).
Fig. 1.
Active ingredients and pharmacology of licorice. a. The main active ingredients in licorice and their pharmacological effects. b. Other typical triterpenoids in licorice. c. Other typical flavonoids in licorice.
Transcriptomic and genomic study of licorice
Genomics and transcriptomics serve as valuable tools in understanding species evolution and genetic diversity and in deciphering the biosynthesis of bioactive compounds in medicinal plants. Before the genome sequencing of Glycyrrhiza species, the transcriptomes of various tissues and their treatments were extensively used to identify the genes related to the biosynthesis of triterpenoids and flavonoids and those associated with plant development. A total of 814 transcriptome datasets are currently available for various Glycyrrhiza species in NCBI’s SRA database. The majority of these datasets pertain to medicinal species. Among them, G. uralensis has the highest number of datasets with 402, followed by G. inflata with 48 datasets and G. glabra with 15 datasets. These datasets provide insights into gene expression and transcription patterns in the roots, stems, and leaves and gene expression profiles under various treatments or conditions. They offer significant support for further research and analysis on these species, helping deepen our understanding of their molecular biology, responses to different treatments, and potential applications.
With the rapid development of sequencing technologies, the application of molecular genetics to nonmodel species for genomic research has become possible. In this context, we summarize the genomic research on Glycyrrhiza species. Among Glycyrrhiza plants, only G. uralensis has had its genome published, with two versions currently available (Table 1). Mochida et al. [112] first sequenced and assembled the whole genome of G. uralensis, creating a reference genome. Initially, high-quality genomic DNA was extracted from G. uralensis for genome sequencing. Illumina sequencing platform was used to generate 380.53 Gb of raw data from three different insert sizes (190 bp, 3 kb, and 10 kb) of sequencing libraries. After short and low-quality reads were filtered out, 327.53 Gb of high-quality sequences were obtained. Flow cytometry analysis estimated the genome size to be 400.95 Mb (±4 Mb), with a sequencing depth of 817 × . However, second-generation sequencing is constrained in assembling short reads into long sequences. Therefore, a third-generation sequencing technology, PacBio RS II, was further employed to obtain 6.76 Gb of data with a sequencing depth of 16.86 × . The low-quality PacBio data were corrected using high-quality Illumina data, resulting in 4.52 Gb of error-corrected long sequences. Compared with the estimated genome size, the dataset had a coverage depth of 11.27 × . Finally, the assembly results from the three sequencing approaches were combined, yielding a G. uralensis genome sequence of 379 Mb that accounted for approximately 94.5 % of the initially estimated genome size. A total of 34,445 protein-coding genes were predicted. Comparative analysis revealed that the G. uralensis genome exhibited conservation in genome composition and gene loci with other Fabaceae plants such as Medicago truncatula and Cicer arietinum. Three genes involved in isoflavones biosynthesis (CYP93C, HI4OMT, and 7-IOMT) were identified with the G. uralensis genome and formed gene clusters. On the basis of genome annotation, several genes involved in triterpenoid saponin biosynthesis were predicted within the cytochrome P450 (CYP450) and UDP-glucuronosyltransferase (UGT) superfamilies, and their gene expression was characterized using the reference genome sequence.
Table 1.
Comparison of the two versions of G. uralensis genome.
| Rai et al [113] | Mochida et al [112] | |
|---|---|---|
| Number of contigs | 54 | 72,148 |
| Total length (contigs) | 459.19 Mb | — |
| N50 (contigs) | 36.03 Mb | 7.32 kb |
| Number of scaffolds | 89 | 12,528 |
| Total length (scaffolds) | 459.21 Mb | 378.86 Mb |
| N50 (scaffolds) | 60.2 Mb | 109.27 kb |
| GC content (scaffolds) | 37.78 % | 35.34 % |
| Number of chromosomes | 8 | — |
| Total length (chromosomes) | 429.07 Mb | — |
| N50 (chromosomes) | 58.56 Mb | — |
| GC content (chromosomes) | 37.13 % | — |
| Anchored | 93.44 % | — |
| Unplaced scaffolds | 81 | — |
| Total gene number | 32,941 | 34,445 |
| Number of chromosome genes | 32,454 | — |
| BUSCO completeness (chromosomes) | 98.6 % | — |
| BUSCO completeness (annotation) | 96.5 % | — |
Obtaining a high-quality genome assembly is crucial to explore the evolutionary basis for defining chemical-type specific traits and provide essential resources for molecular breeding strategies for enhancing the production of medicinal metabolites. To achieve a high-quality G. uralensis genome, Rai et al. [113] utilized single-molecule high-fidelity (HiFi) sequencing and reported a chromosome-level genome for G. uralensis. Their analysis of heterozygosity using HiFi reads revealed a heterozygosity rate of 1.65 %, estimating a genome size of 391.9 Mb. A total of 15.43 Gb of raw PacBio HiFi reads were obtained, equivalent to approximately 38.9 times the estimated genome size. The final assembled genome consisted of 54 contigs with a total size of approximately 459 Mb and an N50 of 36.02 Mb. After scaffolding, 89 scaffolds with an N50 of 60.2 Mb were acquired, and 93.44 % of these scaffolds were anchored to eight chromosomes, totaling approximately 429 Mb. A total of 32,941 genes were annotated, with 32,454 genes located on the eight chromosomes. Whole-genome duplication (WGD) analysis showed that G. uralensis experienced a gamma (γ) WGD event approximately 59.13 million years ago (MYA), followed by a shared WGD event with other Papilionoideae species around 58 MYA. No additional WGD events were observed in the G. uralensis genome. Furthermore, metabolic gene cluster analysis identified 355 gene clusters, including the entire biosynthetic pathway of glycyrrhizic acid.
Biosynthesis and transcriptional regulation of flavonoids and triterpenoids in licorice
Biosynthesis of glycyrrhizic acid
The biosynthesis pathway of glycyrrhizinic acid mainly relies on the mevalonate (MVA) pathway, which catalyzes the direct production of the precursors of glycyrrhizinic acid, isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP) [114]. In the MVA pathway, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) is a key rate-limiting enzyme that catalyzes the irreversible conversion of HMG-CoA into MVA. It is also a key rate-limiting enzyme in the synthesis of glycyrrhizic acid and terpenes, and its expression is significantly positively correlated with the production of glycyrrhizic acid [114]. According to research, the expression level of HMGR in the hairy roots of G. uralensis is significantly positively correlated with the content of glycyrrhizic acid [115].
The catalytic synthesis of glycyrrhizic acid from the precursor IPP involves several key enzymes. In the first stage, IPP is catalyzed sequentially by geranyl diphosphate synthase (GPS), farnesyl diphosphate synthase (FPS), squalene synthase (SQS), and squalene epoxidase (SQE) to produce 2,3-oxidosqualene (2,3-OSC) [114]. 2,3-oxidosqualene is a precursor of triterpenoids and sterols, and its conversion into these compounds is mediated by various oxidosqualene cyclases (OSCs). β-amyrin synthase (β-AS) catalyzes the cyclization of 2,3-oxidosqualene to produce β-amyrin. The expression of β-AS is only observed in the underground parts, which corresponds to the accumulation of glycyrrhizic acid in the roots [114]. CYP450 are key enzymes in the second stage of glycyrrhizic acid biosynthesis. CYP88D6 is a CYP450 monooxygenase that catalyzes a consecutive two-step oxidation at the C-11 position of β-amyrin, leading to the formation of 11-oxo-β-amyrin [116]. The expression pattern of CYP88D6 is consistent with the β-AS gene, as it is exclusively expressed in the underground parts of the plant [116]. G. uralensis has a homologous gene to CYP88D6, known as Uni25647, which shares 97 % amino acid similarity with CYP88D6 and exhibits higher C-11 oxidation enzyme activity than CYP88D6 [117]. Another crucial CYP450 monooxygenase in this pathway is CYP72A154, which is responsible for three consecutive oxidations at the C-30 position of 11-oxo-β-amyrin. This enzymatic process catalyzes the production of glycyrrhizin from 11-oxo-β-amyrin [118]. Cytochrome P450 reductases (CPR) are essential enzymes in the oxidations catalyzed by CYP450 monooxygenases. In the biosynthesis pathway of glycyrrhizic acid, GuCPR1 is also involved in the oxidations at the C-11 and C-30 positions of β-amyrin [117]. Within a yeast synthetic system, GuCPR1 significantly enhances the synthesis efficiency of glycyrrhetinic acid [117].
Glycosylation modification is the final stage in the biosynthesis pathway of glycyrrhizic acid and is catalyzed by UGTs, which add a glucuronosyl group to the C-3 position of glycyrrhetinic acid. GuUGAT (also known as GuUGT3) catalyzes the glucuronosylation of glycyrrhetinic acid, directly producing glycyrrhizic acid through two consecutive glucuronosylation steps [119]. Experiments on amino acid residue mutation for GuUGAT suggested that Gln-352 plays a crucial role in the initial step of glucuronosylation, and His-22, Trp-370, Glu-375, and Gln-392 are essential in the subsequent step of glucuronosylation [119]. Expression profile analysis indicated that GuUGAT is expressed in the roots, stems, and leaves, with the highest expression found in the roots. Salt and drought stress significantly induce the expression of this gene [119].
Along with members of the cellulose synthase superfamily GuCSyGT and GuCsl, O-glycosyltransferase GuGT14 also exhibits triterpenoid glycosyltransferase activity, catalyzing the formation of 3-O-monoglucuronosyl glycyrrhetinic acid from glycyrrhetinic acid [120], [121], [122]. GuUGT73P12 is responsible for catalyzing the second step of glucuronic acid conjugation in the glycyrrhizic acid biosynthesis pathway, thereby converting 3-O-monoglucuronosyl glycyrrhetinic acid into glycyrrhizic acid [7] (Fig. 2).
Fig. 2.
Synthesis pathways of glycyrrhizic acid. AACT: Acetyl-CoA C-acetyltransferase, HMGS: Hydroxymethylglutaryl-CoA synthase, HMGR: 3-hydroxy-3-methylglutaryl-CoA reductase, MK: Mevalonate kinase, MPK: Mevalonate phosphate kinase, MPD: Mevalonate pyrophosphate decarboxylase, GPS: Geranylgeranyl diphosphate synthase, FPS: Farnesyl pyrophosphate synthase, SQS: Squalene synthase, SQE: Squalene epoxidase, β-AS: β-amyrin synthase.
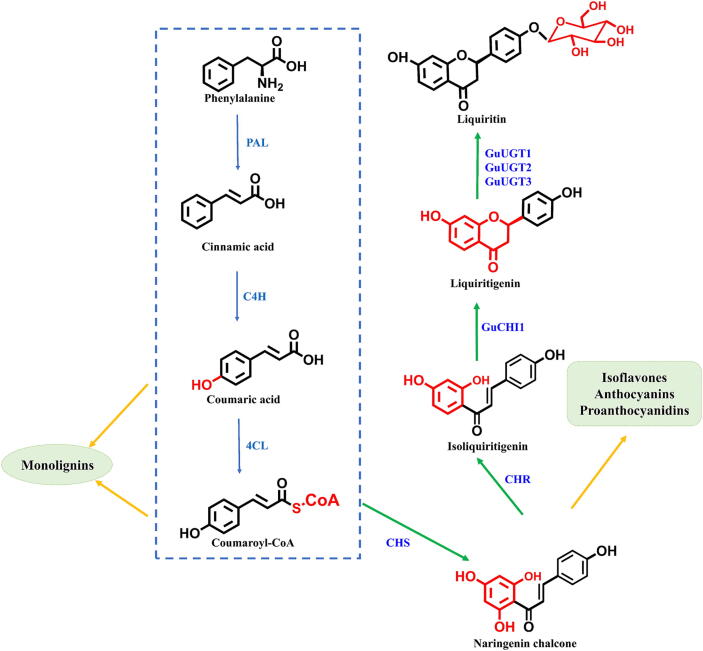
Biosynthesis of liquiritin
The biosynthesis pathway of liquiritin has been successfully elucidated (Fig. 3). Phenylalanine serves as the precursor for liquiritin. The initial three steps of its biosynthesis pathway are identical to those of other flavonoid compounds, collectively known as the common phenylalanine pathway [123]. Sequentially catalyzed by phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate-CoA ligase (4CL), phenylalanine is converted into coumaroyl-CoA. Chalcone synthase (CHS) transforms coumaroyl-CoA into naringenin chalcone, which represents a rate-limiting step in the flavonoid biosynthesis pathway. Chalcone reductase (CHR) then catalyzes the conversion of naringenin chalcone into isoliquiritigenin [123], [124]. Isoliquiritigenin undergoes further catalysis by type II chalcone isomerase GuCHI1 to produce liquiritigenin, which is then subjected to 4ʹ-O-glycosylation modification by GuUGT1, GuUGT2, and GuUGT3 in the final step of liquiritin biosynthesis, resulting in the formation of the end product liquiritin [125].
Fig. 3.
Synthesis pathways of liquiritin. PAL: Phenylalanine ammonla lyase, C4H: Cinnamate 4-hydroxylase, 4CL: 4-coumarate-CoA ligase, CHS: Chalcone synthase, CHR: Chalcone reductase, CHI: Chalcone isomerase.
Transcriptional regulation of triterpenoid and flavonoid biosynthesis in licorice
Transcription factors (TFs) are a class of proteins with unique molecular structures, typically encompassing functional domains such as a DNA-binding domain, transcriptional regulatory domains, oligomerization sites, and nuclear localization signals. These functional domains determine the characteristics and functions of TFs [126]. In plants, TFs recognize and bind to cis-acting elements within the gene promoter regions when stimulated by external factors or during plant growth. This interaction enables them to initiate and regulate gene expression, thereby facilitating the production of gene products [127] (Fig. 4). TFs can also activate or inhibit gene transcription through their interactions with each other and with other related proteins [128], [129]. According to their distinct structure and function, various types of TFs have been categorized in medicinal plant research. Some notable families being studied include MYB, bHLH, WRKY, and bZIP.
Fig. 4.
Transcription factors (TFs) regulating the biosynthesis of triterpenoids and flavonoids in licorice by transmitting signals under abiotic and biotic factors. When licorice is stimulated by external factors, stress signals are transmitted to TFs through signal transmission pathways, such as Ca2+, ROS, and systemic signalling. Then, TFs recognize and bind to cis-acting elements within the gene promoter regions, which enables TFs to initiate and regulate gene expression, thereby facilitating the production of gene products. Some TFs and genes involved in these transcriptional regulation processes have not yet been identified.
The secondary metabolites of plants are the result of the interaction between plants and biotic or abiotic factors during long-term evolution, exhibiting a diverse range of intricate biological functions [130]. The biosynthesis pathway of secondary metabolites in plants is complex and involves multiple enzymes catalysis. However, TFs can activate the coexpression of the multiple synthase genes of secondary metabolites, thereby effectively regulating the secondary metabolic pathway. Hence, the comprehensive analysis of key enzyme genes involved in plant secondary metabolite synthesis and the identification of TFs that regulate their expression have become a prominent research focus in this field.
The plant kingdom harbors a plethora of secondary metabolites with diverse structures; among which, triterpenoids and flavonoids are the predominant constituents in licorice. In this context, we elucidated the current advancements in understanding various types of TFs that govern the biosynthesis of these two classes of secondary metabolites.
