Abstract
The type III secretion system (TTSS) is an essential requirement for the virulence of many Gram-negative bacteria infecting plants, animals and man. Pathogens use the TTSS to deliver effector proteins from the bacterial cytoplasm to the eukaryotic host cell, where the effectors subvert host defences. Plant pathogens have to translocate their effector proteins through the plant cell wall barrier. The best candidates for directing effector protein traffic are bacterial appendages attached to the membrane-bound components of the TTSS. We have investigated the protein secretion route in relation to the TTSS appendage, termed the Hrp pilus, of the plant pathogen Pseudomonas syringae pv. tomato. By pulse expression of proteins combined with immunoelectron microscopy, we show that the Hrp pilus elongates by the addition of HrpA pilin subunits at the distal end, and that the effector protein HrpZ is secreted only from the pilus tip. Our results indicate that both HrpA and HrpZ travel through the Hrp pilus, which functions as a conduit for the long-distance translocation of effector proteins.
Keywords: pathogenesis/pilus assembly/protein translocation/Pseudomonas syringae/type III secretion
Introduction
The type III secretion system (TTSS) is a virulence mechanism shared by many unrelated Gram-negative bacteria that cause diseases in plants and animals (reviewed by Hueck, 1998). Diverse examples include Yersinia pestis, the cause of bubonic plague, and pathovars of Pseudomonas syringae, which attack several crop plants. The conservation of common virulence determinants in a wide range of bacteria infecting phylogenetically distant eukaryotic hosts is intriguing. The export system enables bacteria to deliver proteins, termed effectors, directly to the eukaryotic target cell (Galan and Collmer, 1999). In plant pathogenic bacteria, the components of the TTSS are encoded by a cluster of hrp genes. Mutations in hrp genes lead to pleiotropic loss of the ability to elicit the hypersensitive reaction (HR) in resistant plants and pathogenicity towards hosts (Alfano and Collmer, 1997). Two classes of effector proteins are secreted by the TTSS in plant pathogens: harpin proteins, such as HrpZ, which are readily exported in vitro and target cell membranes (Lee et al., 2001a,b); and avirulence (Avr) and virulence (Vir) proteins, such as AvrPto and VirPphA, which are at best poorly secreted in culture, but are believed to be translocated directly to the plant cell cytoplasm (Jackson et al., 1999; Jin et al., 2001). Expression of avr genes in resistant plants activates the HR via interaction with the protein products of resistance (R) genes (Stevens et al., 1998). The precise role of the effectors in plant pathogenesis is unknown. In the absence of matching R genes, both Avr and Vir proteins are thought, like effectors from animal pathogens, to suppress host defences by interaction with intracellular targets (Leach and White, 1996; Tsiamis et al., 2000). The HrpZ harpin from pathovars of P.syringae has been demonstrated to bind to lipid bilayers and to form an ion-conducting pore in vitro (Lee et al., 2001a).
The common components of the TTSS architecture found in both plant and animal pathogens are: a structure genetically homologous to the flagellar basal body, which traverses the bacterial inner membrane (Hueck, 1998; Kubori et al., 1998); an outer membrane-puncturing pore composed of a protein belonging to the secretin family (Hobbs and Mattick, 1993); and a genetically and morphologically varying extracellular appendage (Ginocchio et al., 1994; Roine et al., 1997a; Ebel et al., 1998; Knutton et al., 1998; Kubori et al., 1998; Blocker et al., 1999; Hoiczyk and Blobel, 2001). The Hrp pilus of the plant pathogen P.syringae pv. tomato DC3000 is the best characterized example of a TTSS-associated appendage (Roine et al., 1997a,b; Taira et al., 1999; Brown et al., 2001). Mutations in the major pilin subunit gene hrpA lead to the Hrp minus phenotype and failure to secrete Avr and harpin proteins such as AvrPto and HrpW in vitro (Wei et al., 2000). The Hrp pilus is, therefore, an indispensable component of the functional TTSS. The HrpA pilin is itself translocated to the bacterial surface by the internal components of the secretion apparatus.
