Abstract
Recombination between homologous DNA molecules is essential for the proper maintenance and duplication of the genome, and for the repair of exogenously induced DNA damage such as double-strand breaks. Homologous recombination requires the RAD52 group proteins, including Rad51, Rad52 and Rad54. Upon treatment of mammalian cells with ionizing radiation, these proteins accumulate into foci at sites of DNA damage induction. We show that these foci are dynamic structures of which Rad51 is a stably associated core component, whereas Rad52 and Rad54 rapidly and reversibly interact with the structure. Furthermore, we show that the majority of the proteins are not part of the same multi-protein complex in the absence of DNA damage. Executing DNA transactions through dynamic multi-protein complexes, rather than stable holo-complexes, allows flexibility. In the case of DNA repair, for example, it will facilitate cross-talk between different DNA repair pathways and coupling to other DNA transactions, such as replication.
Keywords: DNA double-strand break repair/genome instability/green fluorescent protein/photobleaching/Rad51
Introduction
Homologous recombination plays a pivotal role in genome duplication and in providing genome stability. Further more, it is involved in the repair of exogenously induced DNA damage such as double-strand breaks (DSBs) (Flores-Rozas and Kolodner, 2000). Homologous recombination requires, among others, the RAD52 group proteins, including Rad51, Rad52 and Rad54, and the breast cancer susceptibility proteins Brca1 and Brca2 (Modesti and Kanaar, 2001). Immunofluorescence experiments with fixed mammalian cells have revealed that, in response to DNA damage, RAD52 group proteins appear in subnuclear structures referred to as foci (Haaf et al., 1995; Tan et al., 1999; Liu and Maizels, 2000). These foci form at the site of DNA damage (Tashiro et al., 2000) and contain, in addition to homologous recombination proteins, proteins involved in DNA metabolism in general, such as the single-stranded DNA binding protein RPA (Raderschall et al., 1999). Mutant cell lines defective in the formation of Rad51-containing DNA damage-induced foci are sensitive to DNA-damaging agents and display chromosomal instability (Bishop et al., 1998; Takata et al., 2000<//em>, 2001; Yu et al., 2000; O’Regan et al., 2001).
A critical intermediate in the repair of DSBs is a joint molecule between the broken DNA and a homologous double-strand repair template. Biochemical analyses have revealed that joint molecule formation requires close cooperation between the RAD52 group proteins (Baumann and West, 1998; Paques and Haber, 1999; Sung et al., 2000). Rad51 assembles into a nucleoprotein filament on the processed broken DNA, which subsequently pairs with homologous DNA, aided by the Rad52 and Rad54 proteins. Physical interactions among the RAD52 group proteins have been demonstrated biochemically and with the use of yeast two-hybrid and co-immunoprecipitation experiments (Hays et al., 1995; Johnson and Symington, 1995; Golub et al., 1997; Baumann and West, 1998; Chen et al., 1999; Paques and Haber, 1999; Sung et al., 2000). These interactions have led to the suggestion that RAD52 group proteins exist in a multi-protein complex, referred to as a ‘recombinosome’ (Hays et al., 1995). Here we explore the spatio-temporal association between human Rad51, Rad52 and Rad54 in living cells where the proteins have to mediate homologous recombination in the context of chromatin and amid other nuclear structures and processes.
Results
DNA damage response of RAD52 group proteins in living cells
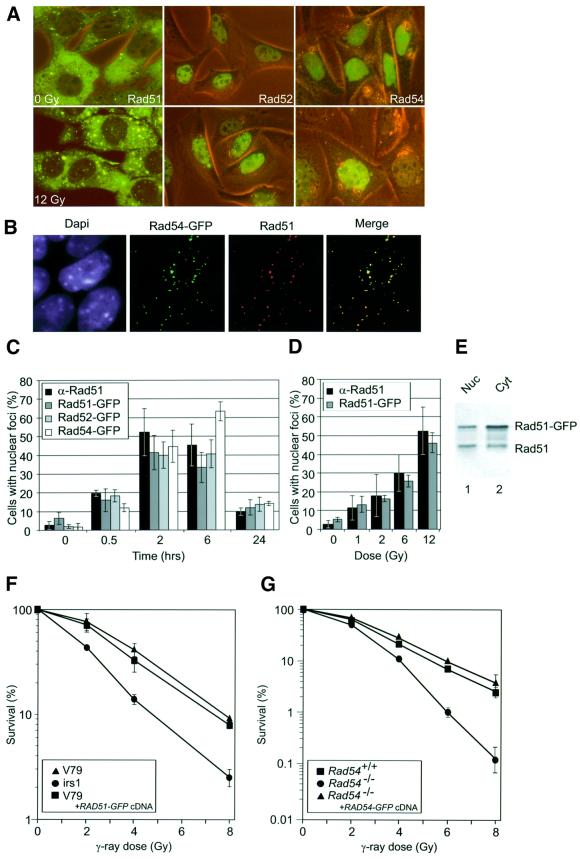