Several TFs have been identified in licorice based on genomics and transcriptomics. Li et al. [131] added methyl jasmonate (MeJA) to G. uralensis cell suspension and identified differentially expressed genes through RNA-seq analysis. The results showed that the MYB TFs GuMYB4 and GuMYB88 were significantly responsive to MeJA induction. Subsequent investigation revealed the nuclear localization of GuMYB4 and GuMYB88 proteins, confirming their positive regulatory role in flavonoid synthesis in G. uralensis cells. Tamura et al. [132] identified a licorice bHLH TF called GubHLH3. Its overexpression in transgenic licorice hairy roots resulted in the increased expression of soyasaponin biosynthetic genes β-AS, CYP93E3, and CYP72A566, leading to the elevated levels of soyasapogenol B and sophoradiol. This finding suggested that GubHLH3 can positively regulate the expression of soyasaponin biosynthetic genes. Furthermore, the soyasaponin biosynthetic genes and GubHLH3 were coexpressed in response to MeJA induction. In a transcriptome analysis conducted by Goyal et al. [133], 147 full-length genes encoding WRKY TFs were identified in G. glabra. They performed a comparative structural analysis of these WRKY genes and identified 23 abiotic stress-responsive GgWRKYs and 17 hormone signal-induced GgWRKYs. In our previous work, 69 members of the bZIP TF gene family were identified in G. uralensis based on the whole-genome data [134]. From these genes, five candidates were upregulated in response to ABA stress. Overexpression and RNAi vectors were then constructed for these candidate genes and successfully performed the genetic transformation of G. uralensis hairy roots. The findings demonstrated a positive correlation between the expression levels of certain genes involved in the biosynthesis of active licorice compounds (such as CHI, CHS, HMGS, SQS, UGT, and β-AS) and the expression levels of the candidate GubZIP genes (GubZIP5, GubZIP33, and GubZIP56) in the overexpression lines and their corresponding RNAi counterparts. Furthermore, the overall change trends of the contents of total flavonoids, total triterpenoids, liquiritin, liquiritigenin, and isoliquiritigenin in the transgenic hairy roots were positively correlated with the expression levels of the candidate GubZIP genes.
Currently, the research on the transcriptional regulation of licorice is limited. Although the enzymes involved in the biosynthesis of triterpenoids and flavonoids in licorice have been studied, the regulatory factors associated with species phenotype formation, development, and bioactive compound synthesis remain unknown. Research on the transcriptional regulation of licorice primarily focuses on the functional validation of TFs and their responses to abiotic stress. Some studies have only examined the effects of biotic or abiotic factors on the content of active compounds, without identifying the specific TFs involved or which genes they regulate (Fig. 4) [135], [136], [137], [138], [139], [140].
In addition to licorice, the TFs regulating terpene and flavonoid biosynthesis in numerous other plants have been identified and extensively studied (Table 2). These studies have a positive impact on guiding the study of transcriptional regulation in licorice. Yu et al. [141] isolated two AP2 TFs, AaERF1 and AaERF2, from Artemisia annua by yeast one-hybrid and confirmed by electrophoretic mobility shift assay that they could bind CBF2 and RAA elements in ADS and CYP71AV1 promoter regions, respectively, which are the key enzymes in artemisinin synthesis pathway. The transient expression of AaERF1 and AaERF2 in Nicotiana tabacum can increase the transcription level of ADS and CYP71AV1 and the production of artemisinin and artemisinin acid. Conversely, the inhibition of AaERF1 and AaERF2 expression by RNAi technology resulted in a decrease in artemisinin and artemisinin acid contents. These findings indicated that AaERF1 and AaERF2 can positively regulate artemisinin synthesis. Zhang et al. [142] first identified six members from 64 highly expressed bZIP TFs in the glandular trichomes of A. annua. Along with ADS and CYP71AVI genes, these six TFs were then introduced into N. tabacum leaves through transformation. Dual-luciferase assay results predicted that AabZIP1 could activate the promoters of these two key enzyme genes. Furthermore, yeast one-hybrid experiments demonstrated that AabZIP1 could directly bind to the ABRE promoter element of ADS and CYP71AV1, thereby activating their expression. The overexpression vector AabZIP1 was then constructed and transformed into A. annua, which significantly increased the content of artemisinin, indicating that the TF AabZIP1 positively regulates artemisinin synthesis. The mechanism of ABA-mediated artemisinin biosynthesis was also elucidated through experiments. The results showed that ABA enhanced the transcriptional activity of AabZIP1 towards the promoters of ADS and CYP71AV1 genes. Moreover, the ABA receptor protein AaPYL9 was isolated from A. annua plants with overexpressed AabZIP1 gene and significantly increased the artemisinin content, suggesting that AabZIP1 plays a key role in ABA-induced artemisinin synthesis. The TFs involved in ABA signal transduction pathway have also been studied in G. uralensis[143], [144], [145], [146]. WRKY (AaWRKY1) [147], [148], bHLH (AabHLH1) [149], NAC (AaNAC1) [150] and MYB (AaMYB1) [151] TFs also regulate artemisinin synthesis. The regulation of terpenoid synthesis by TFs has also been studied in other medicinal plants. The MYB36 and MYB9b of Salvia miltiorrhiza were involved in the regulation of tanshinone synthesis [152], [153]. JAZ is an inhibitor of jasmonic acid signal transduction, and the overexpression of SMJAZ1/2/3/9 significantly promoted the accumulation of tanshinone in the fibrous roots of S. miltiorrhiza [154], [155]. In the glandular trichomes of Mentha spicata, MsMYB directly binds to the cis-acting elements of the large subunit of GPP synthase (MsGPPS.LSU), inhibits gene expression, and negatively regulates terpenoid synthesis [156].
Table 2.
Identified transcription factors involved in the terpenoid and flavonoid biosynthesis of plants.
| Type of compositions | Type of TFs | Transcription factors |
Species | Reference |
|---|---|---|---|---|
| Terpenoids | AP2/ERF | AaORA | Artemisia annua | [168] |
| TAR1 | Artemisia annua | [169] | ||
| TcAP2 | Taxus cuspidata | [170] | ||
| bHLH | AabHLH1 | Artemisia annua | [171] | |
| AabHLH112 | Artemisia annua | [172] | ||
| AaMYC2 | Artemisia annua | [173] | ||
| GubHLH3 | Glycyrrhiza uralensis | [132] | ||
| GubHLHs | Glycyrrhiza uralensis | [174] | ||
| TcJAMYC | Taxus cuspidata | [175] | ||
| SmMYC2a | Salvia miltiorrhiza | [176] | ||
| SmbHLH10 | Salvia miltiorrhiza | [177] | ||
| SmbHLH148 | Salvia miltiorrhiza | [178] | ||
| SmbHLH3 | Salvia miltiorrhiza | [179] | ||
| BpbHLH9 | Betula platyphylla | [180] | ||
| TSAR1 | Medicago truncatula | [181] | ||
| TSAR2 | Medicago truncatula | [181] | ||
| bZIP | AabZIP1 | Artemisia annua | [142] | |
| AabZIP9 | Artemisia annua | [182] | ||
| GubZIP5 | Glycyrrhiza uralensis | [134] | ||
| GubZIP33 | Glycyrrhiza uralensis | [134] | ||
| GubZIP56 | Glycyrrhiza uralensis | [134] | ||
| EIN3 | AaEIN3 | Artemisia annua | [183] | |
| MYB | GuMYBs | Glycyrrhiza uralensis | [174], [184] | |
| GmMYBZ2 | Glycine max | [185] | ||
| PatSWC4 | Pogostemon cablin | [186] | ||
| SmMYB4 | Salvia miltiorrhiza | [187] | ||
| SmMYB36 | Salvia miltiorrhiza | [152] | ||
| SmMYB52 | Salvia miltiorrhiza | [188] | ||
| SmMYB98 | Salvia miltiorrhiza | [189] | ||
| SmMYB111 | Salvia miltiorrhiza | [190] | ||
| TH | PatGT-1 | Pogostemon cablin | [191] | |
| WRKY | AaWRKY1 | Artemisia annua | [147], [148] | |
| AaWRKY9 | Artemisia annua | [192] | ||
| AaGSW1 | Artemisia annua | [193] | ||
| AaGSW2 | Artemisia annua | [194] | ||
| YABBY | AaYABBY5 | Artemisia annua | [195] | |
| MsYABBY5 | Mentha spicata | [196] | ||
| Flavonoids | AP2/ERF | CitERF32 | Citrus reticulata cv. Suavissima | [197] |
| CitERF33 | Citrus reticulata cv. Suavissima | [197] | ||
| bHLH | GubHLHs | Glycyrrhiza uralensis | [174] | |
| GibHLH7 | Glycyrrhiza inflata | [198] | ||
| AmPerilla | Antirrhinum majus | [199] | ||
| AmDelila | Antirrhinum majus | [200] | ||
| AmbHLH-1 | Antirrhinum majus | [201] | ||
| AmbHLH-2 | Antirrhinum majus | [201] | ||
| GtbHLH1 | Gentiana triflora | [202] | ||
| LmbHLH113 | Lonicera macranthoides | [203] | ||
| bZIP | GubZIP5 | Glycyrrhiza uralensis | [134] | |
| GubZIP33 | Glycyrrhiza uralensis | [134] | ||
| GubZIP56 | Glycyrrhiza uralensis | [134] | ||
| MYB | GlMYB4 | Glycyrrhiza uralensis | [131] | |
| GlMYB88 | Glycyrrhiza uralensis | [131] | ||
| GlMYB84 | Glycyrrhiza uralensis | [174] | ||
| GuMYBs | Glycyrrhiza uralensis | [174] | ||
| LmMYB12 | Lonicera macranthoides | [203] | ||
| LmMYB75 | Lonicera macranthoides | [203] | ||
| SmMYB39 | Salvia miltiorrhiza | [204] | ||
| AmRosea1 | Antirrhinum majus | [200], [205] | ||
| AmRosea2 | Antirrhinum majus | [205] | ||
| GtMYB3 | Gentiana triflora | [202] | ||
| DwMYB9 | Dendrobium hybrid Woo Leng | [206] | ||
| InMYB1 | Ipomoea nil | [207] | ||
| GhMYB1 | Gerbera hybrida | [208] | ||
| WDR | InWDR1 | Ipomoea nil | [207] |
The TFs regulating flavonoid synthesis have been extensively investigated across various plant species. In Epimedium sagittatum, EsMYBF1 can activate the promoters of EsF3H and EsFLS genes. Its overexpression in N. tabacum resulted in a significant upregulation of the key genes involved in flavonol biosynthesis including CHS, CHI, F3H, F3′H, and FLS, leading to a substantial increase in flavonol content [157].
Similarly, CsMYBF1 was found to bind and activate Cs4CL, CsCHS and CsFLS promoters in Citrus reticulata. Its inhibition using RNAi technology in C. reticulata significantly reduced flavonol content [158]. However, Xu et al. [159] observed that the overexpression of GbMYBF2 from Ginkgo biloba in Arabidopsis thaliana led to a notable reduction in transcription levels of CHS, F3H, FLS, and ANS genes and the inhibition of anthocyanin and other flavonoid synthesis. This finding suggested that GbMYBF2 plays a negative regulatory role. The GtMYBP3 and GtMYBP4 of Gentiana triflora can activate the expression of genes related to flavonol synthesis, and their overexpression in A. thaliana can significantly increase the flavonol content in seedlings [160]. The color of Rubus idaeus fruit could be red, gold, or black. Anthocyanin, a secondary metabolite of flavonoids, serves as an important factor influencing fruit coloration. bZIP TFs can affect anthocyanin synthesis in red R. idaeus [161]. After the mutation of MTWD40-1 gene in Medicago truncatula, a decrease in isoflavone synthesis within roots was observed, suggesting the potential involvement of MTWD40-1 TF in plant flavonoid biosynthesis [162].
A variety of TFs coregulate the synthesis of metabolic components. Certain plant species have a complex structure known as the MBW complex, which is composed of MYB, bHLH, and WD403 proteins. This complex participates in the regulation of flavonoid biosynthesis. For instance, the MBW complex TT2/TT8/TTG1 in A. thaliana regulates the synthesis of proanthocyanidins in the seed coat [163], [164], [165]. In the leaves of Petunia hybrida, AN1 (bHLH) and AN2 (MYB) proteins coregulate the synthesis of pigments. In anthers, AN1 relies on AN4 (MYB) to activate the structural gene DFR of anthocyanin synthesis pathway [166]. In their study on the regulation of anthocyanin synthesis in Myrica rubra fruits, Liu et al. [167] found that MrbHLH1 could form a transcription complex with MrMYB1 and promote anthocyanin accumulation by selectively activating structural genes.
Synthetic biology of glycyrrhizic acid and liquiritin
Licorice has medicinal value and dietary importance. However, it has low concentrations of specific active ingredients, such as glycyrrhizic acid and liquiritin. The extraction of these natural active components from plants cannot meet the increasing demand. Microbial metabolic engineering has been developed and applied to efficiently produce plant-specific active ingredients. Various strategies have been developed and introduced for the biosynthesis of licorice triterpenoids and flavonoids. One approach is genetic manipulation and fermentation engineering with the use of microbial cell factories for in vitro synthesis.
Fine-tuning the expression ratio of CYP450 enzymes and CPR can optimize the catalytic activity of CYP88D6 oxidation–reduction system. This optimization enables the efficient production of 11-oxo-β-amyrin, the direct precursor of glycyrrhetinic acid, in brewing yeast [209]. Initially, CRISPR-Cas9 technology was employed to knock out the ERG27 and BTS1 genes in brewing yeast, increasing the yield of squalene by 6.91 times. A precursor β-amyrin synthesis pathway was then constructed in this engineered strain, and metabolic engineering strategies were employed to optimize the yield. Finally, a CYP88D6-CPR fusion protein was constructed, and different CPRs from various plant sources were investigated to optimize the CYP88D6 oxidation system, ultimately leading to the improved yield of 11-oxo-β-amyrin.
Sun et al. [209] focused on studying the expression levels of cytochrome CYP88D6 and CPR, and their ratio’s effect on 11-oxo-β-amyrin yield. The results revealed that high CPR expression levels led to growth defects. Moreover, an increased CPR:CYP88D6 expression ratio decreased the concentration of 11-oxo-β-amyrin catalyzed by CYP88D6. Conversely, high CYP88D6:CPR expression ratios improved the efficiency of electron transfer between CYP88D6 and CPR, thereby increasing 11-oxo-β-amyrin yield. With these optimizations, a conversion rate of 91.2 % from β-amyrin to 11-oxo-β-amyrin was achieved in brewing yeast Y321. During fed-batch fermentation, the yield of 11-oxo-β-amyrin was further increased to 810.6 mg/L, laying the foundation for the construction of high-yield glycyrrhetinic acid strains.
Zhu et al. [117] constructed a glycyrrhetinic acid biosynthesis pathway in yeast and demonstrated that CPR can significantly enhance the efficiency of glycyrrhetinic acid synthesis. Previous studies have demonstrated that CYP72A63 is capable of utilizing 11-oxo-β-amyrin as a substrate to generate glycyrrhetinic acid in yeast-engineered strains [117]. However, due to low selectivity, the products consist of a mixture of rare licorice triterpenoids, including glycyrrhetol, glycyrrhetaldehyde, glycyrrhetinic acid, and an isomer of glycyrrhetol, 29-OH-11-oxo-β-amyrin, in a ratio of 45.2:21.4:29.0:4.4. The difficulty in separating and purifying these compounds has limited the production of glycyrrhetinic acid. Sun et al. [210] engineered CYP72A63 mutants to selectively produce each of the four compounds through a combination of homology modeling, molecular docking, and mutagenesis experiments. Model analysis indicated that the typical proton transfer pathway is essential for P450 activity, and that restrictions in proton transfer hinder catalytic performance. By identifying key amino acids involved in proton transfer and conducting further mutagenesis experiments, the catalytic activity of CYP72A63 mutants was improved. Then Sun et al. [211] regulated the genes NHMGR, SIP4, and GPP1 to establish metabolic and microenvironmental compatibility between the licorice triterpenoid biosynthesis pathway and the yeast host, thereby achieving efficient synthesis of licorice triterpenoids in yeast. In a 5L fermenter, the production of licorice triterpenoids reached 4.92 g/L, including 0.96 g/L of 11-oxo-β-amyrin, 2.94 g/L of glycyrrhetaldehyde, and 1.02 g/L of glycyrrhetinic acid. After successfully accomplishing the complete biosynthesis of β-amyrin and glycyrrhetinic acid, the two pivotal precursors of glycyrrhizic acid and 3-O-monoglucuronosyl glycyrrhetinic acid in Saccharomyces cerevisiae, Xu et al. [212] pioneered the whole-cell factory synthesis of glycyrrhizic acid and 3-O-monoglucuronosyl glycyrrhetinic acid in yeast. They achieved this goal through the exploration, characterization, and introduction of glycosyltransferases and UGTs from mammals. The yields reached 5.98 and 2.31 mg/L, respectively. This innovative approach offers a new idea to addressing resource scarcity issues in the production of glycyrrhizic acid and other scarce natural products.