The first TTSS appendage was found in Salmonella typhimurium by Ginocchio et al. (1994), who reported that surface appendages are formed upon contact with epithelial cells. Later, similar structures were observed on shiga toxin-producing Escherichia coli, and the appendages were associated with TTSSs (Ebel et al., 1998). The Hrp pilus of P.syringae was the first TTSS appendage observed in plant pathogenic bacteria (Roine et al., 1997a). Similar structures have now been reported in Erwinia amylovora (Jin et al., 2001) and Ralstonia solanacearum (Van Gijsegem et al., 2000). So-called needle complexes have been isolated from TTSS-expressing Salmonella, Shigella, Yersinia and E.coli (Kubori et al., 1998; Blocker et al., 1999; Hoiczyk and Blobel, 2001; Sekiya et al., 2001). Needle complexes are isolated type III secretion apparatuses that resemble morphologically the flagellar basal body with a needle-like extension attached at the distal end. Structures puncturing the bacterial inner membrane, the periplasm and the outer membrane of the bacterium were distinguished by electron microscopy. The needles observed in all four bacteria were thinner than the appendages observed by Ginocchio et al. (1994) and Ebel et al. (1998). In E.coli, a thick sheath-like structure was shown to be attached onto the core needle complex (Sekiya et al., 2001). The filament resembled in size the appendage observed by Ebel et al. (1998), and indeed it was shown, by immunoelectron microscopy, to be the same structure composed of EspA protein. The EspA filament has been shown to be involved in long-distance protein translocation to eukaryotic cells, allowing protein transfer without direct cell-to-cell contact between E.coli and its host (Shaw et al., 2001).
The Hrp pilus of P.syringae differs morphologically from the appendages found in animal pathogens. It is ∼6–8 nm in diameter (Roine et al., 1997a), thinner than the EspA filament, but similar to the needle of Salmonella. Importantly, the Hrp pilus can extend to a length of several micrometres (Brown et al., 2001), whereas the needle of Yersinia is only 60–80 nm long (Hoiczyk and Blobel, 2001). Whether the Hrp pilus is an extended version of the needle or a thinner variation of the EspA filament is not known. The genes encoding the pilin subunits are not conserved between plant pathogens and animal pathogens, albeit they all encode for small, α-helical proteins (Ebel et al., 1998; Delahay et al., 1999). In all interactions studied, the pili/filaments/needles have been suggested to form a channel between the bacterium and its eukaryotic host, although direct evidence for this has not been found. However, the appendages have been shown to form physical links between bacteria and their animal host cell (Ebel et al., 1998), and to penetrate the plant cell wall (Brown et al., 2001; Hu et al., 2001).
Immunocytochemical studies have demonstrated that secreted harpins and Avr proteins are associated with the Hrp pilus both in vitro and in vivo (Brown et al., 2001; Jin et al., 2001). The patterns of immunogold labelling observed in the earlier work did not distinguish between the secretion of proteins from the base or tip of the appendage. We have addressed the question of the movement of proteins through the pilus by investigating the polarity of pilus elongation and the site of secretion of the HrpZ harpin. Our experiments involved placing both genes under the control of a strictly induced mercury-responsive promoter. Growing bacteria under conditions for pilus development allowed a small appendage to form; next, the addition of HgCl2 induced production of either HrpA or HrpZ, which were identified by immunocytochemistry. By using this inducible system we were able to determine that the Hrp pilus of P.syringae pv. tomato elongates at the distal end and that the harpin protein HrpZ is secreted only from the pilus tip.
Results
FLAG tagging of the Hrp pilus
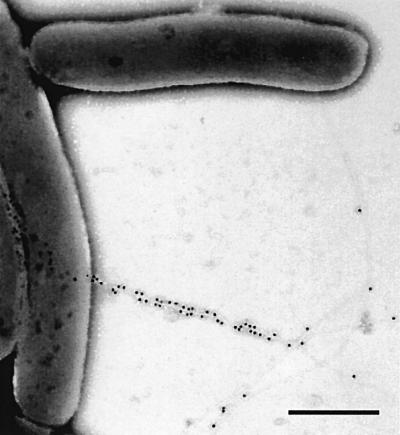
In order to distinguish newly expressed HrpA from the earlier synthesized pilins, we epitope tagged the HrpA pilin subunit. The C-terminal part of the HrpA pilin is responsible for pilus assembly, whereas the N-terminus tolerates insertions and large deletions without affecting pilus formation or function in planta (Taira et al., 1999). A FLAG epitope was cloned at four different N-terminus-encoding sites in the hrpA gene. The cloning sites after codons 15, 23, 24 and 48 were linker insertion sites created earlier by scanning mutagenesis (Taira et al., 1999). Complementation of P.syringae DC3000 hrpA– with pDN18 derivatives encoding the tagged pilins showed that FLAG insertion at all four sites permitted pilin secretion, pilus assembly and function in planta (data not shown). Immunocytochemistry with monoclonal anti-FLAG antiserum revealed that the epitope was exposed on the pilus surface when inserted after codons 23 (Figure 1) or 24, as recognized by immunogold labelling. The anti-FLAG antiserum did not label the pilus with a FLAG insertion after amino acid 48 of the pilin, and very few immunogold particles were observed on pili carrying the FLAG tag after amino acid 15. The pilin FLAG-tagged at amino acid 23 was chosen for further studies and is referred to as FLAG-HrpA in the following text.