The characteristic DNA damage response of RAD52 group proteins observed in fixed cells was reproduced in living cells using the human Rad51, Rad52 and Rad54 proteins tagged with the green fluorescent protein (GFP). After treatment of the cells with ionizing radiation, nuclear foci containing Rad51–GFP, Rad52–GFP and Rad54–GFP were observed (Figure 1A). We infer biologically relevant behavior of the GFP-tagged RAD52 group proteins from the following experiments. By combining the detection of GFP fluorescence with immunofluorescence, we showed pairwise colocalization of the three proteins in the DNA damage-induced foci (Figure 1B; data not shown). The example shown in Figure 1B revealed quantitative colocalization of Rad54–GFP and endogenous Rad51 after irradiation. Furthermore, the kinetics and dose response of the formation of DNA damage-induced Rad51–GFP foci were similar to those of endogenous Rad51, as detected by immunofluorescence (Figure 1C and D). Similar kinetics to those observed for the formation of Rad51 foci were observed for Rad52–GFP and Rad54–GFP foci upon irradiation (Figure 1C). Recently, the presence of Rad51 in the nucleus, as well as in the cytoplasm, has been demonstrated (Davies et al., 2001; Kraakman-van der Zwet et al., 2002). We observed a similar subcellular localization for Rad51–GFP (Figure 1A). Importantly, immunoblot analysis of nuclear and cytoplasmic fractions of the cells expressing Rad51– GFP showed that the expression levels of nuclear endogenous and Rad51–GFP were similar (Figure 1E). Significantly, the presence of Rad51–GFP did not have a negative effect on the survival of cells with respect to irradiation (Figure 1F). The biological activity of Rad52–GFP has recently been revealed by the demonstration that the fusion protein increases the resistance of cells towards DNA-damaging agents (Liu and Maizels, 2000). Furthermore, the Saccharomyces cerevisiae Rad52– GFP protein is fully functional in DNA repair and recombination (Lisby et al., 2001). Finally, Rad54–GFP corrected the ionizing radiation sensitivity of Rad54 knockout mouse embryonic stem cells (Figure 1G).
Fig. 1. DNA damage response of the human RAD52 group proteins in living cells. (A) Detection of Rad51–GFP, Rad52–GFP and Rad54–GFP in living CHO cells before treatment with ionizing radiation by a combination of fluorescence and phase-contrast microscopy (0 Gy; upper panels). All three proteins formed nuclear foci upon treatment with ionizing radiation (12 Gy; lower panels). Images were taken 2 h after irradiation. (B) DNA damage-induced colocalization of Rad51 and Rad54–GFP. Cells expressing Rad54–GFP were fixed 2 h after treatment with ionizing radiation (12 Gy). Nuclei were visualized by DAPI staining. The signal from Rad54–GFP (shown in green) was observed directly by fluorescence microscopy. Endogenous Rad51 (shown in red) was detected by indirect immunofluorescence using antibodies against Rad51. Colocalization of Rad51 and Rad54–GFP (shown in yellow) is evident in the merged image. (C) Kinetics of endogenous Rad51, Rad51–GFP, Rad52–GFP and Rad54–GFP DNA damage-induced foci formation. CHO cells or their derivatives expressing the indicated GFP-tagged RAD52 group proteins were fixed at the indicated times after treatment with ionizing radiation (12 Gy). Detection of foci was performed as described in (B). The percentage of cells containing nuclear foci of endogenous Rad51, Rad51–GFP, Rad52–GFP or Rad54–GFP was determined in three independent experiments. (D) Dose response of endogenous and Rad51–GFP foci. CHO cells and their derivative expressing Rad51–GFP were fixed 2 h after treatment with the indicated doses of ionizing radiation. Detection of foci was performed as described above. (E) Immunoblot of endogenous and Rad51–GFP. Cell-free extracts prepared from V79 cells stably expressing Rad51–GFP were fractionated into nuclear (Nuc) and cytoplasmic (Cyt) fractions, which were analyzed for the presence of endogenous Rad51 and Rad51–GFP by immunoblotting using antibodies against Rad51. (F) Clonogenic survival assays of V79 cells and their indicated derivatives. V79 cells and V79 cells expressing Rad51–GFP were equally sensitive to ionizing radiation as measured by colony-forming ability after irradiation. A derivative of V79 (irs1), defective in the Rad51 paralogue Xrcc2, served as a control for irradiation. (G) Clonogenic survival assays of the Rad54-proficient and -deficient cells after ionizing radiation. The ionizing radiation sensitivity of Rad54–/– mouse embryonic stem cells was corrected to wild-type levels by the expression of Rad54–GFP.