Significant advances have also been achieved in the synthetic biology of flavonoids in licorice. One research searched the G. uralensis genome and comparative transcriptome databases, analyzed and selected homologous genes in known flavonoid biosynthesis pathways, and reconstructed the entire liquiritin biosynthesis pathway in yeast [213]. This study used in vitro and in vivo methods to validate the catalytic functions of candidate genes and achieved the de novo synthesis of liquiritigenin in S. cerevisiae for the first time, utilizing endogenous yeast metabolites as precursors and cofactors. GuPAL1 and GuC4H1 were also simultaneously introduced into the yeast strain WAT11, resulting in a recombinant yeast called WM1 that could produce p-coumaric acid under galactose induction. After 36 h of cultivation, the p-coumalic acid content reached 7.59 mmol/L. When the vectors carrying Gu4CL1 and GuCHS1::GuCHR1 were introduced into WM1, a yeast strain named WM2-1 capable of producing small amounts of isoliquiritigenin was obtained. No isoliquiritigenin was detected when GuCHS1 and GuCHR1 were coexpressed with substrates in yeast strain WM1. GuCHI1 and GuUGT1 were subsequently introduced into WM2-1, resulting in the yeast strain WM3-1, which is capable of producing liquiritigenin, and WM4-1, which is capable of producing liquiritin. In WM4-1, a higher proportion of isoliquiritigenin was glycosylated before isomerization, leading to a 4.7-fold higher yield of isoliquiritoside compared with liquiritin.
To enhance the conversion of upstream metabolites toward downstream flavonoid structures along the liquiritin pathway in the described recombinant strains, this study attempted to overexpress the key genes involved in isoliquiritigenin biosynthesis [213]. The results showed that yeast WM2-2, which overexpressed GuCHS1::GuCHR1, significantly increased isoliquiritigenin production. The isoliquiritigenin yield in WM2-2 was 18 times higher than that in WM2-1. GuCHI1 was overexpressed in WM3-1 and WM4-1 to address the issue of significantly lower liquiritin production compared with isoliquiritoside in WM4-1. This process led to a 1.3-fold increase in liquiritigenin and a 2.1-fold increase in liquiritin production. Furthermore, the simultaneous overexpression of GuCHS1::GuCHR1 and GuCHI1 resulted in a 5.3-fold increase in liquiritigenin production and a 6.4-fold increase in liquiritin production.
Yin et al. [213] also found that overexpressing GuCHI1 and GuCHS1::GuCHR1 in the yeast system increased the yield of downstream products and significantly increased the accumulation of upstream precursor compounds such as p-coumaric acid. This phenomenon suggested interactions between different genes in the liquiritin biosynthesis pathway. In addition, the downstream products of the liquiritin biosynthesis pathway might play a role in the positive or negative feedback on the upstream pathway. This type of cross-talk and regulation is common in metabolic pathways, where the production of one compound can influence the synthesis of another at different points in the pathway. Understanding these interactions is crucial for optimizing the production of specific compounds through metabolic engineering.
In addition to liquiritin, synthetic biology research on other flavonoid compounds has also made breakthroughs. Wang et al. [214] identified and functionally characterized the apiosyltransferase GuApiGT in G. uralensis. They reconstructed the de novo synthesis pathway of liquiritin apioside and isoliquiritin apioside in Nicotiana benthamiana by introducing 12 genes. The pathway was divided into three modules. For flavonoid aglycone module, AtPAL, AtC4H, At4CL, AtCHS and GuCHR were used to synthesize isoliquiritigenin. Pgm, GalU, CalS8 and UAXS were used for UDP-donor module to produce UDP-Glc and UDP-Api. For glycosyltransferase module, GuApiGT and GuGT14 were used as glycosyltransferases. This approach successfully achieved the total synthesis of liquiritin apioside (5.46 mg/g) and isoliquiritin apioside (4.73 mg/g) in N. benthamiana, with yields comparable to those found in licorice root. Additionally, by altering the gene combination in the flavonoid module, the authors achieved the total synthesis of eight other apiosides.
Conclusions and future prospects
Glycyrrhiza plants and their active compounds have extensive medicinal prospects. These medicinal plants have a history of thousands of years of use in traditional Chinese medicine and have been cultivated to meet the demands for herbal medicine and chemical extraction. Molecular design approaches for producing improved licorice varieties and efficiently producing triterpenoids and flavonoids have been widely researched. Furthermore, the utilization of resources from other licorice species, in addition to those defined in pharmacopeias, is important for pharmaceutical and healthcare products. A forward-looking strategy based on multiomic approaches has been proposed and involves exploring the genetic resources of the Glycyrrhiza genus and improving the quality, biosynthesis, and regulation of active compounds and the efficiency of in vitro production (Fig. 5).
Fig. 5.
Strategies for the development and utilization of resources of Glycyrrhiza plants. Firstly, the resources of Glycyrrhiza species are collected to obtain their multi-omics information, such as genome, transcriptome and metabolome. Through these genetic information, the unknown biosynthesis pathway of active ingredients in licorice could be analyzed. The target medicinal ingredients are then produced industrially through molecular breeding and synthetic biology. Molecular breeding methods mainly include molecular marker-assisted breeding, genetic engineering breeding, and molecular designing breeding. The synthetic biology of licorice could be improved in terms of biological elements, metabolic pathways, chassis cells and fermentation process. Finally, these medicinal ingredients are used for drug development and clinical treatment of diseases.
Population genomics of Glycyrrhiza
Conducting large-scale comparative analyses of metabolism and genome is an effective strategy for elucidating the mechanisms behind the metabolic diversity of certain compounds within a taxonomic group. Medicinal plants contain a wealth of secondary metabolites closely related to their pharmacological activities. However, the synthesis of these secondary metabolites in plants is a complex process, with significant variation in both the structure and content of these metabolites [215]. Over the past two decades, a considerable number of species genomes have been sequenced, providing comprehensive support for advancements in medicinal plant genomics and establishing a theoretical foundation for revealing the biosynthetic pathways of secondary metabolites [216].
Recent studies have explored the metabolic diversity of the same type of compounds within a taxonomic group by focusing on functional validation, structural analysis, and comparative genomics. For instance, in the genus Papaver, species like P. rhoeas, P. setigerum, and P. somniferum exhibit varying levels of noscapine and morphinan compounds, with P. somniferum containing the highest amounts [217]. Yang et al. [217] constructed high-quality genomes for these three species and analyzed the relationship among copy numbers of key genes in the morphinan synthesis pathway, gene expression, and morphinan content. They found that the CODM, T6ODM, and COR genes are under positive selection in P. somniferum, providing new insights into the evolution of the benzylisoquinoline alkaloid biosynthetic pathway. In the case of abietane-type diterpenes (ATDs), specific to the genus Salvia, Hu et al. [218] demonstrated that the chemical diversity of ATDs is driven by the functional diversification of the cytochrome P450 subfamily CYP76AK, resulting in oxidation differences at the C-20 position of the terpene skeleton. This diversification results in three distinct catalytic features with noticeable phylogenetic divergence, shaping the chemical diversity of ATDs within Salvia plants. An et al. [219] conducted comparative genomic studies on Menispermaceae species, showing that stereoselective differences and functional diversification (C-C linkage differences) among CYP80G members in different species have led to the structural diversity of benzylisoquinoline alkaloids.
Research on Glycyrrhiza species is somewhat limited. Molecular biology and pharmacological studies have predominantly focused on G. uralensis, G. glabra, and G. inflata. Research on other licorice species within this genus has lagged behind, especially in terms of multiomics. Although the genome of G. uralensis has been published, an ongoing research aims to sequence and assemble the genomes of the other two species mentioned in pharmacopeias, namely, G. glabra, and G. inflata. In addition to the species used for medicinal purposes, genomic studies must also be conducted on nonmedicinal species. Such research can explore the evolution of enzyme genes in the glycyrrhizic acid biosynthesis pathway, trace the origins of these genes, and investigate why some species produce glycyrrhizic acid and others do not. This information is of great significance for breeding high-yield licorice varieties that produce glycyrrhizic acid and for the field of synthetic biology related to glycyrrhizic acid biosynthesis.
Biosynthetic pathway elucidation of active compounds in licorice
Licorice contains numerous active compounds that can be used for medicinal purposes, but only the biosynthetic pathways of glycyrrhizic acid and liquiritin have been fully elucidated. Most natural products share common upstream metabolic pathways that have been relatively well-studied, including the mevalonate pathway, methylerythritol phosphate pathway, shikimate pathway, and malonic acid pathway [220]. Therefore, elucidating the biosynthetic pathway of a specific natural product, or a particular class of them, typically refers to uncovering its downstream, specialized branch metabolic pathways. To investigate the unknown biosynthetic pathways of active compounds in licorice, the following strategies can be employed.
Firstly, hypothesize potential biosynthetic pathways based on isolated and identified intermediate compounds, and further confirm these pathways through isotope tracing. Hypothesizing a possible metabolic pathway based on existing knowledge is the first step in elucidating natural product biosynthetic pathways [221]. The principles of chemical reactions and the structures of intermediate compounds can both provide clues for predicting the synthesis pathway. Isotope tracing is commonly used for pathway prediction and validation. Isotope-labeled precursor compounds have the same biological and chemical properties as unlabeled compounds but differ in molecular mass, allowing them to be distinguished through mass spectrometry and nuclear magnetic resonance instruments. Thus, after feeding isotope-labeled precursors, any newly isotope-labeled compounds detected are considered intermediates or end products of the metabolic pathway [222].
Next, use transcriptome analysis to obtain co-expression data of related genes or conduct genome scans to identify gene clusters associated with secondary metabolism, thus narrowing down candidate genes. The rapid development of high-throughput sequencing technology has made it possible to quickly and affordably obtain the genetic information of a species. However, selecting candidate genes potentially involved in the biosynthesis of specific natural products from so many genes remains a highly challenging task. Currently, co-expression analysis based on transcriptome sequencing and gene cluster mining based on genome sequencing are the two main methods for identifying candidate genes [223], [224].
Finally, heterologously express the candidate genes and test enzyme activity to confirm their catalytic functions, and develop a licorice genetic transformation system to study the inhibition or overexpression of relevant enzyme-coding genes in the original species, further verifying enzyme function within the species. By identifying and validating the functions of all enzymes in the pathway, the goal of pathway elucidation can ultimately be achieved. Gene co-expression analysis and gene cluster mining can effectively narrow down the range of candidate genes, reducing the workload for functional validation. Due to the structural diversity of natural products, the enzymes involved in their biosynthetic pathways also exhibit great diversity, meaning that each enzyme’s functional validation requires a unique approach. Functional verification of candidate genes involves a combination of experimental techniques in molecular biology and analytical chemistry, such as heterologous expression and enzyme activity assays, gene suppression, and gene overexpression [225], [226], [227]. This is the most critical and challenging step in elucidating natural product biosynthetic pathways.
The elucidation of the biosynthetic pathways of natural products in licorice will provide a wealth of components for synthetic biology research on active compounds in licorice, advancing the development of this field and offering new sources for the production and research of natural drugs derived from licorice. Additionally, pathway elucidation will provide functional molecular markers for molecular breeding research in licorice, accelerating the selection and cultivation of high-quality varieties and promoting the growth of the licorice industry.
Breeding of licorice
The wild resources of licorice are scarce, and the active compound content in cultivated licorice is generally low, failing to meet pharmacopoeia standards [228]. There is an urgent need to improve important traits and quality in licorice breeding. Licorice breeding has a long history. As early as the 1990 s, researchers reported studies on radiation breeding of licorice [229]. The results showed that plants selected from treatments with 60,000 roentgen of gamma-ray irradiation exhibited excellent medicinal traits, including straight stems, dense texture, rich sweetness, reddish-brown skin, and thicker bark. Additionally, these plants had desirable qualities such as higher dry weight and increased glycyrrhizic acid content. These are the main traits and internal qualities that licorice breeding research aims to acquire and continuously improve. Unfortunately, no new variety was developed from this work. In 2014, the first new licorice variety in China, “Guogan No. 1” was successfully bred. Compared to other licorice sources, the seeds of “Guogan No. 1” have lower values for seed traits such as thousand-seed weight, length, width, and height. However, “Guogan No. 1” exhibits more vigorous above-ground growth and higher levels of key medicinal components [230].
Traditional breeding methods include direct selection of naturally superior varieties, hybrid breeding, space breeding, and artificial induction of polyploidy breeding [231]. As a perennial herbaceous plant with a long growing cycle, licorice typically takes 3 to 5 years before it can be harvested, making the breeding process quite lengthy. The breeding of “Guogan No. 1” took 13 years [230]. As early as the late 20th century, farmers began selecting high-quality licorice during the transition from wild to cultivated licorice. This was primarily done by collecting and planting seeds based on their judgment of the quality of licorice from traditional regions. However, due to the sparse distribution of wild licorice and low seed yield, large-scale cultivation and the development of new varieties did not occur [230]. Given the long breeding cycles, high technical difficulty of seed propagation, and slow reproduction rate, the production cost of seeds for new licorice varieties is higher than collecting wild seeds, which makes it challenging for average growers to accept during promotion. As quality control for medicinal materials and traceability in the production process of raw herbs become increasingly stringent, high-quality, high-yield, and disease-resistant new varieties are expected to dominate the market in the future. However, the existing new varieties are far from meeting market demands, so the breeding of new licorice varieties must continue to advance.
Therefore, there is an urgent need to shorten the breeding cycle through effective breeding methods in order to rapidly and efficiently cultivate new licorice varieties with high active compound content, superior quality, and stress resistance. This will help improve the quality and yield of licorice raw materials, reduce production costs, and enhance the supply of high-quality raw materials. Plant molecular breeding is an important approach to improving the quality of medicinal plants [232]. By using molecular breeding techniques, combining the phenotypes and genotypes of licorice, it is possible to increase breeding efficiency, shorten the breeding cycle, and select new licorice varieties with superior quality, high yield, and strong resistance.
With the establishment of a licorice germplasm resource bank and the publication of genetic information such as genomes, transcriptomes, and genetic maps, the future direction of licorice breeding will focus on rapidly cultivating superior varieties by utilizing molecular breeding methods, including molecular marker-assisted breeding, genetic engineering breeding, and molecular designing breeding. Based on the genetic background and biological characteristics of licorice, as well as the current state of plant molecular breeding technology, a licorice molecular breeding strategy is proposed: 1) Collecting germplasm resources of licorice and selecting superior traits. 2) Screening high-quality molecular markers and associating them with desirable traits to enable rapid selection of germplasm. 3) Identifying functional genes related to high content and resistance for use in genetic engineering breeding. 4) Integrating omics data and breeding technologies to implement molecular designing breeding from gene design to whole-system breeding.
The first step involves collecting licorice germplasm resources and recording traits related to active ingredient content, yield of medicinal parts, and resistance. There are significant differences in the levels of medicinal components between the roots and aerial parts of different licorice plants. Additionally, there are varying degrees of correlation between licorice growth characteristics and the levels of active components [26]. One study conducted morphological observations and quantitative measurements of key medicinal active ingredients, while also examining three aspects: the sexual reproductive characteristics of licorice, germplasm resource screening, and the cloning and expression patterns of the key enzyme genes in the glycyrrhizic acid metabolic pathway. It was found that white-flowered types of licorice have better medicinal quality [233].