Fig. 1. Immunogold localization of the FLAG epitope (20 nm gold particles) in a Hrp pilus composed entirely of FLAG-HrpA. The FLAG tag was inserted after amino acid 23 of HrpA and cloned into pDN18. The hrpA mutant of DC3000 harbouring the FLAG-HrpA-encoding pDN18 derivative was used to examine pilus production after 18 h incubation in Hrp-inducing minimal medium. Bar = 0.5 µm.
FLAG-HrpA produced under the control of a mercury-inducible promoter
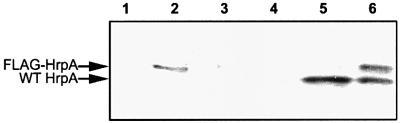
In order to express FLAG-HrpA independently of the rest of the TTSS, we cloned the gene encoding FLAG-HrpA under the control of a mercury-inducible promoter (see Materials and methods). The fusion construct, termed pMerFLAGHrpA, was introduced to wild-type P.syringae DC3000 together with the mer repressor-harbouring plasmid pTPT11 (Petänen et al., 2001). Immunoblotting with anti-FLAG and anti-HrpA antisera showed that the expression of FLAG-HrpA was completely turned off in the absence of mercury and strongly induced by 100 nM HgCl2, both in the minimal medium, which induces all hrp promoters (Huynh et al., 1989), and in a rich medium, where hrp genes are normally silent. Wild-type DC3000 harbouring pMerFLAGHrpA expressed and secreted both FLAG-HrpA and wild-type HrpA encoded by the chromosomal hrpA locus when grown in minimal medium in the presence of 100 nM HgCl2 (Figure 2).
Fig. 2. Immunoblot showing production and secretion of FLAG-HrpA by DC3000 harbouring pMerFLAGHrpA with anti-HrpA antiserum. Bacteria were grown for 18 h before extraction of proteins, which were separated by 16% SDS–PAGE and probed with anti-HrpA antibodies. Lanes 1 and 2 are cellular fractions; lanes 3–6 are extracellular fractions. Lanes 1 and 3, King’s B medium; lanes 2 and 4, King’s B with HgCl2; lane 5, Hrp-inducing medium; lane 6, Hrp-inducing medium with HgCl2.
Immunoelectron microscopy of the growing pilus
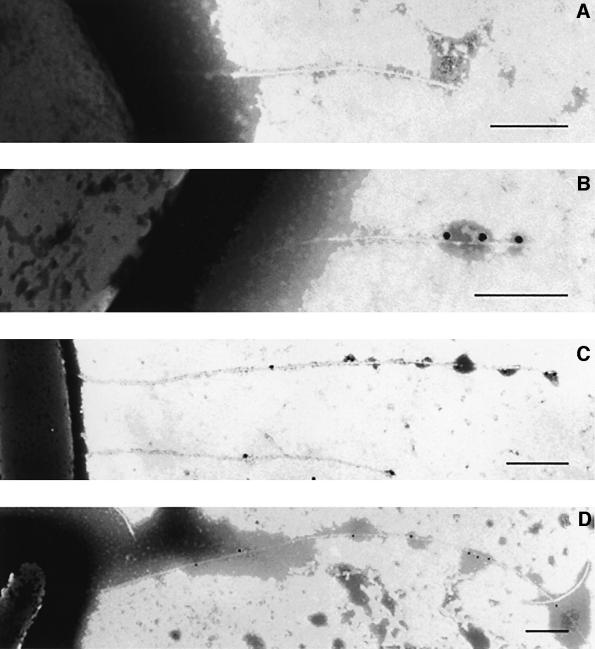
The development of Hrp pili was examined using modifications of the method devised by Brown et al. (2001), in which bacteria are grown on electron microscopy grids to avoid disrupting their fragile appendages. DC3000 (pMerFLAGHrpA) was grown for 8 h in hrp-inducing minimal medium before induction of FLAG-HrpA by addition of HgCl2. Samples were examined 15, 30 and 60 min after mercury induction. By the first time point, only 15 min after mercury induction, a few pilus-associated gold particles were observed at the distal end of the appendage (Figure 3B). At later time points, sites of immunogold labelling increased, but only at the distal end of the pilus (Figure 3C and D). The density of labelling of the Hrp pilus with anti-FLAG antibody was lower in the DC3000 wild-type background than in the hrpA– mutant (compare Figures 1 and 3). Such a reduced level of FLAG-HrpA incorporation would be expected if the pili produced were a mixture of wild-type and epitope-tagged HrpA, and indicates that there is no absolute selection in recruitment of HrpA or FLAG-HrpA for pilus assembly.