Mobility of RAD52 group proteins in the nucleus
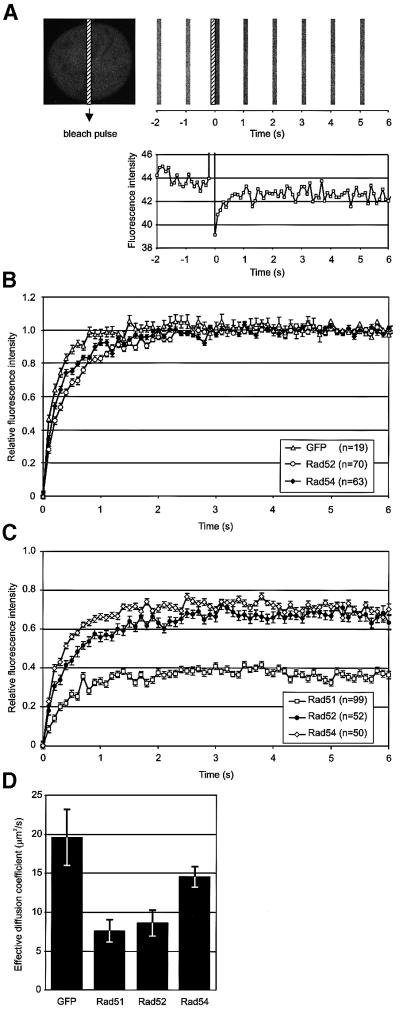
To ascertain whether the RAD52 group proteins are constituents of the same pre-assembled DNA repair complex in living cells, we analyzed the dynamic behavior of the proteins using fluorescence redistribution after photobleaching (FRAP) (White and Stelzer, 1999; Houtsmuller and Vermeulen, 2001; Misteli, 2001). The effective diffusion coefficient (Deff) of the proteins and the fraction of the proteins that was mobile were determined by measuring the kinetics of fluorescence recovery in a nuclear area that had been photobleached (Figure 2). The fluorescence in a small strip spanning the width of the entire nucleus was bleached using a 200 ms high-intensity laser pulse (Ellenberg et al., 1997; Houtsmuller et al., 1999; Houtsmuller and Vermeulen, 2001). Subsequently, the recovery of fluorescence in the strip was monitored at intervals of 100 ms. Figure 2A shows the primary data for a single cell containing Rad54–GFP. Measurements were performed on >60 cells for Rad52 and Rad54. For estimation of Deff, the final post-bleach pulse fluorescence intensity measured was set to 1, and the fluorescence intensity immediately after the bleach pulse was set to 0 (see Materials and methods; Figure 2D). The normalized data are shown in Figure 2B.
Fig. 2. Fluorescence redistribution after photobleaching (FRAP) analyses of RAD52 group proteins in CHO cells. Cells stably expressing Rad51–GFP, Rad52–GFP and Rad54–GFP were subjected to a local bleach pulse, and the kinetics of fluorescence recovery in the bleached area was determined. (A) An example of the primary data obtained using the photobleaching protocol on a cell nucleus (shown left) containing Rad54–GFP. The fluorescence in a small strip (indicated by the hatched rectangle) spanning the entire nucleus was bleached with a 200 ms high-intensity laser pulse. The recovery of fluorescence in the strip was monitored at intervals of 100 ms. For clarity, only strips obtained every second are shown above the time scale of the experiment. The measured fluorescence intensities over time are plotted below. (B) The photobleaching protocol was applied to a number (n) of cells containing GFP, Rad52–GFP and Rad54–GFP. The fluorescence intensity immediately after bleaching was set to 0, and the final post-bleach pulse fluorescence intensity measured was set to 1. The normalized data are plotted. (C) The photobleaching protocol was applied to a number (n) of cells containing Rad51–GFP, Rad52–GFP and Rad54–GFP. In this case, the final measured fluorescence intensity was normalized to the pre-bleach pulse fluorescence intensity. (D) The effective diffusion coefficients of the RAD52 group proteins were determined by fitting the experimentally obtained curves [shown in (B) and (C)] to a mathematical model describing diffusion (see Materials and methods). Measurements were performed in triplicate, and consistent results were obtained among different sets of experiments. Error bars indicate twice the standard error of the mean.
The bleaching protocol employed above led to the irreversible bleaching of ∼30% of all fluorescent proteins in the cell, due to the ratio between the nuclear volume irradiated with the laser and the total nuclear volume, and the diffusion of the proteins during the laser pulse. In the experiment shown in Figure 2C, the final measured fluorescence intensity was normalized to the pre-bleach pulse fluorescence intensity. The fluorescence in the bleached area recovered to ∼70% for both the Rad52 and Rad54 proteins. This behavior was similar to that of free GFP in our experimental set-up, and indicates that all of the detected Rad52 and Rad54 molecules in the cell were mobile (Ellenberg et al., 1997; White and Stelzer, 1999; Houtsmuller and Vermeulen, 2001; Misteli, 2001; Pederson, 2001). In contrast, Rad51 behaved differently, in that it was present in two distinct kinetic pools. Approximately half of the Rad51 proteins were immobile within the time scale of the measurements.
The fluorescence recovery curves were used to calculate the Deff of the mobile fraction of the RAD52 group proteins (Figure 2D). All three recombination proteins had a lower mobility than free GFP and, in turn, Rad51 and Rad52 had a lower mobility than Rad54. The observed differences in the dynamic behavior of the RAD52 group proteins in the absence of DNA damage indicate that even though they colocalize in DNA damage-induced foci, the majority of the proteins are not constituents of the same pre-assembled multi-protein complex in undamaged cells. These observations are consistent with the inability to co-immunoprecipitate RAD52 group proteins under physiological conditions in the absence of DNA damage (Tan et al., 1999; data not shown). Interestingly, in irradiated cells, the diffusion rates of the Rad52 and Rad54 proteins in the nucleoplasm did not differ from those in unirradiated cells (data not shown).