Trait-linked genes and molecular markers (e.g., RAPD, SRAP, SSR, SNP, Indel, CNV) can be identified by constructing genetic populations and conducting resequencing for marker-assisted breeding. An SSR molecular marker analysis showed that 64 of 69 randomly designed and synthesized SSR pairs were successfully amplified, and 33 pairs of primers exhibited polymorphism in G. uralensis, G. inflata, G. glabra. and G. pallidiflora [234]. Cluster analysis effectively distinguished cultivated licorice from different traditional regions, but licorice grown from seeds and seedlings in the same region did not cluster well. The genetic diversity of cultivated licorice from different regions was diverse, and those within the same region, genetic diversity varied due to different cultivation generations. This suggests that some genetic diversity may be lost after multiple generations of sexual reproduction, possibly due to environmental selection, providing a theoretical basis for the breeding and cultivation of high-quality licorice.
By exploring the published genetic information of licorice, including its whole genome, chloroplast genome, transcriptome, and genetic maps, TFs and key genes related to the yield of medicinal parts, active ingredient content, and resistance can be identified. As mentioned earlier, several versions of licorice genome and transcriptome have been published [112], [113], providing a rich database and genetic background for analyzing the biosynthetic pathways of licorice's active components. Additionally, based on these data, TFs associated with high-quality traits in licorice have also been discovered [131], [132], [134], [174]. TFs play a crucial regulatory role in the growth and development of plants, enabling them to respond to biotic and abiotic stresses. The accumulation of specialized metabolites in plants is closely associated with the regulation of TFs and their adaptability to the environment [235]. The molecular mechanisms by which TFs regulate the synthesis of licorice secondary metabolites by participating in abiotic stress signaling pathways remain largely unexplored [127]. A significant gap exists in our understanding of this field, and further research is needed to uncover these molecular mechanisms. Genome-wide association study (GWAS) is a powerful tool that helps us understand the relationship between genetic variations and various important traits in natural populations. Through GWAS, regulatory factors or functional genes can be accurately identified, contributing to a deep understanding of plant biological processes, including growth, development, and metabolism. In addition, the discovery of new genes and TFs provides valuable resources for molecular plant breeding, aiding in the improvement of plant traits and adaptability and thereby enhancing medicinal plant yield, quality, and stress resistance. The application of GWAS has already achieved numerous significant breakthroughs in the fields of plant genetics and molecular breeding, offering endless possibilities for future research and medicinal plant improvement [236], [237], [238].
Genetic engineering breeding is a breeding method that utilizes genetic engineering technology to transfer foreign genes into a specific plant, enabling it to express a desired trait through regulation. Genetic engineering breeding can overcome gene exchange barriers between different species, addressing challenges that traditional breeding methods cannot easily solve [232]. Currently, Agrobacterium-mediated transformation is commonly used for genetic engineering in licorice. This method is widely applied in plant genetic engineering due to its high transformation efficiency, precise insertion of large DNA fragments, stable genetic expression, and relatively simple technical and equipment requirements [131]. The biosynthetic pathways of two medicinal components in licorice, glycyrrhizic acid and liquiritin, have been fully elucidated. Research has shown that overexpressing key enzyme genes (e.g., CHS, CHI, HMGR, SQS, β-AS) in the pathways or TFs (e.g., GubHLH3, GuMYB88, GubZIPs) that control these key genes can increase the content of glycyrrhizic acid or liquiritin [131], [132], [239], [240], [241]. The CRISPR-Cas9 system has been successfully applied in licorice. The study by Wang et al. [242] showed that hair roots with ARPI or UGE knockout quickly died after induction, indicating that these genes are essential for the growth of licorice hair roots, while hair roots with β-AS knockout grew healthily but were unable to produce glycyrrhizic acid. Gene knockout and base editing technologies based on CRISPR-Cas will aid in studying gene functions in licorice and enable precise editing of key sites in enzymes involved in the biosynthesis of active compounds, which will help identify and develop enzymes with higher catalytic activity, effectively enhancing the content of active compounds in licorice [7]. This strategy not only directly creates high-value licorice germplasm with elevated active compound content for industrial applications but also efficiently identifies key active residues of catalytic enzymes, promoting the advancement of basic research on enzyme catalysis mechanisms.
Additionally, molecular designing breeding can also be applied to licorice breeding. It is a method based on whole-genome sequencing that involves screening and optimizing various factors in the breeding process to design a target genotype with desirable traits, enabling the selection of parental combinations and offspring screening for the target genotype [243]. Molecular design breeding represents the future direction of licorice molecular breeding, allowing a shift from traditional breeding to efficient, targeted precision breeding. Wang et al. [244] defined three steps involved in molecular designing breeding: 1) Identifying trait loci associated with breeding targets through genes/quantitative trait locis (QTLs) or their closely linked markers. 2) Selecting new varieties with target genotypes that meet the conditions and requirements of different ecotypes and breeding objectives. 3) Implementing a breeding plan based on established goals, utilizing techniques such as marker-assisted selection, association analysis, QTL analysis, genetic engineering, genome-wide selection, and mutagenesis to carry out design breeding. The main focus of molecular designing breeding research is the efficient discovery of QTLs associated with important agronomic traits and the establishment of genetic information support for core germplasm resources and key parental lines. Molecular designing breeding leverages elite germplasm resources identified during the breeding process, selecting suitable design elements based on the target genotype to enable multi-gene assembly breeding, significantly shortening the breeding cycle. Additionally, molecular designing breeding requires careful selection of parental combinations, thereby reducing the workload of field trials and effectively improving breeding efficiency. Based on published genome, metabolome, and transcriptome data of licorice, target genotypes aligned with breeding goals are identified, along with relevant metabolic pathways and transcriptional regulatory pathways. This enables efficient and precise molecular designing breeding of licorice through comprehensive design.
Application of synthetic biology in Glycyrrhiza
Glycyrrhizic acid and liquiritin are the main medicinal components of licorice. Although their biosynthetic pathways have been fully elucidated and their synthetic pathways have been constructed in yeast expression systems, their in vitro production remains very low. Achieving industrial production levels for the desired products remains a highly challenging task. To enhance the yield of target products in licorice synthetic biology, the following optimization strategies can be implemented.
Biological elements include ribosome-binding site, origins of replication, promoters, terminators, functional proteins. Adjustments to individual elements, such as increasing gene copy number, promoter modification, and codon optimization, can effectively increase the expression levels of pathway enzyme genes and improve biochemical reaction efficiency [245]. It can significantly boost the production of important precursor substances and target products in engineered microorganisms.
Modifying individual elements within a biosynthetic pathway can improve the yield of the target product to some extent. However, local optimizations often lead to the accumulation of intermediates, increasing cellular burden and potentially causing growth-inhibitory toxicity, which hinders the efficient production of the desired product. Therefore, enhancing target product yield through systematic optimization of metabolic pathways is also a critical aspect of licorice synthetic biology [246]. The systematic optimization of metabolic pathway mainly includes multiple module metabolic engineering, multiplex automated genome engineering, multiplex iterative plasmid engineering, customized optimization of metabolic pathways by combinatorial transcriptional engineering, and tunable intergenic regions [220].
Differences in chassis cell properties can significantly impact the yield of natural products. Selecting the right chassis cells is a key step in synthetic biology. Chassis cells currently used include Escherichia coli, Saccharomyces cerevisiae, plant cells, and other microorganisms. Primary or secondary metabolites from these chassis cells provide precursors for the heterologous biosynthesis of natural products. Optimizing the chassis system can enhance substrate supply for target metabolic pathways, redirect pathway flux, and increase the yield of natural products. Firstly, modifying or optimizing genes in the metabolic pathway can enhance precursor supply and improve substrate utilization efficiency. Secondly, directing more synthesis into the specific pathways for glycyrrhizic acid and liquiritin allows more precursors to flow into these pathways, thus increasing their yield. Yin et al. [213] enhanced the biosynthesis of liquiritin in yeast by overexpressing pathway genes CHI, CHS, and CHR. At the same time, downregulation or blocking of competing branch pathway genes reduces diversion into other pathways. Sun et al. [209] constructed a yeast strain by deleting the BTS1 and ERG27 genes, which decreases metabolic flux toward branch pathways in the MVA pathway, directing more flux toward glycyrrhizic acid synthesis. Finally, reconstructing metabolic pathways in heterologous chassis cells, using more active isozymes to replace native enzymes, increases the catalytic efficiency of enzymes in the reconstructed pathway, effectively enhancing target metabolite production in engineered strains. The whole-cell factory synthesis of glycyrrhizic acid and 3-O-monoglucuronosyl glycyrrhetinic acid in yeast was realized through the exploration, characterization, and introduction of glycosyltransferases and UGTs from mammals [212]. These strategies effectively increase substrate utilization efficiency and balance the entire metabolic pathway, directly impacting the final product yield.
Target product yield can also be improved by optimizing cultivation and production conditions. Culture conditions significantly impact the production of glycyrrhizic acid, liquiritin, and their precursor substances in engineered strains. Selecting appropriate conditions can allow for tailored optimizations for different production targets. Sun et al. [209] constructed a high-yield β-amyrin yeast strain using yeast’s own compounds as starting substrates, which is directly linked to yeast growth state. Therefore, high-density fermentation with precise control of environmental factors such as temperature, glucose concentration, airflow, and stirring rate enables rapid production of large amounts of cells and target products in a shorter time.
By modifying host cells and precisely controlling the expression of exogenous genes, it is possible to achieve coordinated expression of multiple genes, maximize the efficiency of licorice biosynthetic system, and increase the yield of target products. This provides new lead compounds for the development of natural licorice-based drugs.
Glycyrrhiza species have a history of clinical use as medicinal herbs for thousands of years, and their highly diverse morphological and chemical properties have garnered significant research attention. Many resources of Glycyrrhiza species remain largely untapped, and multiomic studies, breeding, and synthetic biology of the Glycyrrhiza lineage are expected to facilitate new discoveries in the fields of medicine and human health.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 32070368), National Key R&D Program of China (No. 2023YFC3504104), and the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS) (No. 2021-I2M-1-071). Fig. 4, Fig. 5 in the review were drawn by BioRender (https://www.biorender.com/).
Biographies

Liwei Wu PhD candidate. Affiliation: Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China. Research Directions: Molecular identification of traditional Chinese medicine; Functional genes of medicinal plants.

Tingyu Ma PhD / Research assistant. Affiliation: Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China. Research Directions: Biosynthesis of natural products; Molecular-assisted breeding of medicinal plants.

Chenxi Zang PhD candidate. Affiliation: Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China. Research Directions: Molecular identification of traditional Chinese medicine; Functional genes of medicinal plants.

Zhichao Xu PhD / Professor. Affiliation: College of Life Science, Northeast Forestry University, Harbin, China. Research Directions: Biosynthesis; Metabolic engineering; Genomic evolution.

Wei Sun PhD / Professor. Affiliation: Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China. Research Directions: Medicinal resources; Metabolic engineering; Molecular identification of traditional Chinese medicine.

Hongmei Luo PhD / Professor. Affiliation: Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China. Research Directions: Functional genomics of medicinal plants; Analysis of secondary metabolic pathway.

Meihua Yang PhD / Professor. Affiliation: Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China. Research Directions: Quality control and analysis of Chinese medicine; Exogenous pollutants and safety of Chinese medicine.

Jingyuan Song PhD / Professor. Affiliation: Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China. Research Directions: Medicinal resources; Herbgenomics.

Shilin Chen PhD / Professor. Affiliation: Institute of Herbgenomics, Chengdu University of Traditional Chinese Medicine, Chengdu, China Research Directions: Medicinal resources; Herbgenomics. Achievement: Shilin Chen works as the Academician of Chinese Academy of Engineering and director of the World Health Organization Collaborating Center for Traditional Medicine.

Hui Yao PhD / Professor. Affiliation: Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China. Research Directions: Molecular identification of traditional Chinese medicine; Functional genes of medicinal plants. Achievement: Hui Yao has published more than 150 papers, including internationally renowned journals, and was listed in the “Top 2% of Global Scientists - Annual Scientific Influence List”.
Contributor Information
Shilin Chen, Email: slchen@cdutcm.edu.cn.
Hui Yao, Email: scauyaoh@sina.com.