Fig. 3. Pili incorporating induced FLAG-HrpA are decorated by immunogold labelling (20 nm gold particles) of the FLAG epitope at the distal end of the appendage. Bacteria (DC3000 pMerFLAGHrpA) were grown for 8 h in minimal medium to allow pilus formation and then examined after the addition of HgCl2 to induce FLAG-HrpA. (A) Pilus before addition of HgCl2; note the absence of immunogold label; (B–D) 15, 30 and 60 min, respectively, after FLAG-HrpA induction. Bars: (A) and (B) = 0.25 µm; (C) and (D) = 0.5 µm.
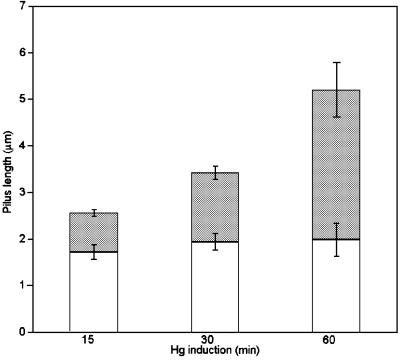
Quantitative analysis of the length of pilus containing incorporated FLAG-HrpA demonstrated the presence of an unlabelled basal (proximal) portion of the pilus and an expanding, FLAG-tagged distal region (Figure 4). The time course also allowed the rate of extension to be determined as ∼50 nm/min. Our results clearly demonstrated rapid growth of the pilus by the incorporation of HrpA subunits at the tip of the filament and suggest that HrpA monomers are translocated acropetally through the growing pilus. The mean numbers of particles of gold labels recorded per pilus 15, 30 and 60 min after induction were 3.9 ± 0.3, 5.1 ± 0.9 and 8.7 ± 0.7, respectively. An additional feature of the quantitative study was the finding that no short pili labelled all along their filament were observed. The absence of such appendages suggests that no new TTSS or pili were formed during the incubation periods after the addition of HgCl2 to induce FLAG-HrpA.
Fig. 4. Quantitative analysis of the development of the Hrp pilus. The length of the unlabelled proximal (open column) and immunogold- labelled distal (shaded column) portion of the Hrp pilus incorporating the FLAG-HrpA subunit was recorded from at least 30 pili at each time point after induction of FLAG-HrpA (means ± SEM are given). Immunogold labelling was achieved using anti-FLAG monoclonal antibodies.
HrpZ secretion occurs at the pilus tip
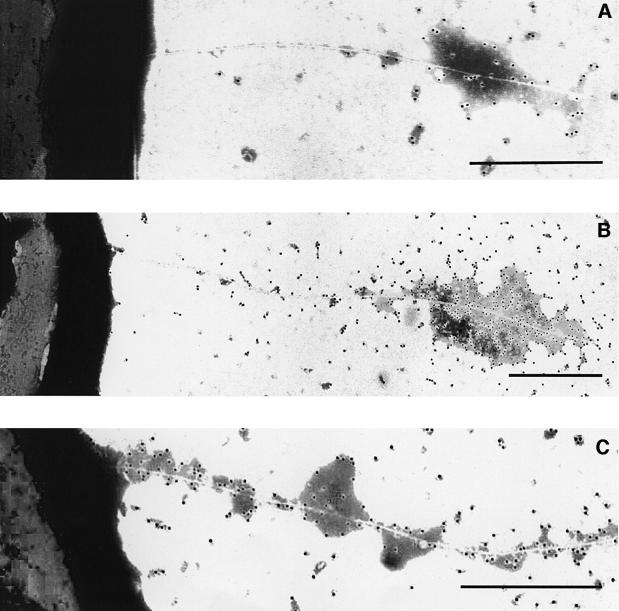
We next addressed the possibility that effector proteins may also travel through the pilus. As Avr and Vir proteins are secreted in vitro with low efficiency, we used the harpin protein HrpZ as a model. We cloned the hrpZ gene from P.syringae pv. phaseolicola under the control of the mer promoter, as described in Materials and methods, and transferred the resulting plasmid, pMerHrpZ, into DC3000ΔhrpZ. As the translocation of HrpZ was examined in an hrpZ deletion mutant, an epitope tag was unnecessary for this experiment, and secretion could easily be followed using polyclonal antiserum to HrpZ. DC3000ΔhrpZ(pMerHrpZ) was grown on electron microscopy grids as above. When the mercury induction was performed 8 h after Hrp induction, newly made HrpZ was first detected at the tip of the pilus, and in time coated the distal portion of the growing appendage (Figure 5A and B). If bacteria were exposed to HgCl2 throughout the incubation, all parts of the emerging pilus were decorated by the HrpZ antiserum (Figure 5C). As observed with HrpA, a secretion route through the pilus was demonstrated.