Turnover of RAD52 group proteins in DNA damage-induced foci
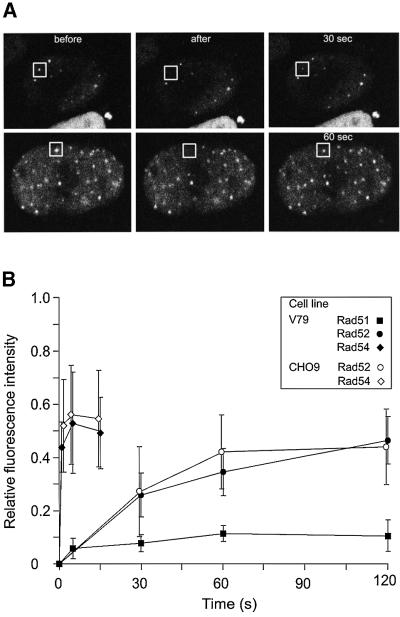
Next we addressed the nature of the DNA damage-induced foci formed by the RAD52 group proteins. It is not known whether these subnuclear structures form due to long-lived protein–protein interactions between their constituents or whether they are dynamic structures in which the RAD52 group proteins turnover. Therefore, we photobleached a single Rad52-containing focus in cells treated with ionizing radiation (Figure 3A). Interestingly, the fluorescence of the focus recovered over time, indicating that unbleached Rad52 molecules from the nucleoplasm were exchanging with bleached Rad52 molecules in the DNA damage-induced structure. We quantitated the fluorescence recovery of individual foci for the Rad51, Rad52 and Rad54 proteins. Intriguingly, the different RAD52 group proteins displayed very different residence times in the DNA damage-induced structures (Figure 3B). Half the original fluorescence intensity recovered in 0.5 s for Rad54 and 26 s for Rad52. In contrast to Rad52 and Rad54, Rad51 fluorescence hardly recovered over time, implying that it resides in the DNA damage-induced structures for much longer.
Fig. 3. Different residence times of RAD52 group proteins in DNA damage-induced foci. (A) Individual DNA damage-induced foci (indicated by squares) in cells stably expressing Rad52–GFP were photobleached. Images were collected before, immediately after and at the indicated times after the bleach pulse. (B) Quantitative FRAP analysis of DNA damage-induced Rad51–GFP, Rad52–GFP and Rad54–GFP foci. Recovery of fluorescence was measured at the indicated time points after the bleach pulse. All data points represent the mean of at least 10 different measurements, and the error bars indicate twice the standard error of the mean. The results were independent of the cell line used, because similar results were obtained with V79 and CHO9 cells. The major cause of fluctuations in fluorescence intensity of the foci was cellular movement.
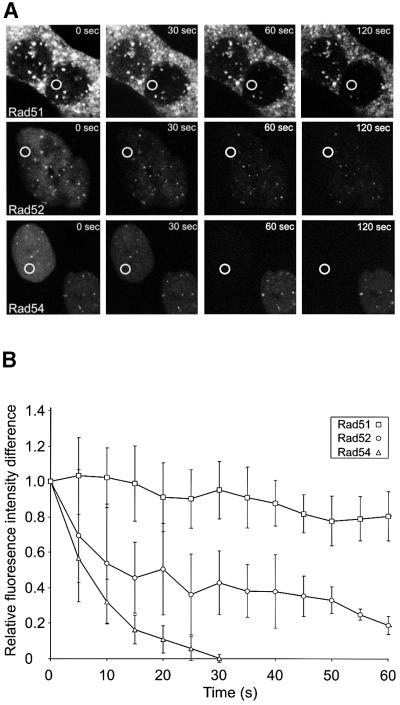
To obtain independent confirmation of the dynamic behavior of the RAD52 group proteins in the DNA damage-induced structures, we examined them using fluorescence loss in photobleaching (FLIP) (White and Stelzer, 1999; Houtsmuller and Vermeulen, 2001; Misteli, 2001; Pederson, 2001). In these experiments, the laser pulse used for bleaching fluorescence was not aimed at the structures. Instead, an area in the nucleoplasm, devoid of foci, was repeatedly bleached. The fluorescence intensity of the nucleoplasm, in a region away from the bleached area, and of the DNA damage-induced structures in the cells was measured after every bleach pulse (Figure 4). The observed relative loss of fluorescence in the structures demonstrated their dynamic nature, because it is due to the equilibrium between dissociation of unbleached proteins (and later also of bleached proteins) and association of (un)bleached proteins. Bleaching was specific for the cells on which the laser was aimed, as can be seen from the fluorescence signal in the control cells shown for Rad51 and Rad54, which hardly changed over time except for a low amount of bleaching due to monitoring of the cells (Figure 4A). Quantitation of these FLIP experiments revealed a qualitatively similar result to that found in the FRAP experiment (Figure 4B). The residence time of Rad54 in the DNA damage-induced structures was shorter than that of Rad52, whereas Rad51 was a much more long-lived component of the structures with little turnover and therefore a long residence time.