References
- 1.Zheng Y., Wei J., Leng K., Tao W., Fang S., Peng G., et al. Research advances in resource chemistry and utilization of genus Glycyrrhiza. Mod Chin Med. 2015;17:1096–1108. [Google Scholar]
- 2.Yang R., Wang L., Liu Y. Research progress on tissue culture of Glycyrrhizae Radix et Rhizoma, Chin Tradit Herb. Drugs. 2014;45:1796–1802. [Google Scholar]
- 3.Commission C.P. China Medical Science Press; Beijing: 2020. Pharmacopoeia of the People's Republic of China. [Google Scholar]
- 4.Yang B., Fang Y. Application of Radix Glycyrrhizae in harmonizing Chinese medicine, Shanghai. J Tradit Chin Med. 2012;46:91–94. [Google Scholar]
- 5.Zhao J., Liu F., Lin H. Effects of the inducement on callus with different explants of Glycyrrhiza uralensis. Guangdong Agric Sci. 2011;38:36–38. [Google Scholar]
- 6.Deng T., Peng C., Peng D., Yu N., Chen W., Wang L. Research progress on chemical constituents and pharmacological effects of Glycyrrhizae Radix et Rhizoma and discussion of Q-markers. China J Chin Mater Med. 2021;46:2660–2676. doi: 10.19540/j.cnki.cjcmm.20210304.201. [DOI] [PubMed] [Google Scholar]
- 7.Zhou D., Li H., Tong X., Yu F., Kong D., Liu Y. Research progress in the biosynthesis and regulation of glycyrrhizic acid and liquiritin, Biotechnol. Bulletin of the Georgian Academy of Sciences. 2023;39:65–74. [Google Scholar]
- 8.Shin Y.W., Bae E.A., Lee B., Lee S.H., Kim J.A., Kim Y.S., et al. In vitro and in vivo antiallergic effects of Glycyrrhiza glabra and its components. Planta Med. 2007;73:257–261. doi: 10.1055/s-2007-967126. [DOI] [PubMed] [Google Scholar]
- 9.Shin E.M., Zhou H.Y., Guo L.Y., Kim J.A., Lee S.H., Merfort I., et al. Anti-inflammatory effects of glycyrol isolated from Glycyrrhiza uralensis in LPS-stimulated RAW264.7 macrophages. Int Immunopharmacol. 2008;8:1524–1532. doi: 10.1016/j.intimp.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Hawthorne S., Gallagher S. Effects of glycyrrhetinic acid and liquorice extract on cell proliferation and prostate-specific antigen secretion in LNCaP prostate cancer cells. J Pharm Pharmacol. 2008;60:661–666. doi: 10.1211/jpp.60.5.0013. [DOI] [PubMed] [Google Scholar]
- 11.Ming L.J., Yin A.C. Therapeutic effects of glycyrrhizic acid. Nat Prod Commun. 2013;8:415–418. [PubMed] [Google Scholar]
- 12.Li W., Song X., Zhang L., Li W., Yu H., An J. Advances in research on chemical constituents of Radix Glycyrrhizae. J Liaoning Univ Tradit Chin Med. 2012;14:40–44. [Google Scholar]
- 13.Li G., Zhou C., Jiang X., He Z. Research progress on cultivation of licorice and glycyrrhizin biosynthesis and regulation. J Chin Med Mater. 2004;27:462–465. [Google Scholar]
- 14.Yao H., Zhao Y., Chen D.F., Chen J.K., Zhou T.S. ISSR primer screening and preliminary evaluation of genetic diversity in wild populations of Glycyrrhiza uralensis. Biol Plant. 2008;52:117–120. [Google Scholar]
- 15.Ye X., Xue J., Xie X., Huang Z. Effects of different disturbances on plant growth and content of main medicinal ingredients of rhizomatous clonal plant Glycyrrhiza uralensis in a natural population, Chin. J Plant Ecol. 2020;44:951–961. [Google Scholar]
- 16.Xiang Y. Beijing University of Chinese Medicine; Beijing: 2015. Study on the relationship between secondary metabolites of licorice based on glycyrrhizic acid content model, in. [Google Scholar]
- 17.Huang M., Wang W., Wei S. Investigation on medicinal plant resources of Glycyrrhiza uralensis in China and chemical assessment of its underground part. China J Chin Mater Med. 2010;35:947–952. doi: 10.4268/cjcmm20100802. [DOI] [PubMed] [Google Scholar]
- 18.Wei S., Wang W., Wang J., Sun Z., Liu C., Wang H., et al. Preliminary study in glycyrrhizin content and its influencing factors of wild and cultivated in different region of China. China J Chin Mater Med. 2012;37:1341–1345. [PubMed] [Google Scholar]
- 19.Li X., Lu J. Fudan University Press; Shanghai: 2015. Taxonomy and experimental biology of the genus Glycyrrhiza L. [Google Scholar]
- 20.Kruganova E. A review of species from the genera Glycyrrhiza L. and Meristotropis Fish. et CA Mey. Acta Inst Bot Academ Sci Slovacae. 1955;11:161–197. [Google Scholar]
- 21.Meng L. Institute of Botany, Chinese Academy of Sciences; Beijing: 2005. Systematics of Glycyrrhiza L. (Fabaceae)—With a special reference to its relationship to Glycyrrhizopsis Boiss. [Google Scholar]
- 22.Duan L., Han L., Liu B., Leostrin A., HarrisHarris A., Wang L., et al. Species delimitation of the liquorice tribe (Leguminosae: Glycyrrhizeae) based on phylogenomic and machine learning analyses. J Syst Evol. 2023;61:22–41. [Google Scholar]
- 23.Wu L., Fan P., Cai J., Zang C., Lin Y., Xu Z., et al. Comparative genomics and phylogenomics of the genus Glycyrrhiza (Fabaceae) based on chloroplast genomes. Front Pharmacol. 2024;15:1371390. doi: 10.3389/fphar.2024.1371390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi H., Miwa E., Inoue K. Phylogenetic relationship of Glycyrrhiza lepidota, American licorice, in genus Glycyrrhiza based on rbcL sequences and chemical constituents. Biol Pharm Bull. 2005;28:161–164. doi: 10.1248/bpb.28.161. [DOI] [PubMed] [Google Scholar]
- 25.Duan L., Harris A.J., Su C., Zhang Z.R., Arslan E., Ertuğrul K., et al. Chloroplast phylogenomics reveals the intercontinental biogeographic history of the liquorice genus (Leguminosae: Glycyrrhiza) Front Plant Sci. 2020;11:793. doi: 10.3389/fpls.2020.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q., Ye M. Chemical analysis of the Chinese herbal medicine Gan-Cao. licoriceJ Chromatogr A. 2009;1216:1954–1969. doi: 10.1016/j.chroma.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y., Li Z., Ma L., Guo H., Chen Y., Fu X., et al. Simultaneous determination of 4 flavonoid glycosides in licorice and application in original plants identification. Chin Pharm J. 2011;46:1112–1116. [Google Scholar]
- 28.Yang R., Li W., Yuan B., Ma Y., Zhou S., Liu C., et al. Simultaneous determination of 18α-glycyrrhizic acid and 18β-glycyrrhizic acid in three licorice samples from different origin by HPLC. Chin J Pharm Anal. 2016;36:1065–1071. [Google Scholar]
- 29.Liu Q., Guo S., Zheng X., Shen X., Zhang T., Liao B., et al. Licorice germplasm resources identification using DNA barcodes inner-variants. Plants (Basel) 2021;10:2036. doi: 10.3390/plants10102036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang R., Li W., Ma Y., Zhou S., Xue Y., Lin R., et al. The molecular identification of licorice species and the quality evaluation of licorice slices. Acta Pharm Sin. 2017;52:318–326. [PubMed] [Google Scholar]
- 31.Zhang J., Hu D., Liu S., Wu J. Research progress on active components and pharmacological effects of common glycyrrhiza varieties. J Tradit Chin Vet Med. 2012;31:23–27. [Google Scholar]
- 32.Kitagawa I., Zhou J., Sakagami M., Uchida E., Yoshikawa M. Licorice-saponins F3, G2, H2, J2, and K2, five new oleanene-triterpene oligoglycosides from the root of Glycyrrhiza uralensis. Chem Pharm Bull. 1991;39:244–246. [Google Scholar]
- 33.Astaf'eva O.V., Sukhenko L.T. Comparative analysis of antibacterial properties and chemical composition of Glycyrrhiza glabra L. from Astrakhan region (Russia) and Calabria region (Italy) Bull Exp Biol Med. 2014;156:829–832. doi: 10.1007/s10517-014-2462-8. [DOI] [PubMed] [Google Scholar]
- 34.Montoro P., Maldini M., Russo M., Postorino S., Piacente S., Pizza C. Metabolic profiling of roots of liquorice (Glycyrrhiza glabra) from different geographical areas by ESI/MS/MS and determination of major metabolites by LC-ESI/MS and LC-ESI/MS/MS. J Pharm Biomed Anal. 2011;54:535–544. doi: 10.1016/j.jpba.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Peng L., Hu Z. Research progress on biological and chemical constituents of licorice, Chin Tradit Herb. Drugs. 2005;36:1744–1747. [Google Scholar]
- 36.Zhao Y., Zhang R., Liu M. Study on chemical constituents of Glycyrrhiza inflata (Ⅰ) J Peking Univ (Health Sciences) 1990;22:283–284. [Google Scholar]
- 37.Sun L., Yang Y. Isolation and identification of chemical constituents from Glycyrrhiza inflata Bat. Anhui Med Pharm J. 2013;17:1121–1123. [Google Scholar]
- 38.Zhang J., Liu F., Li N., Qing D., Sun Y., Jia X., et al. Identification of flavonoids gruffs of Glycyrrhiza inflate waste by UPLC-TOF-MS. Drugs & Clinic. 2012;27:558–561. [Google Scholar]
- 39.Liu F., Ni H., Qing D. Chemical constituents from the residues of Glycyrrhiza inflata Batal., Chin. J Med Chem. 2011;21:312–314. [Google Scholar]
- 40.Li N., Liu F., Ni H., Meng D., Zhang J., Jia X. Isolation and identification on chemical constituents of residue of Glycyrrhiza inflata Batal. J Shenyang Pharm Univ. 2011;28:368–370. [Google Scholar]
- 41.Li W., Asada Y., Yoshikawa T. Flavonoid constituents from Glycyrrhiza glabra hairy root cultures. Phytochemistry. 2000;55:447–456. doi: 10.1016/s0031-9422(00)00337-x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang B., Guan Y., Tian Q., Liu H. Research progress on chemical constituents of flavonoids in Glycyrrhiza uralensis. Chin Tradit Pat Med. 2006;28:1362–1365. [Google Scholar]
- 43.Yang L., Liu Y., Lin S. Analysis of flavonoids in the roots of six species of Glycyrrhiza by high performance liquid chromatography. Acta Pharm Sin. 1990;25:840–848. [PubMed] [Google Scholar]
- 44.Li N., Zhang C., Zhong G., Xiu L., Liu H., Chen S., et al. Research progress on chemical constituents and pharmacological effects of different varieties of Glycyrrhizae Radix et Rhizoma and predictive analysis of quality markers, Chin Tradit Herb. Drugs. 2021;52:7680–7692. [Google Scholar]
- 45.Gupta V.K., Fatima A., Faridi U., Negi A.S., Shanker K., Kumar J.K., et al. Antimicrobial potential of Glycyrrhiza glabra roots. J Ethnopharmacol. 2008;116:377–380. doi: 10.1016/j.jep.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Zhang H., Shi Y., Wang W. Inhibition of clycyrrhize polysaccharide on virus. Acta Sci Nat Univ Nankai. 2001;34:126–128. [Google Scholar]
- 47.Wang Y., Zhang H., Lv J., Shi Y., He M., Wang S., et al. Research on the biological activities of a polysaccharide extracted from residue of Glycyrrhiza uralensis Fisch. Acta Sci Nat Univ Nankai. 2000;33:46–48. [Google Scholar]
- 48.Cong Y. Xinjiang Medical University; Urumchi: 2008. Studies on isolation, purification, structural elucidation and biological activities of polysaccharides from Glycyrrhiza inflata Bat. [Google Scholar]
- 49.Li W., Song X., Sun C., Xia Q. Content determination of polysaccharides in Radix Glycyrrhizae from three different species, Tianjin. J Tradit Chin Med. 2013;30:47–49. [Google Scholar]
- 50.Li W., Xia Q., Sun C., Song X. Analysis of monosaccharide compositions of various kinds of liquorice polysaccharide by HPLC precolumn derivatization. J Liaoning Univ Tradit Chin Med. 2014;16:56–58. [Google Scholar]
- 51.Lu J., Cao J., Li W., Niu X., Zhao Y. A new coumarins from residue of Glycyrrhiza uralensis, Chin Tradit Herb. Drugs. 2015;46:174–177. [Google Scholar]
- 52.Liu Y. Beijing University of Chinese Medicine; Beijing: 2011. Establishment of multi-index detection method for quality evaluation of licorice and its application in identification of different sources of licorice, in. [Google Scholar]
- 53.Yang X., Zou K., Yao B., Zhang R. NMR study on coumarin A of Glycyrrhiza inflata, Chin. J Magn Reson. 1994;11:211–214. [Google Scholar]
- 54.Zhang J., Yao J., Yang Y., Wang Y., Gu L., Wang Y. Analysis and contents mensurate of alkaloid in liquorice. Acta Bot Boreali Occidentalia Sin. 2001;21:211–214. [Google Scholar]
- 55.Ma J., Zhang J., Wang Y., Yao J., Yang Y. Studies on the composition of the volatile oil from the roots of Glycyrrhiza uralensis. Acta Pharm Sin. 2006;15:120–123. [Google Scholar]
- 56.Peng L. Jilin Agricultural University; Changchun: 2016. Study glycyrrhetinic acid derivatives and screening of biological activity, in. [Google Scholar]
- 57.Liang X., Yin X., Li M. Distribution of micro-elements in Glycyrrhiza uralensis Fisch. and its effect on the quality of medicinal materials. J Anhui Agric Sci. 2008;36:11411–11412. [Google Scholar]
- 58.Song W., Qiao X., Chen K., Wang Y., Ji S., Feng J., et al. Biosynthesis-based quantitative analysis of 151 secondary metabolites of licorice to differentiate medicinal Glycyrrhiza species and their hybrids. Anal Chem. 2017;89:3146–3153. doi: 10.1021/acs.analchem.6b04919. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y. Yanbian University; Yanji: 2013. Study on antibacterial activity of extractive and active constituent of Glycyrrhizae, in. [Google Scholar]
- 60.Tsukiyama R., Katsura H., Tokuriki N., Kobayashi M. Antibacterial activity of licochalcone A against spore-forming bacteria. Antimicrob Agents Chemother. 2002;46:1226–1230. doi: 10.1128/AAC.46.5.1226-1230.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu D. Peking Union Medical College; Beijing: 2016. The therapeutic effect of licorice flavonoids and licochalcone A on ulcerative colitis and its molecular mechanism, in. [Google Scholar]
- 62.Gangwar B., Kumar S., Darokar M.P. Glabridin averts biofilms formation in methicillin-resistant Staphylococcus aureus by modulation of the surfaceome. Front Microbiol. 2020;11:1779. doi: 10.3389/fmicb.2020.01779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chandrasekaran C.V., Deepak H.B., Thiyagarajan P., Kathiresan S., Sangli G.K., Deepak M., et al. Dual inhibitory effect of Glycyrrhiza glabra (GutGard™) on COX and LOX products. Phytomedicine. 2011;18:278–284. doi: 10.1016/j.phymed.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 64.Kim J.Y., Kang J.S., Kim H.M., Ryu H.S., Kim H.S., Lee H.K., et al. Inhibition of bone marrow-derived dendritic cell maturation by glabridin. Int Immunopharmacol. 2010;10:1185–1193. doi: 10.1016/j.intimp.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 65.Chakotiya A.S., Tanwar A., Narula A., Sharma R.K. Alternative to antibiotics against Pseudomonas aeruginosa: Effects of Glycyrrhiza glabra on membrane permeability and inhibition of efflux activity and biofilm formation in Pseudomonas aeruginosa and its in vitro time-kill activity. Microb Pathog. 2016;98:98–105. doi: 10.1016/j.micpath.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y., Wang L., Luo R., Sun Y., Zou M., Wang T., et al. Glycyrrhizic acid against Mycoplasma gallisepticum-induced inflammation and apoptosis through suppressing the MAPK pathway in chickens. J Agric Food Chem. 2022;70:1996–2009. doi: 10.1021/acs.jafc.1c07848. [DOI] [PubMed] [Google Scholar]