Fig. 5. Immunogold localization of HrpZ in DC3000ΔhrpZ harbouring pMerHrpZ, which allows mercury-driven HrpZ expression. (A and B) Emergence of HrpZ (immunogold label) at the tip of the pilus 30 and 60 min, respectively, after the addition of HgCl2 to induce HrpZ following 8 h growth in minimal medium to allow pilus formation. (C) Labelling along the proximal base of the pilus if bacteria were grown continuously in minimal medium + HgCl2 for 8 h. Bars = 0.5 µm.
Discussion
The molecular mechanism by which TTSS appendages contribute to protein delivery has until now been elusive. The results of our pulse-expression experiments with P.syringae demonstrated that secreted proteins emerge in the extracellular space at the pilus tip. The simplest explanation of our immunocytochemical data is that the pilus functions as a conduit through which travel pilin subunits and TTSS-dependent effectors. In P.syringae, the long Hrp pilus potentially allows the long-distance delivery of proteins directly to the plant cell membrane by crossing the plant cell wall barrier (Brown et al., 2001). The protein traffic from animal pathogens such as Shigella or Salmonella to their hosts occurs between membranes that are in close contact. Perhaps in consequence, the needle extensions observed in these bacteria are considerably shorter than the Hrp pili.
The Hrp pilus of P.syringae and the needles of animal pathogens have an external diameter of 6–8 nm (Roine et al., 1997b; Kubori et al., 1998; Blocker et al., 1999; Brown et al., 2001). The internal diameter of the Yersinia needle has been estimated to be 2 nm (Hoiczyk and Blobel, 2001). Considering pilus dimensions, proteins using this secretion route would be expected to be at least partially unfolded for transport. Many of the effector proteins translocated by TTSSs have been shown to be dependent on chaperones, which keep the effector in a partially unfolded form in the bacterial cytoplasm (Stebbins and Galan, 2001). It is envisaged that, in P.syringae, the effectors would therefore enter the secretion channel unfolded, travel the remarkably long intra-pilus route across the plant cell wall, and take their active conformation at their destination following secretion. Such a scenario is probably the case for the Hrp pilin itself. Hrp pilin can auto-assemble to form filaments in vitro (Roine et al., 1997b), and may therefore be able to fold and combine to form the pilus structure at the tip of the growing appendage. The mode of pilus extension also explains how the long and flexible pilus may wriggle through the porous cellulose matrix that comprises the plant cell wall. As growth occurs at the tip, pilus assembly must occur inside the plant cell wall rather than the structure being driven into the wall by basal extension, as proposed by Brown et al. (2001).
The next interesting question concerns the fate of the pilus when it reaches the plant cell membrane. It is possible that the emerging pilin monomers are dispersed within the lipid membrane. Alternatively, the pilus may puncture the membrane, as suggested for the Yersinia needle (Hoiczyk and Blobel, 2001). Whether pilus assembly provides enough energy to force the pilus through the membrane or if a plant membrane protein is involved in this interaction is not known. HrpZ would be an excellent candidate to serve as a mediator in the pilus–membrane interaction, since it has been shown to insert in lipid membranes (Lee et al., 2001a) and, as it is secreted at the pilus tip, it is at the right place at a high local concentration.
Clearly, the assembly of the Hrp pilus diverges from the basal-growth pattern of type I and type IV pili (Hultgren and Normark, 1991). Our result, showing distal extension of the Hrp pilus, indicates similarity with the bacterial flagellum, since newly synthesized flagellin monomers are also added to the tip of the filament (Emerson et al., 1970). It has been suggested, based on protein homology (Macnab, 1999), that the flagellar apparatus is a member of the TTSS family. Our observation supports the idea of a common evolutionary origin of the flagellum and the TTSS by revealing functional and constructional similarities. The flagellum is a hollow structure (Namba et al., 1989) through which flagellin monomers reach the distal end of the filament. Our observations indicate that the needle/pilus part of the TTSS is also a hollow tube, through which the secreted proteins travel to the tip.
Our experiments have confirmed the route for TTSS-dependent protein delivery. Most significantly, they raise fundamental questions for further study, including the energy requirements for movement through the long pilus, and the possible need for regulation of protein traffic in the narrow and presumably crowded pilus tunnel.