Fig. 4. FLIP in DNA damage-induced Rad51–GFP, Rad52–GFP and Rad54–GFP foci. (A) A region (indicated by circles) in the nucleoplasm of ionizing radiation-treated (12 Gy) cells expressing Rad51–GFP, Rad52–GFP or Rad54–GFP was repeatedly bleached 2 h after the irradiation. Cells were imaged between bleach pulses at the indicated times after the initial bleach pulse. (B) Quantitative FLIP analysis of DNA damage-induced foci containing Rad51–GFP, Rad52–GFP and Rad54–GFP. The difference between the loss in fluorescence of the DNA damage-induced foci and that of the nucleoplasm was determined at the indicated time points after the initial bleach pulse. The resulting curves were corrected for background bleaching due to monitoring of the cells. For each data point, at least five different cells and five foci per cell were analyzed. Error bars indicate twice the standard error of the mean.
Absence of immobile Rad52 and Rad54 in DNA damage-induced foci
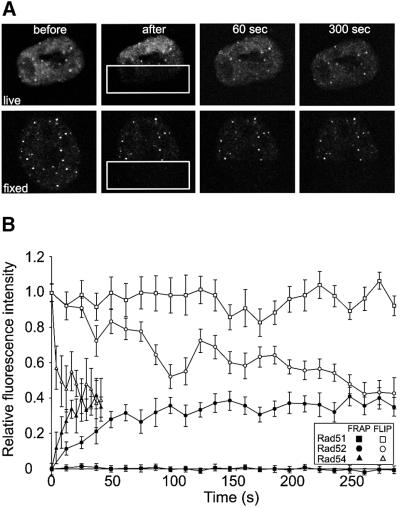
The FRAP and FLIP experiments clearly revealed that the majority of Rad51 molecules are stably associated with the DNA damage-induced structures. In contrast, Rad52 and Rad54 were not stably associated with the structures. However, we could not rule out, from the FRAP and FLIP experiments by themselves, the possibility that a minor fraction of these proteins was stably associated. To address this issue, we performed a set of experiments in which FRAP and FLIP techniques were applied simultaneously in the same cell (Figure 5). Half a cell containing DNA damage-induced structures was bleached (Figure 5A). Subsequently, the recovery of the fluorescence was monitored in a number of foci in the bleached half of the cell, and loss of fluorescence was monitored in a number of foci in the unbleached half of the same cell. The change in fluorescence intensity of foci containing Rad51, Rad52 and Rad54 was quantitated over time (Figure 5B). If a stably associated fraction of the RAD52 group proteins with the DNA damage-induced structures was present, the FRAP and FLIP curves would not converge. This is because both a fraction of the bleached proteins in the bleached DNA damage-induced structures and a fraction of the fluorescent proteins in the unbleached structures would not be replaced. Consistent with a long residence time, the FRAP and FLIP curves for Rad51 hardly changed over time. Significantly, for Rad52 and Rad54, the curves for the FRAP and FLIP experiments converged completely, showing the absence of a long-lived immobile fraction of either of the two proteins in the DNA damage-induced structures. Finally, the simultaneous FRAP/FLIP experiment demonstrated that the bleaching itself did not affect the dynamic behavior of the GFP-tagged RAD52 group proteins, since both the ‘frapped’ and the ‘flipped’ side of the cell returned to the same fluorescence intensity, as did the fluorescence intensity ratio between the nucleoplasm and the DNA damage-induced structures.
Fig. 5. FRAP and FLIP in DNA damage-induced Rad51–GFP, Rad52–GFP and Rad54–GFP foci. (A) A region (indicated by rectangles) of a cell containing Rad52–GFP foci was bleached by a single laser pulse (upper panels). The cell was imaged at the indicated times after bleaching. FLIP was measured in foci in the unbleached half of the cell, and FRAP was measured in foci in the bleached half of the same cell. The same experimental protocol applied to a fixed cell demonstrates the requirement for protein mobility to observe FRAP and FLIP (lower panels). (B) Quantitation of the simultaneous FRAP/FLIP experiment on DNA damage-induced Rad51–GFP, Rad52–GFP and Rad54–GFP foci. Error bars indicate twice the standard error of the mean.
Discussion
Different mobilities of RAD52 group proteins in the nucleus
The results of our experiments suggest that the major fraction of the RAD52 group proteins is not part of the same pre-assembled holo-complex in the absence of DNA damage. Instead, the majority of the proteins are diffusing through the nucleus independently. Once a DSB arises, it might represent a site with a slightly increased affinity for one of the RAD52 group proteins compared with intact DNA. In this regard, Rad52 itself is a good candidate protein, because it preferentially binds to DNA ends (Van Dyck et al., 1999). The difference in affinity ensures that Rad52 will be immobilized for a longer time at the DSB site than at other sites in the genome. Therefore, on average, Rad52 will accumulate at the DSB site. From the observed fluorescence intensity of the Rad52 protein in the DNA damage-induced structures, we suspect that these structures do not represent a single Rad52 heptamer bound to a single DSB end (Stasiak et al., 2000). Because fewer DNA damage-induced structures are observed compared with the number of DSBs generated by a given dose of ionizing radiation, it is possible that these structures represent sites where multiple DSBs are processed. Alternatively, or in addition, multiple Rad52 heptamers might be required to process a DNA lesion. Accumulation of Rad52 at the DSB sites might, in turn, generate sites of increased affinity for the other RAD52 group proteins, such as Rad51. The reason why Rad51 is the most stable component of DNA damage-induced structures could be due to the fact that this protein is part of a higher order structure, the nucleoprotein filament, in which it could be kept through cooperative interactions (De Zutter and Knight, 1999). Although we cannot rule out the possibility that the GFP tag influences the residence time of Rad51 in the DNA damage-induced structures, we believe this to be unlikely because the dose response and the kinetics of appearance and disappearance of DNA damage-induced structures containing endogenous Rad51 and Rad51–GFP are the same (Figure 1C and D).