- 67.Ma C., Ma Z., Liao X.L., Liu J., Fu Q., Ma S. Immunoregulatory effects of glycyrrhizic acid exerts anti-asthmatic effects via modulation of Th1/Th2 cytokines and enhancement of CD4(+)CD25(+)Foxp3+ regulatory T cells in ovalbumin-sensitized mice. J Ethnopharmacol. 2013;148:755–762. doi: 10.1016/j.jep.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 68.Adianti M., Aoki C., Komoto M., Deng L., Shoji I., Wahyuni T.S., et al. Anti-hepatitis C virus compounds obtained from Glycyrrhiza uralensis and other Glycyrrhiza species. Microbiol Immunol. 2014;58:180–187. doi: 10.1111/1348-0421.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Chen X., Wang W., Zhang Y., Yang Z., Jin Y., et al. Glycyrrhizic acid as the antiviral component of Glycyrrhiza uralensis Fisch. against coxsackievirus A16 and enterovirus 71 of hand foot and mouth disease. J Ethnopharmacol. 2013;147:114–121. doi: 10.1016/j.jep.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soufy H., Yassein S., Ahmed A.R., Khodier M.H., Kutkat M.A., Nasr S.M., et al. Antiviral and immune stimulant activities of glycyrrhizin against duck hepatitis virus. Afr J Tradit Complement Altern Med. 2012;9:389–395. doi: 10.4314/ajtcam.v9i3.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun Z.G., Zhao T.T., Lu N., Yang Y.A., Zhu H.L. Research progress of glycyrrhizic acid on antiviral activity. Mini Rev Med Chem. 2019;19:826–832. doi: 10.2174/1389557519666190119111125. [DOI] [PubMed] [Google Scholar]
- 72.Zuo J., Meng T., Wang Y., Tang W. A review of the antiviral activities of glycyrrhizic acid, glycyrrhetinic acid and glycyrrhetinic acid monoglucuronide. Pharmaceuticals. 2023;16:641. doi: 10.3390/ph16050641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu S., Zhu Y., Xu J., Yao G., Zhang P., Wang M., et al. Glycyrrhizic acid exerts inhibitory activity against the spike protein of SARS-CoV-2. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao Z., Xiao Y., Xu L., Liu Y., Jiang G., Wang W., et al. Glycyrrhizic acid nanoparticles as antiviral and anti-inflammatory agents for COVID-19 treatment. ACS Appl Mater Interfaces. 2021;13:20995–21006. doi: 10.1021/acsami.1c02755. [DOI] [PubMed] [Google Scholar]
- 75.Xu H., Li S., Liu J., Cheng J., Kang L., Li W., et al. Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19. Proc Natl Acad Sci USA. 2023;120 doi: 10.1073/pnas.2301775120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin J.C., Cherng J.M., Hung M.S., Baltina L.A., Baltina L., Kondratenko R. Inhibitory effects of some derivatives of glycyrrhizic acid against Epstein-Barr virus infection: structure-activity relationships. Antiviral Res. 2008;79:6–11. doi: 10.1016/j.antiviral.2008.01.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang M., Fan J., Zhu W., Li X. Inhibition effect of extracts of G.inflata Bat. on mushroom tyrosinase. Chin J Appl Chem. 2012;29:898–899. [Google Scholar]
- 78.Xu C., Liang C., Sun W., Chen J., Chen X. Glycyrrhizic acid ameliorates myocardial ischemic injury by the regulation of inflammation and oxidative state. Drug Des Devel Ther. 2018;12:1311–1319. doi: 10.2147/DDDT.S165225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen J., Li W.C., Gu X.L. Optimized Extraction, Preliminary Characterization, and In Vitro Antioxidant Activity of Polysaccharides from Glycyrrhiza Uralensis Fisch. Med Sci Monit. 2017;23:1783–1791. doi: 10.12659/MSM.900471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hong Y.K., Wu H.T., Ma T., Liu W.J., He X.J. Effects of Glycyrrhiza glabra polysaccharides on immune and antioxidant activities in high-fat mice. Int J Biol Macromol. 2009;45:61–64. doi: 10.1016/j.ijbiomac.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 81.Maocheng X., Ding J., Li G. A comparative study on scavenging reactive oxygen species of Glycyrrhiza glabra and G. uralensis. J Plant Res Env. 2003;12:29–31. [Google Scholar]
- 82.Ojha S., Golechha M., Kumari S., Bhatia J., Arya D.S. Glycyrrhiza glabra protects from myocardial ischemia-reperfusion injury by improving hemodynamic, biochemical, histopathological and ventricular function. Exp Toxicol Pathol. 2013;65:219–227. doi: 10.1016/j.etp.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 83.Cong Y., Kasmu J., Abudzi P., Chen J., Wang X. Extraction and in vitro antioxidant activity of Glycyrrhiza inflata Bat. Xinjiang, J Chin Med Mater. 2009;32:1435–1438. [Google Scholar]
- 84.Pan L.C., Zhu Y.M., Zhu Z.Y., Xue W., Liu C.Y., Sun H.Q., et al. Chemical structure and effects of antioxidation and against α-glucosidase of natural polysaccharide from Glycyrrhiza inflata Batalin. Int J Biol Macromol. 2020;155:560–571. doi: 10.1016/j.ijbiomac.2020.03.192. [DOI] [PubMed] [Google Scholar]
- 85.Sun G., Luo Z. Anti-aging effect of liquiritin on aging model rats. Chin J Gerontol. 2014;34:1895. [Google Scholar]
- 86.Wei F., Jiang X., Gao H.Y., Gao S.H. Liquiritin induces apoptosis and autophagy in cisplatin (DDP)-resistant gastric cancer cells in vitro and xenograft nude mice in vivo. Int J Oncol. 2017;51:1383–1394. doi: 10.3892/ijo.2017.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma M., Zhou X., Hu Y., Xiao H., Li X. Comparison in anti-proliferation effects of active components from Glycyrrhiza uralensis on four kinds of human cancer cells. Lishizhen Med Mater Med Res. 2008;19:9–11. [Google Scholar]
- 88.Park S.Y., Lim S.S., Kim J.K., Kang I.J., Kim J.S., Lee C., et al. Hexane-ethanol extract of Glycyrrhiza uralensis containing licoricidin inhibits the metastatic capacity of DU145 human prostate cancer cells. Br J Nutr. 2010;104:1272–1282. doi: 10.1017/S0007114510002114. [DOI] [PubMed] [Google Scholar]
- 89.Liao B., Ablat S., Ablish M., Ren B., Yang X. Study on the effect of crude extract and purified extract of Xinjiang Glycyrrhiza inflata Batalin on cervical cancer cells. Northwest Pharm J. 2017;32:472–476. [Google Scholar]
- 90.Cho J.J., Chae J.I., Yoon G., Kim K.H., Cho J.H., Cho S.S., et al. Licochalcone A, a natural chalconoid isolated from Glycyrrhiza inflata root, induces apoptosis via Sp1 and Sp1 regulatory proteins in oral squamous cell carcinoma. Int J Oncol. 2014;45:667–674. doi: 10.3892/ijo.2014.2461. [DOI] [PubMed] [Google Scholar]
- 91.Cai S., Bi Z., Bai Y., Zhang H., Zhai D., Xiao C., et al. Glycyrrhizic acid-induced differentiation repressed stemness in hepatocellular carcinoma by targeting c-Jun N-Terminal Kinase 1. Front Oncol. 2019;9:1431. doi: 10.3389/fonc.2019.01431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cai Y., Zhao B., Liang Q., Zhang Y., Cai J., Li G. The selective effect of glycyrrhizin and glycyrrhetinic acid on topoisomerase IIα and apoptosis in combination with etoposide on triple negative breast cancer MDA-MB-231 cells. Eur J Pharmacol. 2017;809:87–97. doi: 10.1016/j.ejphar.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 93.Farooqui A., Khan F., Khan I., Ansari I.A. Glycyrrhizin induces reactive oxygen species-dependent apoptosis and cell cycle arrest at G(0)/G(1) in HPV18(+) human cervical cancer HeLa cell line. Biomed Pharmacother. 2018;97:752–764. doi: 10.1016/j.biopha.2017.10.147. [DOI] [PubMed] [Google Scholar]
- 94.Zhang X., Zhao S., Song X., Jia J., Zhang Z., Zhou H., et al. Inhibition effect of glycyrrhiza polysaccharide (GCP) on tumor growth through regulation of the gut microbiota composition. J Pharmacol Sci. 2018;137:324–332. doi: 10.1016/j.jphs.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 95.Gou S.H., Liu J., He M., Qiang Y., Ni J.M. Quantification and bio-assay of α-glucosidase inhibitors from the roots of Glycyrrhiza uralensis Fisch. Nat Prod Res. 2016;30:2130–2134. doi: 10.1080/14786419.2015.1114940. [DOI] [PubMed] [Google Scholar]
- 96.Kent U.M., Aviram M., Rosenblat M., Hollenberg P.F. The licorice root derived isoflavan glabridin inhibits the activities of human cytochrome P450S 3A4, 2B6, and 2C9. Drug Metab Dispos. 2002;30:709–715. doi: 10.1124/dmd.30.6.709. [DOI] [PubMed] [Google Scholar]
- 97.Carmeli E., Fogelman Y. Antioxidant effect of polyphenolic glabridin on LDL oxidation. Toxicol Ind Health. 2009;25:321–324. doi: 10.1177/0748233709103034. [DOI] [PubMed] [Google Scholar]
- 98.Lai T., Shen Y., Chen C., Huang B., Deng T., Zhao Z., et al. Glycyrrhizic acid ameliorates myocardial ischemia-reperfusion injury in rats through inhibiting endoplasmic reticulum stress. Eur J Pharmacol. 2021;908 doi: 10.1016/j.ejphar.2021.174353. [DOI] [PubMed] [Google Scholar]
- 99.Cao Z.Y., Liu Y.Z., Li J.M., Ruan Y.M., Yan W.J., Zhong S.Y., et al. Glycyrrhizic acid as an adjunctive treatment for depression through anti-inflammation: A randomized placebo-controlled clinical trial. J Affect Disord. 2020;265:247–254. doi: 10.1016/j.jad.2020.01.048. [DOI] [PubMed] [Google Scholar]
- 100.Murck H., Lehr L., Hahn J., Braunisch M.C., Jezova D., Zavorotnyy M. Adjunct therapy with Glycyrrhiza glabra rapidly improves outcome in depression-A pilot study to support 11-beta-hydroxysteroid dehydrogenase type 2 inhibition as a new target. Front Psychiatry. 2020;11 doi: 10.3389/fpsyt.2020.605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paudel Y.N., Angelopoulou E., Semple B., Piperi C., Othman I., Shaikh M.F. Potential neuroprotective effect of the HMGB1 inhibitor glycyrrhizin in neurological disorders. ACS Chem Neurosci. 2020;11:485–500. doi: 10.1021/acschemneuro.9b00640. [DOI] [PubMed] [Google Scholar]
- 102.Rashedinia M., Saberzadeh J., Khosravi Bakhtiari T., Hozhabri S., Arabsolghar R. Glycyrrhizic acid ameliorates mitochondrial function and biogenesis against aluminum toxicity in PC12 cells. Neurotox Res. 2019;35:584–593. doi: 10.1007/s12640-018-9967-2. [DOI] [PubMed] [Google Scholar]
- 103.Han S., Sun L., He F., Che H. Anti-allergic activity of glycyrrhizic acid on IgE-mediated allergic reaction by regulation of allergy-related immune cells. Sci Rep. 2017;7:7222. doi: 10.1038/s41598-017-07833-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Y., Zhang L., Zhang Y., Xu J.J., Sun L.L., Li S.Z. The protective role of liquiritin in high fructose-induced myocardial fibrosis via inhibiting NF-κB and MAPK signaling pathway. Biomed Pharmacother. 2016;84:1337–1349. doi: 10.1016/j.biopha.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 105.Wang W., Hu X., Zhao Z., Liu P., Hu Y., Zhou J., et al. Antidepressant-like effects of liquiritin and isoliquiritin from Glycyrrhiza uralensis in the forced swimming test and tail suspension test in mice. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1179–1184. doi: 10.1016/j.pnpbp.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 106.Sun S., He X., Chai W., Lv C., Lv A., Yu C. Immunoregulation function of Glycyrrhizae Radix et Rhizoma polysaccharide to γδT cells from human peripheral blood. Chin J Exp Tradit Med Form. 2013;19:242–245. [Google Scholar]
- 107.Ayeka P.A., Bian Y., Githaiga P.M., Zhao Y. The immunomodulatory activities of licorice polysaccharides (Glycyrrhiza uralensis Fisch.) in CT 26 tumor-bearing mice. BMC Complement Altern Med. 2017;17:536. doi: 10.1186/s12906-017-2030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ahn J., Um M., Choi W., Kim S., Ha T. Protective effects of Glycyrrhiza uralensis Fisch. on the cognitive deficits caused by beta-amyloid peptide 25–35 in young mice. Biogerontology. 2006;7:239–247. doi: 10.1007/s10522-006-9023-0. [DOI] [PubMed] [Google Scholar]
- 109.Park S.H., Kang J.S., Yoon Y.D., Lee K., Kim K.J., Lee K.H., et al. Glabridin inhibits lipopolysaccharide-induced activation of a microglial cell line, BV-2, by blocking NF-kappaB and AP-1. Phytother Res. 2010;24(Suppl 1):S29–S34. doi: 10.1002/ptr.2872. [DOI] [PubMed] [Google Scholar]
- 110.Somjen D., Katzburg S., Vaya J., Kaye A.M., Hendel D., Posner G.H., et al. Estrogenic activity of glabridin and glabrene from licorice roots on human osteoblasts and prepubertal rat skeletal tissues. J Steroid Biochem Mol Biol. 2004;91:241–246. doi: 10.1016/j.jsbmb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 111.Kim Y.H., Shin E.K., Kim D.H., Lee H.H., Park J.H., Kim J.K. Antiangiogenic effect of licochalcone A. Biochem Pharmacol. 2010;80:1152–1159. doi: 10.1016/j.bcp.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 112.Mochida K., Sakurai T., Seki H., Yoshida T., Takahagi K., Sawai S., et al. Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. 2017;89:181–194. doi: 10.1111/tpj.13385. [DOI] [PubMed] [Google Scholar]
- 113.Rai A., Hirakawa H., Rai M., Shimizu Y., Shirasawa K., Kikuchi S., et al. Chromosome-scale genome assembly of Glycyrrhiza uralensis revealed metabolic gene cluster centred specialized metabolites biosynthesis. DNA Res. 2022;29:dsac043. doi: 10.1093/dnares/dsac043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han Y.X., Jia Q.J., Yang D.F., Chai W.G., Zhang X.M., He Q.L., et al. Current advances in environmental stimuli regulating the glycyrrhizic acid biosynthesis pathway. Fitoterapia. 2021;151 doi: 10.1016/j.fitote.2021.104860. [DOI] [PubMed] [Google Scholar]
- 115.Liu Y., Zhan X., Li W., Gao Y., Wen H., Chen H., et al. Copy number variations of functional genes influence contents of glycyrrhizic acid in Glycyrrhiza uralensis. Acta Physiol Plant. 2014;36:1433–1440. [Google Scholar]
- 116.Seki H., Ohyama K., Sawai S., Mizutani M., Ohnishi T., Sudo H., et al. Licorice beta-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA. 2008;105:14204–14209. doi: 10.1073/pnas.0803876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu M., Wang C., Sun W., Zhou A., Wang Y., Zhang G., et al. Boosting 11-oxo-β-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants. Metab Eng. 2018;45:43–50. doi: 10.1016/j.ymben.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 118.Seki H., Sawai S., Ohyama K., Mizutani M., Ohnishi T., Sudo H., et al. Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell. 2011;23:4112–4123. doi: 10.1105/tpc.110.082685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu G., Cai W., Gao W., Liu C. A novel glucuronosyltransferase has an unprecedented ability to catalyse continuous two-step glucuronosylation of glycyrrhetinic acid to yield glycyrrhizin. New Phytol. 2016;212:123–135. doi: 10.1111/nph.14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chung S.Y., Seki H., Fujisawa Y., Shimoda Y., Hiraga S., Nomura Y., et al. A cellulose synthase-derived enzyme catalyses 3-O-glucuronosylation in saponin biosynthesis. Nat Commun. 2020;11:5664. doi: 10.1038/s41467-020-19399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen K., Hu Z.M., Song W., Wang Z.L., He J.B., Shi X.M., et al. Diversity of O-Glycosyltransferases contributes to the biosynthesis of flavonoid and triterpenoid glycosides in Glycyrrhiza uralensis. ACS Synth Biol. 2019;8:1858–1866. doi: 10.1021/acssynbio.9b00171. [DOI] [PubMed] [Google Scholar]
- 122.Jozwiak A., Sonawane P.D., Panda S., Garagounis C., Papadopoulou K.K., Abebie B., et al. Plant terpenoid metabolism co-opts a component of the cell wall biosynthesis machinery. Nat Chem Biol. 2020;16:740–748. doi: 10.1038/s41589-020-0541-x. [DOI] [PubMed] [Google Scholar]
- 123.Liu W., Feng Y., Yu S., Fan Z., Li X., Li J., et al. The flavonoid biosynthesis network in plants. Int J Mol Sci. 2021;22:12824. doi: 10.3390/ijms222312824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bomati E.K., Austin M.B., Bowman M.E., Dixon R.A., Noel J.P. Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis. J Biol Chem. 2005;280:30496–30503. doi: 10.1074/jbc.M502239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jiang D., Li P., Yin Y., Ren G., Liu C. Molecular cloning and functional characterization of UGTs from Glycyrrhiza uralensis flavonoid pathway. Int J Biol Macromol. 2021;192:1108–1116. doi: 10.1016/j.ijbiomac.2021.09.136. [DOI] [PubMed] [Google Scholar]