Materials and methods
Strains and growth media
Pseudomonas syringae DC3000 (D.Cuppels, London, Ontario, Canada) and its hrpA– derivative (Roine et al., 1997b) or hrpZ– derivative (A.Collmer, Ithaca, NY) were used in all experiments. Hrp induction was carried out in minimal medium with fructose as the sole carbon source (Huynh et al., 1989), and antibiotics used were rifampicin (75 µg/l), tetracycline (10 µg/l) and chloramphenicol (35 µg/l). King’s B (King et al., 1954) was used as the Hrp-repressing medium.
Immunoblotting
DC3000 harbouring pMerFLAGHrpA was grown overnight in King’s B or Hrp-inducing medium in the presence or absence of 100 nM HgCl2. Cell density was adjusted spectrophotometrically to be the same in all samples. Cellular and extracellular fractions were separated by centrifugation at 16 000 g for 15 min. Supernatant representing secreted and detached pili, and cell pellet representing cell-associated pili, were boiled in sample buffer and an equal amount of each sample was loaded in the gel. The wild-type HrpA and FLAG-tagged HrpA were separated in 16% SDS–polyacrylamide gel, and transferred on PVDF membranes (Schleicher & Schuell, USA). HrpA antiserum (Roine et al., 1997a), anti-rabbit alkaline phosphatase conjugate (Sigma) and standard colour reaction were used to visualize the pilins.
FLAG tagging the Hrp pilus
A FLAG epitope-encoding linker was made by annealing oligonucleotides GGCCGCGACTACAAGGACGATGACGATAAG and GGCCCTTATCGTCATCGTCCTTGTAGTCGC together. The linker contains NotI-compatible cohesive ends. The FLAG linker was cloned into NotI sites in hrpA created earlier by linker scanning mutagenesis (Taira et al., 1999).
Expression of FLAG-HrpA and HrpZ under the control of a mercury-inducible promoter
The gene encoding FLAG-HrpA without its natural promoter was fused with the mer promoter of Tn21 (Misra et al., 1984) at the transcription start point of mer and –42 bp upstream of the hrpA start codon (transcriptional start of hrpA), using standard PCR protocols. The oligonucleotides used were: (1) hrpA 3′ region-specific GTACCTTCTAGAGGTAGCGGCC, introducing a XbaI site; (2) mer 5′ promoter region-specific AATTAAGCTTCGAGCTAAGGCATAGC, introducing a HindIII site; (3) fusion oligonucleotide GAAGTAAGGTTACGCTATACCAAGCAATCACGCTG, containing sequence specific to the mer promoter immediately upstream of the transcription start site, and sequence specific to hrpA immediately after the transcription site; and (4) the complement of oligonucleotide 3. The mer promoter was amplified with oligonucleotides 2 and 4, and hrpA with oligonucleotides 1 and 3. The resulting PCR fragments were mixed and used as templates in PCR with oligonucleotides 1 and 2. The final PCR product was cloned into a broad host range plasmid pBBR1MCS (Kanter-Smoler et al., 1994) as a HindIII–XbaI fragment. Since the fusion disrupts the merR promoter required for mer repressor production, plasmid pTPT11, which harbours the intact gene for the mer repressor, was used to provide the repressor in trans. For mer promoter-driven HrpZ, we PCR amplified the mer promoter and the entire merR gene with oligonucleotide 2 above and oligonucleotide TCGGATCCTTTTGAATTTGGATTG, which introduce HindIII and BamHI sites, respectively, and cloned the resulting fragment in pDN18. The gene encoding P.syringae pv. phaseolicola hrpZ was amplified with oligonucleotides AAGGATCCTACTTTGAGGAGGTTG and TTGAATTCTCCGTCAGGCGG, which introduce BamHI and EcoRI sites, and the resulting fragment was cloned after the mer promoter in pDN18. In pMerHrpZ, 19 bp were included upstream of the hrpZ start codon, hence the ribosome-binding site of the original mer gene was replaced by the ribosome-binding site of hrpZ.
Electron microscopy
Methods were essentially as described by Brown et al. (2001) apart from the following modifications. Carbon/formvar-coated gold grids (300 mesh) were floated carbon side down on 18 µl drops of bacterial suspension in Hrp-inducing minimal medium adjusted to an OD of 0.3 at 600 nm. For the pulse-expression experiments, grids were incubated at 20°C for 8 h when 2 µl of HgCl2 stock solution were added to the drop, giving a final concentration of 100 nM. Grids were further incubated for 15, 30 and 60 min, after which they were fixed by addition of 80 µl of 2% formaldehyde and 0.5% glutaraldehyde in 50 mM sodium cacodylate buffer pH 7.2 to the 20 µl droplet. Grids were left to fix this way for 15 min before being floated on a fresh drop of fixative at 4°C overnight. Anti-FLAG M2 monoclonal antibody (Sigma, UK) was used at a concentration of 50 µg/ml. Grids were washed by passing them through six 20 µl drops, rather than in a flow of buffer from a wash bottle, in order to preserve the delicate pili. HrpZ was labelled with a rabbit polyclonal antibody as described previously (Brown et al., 2001). Second antibodies were goat anti-mouse IgG–20 nm gold and goat anti-rabbit IgG–10 nm gold (British Biocell International, Cardiff, UK) diluted 1:50, and were used to visualize FLAG and HrpZ, respectively. Grids were negatively stained in 1% phosphotungstic acid/KOH at pH 6.5 for 10 s and were observed at an accelerating voltage of 75 kV in an Hitachi H-7000 transmission electron microscope.