A scenario in which the DNA repair proteins diffuse through the nucleus in relatively small complexes, and assemble ‘on-the-spot’ in DNA repair complexes, may be favorable to holo-complex formation prior to binding to damage, because small complexes have more efficient access than bulky holo-complexes to DNA damage located in condensed (hetero)chromatin regions (Cremer and Cremer, 2001; Houtsmuller and Vermeulen, 2001). Moreover, the observed homogeneous distribution of freely mobile DNA repair proteins, probably due to free diffusion (Phair and Misteli, 2000), ensures that all required factors are always present in the vicinity of DNA lesions wherever they occur, allowing rapid and efficient detection and subsequent repair. The observation that the induction of DNA damage does not influence the diffusion rates of the Rad52 and Rad54 proteins that are located in the nucleoplasm (as opposed to the proteins in the DNA damage-induced structures) argues that potential complexes between Rad52 and Rad54 do not pre-assemble away from the DNA damage-induced structures.
Cross-talk between DNA repair pathways
Performing the repair of DNA lesions by freely diffusing proteins that are temporarily immobilized due to encountering sites of increased affinity has an additional important advantage over a mechanism involving pre-assembled holo-complexes (Kowalczykowski, 2000). In situ assembly allows a greater flexibility in the composition of a DNA repair complex. Because different components can rapidly and reversibly interact with the DNA damage-induced structure, the specific components required for the repair of a particular lesion can be selected. For example, although both the repair of a DSB and an interstrand DNA cross-link through homologous recombination require Rad51, repair of an interstrand DNA cross-link requires, in addition, structure-specific endonucleases, such as ERCC1/XPF (Dronkert and Kanaar, 2001). ERCC1/XPF is also involved in nucleotide excision repair, but the other components of this DNA repair pathway do not play a major role in mammalian interstrand DNA cross-link repair. The rapid and reversible interaction of proteins with DNA damage-induced structures alleviates the necessity of having to dissemble a DNA repair holo-complex that does not contain all the specialized components required to repair the lesion with which it is associated. Furthermore, in situ assembly allows exchange of components between different multi-step DNA repair pathways. This cross-talk is biologically significant because it will lead to an increase in the diversity of DNA lesions that can be repaired and provides a mechanism to link DNA repair with other DNA transactions, such as replication.
Materials and methods
DNA constructs and cell lines
Plasmids EGFP-Rad51, EGFP-Rad52 and EGFP-Rad54 were generated by inserting cDNAs encoding the respective human RAD52 group proteins into pEGFP-C1, pEGFP-C3 and pEGFP-N1 (Clontech). The constructs were transfected into CHO9 and V79 Chinese hamster ovary cells and mouse embryonic stem cells. Stable clones were selected using G418 or puromycin. Upon immunoblot analysis, the nuclear expression levels of Rad51–GFP (Figure 1E) and Rad54–GFP (data not shown) were found to be similar to those of the endogenous proteins. Quantitation of the nuclear fluorescence intensity of cells expressing Rad52–GFP and Rad54–GFP showed that both proteins were expressed to similar levels. Taken together, these data indicate that none of the nuclear GFP-tagged RAD52 group proteins is overexpressed.
Epifluorescence microscopy, cell survival assays and immunoblotting
Cells were treated with ionizing radiation using a 137Cs source and fixed with 2% paraformaldehyde. Endogenous Rad51 was detected using indirect immunofluorescence with a polyclonal antibody raised against human Rad51 in rabbits (Tan et al., 1999). The signal from GFP-tagged proteins was observed directly by fluorescence microscopy. Quantitation of DNA damage-induced foci and cell survival assays were performed as described previously (Essers et al., 1997; Tan et al., 1999). Control cell lines used in the survival assays were irs1 (Cartwright et al., 1998; Liu et al., 1998) and Rad54–/– (Essers et al., 1997). Cell lines stably expressing Rad51–GFP were analyzed for the presence of Rad51 using immunoblotting after cellular fractionation into nuclear and cytoplasmic fractions (Davies et al, 2001; Kraakman-van der Zwet et al., 2002). Five-fold more nuclear fraction than cytoplasmic fraction was transferred to the blots. We believe that the high local concentrations of Rad51 that could be observed in the cytoplasm (Figure 1A) reflect the presence of Rad51 in a cytoplasmic organelle. The structures were present before irradiation and did not increase upon irradiation.
Confocal microscopy
Cells were treated with ionizing radiation and subjected to photobleaching experiments 2 h after irradiation. Confocal images of living cells expressing GFP-tagged RAD52 group proteins were obtained using a Zeiss LSM 410 microscope equipped with a 200 mW Ar laser at 488 nm and a 40× 1.3 n.a. oil immersion lens. Images of single nuclei were taken at a lateral sample interval of 100 nm. GFP fluorescence was detected using a dichroic beamsplitter (488/543 nm) and an additional 515–540 nm bandpass emission filter placed in front of the photomultiplier tube.