- 126.Dong D. Hunan University; Changsha: 2011. Stable expression of transcription factor FoxA1 or FoxM1 effects cell pluripotency and differentiation potential of stem cells, in. [Google Scholar]
- 127.Zhang H., Zhu J., Gong Z., Zhu J.K. Abiotic stress responses in plants. Nat Rev Genet. 2022;23:104–119. doi: 10.1038/s41576-021-00413-0. [DOI] [PubMed] [Google Scholar]
- 128.Doebley J., Lukens L. Transcriptional regulators and the evolution of plant form. Plant Cell. 1998;10:1075–1082. doi: 10.1105/tpc.10.7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gao K., Wang L., Wu S., Sui C. Advances in studies on transcriptional factor regulation of biosynthesis of active components in medicinal plants, Chin Tradit Herb. Drugs. 2015;46:3100–3108. [Google Scholar]
- 130.Tian L. Using hairy roots for production of valuable plant secondary metabolites. Adv Biochem Eng Biotechnol. 2015;149:275–324. doi: 10.1007/10_2014_298. [DOI] [PubMed] [Google Scholar]
- 131.Li Y., Chen X., Wang J., Zou G., Wang L., Li X. Two responses to MeJA induction of R2R3-MYB transcription factors regulate flavonoid accumulation in Glycyrrhiza uralensis Fisch. PLoS One. 2020;15:e0236565. doi: 10.1371/journal.pone.0236565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tamura K., Yoshida K., Hiraoka Y., Sakaguchi D., Chikugo A., Mochida K., et al. The basic helix-loop-helix transcription factor GubHLH3 positively regulates soyasaponin biosynthetic genes in Glycyrrhiza uralensis. Plant Cell Physiol. 2018;59:778–791. doi: 10.1093/pcp/pcy046. [DOI] [PubMed] [Google Scholar]
- 133.Goyal P., Manzoor M.M., Vishwakarma R.A., Sharma D., Dhar M.K., Gupta S. A comprehensive transcriptome-wide identification and screening of WRKY gene family engaged in abiotic stress in Glycyrrhiza glabra. Sci Rep. 2020;10:373. doi: 10.1038/s41598-019-57232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wu L. Peking Union Medical College; Beijing: 2022. Functional identification of ABA-responsive bZIP transcription factors involved in the regulation of active ingredient biosynthesis in Glycyrrhiza uralensis, in. [Google Scholar]
- 135.Afreen F., Zobayed S.M., Kozai T. Spectral quality and UV-B stress stimulate glycyrrhizin concentration of Glycyrrhiza uralensis in hydroponic and pot system. Plant Physiol Biochem. 2005;43:1074–1081. doi: 10.1016/j.plaphy.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 136.Marui A., Nagafuchi T., Shinogi Y., Yasufuku N., Omine K., Kobayashi T., et al. Cultivation research for high-glycyrrhizin licorice by applying low temperature and Ca2+ ion as environmental stress based on field investigation. J Fac Agric Kyushu Univ. 2011;56:367–371. [Google Scholar]
- 137.Hou J., Li W., Zheng Q., Wang W., Xiao B., Xing D. Effect of low light intensity on growth and accumulation of secondary metabolites in roots of Glycyrrhiza uralensis Fisch. Biochem Syst Ecol. 2010;38:160–168. [Google Scholar]
- 138.Behdad A., Mohsenzadeh S., Azizi M., Moshtaghi N. Salinity effects on physiological and phytochemical characteristics and gene expression of two Glycyrrhiza glabra L. populations. Phytochemistry. 2020;171 doi: 10.1016/j.phytochem.2019.112236. [DOI] [PubMed] [Google Scholar]
- 139.Xie W., Hao Z., Zhou X., Jiang X., Xu L., Wu S., et al. Arbuscular mycorrhiza facilitates the accumulation of glycyrrhizin and liquiritin in Glycyrrhiza uralensis under drought stress. Mycorrhiza. 2018;28:285–300. doi: 10.1007/s00572-018-0827-y. [DOI] [PubMed] [Google Scholar]
- 140.Han Y., Hou Z., Zhang X., He Q., Liang Z. Multi-dimensional “projection” - the impact of abiotic stresses on the content of seven active compounds and expression of related genes in Glycyrrhiza uralensis Fisch. Environ Exp Bot. 2022;197 [Google Scholar]
- 141.Yu Z.X., Li J.X., Yang C.Q., Hu W.L., Wang L.J., Chen X.Y. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant. 2012;5:353–365. doi: 10.1093/mp/ssr087. [DOI] [PubMed] [Google Scholar]
- 142.Zhang F., Fu X., Lv Z., Lu X., Shen Q., Zhang L., et al. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol Plant. 2015;8:163–175. doi: 10.1016/j.molp.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 143.Wu L., Xu Z., Wang Q., Nie L., Cui Y., Wang Y., et al. Systematic screening and analysis of bZIP transcription factors in Glycyrrhiza uralensis and their response to ABA stress. Acta Pharm Sin. 2022;57:818–830. [Google Scholar]
- 144.Han Y., Hou Z., He Q., Zhang X., Yan K., Han R., et al. Genome-wide characterization and expression analysis of bZIP gene family under abiotic stress in Glycyrrhiza uralensis. Front Genet. 2021;12 doi: 10.3389/fgene.2021.754237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cui Y.X., Xu Z.C., Chen X.L., Nie L.P., Wu L.W., Wang Y., et al. Genome-wide identification of abscisic acid (ABA) receptor pyrabactin resistance 1-like protein (PYL) family members and expression analysis of PYL genes in response to different concentrations of ABA stress in Glycyrrhiza uralensis, Chin. J Nat Med. 2020;18:606–611. doi: 10.1016/S1875-5364(20)30072-8. [DOI] [PubMed] [Google Scholar]
- 146.Nie L., Xu Z., Wu L., Chen X., Cui Y., Wang Y., et al. Genome-wide identification of protein phosphatase 2C family members in Glycyrrhiza uralensis Fisch. and their response to abscisic acid and polyethylene glycol stress. J Taibah Univ Sci. 2021;15:1260–1268. [Google Scholar]
- 147.Ma D., Pu G., Lei C., Ma L., Wang H., Guo Y., et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009;50:2146–2161. doi: 10.1093/pcp/pcp149. [DOI] [PubMed] [Google Scholar]
- 148.Jiang W., Fu X., Pan Q., Tang Y., Shen Q., Lv Z., et al. Overexpression of AaWRKY1 leads to an enhanced content of artemisinin in Artemisia annua. Biomed Res Int. 2016;2016:7314971. doi: 10.1155/2016/7314971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ji Y., Xiao J., Shen Y., Ma D., Li Z., Pu G., et al. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 2014;55:1592–1604. doi: 10.1093/pcp/pcu090. [DOI] [PubMed] [Google Scholar]
- 150.Lv Z., Wang S., Zhang F., Chen L., Hao X., Pan Q., et al. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and botrytis cinerea in Artemisia annua. Plant Cell Physiol. 2016;57:1961–1971. doi: 10.1093/pcp/pcw118. [DOI] [PubMed] [Google Scholar]
- 151.Matías-Hernández L., Jiang W., Yang K., Tang K., Brodelius P.E., Pelaz S. AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 2017;90:520–534. doi: 10.1111/tpj.13509. [DOI] [PubMed] [Google Scholar]
- 152.Ding K., Pei T., Bai Z., Jia Y., Ma P., Liang Z. SmMYB36, a novel R2R3-MYB transcription factor, enhances tanshinone accumulation and decreases phenolic acid content in Salvia miltiorrhiza hairy roots. Sci Rep. 2017;7:5104. doi: 10.1038/s41598-017-04909-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang J., Zhou L., Zheng X., Zhang J., Yang L., Tan R., et al. Overexpression of SmMYB9b enhances tanshinone concentration in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2017;36:1297–1309. doi: 10.1007/s00299-017-2154-8. [DOI] [PubMed] [Google Scholar]
- 154.Shi M., Zhou W., Zhang J., Huang S., Wang H., Kai G. Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by jasmonate zim-domain repressor proteins. Sci Rep. 2016;6:20919. doi: 10.1038/srep20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhou Y., Zhou X., Li Q., Chen J., Xiao Y., Zhang L., et al. Molecular cloning, bioinformatics analysis, and transcriptional profiling of JAZ1 and JAZ2 from Salvia miltiorrhiza. Biotechnol Appl Biochem. 2017;64:27–34. doi: 10.1002/bab.1454. [DOI] [PubMed] [Google Scholar]
- 156.Reddy V.A., Wang Q., Dhar N., Kumar N., Venkatesh P.N., Rajan C., et al. Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit. MsGPPS.LSUPlant Biotechnol J. 2017;15:1105–1119. doi: 10.1111/pbi.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huang W., Khaldun A.B., Chen J., Zhang C., Lv H., Yuan L., et al. A R2R3-MYB transcription factor regulates the flavonol biosynthetic pathway in a traditional Chinese medicinal plant. Epimedium sagittatum, Front Plant Sci. 2016;7:1089. doi: 10.3389/fpls.2016.01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu C., Long J., Zhu K., Liu L., Yang W., Zhang H., et al. Characterization of a citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Sci Rep. 2016;6:25352. doi: 10.1038/srep25352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu F., Ning Y., Zhang W., Liao Y., Li L., Cheng H., et al. An R2R3-MYB transcription factor as a negative regulator of the flavonoid biosynthesis pathway in Ginkgo biloba. Funct Integr Genomics. 2014;14:177–189. doi: 10.1007/s10142-013-0352-1. [DOI] [PubMed] [Google Scholar]
- 160.Nakatsuka T., Saito M., Yamada E., Fujita K., Kakizaki Y., Nishihara M. Isolation and characterization of GtMYBP3 and GtMYBP4, orthologues of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J Exp Bot. 2012;63:6505–6517. doi: 10.1093/jxb/ers306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.McCallum S., Woodhead M., Hackett C.A., Kassim A., Paterson A., Graham J. Genetic and environmental effects influencing fruit colour and QTL analysis in raspberry. Theor Appl Genet. 2010;121:611–627. doi: 10.1007/s00122-010-1334-5. [DOI] [PubMed] [Google Scholar]
- 162.Pang Y., Wenger J.P., Saathoff K., Peel G.J., Wen J., Huhman D., et al. A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 2009;151:1114–1129. doi: 10.1104/pp.109.144022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., et al. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell. 2003;15:2514–2531. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Baudry A., Caboche M., Lepiniec L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006;46:768–779. doi: 10.1111/j.1365-313X.2006.02733.x. [DOI] [PubMed] [Google Scholar]
- 165.Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of banyuls and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 166.Spelt C., Quattrocchio F., Mol J.N., Koes R. anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell. 2000;12:1619–1632. doi: 10.1105/tpc.12.9.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu X., Yin X., Allan A., Wang K., Shi Y., Huang Y., et al. The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry (Myrica rubra) during anthocyanin biosynthesis. Plant Cell Tiss Organ Cult. 2013;115:285–298. [Google Scholar]
- 168.Lu X., Zhang L., Zhang F., Jiang W., Shen Q., Zhang L., et al. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 2013;198:1191–1202. doi: 10.1111/nph.12207. [DOI] [PubMed] [Google Scholar]
- 169.Tan H., Xiao L., Gao S., Li Q., Chen J., Xiao Y., et al. Trichome and artemisinin regulator 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol Plant. 2015;8:1396–1411. doi: 10.1016/j.molp.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 170.Dai Y., Qin Q., Dai D., Kong L., Li W., Zha X., et al. Isolation and characterization of a novel cDNA encoding methyl jasmonate-responsive transcription factor TcAP2 from Taxus cuspidata. Biotechnol Lett. 2009;31:1801–1809. doi: 10.1007/s10529-009-0068-4. [DOI] [PubMed] [Google Scholar]
- 171.Li L., Hao X., Liu H., Wang W., Fu X., Ma Y., et al. Jasmonic acid-responsive AabHLH1 positively regulates artemisinin biosynthesis in Artemisia annua. Biotechnol Appl Biochem. 2019;66:369–375. doi: 10.1002/bab.1733. [DOI] [PubMed] [Google Scholar]
- 172.Xiang L., Jian D., Zhang F., Yang C., Bai G., Lan X., et al. The cold-induced transcription factor bHLH112 promotes artemisinin biosynthesis indirectly via ERF1 in Artemisia annua. J Exp Bot. 2019;70:4835–4848. doi: 10.1093/jxb/erz220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shen Q., Lu X., Yan T., Fu X., Lv Z., Zhang F., et al. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 2016;210:1269–1281. doi: 10.1111/nph.13874. [DOI] [PubMed] [Google Scholar]
- 174.Wang C., Chen L., Cai Z., Chen C., Liu Z., Liu S., et al. Metabolite profiling and transcriptome analysis explains difference in accumulation of bioactive constituents in licorice (Glycyrrhiza uralensis) under salt stress. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.727882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nims E., Vongpaseuth K., Roberts S.C., Walker E.L. TcJAMYC: a bHLH transcription factor that activates paclitaxel biosynthetic pathway genes in yew. J Biol Chem. 2015;290:20104. doi: 10.1074/jbc.A109.026195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhou Y., Ma Y., Zeng J., Duan L., Xue X., Wang H., et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat Plants. 2016;2:16183. doi: 10.1038/nplants.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xing B., Yang D., Yu H., Zhang B., Yan K., Zhang X., et al. Overexpression of SmbHLH10 enhances tanshinones biosynthesis in Salvia miltiorrhiza hairy roots. Plant Sci. 2018;276:229–238. doi: 10.1016/j.plantsci.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 178.Xing B., Liang L., Liu L., Hou Z., Yang D., Yan K., et al. Overexpression of SmbHLH148 induced biosynthesis of tanshinones as well as phenolic acids in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2018;37:1681–1692. doi: 10.1007/s00299-018-2339-9. [DOI] [PubMed] [Google Scholar]
- 179.Zhang C., Xing B., Yang D., Ren M., Guo H., Yang S., et al. SmbHLH3 acts as a transcription repressor for both phenolic acids and tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. Phytochemistry. 2020;169 doi: 10.1016/j.phytochem.2019.112183. [DOI] [PubMed] [Google Scholar]
- 180.Yin J., Li X., Zhan Y., Li Y., Qu Z., Sun L., et al. Cloning and expression of BpMYC4 and BpbHLH9 genes and the role of BpbHLH9 in triterpenoid synthesis in birch. BMC Plant Biol. 2017;17:214. doi: 10.1186/s12870-017-1150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mertens J., Pollier J., Vanden Bossche R., Lopez-Vidriero I., Franco-Zorrilla J.M., Goossens A. The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 2016;170:194–210. doi: 10.1104/pp.15.01645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shen Q., Huang H., Zhao Y., Xie L., He Q., Zhong Y., et al. The Transcription Factor Aabzip9 Positively Regulates the Biosynthesis of Artemisinin in Artemisia annua. Front Plant Sci. 2019;10:1294. doi: 10.3389/fpls.2019.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tang Y., Li L., Yan T., Fu X., Shi P., Shen Q., et al. AaEIN3 mediates the downregulation of artemisinin biosynthesis by ethylene signaling through promoting leaf senescence in Artemisia annua. Front Plant Sci. 2018;9:413. doi: 10.3389/fpls.2018.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhang N. Lanzhou Jiaotong University; Lanzhou: 2023. Analysis and exploration of MYB gene family regulating glycyrrhizic acid biosynthesis in Glycyrrhiza uralensis. [Google Scholar]
- 185.Zhou M.L., Hou H.L., Zhu X.M., Shao J.R., Wu Y.M., Tang Y.X. Soybean transcription factor GmMYBZ2 represses catharanthine biosynthesis in hairy roots of Catharanthus roseus. Appl Microbiol Biotechnol. 2011;91:1095–1105. doi: 10.1007/s00253-011-3288-1. [DOI] [PubMed] [Google Scholar]