Images were recorded of 30 pili for each time point for the quantitative analysis of pilus elongation using FLAG labelling. Only those pili that could be tracked back unambiguously to a bacterium were sampled. The lengths of the epitope-labelled tip and the unlabelled basal portion were measured from negatives and the numbers of gold particles recorded.
Acknowledgments
Acknowledgements
Tiina Petänen is acknowledged for providing mer-harbouring plasmids and for valuable advice in handling the mercury induction. Ghislain Pras and Alexis Prasit are thanked for their help with Adobe Photoshop. The Academy of Finland (C.-M.L., S.T., M.R.), University of Helsinki (T.B.) and UK BBSRC (I.B., C.S., J.M.) are acknowledged for financial support. Work at Wye was completed under MAFF licence PHL 30A/3755 (4/2001).
Note added in proof
After submission of this manuscript and in support of our findings, extrusion of effector protein AvrPto from the Hrp pilus tip of Pseudomonas syringae was described by Q.Jin and S.Y.He (Science, 294, 2556–2558, 2001).
References
- Alfano J.R. and Collmer,A. (1997) The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins and death. J. Bacteriol., 179, 5655–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A., Gounon,P., Larquet,E., Niebuhr,K., Cabiaux,V., Parsot,C. and Sansonetti,P. (1999) The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol., 147, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown I.R., Mansfield,J.W., Taira,S., Roine,E. and Romantschuk,M. (2001) Immunocytochemical localization of HrpA and HrpZ supports a role for a transfer of effector proteins from Pseudomonas syringae pv. tomato across the plant cell wall. Mol. Plant Microbe Interact., 14, 394–404. [DOI] [PubMed] [Google Scholar]
- Delahay R.M., Knutton,S., Shaw,R.K., Hartland,E.L., Pallen,M.J. and Frankel,G. (1999) The coiled-coil domain of EspA is essential for the assembly of the type III secretion translocon on the surface of enteropathogenic Escherichia coli. J. Biol. Chem., 274, 35969–35974. [DOI] [PubMed] [Google Scholar]
- Ebel F., Podzadel,T., Rohde,M., Kresse,A.U., Kramer,S., Deibel,C., Guzman,C.A. and Chakraborty,T. (1998) Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol. Microbiol., 30, 147–161. [DOI] [PubMed] [Google Scholar]
- Emerson S.U., Tokuyasu,K. and Simon,M. (1970) Bacterial flagella: polarity of elongation. Science, 169, 190–192. [DOI] [PubMed] [Google Scholar]
- Galan J.E. and Collmer,A. (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science, 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Ginocchio C.C., Olmsted,S.B., Wells,C.L. and Galan,J.E. (1994) Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium. Cell, 76, 717–724. [DOI] [PubMed] [Google Scholar]
- Hobbs M. and Mattick,J.S. (1993) Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol., 10, 233–243. [DOI] [PubMed] [Google Scholar]
- Hoiczyk E. and Blobel,G. (2001) Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl Acad. Sci. USA, 98, 4669–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Yuan,J., Jin,Q.-L., Hart,P. and He,S.Y. (2001) Immunogold labeling of Hrp pili of Pseudomonas syringae pv. tomato assembled in minimal medium and in planta. Mol. Plant Microbe Interact., 14, 234–241. [DOI] [PubMed] [Google Scholar]
- Hueck C.J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev., 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultgren S.J. and Normark,S. (1991) Biogenesis of the bacterial pilus. Curr. Opin. Genet. Dev., 1, 313–318. [DOI] [PubMed] [Google Scholar]
- Huynh T.V., Dahlbeck,D. and Staskawicz,B.J. (1989) Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science, 245, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Jackson R.J. et al. (1999) Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl Acad. Sci. USA, 96, 10875–10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Hu,W., Brown,I., McGhee,G., Hart,P., Jones,A.L. and He,S.Y. (2001) Visualization of secreted Hrp and Avr proteins along the Hrp pilus during type III secretion in Erwinia amylovora and Pseudomonas syringae. Mol. Microbiol., 40, 1129–1139. [DOI] [PubMed] [Google Scholar]