Photobleaching experiments
To determine the Deff of freely mobile GFP-labeled RAD52 group proteins, a small region with a width of 2 µm and spanning the entire nucleus was bleached for 200 ms at high laser intensity (100% of the 488 nm line of a 200 mW Ar laser) (Ellenberg et al., 1997; Houtsmuller et al., 1999; Houtsmuller and Vermeulen, 2001). Subsequently, the recovery of fluorescence in the region was monitored at intervals of 100 ms at 3% of the laser intensity applied for bleaching. The Deff was estimated by calculating relative fluorescence at several time points, after small corrections for monitor bleaching: FRdiff(t) = (It – I0)/(I∞ – I0), where I∞ is the fluorescence intensity measured after complete recovery, I0 is the fluorescence intensity immediately after bleaching and It is the measured fluorescence intensity at 100 ms intervals. The Deff was calculated as the value of D in the theoretical equation for one-dimensional diffusion: FT(t) = 1 – [w2 × (w2 + 4πDt)–1]1/2 (Ellenberg et al., 1997), for which Σ[FRdiff(t) – FT(t)]2 is minimal (least-squares fitting). For visualization and estimation of a potentially present immobile fraction, relative fluorescence was calculated, after small corrections for monitor bleaching, as FRimm(t) = (It – I0)/(It<0 – I0), where It<0 is the intensity immediately before bleaching. The immobile fraction was calculated as Nimmobile/Ntot = 1 – FRimm(∞) × (1 – Nmobile,bleached/Ntot)–1, where Nmobile,bleached/Ntot is the fraction of mobile molecules bleached by the pulse.
To determine the residence time of RAD52 group proteins in foci formed upon γ-irradiation, FRAP and FLIP experiments were applied. Foci, induced by treatment of cells with 12 Gy ionizing radiation, were analyzed 2 h after irradiation. In FRAP experiments, the fluorescence recovery of foci bleached for 1 s (at 100% intensity) was monitored (at 3% intensity), with time intervals as indicated. Relative fluorescence in each focus (spot) was calculated as FRspot(t) = (Ispot,t – Ispot,0)/[(Inucl,t<0/nucl,∞) × (Ispot,t<0 – Ispot,0)], where Inucl is the fluorescence intensity in the vicinity of the foci and Ispot is the intensity in the focus after subtraction of Inucl (Ispot = Imeasured – Inucl). The time required for the recovery of half the relative fluorescence intensity was used as a measure for the residence time of individual proteins. In FLIP measurements, the loss of fluorescence was monitored in foci (at 3% laser power) in between repetitive bleach pulses (1 s at 100% laser intensity with 5 s intervals) at a distant region in the same nucleus. Relative fluorescence was calculated, after small corrections for monitor bleaching, as FRspot(t) = (Ispot,t – Inucl,t)/(Ispot,0 – Inucl,0), and the difference in relative fluorescence between the nucleoplasm and the foci was plotted against time. The longer apparent residence time for the proteins measured using the multi-bleach pulse FLIP protocol, compared with FRAP, is due to the time required to diffuse from the bleached area to the DNA damage-induced structure.
In the simultaneous FRAP/FLIP experiment, half the nucleus was bleached for 2 s at 50% laser intensity (Figure 5A). Subsequently, the redistribution of fluorescence in the nucleoplasm and the exchange of bleached and unbleached molecules between foci and nucleoplasm were monitored by taking confocal images at fixed time intervals (3 s for Rad54 and 12 s for Rad51 and Rad52). The relative intensities IR of the foci in the bleached and unbleached halves of the cell were calculated separately as IR = (It – I0)/(It<0 – I0), where It is the intensity of the foci measured at consecutive time points, I0 is the intensity of the bleached foci immediately after bleaching and It<0 is the intensity of the foci before bleaching. If a fraction of the fluorescent molecules is stably associated with the foci, the size of this fraction is given by Fstably bound = |IR,bleached – IR,unbleached|. If the curves reach the same level (IR,bleached = IR,unbleached), there is no stably bound fraction.
Acknowledgments
Acknowledgements
We thank T.Cervelli, K.Mattern and C.Beerens for their contributions. This work was supported by grants from the Netherlands Organization for Scientific Research (NWO), the Dutch Cancer Society (KWF) and the Association for International Cancer Research.