- 186.Chen X., Li J., Liu Y., Wu D., Huang H., Zhan R., et al. PatSWC4, a methyl jasmonate-responsive MYB (v-myb avian myeloblastosis viral oncogene homolog)-related transcription factor, positively regulates patchoulol biosynthesis in Pogostemon cablin. Ind Crops Prod. 2020;154 [Google Scholar]
- 187.Tian Q., Han L., Zhu X., Zhang C., Li Y., Xue X., et al. SmMYB4 Is a R2R3-MYB Transcriptional Repressor Regulating the Biosynthesis of Phenolic Acids and Tanshinones in Salvia miltiorrhiza. Metabolites. 2022;12:968. doi: 10.3390/metabo12100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yang R., Wang S., Zou H., Li L., Li Y., Wang D., et al. R2R3-MYB Transcription Factor SmMYB52 Positively Regulates Biosynthesis of Salvianolic Acid B and Inhibits Root Growth in Salvia miltiorrhiza. Int J Mol Sci. 2021;22:9538. doi: 10.3390/ijms22179538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hao X., Pu Z., Cao G., You D., Zhou Y., Deng C., et al. Tanshinone and salvianolic acid biosynthesis are regulated by SmMYB98 in Salvia miltiorrhiza hairy roots. J Adv Res. 2020;23:1–12. doi: 10.1016/j.jare.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li S., Wu Y., Kuang J., Wang H., Du T., Huang Y., et al. SmMYB111 Is a Key Factor to Phenolic Acid Biosynthesis and Interacts with Both SmTTG1 and SmbHLH51 in Salvia miltiorrhiza. J Agric Food Chem. 2018;66:8069–8078. doi: 10.1021/acs.jafc.8b02548. [DOI] [PubMed] [Google Scholar]
- 191.Li J., Chen X., Zhou X., Huang H., Wu D., Shao J., et al. Identification of trihelix transcription factors in Pogostemon cablin reveals PatGT-1 negatively regulates patchoulol biosynthesis. Ind Crops Prod. 2021;161 [Google Scholar]
- 192.Fu X., Peng B., Hassani D., Xie L., Liu H., Li Y., et al. AaWRKY9 contributes to light- and jasmonate-mediated to regulate the biosynthesis of artemisinin in Artemisia annua. New Phytol. 2021;231:1858–1874. doi: 10.1111/nph.17453. [DOI] [PubMed] [Google Scholar]
- 193.Chen M., Yan T., Shen Q., Lu X., Pan Q., Huang Y., et al. GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 2017;214:304–316. doi: 10.1111/nph.14373. [DOI] [PubMed] [Google Scholar]
- 194.Xie L., Yan T., Li L., Chen M., Ma Y., Hao X., et al. The WRKY transcription factor AaGSW2 promotes glandular trichome initiation in Artemisia annua. J Exp Bot. 2021;72:1691–1701. doi: 10.1093/jxb/eraa523. [DOI] [PubMed] [Google Scholar]
- 195.Kayani S.I., Yanan M., Fu X., Shen Q., Li Y., Rahman S.U., et al. JA-Regulated AaGSW1-AaYABBY5/AaWRKY9 Complex Regulates Artemisinin Biosynthesis in Artemisia annua. Plant Cell Physiol. 2023;64:771–785. doi: 10.1093/pcp/pcad035. [DOI] [PubMed] [Google Scholar]
- 196.Wang Q., Reddy V.A., Panicker D., Mao H.Z., Kumar N., Rajan C., et al. Metabolic engineering of terpene biosynthesis in plants using a trichome-specific transcription factor MsYABBY5 from spearmint. Mentha spicataPlant Biotechnol J. 2016;14:1619–1632. doi: 10.1111/pbi.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhao C., Liu X., Gong Q., Cao J., Shen W., Yin X., et al. Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol J. 2021;19:671–688. doi: 10.1111/pbi.13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jia T. Hebei University; Baoding: 2023. Expression and functional analysis of bHLH differential genes in Glycyrrhiza inflata based on salt stress transcriptome sequencing, in. [Google Scholar]
- 199.Goodrich J., Carpenter R., Coen E.S. A common gene regulates pigmentation pattern in diverse plant species. Cell. 1992;68:955–964. doi: 10.1016/0092-8674(92)90038-e. [DOI] [PubMed] [Google Scholar]
- 200.Sharma S., Holme I.B., Dionisio G., Kodama M., Dzhanfezova T., Joernsgaard B., et al. Cyanidin based anthocyanin biosynthesis in orange carrot is restored by expression of Am Rosea1 and Am Delila, MYB and bHLH transcription factors. Plant Mol Biol. 2020;103:443–456. doi: 10.1007/s11103-020-01002-1. [DOI] [PubMed] [Google Scholar]
- 201.Albert N.W., Butelli E., Moss S.M.A., Piazza P., Waite C.N., Schwinn K.E., et al. Discrete bHLH transcription factors play functionally overlapping roles in pigmentation patterning in flowers of Antirrhinum majus. New Phytol. 2021;231:849–863. doi: 10.1111/nph.17142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nakatsuka T., Haruta K.S., Pitaksutheepong C., Abe Y., Kakizaki Y., Yamamoto K., et al. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol. 2008;49:1818–1829. doi: 10.1093/pcp/pcn163. [DOI] [PubMed] [Google Scholar]
- 203.Chen Z., Liu G., Tang N., Li Z. Transcriptome Analysis Reveals Molecular Signatures of Luteoloside Accumulation in Senescing Leaves of Lonicera macranthoides. Int J Mol Sci. 2018;19:1012. doi: 10.3390/ijms19041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang S., Ma P., Yang D., Li W., Liang Z., Liu Y., et al. Cloning and characterization of a putative R2R3 MYB transcriptional repressor of the rosmarinic acid biosynthetic pathway from Salvia miltiorrhiza. PLoS One. 2013;8:e73259. doi: 10.1371/journal.pone.0073259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schwinn K., Venail J., Shang Y., Mackay S., Alm V., Butelli E., et al. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell. 2006;18:831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wu X.M., Lim S.H., Yang W.C. Characterization, expression and phylogenetic study of R2R3-MYB genes in orchid. Plant Mol Biol. 2003;51:959–972. doi: 10.1023/a:1023050110077. [DOI] [PubMed] [Google Scholar]
- 207.Morita Y., Saitoh M., Hoshino A., Nitasaka E., Iida S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol. 2006;47:457–470. doi: 10.1093/pcp/pcj012. [DOI] [PubMed] [Google Scholar]
- 208.Zhong C., Tang Y., Pang B., Li X., Yang Y., Deng J., et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic Res. 2020;7:78. doi: 10.1038/s41438-020-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sun M., Xin Q., Hou K., Qiu J., Wang L., Chao E., et al. Production of 11-Oxo-β-Amyrin in Saccharomyces cerevisiae at High Efficiency by Fine-Tuning the Expression Ratio of CYP450:CPR. J Agric Food Chem. 2023;71:3766–3776. doi: 10.1021/acs.jafc.2c08261. [DOI] [PubMed] [Google Scholar]
- 210.Sun W., Xue H., Liu H., Lv B., Yu Y., Wang Y., et al. Controlling chemo- and regioselectivity of a plant P450 in yeast cell toward rare licorice triterpenoid biosynthesis. ACS Catal. 2020;10:4253–4260. [Google Scholar]
- 211.Sun W., Wan S., Liu C., Wang R., Zhang H., Qin L., et al. Establishing cell suitability for high-level production of licorice triterpenoids in yeast. Acta Pharm Sin B. 2024;14:4134–4148. doi: 10.1016/j.apsb.2024.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xu K., Zhao Y.J., Ahmad N., Wang J.N., Lv B., Wang Y., et al. O-glycosyltransferases from Homo sapiens contributes to the biosynthesis of Glycyrrhetic Acid 3-O-mono-β-D-glucuronide and Glycyrrhizin in Saccharomyces cerevisiae, Synth Syst. Biotechnol. 2021;6:173–179. doi: 10.1016/j.synbio.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yin Y., Li Y., Jiang D., Zhang X., Gao W., Liu C. De novo biosynthesis of liquiritin in Saccharomyces cerevisiae. Acta Pharm Sin B. 2020;10:711–721. doi: 10.1016/j.apsb.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang H.T., Wang Z.L., Chen K., Yao M.J., Zhang M., Wang R.S., et al. Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants. Nat Commun. 2023;14:6658. doi: 10.1038/s41467-023-42393-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhao C., Zhang Z., Sun L., Bai R., Wang L., Chen S. Genome sequencing provides potential strategies for drug discovery and synthesis. Acupunct Herb Med. 2023;3:244–255. [Google Scholar]
- 216.Sun Y., Shang L., Zhu Q.H., Fan L., Guo L. Twenty years of plant genome sequencing: achievements and challenges. Trends Plant Sci. 2022;27:391–401. doi: 10.1016/j.tplants.2021.10.006. [DOI] [PubMed] [Google Scholar]
- 217.Yang X., Gao S., Guo L., Wang B., Jia Y., Zhou J., et al. Three chromosome-scale Papaver genomes reveal punctuated patchwork evolution of the morphinan and noscapine biosynthesis pathway. Nat Commun. 2021;12:6030. doi: 10.1038/s41467-021-26330-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hu J., Qiu S., Wang F., Li Q., Xiang C.L., Di P., et al. Functional divergence of CYP76AKs shapes the chemodiversity of abietane-type diterpenoids in genus Salvia. Nat Commun. 2023;14:4696. doi: 10.1038/s41467-023-40401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.An Z., Gao R., Chen S., Tian Y., Li Q., Tian L., et al. Lineage-specific CYP80 expansion and benzylisoquinoline alkaloid diversity in early-diverging eudicots. Adv Sci. 2024;11:e2309990. doi: 10.1002/advs.202309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chen S. Science Press; Beijing: 2018. Herbgenomics. [Google Scholar]
- 221.Di P., Zhang L., Chen J., Tan H., Xiao Y., Dong X., et al. 13C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem Biol. 2013;8:1537–1548. doi: 10.1021/cb3006962. [DOI] [PubMed] [Google Scholar]
- 222.Guo J., Zhou Y.J., Hillwig M.L., Shen Y., Yang L., Wang Y., et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci USA. 2013;110:12108–12113. doi: 10.1073/pnas.1218061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Miettinen K., Dong L., Navrot N., Schneider T., Burlat V., Pollier J., et al. The seco-iridoid pathway from Catharanthus roseus. Nat Commun. 2014;5:3606. doi: 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.King A.J., Brown G.D., Gilday A.D., Larson T.R., Graham I.A. Production of bioactive diterpenoids in the euphorbiaceae depends on evolutionarily conserved gene clusters. Plant Cell. 2014;26:3286–3298. doi: 10.1105/tpc.114.129668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Karamat F., Olry A., Doerper S., Vialart G., Ullmann P., Werck-Reichhart D., et al. CYP98A22, a phenolic ester 3'-hydroxylase specialized in the synthesis of chlorogenic acid, as a new tool for enhancing the furanocoumarin concentration in Ruta graveolens. BMC Plant Biol. 2012;12:152. doi: 10.1186/1471-2229-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kumar K., Kumar S.R., Dwivedi V., Rai A., Shukla A.K., Shanker K., et al. Precursor feeding studies and molecular characterization of geraniol synthase establish the limiting role of geraniol in monoterpene indole alkaloid biosynthesis in Catharanthus roseus leaves. Plant Sci. 2015;239:56–66. doi: 10.1016/j.plantsci.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 227.Wang L., Zhao S.J., Liang Y.L., Sun Y., Cao H.J., Han Y. Identification of the protopanaxatriol synthase gene CYP6H for ginsenoside biosynthesis in Panax quinquefolius. Funct Integr Genomics. 2014;14:559–570. doi: 10.1007/s10142-014-0386-z. [DOI] [PubMed] [Google Scholar]
- 228.Wang H., Song W., Tao W., Zhang J., Zhang X., Zhao J., et al. Identification wild and cultivated licorice by multidimensional analysis. Food Chem. 2021;339 doi: 10.1016/j.foodchem.2020.128111. [DOI] [PubMed] [Google Scholar]
- 229.Gou k., Ren q. Studies on radioactive breeding of Glycyrrhiza uralensis. Acta Bot Yunnanica. 1993;15:214–216. [Google Scholar]
- 230.Jiao L., Li X., Wang J., Zeng Y., Liu M., Zheng S., et al. Breeding and popularization of new varieties for six key Chinese herbs including Radix et Rhizoma Glycyrrhizae. Mod Chin Med. 2021;23:1463–1475. [Google Scholar]
- 231.Liu Y., Liu C., Zeng B., Fan B., Li P., Xu D., et al. Research progress on germplasm resources of Glycyrrhizae Radix et Rhizoma, Chin Tradit Herb. Drugs. 2013;44:3593–3598. [Google Scholar]
- 232.Ma T., Xiang L., Zhang D., Shi Y., Ding D., Shen X., et al. Statues and research strategy of molecular breeding in Artemisia annua. China J Chin Mater Med. 2018;43:1403–3050. doi: 10.19540/j.cnki.cjcmm.2018.0093. [DOI] [PubMed] [Google Scholar]
- 233.Ma C. Beijing University of Chinese Medicine; Beijing: 2009. Breeding basis of Glycyrrhiza uralensis and cloning of HMGR gene, in. [Google Scholar]
- 234.Liu Y., Zhang P., Song M., Hou J., Qing M., Wang W., et al. Transcriptome analysis and development of SSR molecular markers in Glycyrrhiza uralensis Fisch. PLoS One. 2015;10:e0143017. doi: 10.1371/journal.pone.0143017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Colinas M., Goossens A. Combinatorial Transcriptional Control of Plant Specialized Metabolism. Trends Plant Sci. 2018;23:324–336. doi: 10.1016/j.tplants.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 236.Alqudah A.M., Sallam A., Stephen Baenziger P., Börner A., Gwas, Fast-forwarding gene identification and characterization in temperate Cereals: lessons from Barley - A review. J Adv Res. 2020;22:119–135. doi: 10.1016/j.jare.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Huang X., Han B. Natural variations and genome-wide association studies in crop plants. Annu Rev Plant Biol. 2014;65:531–551. doi: 10.1146/annurev-arplant-050213-035715. [DOI] [PubMed] [Google Scholar]
- 238.Khan S.U., Saeed S., Khan M.H.U., Fan C., Ahmar S., Arriagada O., et al. Advances and challenges for QTL analysis and GWAS in the plant-breeding of high-yielding: a focus on rapeseed. Biomolecules. 2021;11:1516. doi: 10.3390/biom11101516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lu H., Liu J., Zhang H., Yin T., Gao S. Ri-mediated transformation of Glycyrrhiza uralensis with a squalene synthase gene (GuSQS1) for production of glycyrrhizin. Plant Mol Biol Rep. 2008;26:1–11. [Google Scholar]
- 240.Zhang H.C., Liu J.M., Lu H.Y., Gao S.L. Enhanced flavonoid production in hairy root cultures of Glycyrrhiza uralensis Fisch by combining the over-expression of chalcone isomerase gene with the elicitation treatment. Plant Cell Rep. 2009;28:1205–1213. doi: 10.1007/s00299-009-0721-3. [DOI] [PubMed] [Google Scholar]
- 241.Hou J., Yin Y., Tian S., Zhang Z., Yang L., Li W., et al. Overexpressing of chalcone isomerase(CHI) gene enhances flavonoid accumulation in Glycyrrhiza uralensis hairy roots. Acta Pharm Sin. 2021;56:319–327. [Google Scholar]
- 242.Wang D., Zhang Z., Yang L., Tian S., Liu Y. ARPI, β-AS, and UGE regulate glycyrrhizin biosynthesis in Glycyrrhiza uralensis hairy roots. Plant Cell Rep. 2021;40:1285–1296. doi: 10.1007/s00299-021-02712-6. [DOI] [PubMed] [Google Scholar]
- 243.Zhu H., Liu Z., Fu X., Dai Z., Wang S., Zhang G., et al. Detection and characterization of epistasis between QTLs on plant height in rice using single segment substitution lines. Breed Sci. 2015;65:192–200. doi: 10.1270/jsbbs.65.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wang J., Li H., Zhang X., Yin C., Li Y., Ma Y., et al. Molecular design breeding in crops in China. Acta Agron Sin. 2011;37:191–201. [Google Scholar]
- 245.Jawed K., Mattam A.J., Fatma Z., Wajid S., Abdin M.Z., Yazdani S.S. Engineered production of short chain fatty acid in Escherichia coli using fatty acid synthesis pathway. PLoS One. 2016;11:e0160035. doi: 10.1371/journal.pone.0160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wu J., Du G., Zhou J., Chen J. Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy. Metab Eng. 2013;16:48–55. doi: 10.1016/j.ymben.2012.11.009. [DOI] [PubMed] [Google Scholar]