- Kanter-Smoler G., Dahlqvist,A. and Sunnerhagen,P. (1994) pBBRMCS: a broad-host-range cloning vector. Biotechniques, 16, 800–802. [PubMed] [Google Scholar]
- King E.O., Ward,M.K. and Raney,D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med., 22, 301–307. [PubMed] [Google Scholar]
- Knutton S., Rosenshine,I., Pallen,M.J., Nisan,I., Neves,B.C., Bain,C., Wolff,C., Dougan,G. and Frankel,G. (1998) A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J., 17, 2166–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T., Matsushima,Y., Nakamura,D., Uralil,J., Lara-Tejero,M., Sukhan,A., Galan,J.E. and Aizawa,S.I. (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science, 280, 602–605. [DOI] [PubMed] [Google Scholar]
- Leach J.E and White,F.F. (1996) Bacterial avirulence proteins. Annu. Rev. Phytopathol., 34, 153–179. [DOI] [PubMed] [Google Scholar]
- Lee J. et al. (2001a) HrpZ(Psph) from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc. Natl Acad. Sci. USA, 98, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Klessig,D.F. and Nurnberger,T. (2001b) A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell, 13, 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R.M. (1999) The bacterial flagellum: reversible rotary propeller and type III export apparatus. J. Bacteriol., 181, 7149–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T.K., Brown,N.L., Fritzinger,D., Pridmore,R., Barnes,W. and Silver,S. (1984) Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc. Natl Acad. Sci. USA, 81, 5975–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba K., Yamashita,I. and Vonderviszt,F. (1989) Structure of the core and the central channel of bacterial flagella. Nature, 342, 648–654. [DOI] [PubMed] [Google Scholar]
- Petänen T., Virta,M., Karp,M. and Romantschuk,M. (2001) Construction and use of broad host range mercury and arsenite sensor plasmids in the soil bacterium Pseudomonas fluorescens OS8. Microb. Ecol., 41, 360–368. [DOI] [PubMed] [Google Scholar]
- Roine E., Wei,W., Yuan,J., Nurmiaho-Lassila,E.L., Kalkkinen,N., Romantschuk,M. and He,S.Y. (1997a) Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl Acad. Sci. USA, 94, 3459–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine E., Saarinen,J., Kalkkinen,N. and Romantschuk,M. (1997b) Purified HrpA of Pseudomonas syringae pv. tomato DC3000 reassembles into pili. FEBS Lett., 417, 168–172. [DOI] [PubMed] [Google Scholar]
- Sekiya K., Ohishi,M., Ogino,T., Tamano,K., Sasakawa,C. and Abe,A. (2001) Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl Acad. Sci. USA, 98, 11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R.K., Daniell,S., Ebel,F., Frankel,G. and Knutton,S. (2001) EspA filament-mediated protein translocation into red blood cells. Cell. Microbiol., 3, 213–222. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E. and Galan,J.E. (2001) Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature, 414, 77–81. [DOI] [PubMed] [Google Scholar]
- Stevens C., Bennett,M.A., Athanassopoulos,E., Tsiamis,G., Taylor,J.D. and Mansfield,J.W. (1998) Sequence variations in alleles of the avirulence gene avrPphE.R2 from Pseudomonas syringae pv. phaseolicola lead to loss of recognition of the AvrPphE protein within bean cells and a gain in cultivar-specific virulence. Mol. Microbiol., 29l, 165–177. [DOI] [PubMed] [Google Scholar]
- Taira S., Tuimala,J., Roine,E., Nurmiaho-Lassila,E.L., Savilahti,H. and Romantschuk,M. (1999) Mutational analysis of the Pseudomonas syringae pv. tomato hrpA gene encoding Hrp pilus subunit. Mol. Microbiol., 34, 737–744. [DOI] [PubMed] [Google Scholar]
- Tsiamis G. et al. (2000) Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J., 19, 3204–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gijsegem F., Vasse,J., Camus,J.-C., Marenda,M. and Boucher,C. (2000) Ralstonia solanacearum produces Hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol. Microbiol., 36, 249–260. [DOI] [PubMed] [Google Scholar]
- Wei W., Plovanich-Jones,A., Deng,W.-L., Jin,Q.-L., Collmer,A., Huang,H.-C. and He,S.Y. (2000) The gene coding for the Hrp pilus structural protein is required for type III secretion of Hrp and Avr proteins in Pseudomonas syringae pv. tomato. Proc. Natl Acad. Sci. USA, 97, 2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]