References
- Baumann P. and West,S.C. (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Ear,U., Bhattacharyya,A., Calderone,C., Beckett,M., Weichselbaum,R.R. and Shinohara,A. (1998) Xrcc3 is required for assembly of Rad51 complexes in vivo. J. Biol. Chem., 273, 21482–21488. [DOI] [PubMed] [Google Scholar]
- Cartwright R., Tambini,C.E., Simpson,P.J. and Thacker,J. (1998) The XRCC2 DNA repair gene from human and mouse encodes a novel member of the recA/RAD51 family. Nucleic Acids Res., 26, 3084–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. et al. (1999) Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J. Biol. Chem., 274, 12748–12752. [DOI] [PubMed] [Google Scholar]
- Cremer T. and Cremer,C. (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Rev. Genet., 2, 292–301. [DOI] [PubMed] [Google Scholar]
- Davies A.A., Masson,J., McIlwraith,M.J., Stasiak,A.Z., Stasiak,A., Venkitaraman,A.R. and West,S.C. (2001) Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell, 7, 273–282. [DOI] [PubMed] [Google Scholar]
- De Zutter J.K. and Knight,K.L. (1999) The hRad51 and RecA proteins show significant differences in cooperative binding to single-stranded DNA. J. Mol. Biol., 293, 769–780. [DOI] [PubMed] [Google Scholar]
- Dronkert M.L.G. and Kanaar,R. (2001) Repair of DNA interstrand cross-links. Mutat. Res., 486, 217–247. [DOI] [PubMed] [Google Scholar]
- Ellenberg J., Siggia,E.D., Moreira,J.E., Smith,C.L., Presley,J.F., Worman,H.J. and Lippincott-Schwartz,J. (1997) Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol., 138, 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., Hendriks,R.W., Swagemakers,S.M.A., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H.J. and Kanaar,R. (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H. and Kolodner,R.D. (2000) Links between replication, recombination and genome instability in eukaryotes. Trends Biochem. Sci., 25, 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E.I., Kovalenko,O.V., Gupta,R.C., Ward,D.C. and Radding,C.M. (1997) Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res., 25, 4106–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T., Golub,E.I., Reddy,G., Radding,C.M. and Ward,D.C. (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA, 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays S.L., Firmenich,A.A. and Berg,P. (1995) Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl Acad. Sci. USA, 92, 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtsmuller A.B. and Vermeulen,W. (2001) Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem. Cell Biol., 115, 13–21. [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Rademakers,S., Nigg,A.L., Hoogstraten,D., Hoeijmakers,J.H. and Vermeulen,W. (1999) Action of DNA repair endonuclease ERCC1/XPF in living cells. Science, 284, 958–961. [DOI] [PubMed] [Google Scholar]
- Johnson R.D. and Symington,L.S. (1995) Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol., 15, 4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S.C. (2000) Some assembly required. Nature Struct. Biol., 7, 1087–1089. [DOI] [PubMed] [Google Scholar]
- Kraakman-van der Zwet M. et al. (2002) Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol., 22, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Rothstein,R. and Mortensen,U.H. (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA, 98, 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N. et al. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell, 1, 783–793. [DOI] [PubMed] [Google Scholar]
- Liu Y. and Maizels,N. (2000) Coordinated response of mammalian Rad51 and Rad52 to DNA damage. EMBO rep., 1, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. (2001) Protein dynamics: implications for nuclear architecture and gene expression. Science, 291, 843–847. [DOI] [PubMed] [Google Scholar]
- Modesti M. and Kanaar,R. (2001) Homologous recombination: from model organisms to human disease. Genome Biol., 2, REVIEWS1014.1–1014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan P., Wilson,C., Townsend,S. and Thacker,J. (2001) XRCC2 is a nuclear RAD51-like protein, required for damage-dependent RAD51 focus formation without the need for ATP binding. J. Biol. Chem., 276, 22148–22153. [DOI] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. (2001) Protein mobility within the nucleus—what are the right moves? Cell, 104, 635–638. [DOI] [PubMed] [Google Scholar]
- Phair R.D. and Misteli,T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Raderschall E., Golub,E.I. and Haaf,T. (1999) Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl Acad. Sci. USA, 96, 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak A.Z., Larquet,E., Stasiak,A., Muller,S., Engel,A., Van Dyck,E., West,S.C. and Egelman,E.H. (2000) The human Rad52 protein exists as a heptameric ring. Curr. Biol., 10, 337–340. [DOI] [PubMed] [Google Scholar]
- Sung P., Trujillo,K.M. and Van Komen,S. (2000) Recombination factors of Saccharomyces cerevisiae. Mutat. Res., 451, 257–275. [DOI] [PubMed] [Google Scholar]
- Takata M. et al. (2000) The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol., 20, 6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M., Sasaki,M.S., Tachiiri,S., Fukushima,T., Sonoda,E., Schild,D., Thompson,L.H. and Takeda,S. (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol., 21, 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.L., Essers,J., Citterio,E., Swagemakers,S.M., de Wit,J., Benson,F.E., Hoeijmakers,J.H. and Kanaar,R. (1999) Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol., 9, 325–328. [DOI] [PubMed] [Google Scholar]
- Tashiro S., Walter,J., Shinohara,A., Kamada,N. and Cremer,T. (2000) Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol., 150, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck E., Stasiak,A.Z., Stasiak,A. and West,S.C. (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature, 398, 728–731. [DOI] [PubMed] [Google Scholar]
- White J. and Stelzer,E. (1999) Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol., 9, 61–65. [DOI] [PubMed] [Google Scholar]
- Yu V.P., Koehler,M., Steinlein,C., Schmid,M., Hanakahi,L.A., van Gool,A.J., West,S.C. and Venkitaraman,A.R. (2000) Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev., 14, 1400–1406. [PMC free article] [PubMed] [Google Scholar]