Abstract
Activity-dependent local translation of dendritic mRNAs is one process that underlies synaptic plasticity. Here, we demonstrate that several of the factors known to control polyadenylation-induced translation in early vertebrate development [cytoplasmic polyadenylation element-binding protein (CPEB), maskin, poly(A) polymerase, cleavage and polyadenylation specificity factor (CPSF) and Aurora] also reside at synaptic sites of rat hippocampal neurons. The induction of polyadenylation at synapses is mediated by the N-methyl-d-aspartate (NMDA) receptor, which transduces a signal that results in the activation of Aurora kinase. This kinase in turn phosphorylates CPEB, an essential RNA-binding protein, on a critical residue that is necessary for polyadenylation-induced translation. These data demonstrate a remarkable conservation of the regulatory machinery that controls signal-induced mRNA translation, and elucidates an axis connecting the NMDA receptor to localized protein synthesis at synapses.
Keywords: polyadenylation/protein phosphorylation/synapse/translation
Introduction
The experience-dependent changes in synaptic strength, i.e. synaptic plasticity, may be a causal link to higher order neural activity such as learning and memory. Such changes that are of short duration probably involve the modification of proteins already present at synapses prior to their activation; changes that are longer lasting require the synthesis of new proteins that presumably become resident at the activated synapse. The observation that dendritic shafts contain many components of the translational apparatus has suggested that local mRNA translation could be involved in modulating synaptic plasticity (e.g. Crino and Eberwine, 1996; Martin et al., 2000; Steward and Schuman, 2001; Richter and Lorenz, 2002). This possibility is underscored by three observations: (i) isolated dendritic (Kacharmina et al., 2000) and synaptic (Bagni et al., 2000; Scheetz et al., 2000) compartments can synthesize new protein; (ii) two forms of synaptic plasticity, brain-derived neurotrophic factor (BDNF)-induced long-lasting phase of long-term potentiation (L-LTP) (Kang and Schuman, 1996) and 3,5-dihydroxyphenylglycine (DHPG)-induced long-term depression (LTD) (Huber et al., 2000) occur in dendritic layers severed from nuclei-containing cell bodies but only in the absence of protein synthesis inhibitors; and (iii) a reporter mRNA in transfected hippocampal neurons can undergo BDNF-induced translational induction in dendrites (Aakalu et al., 2001).
While the mechanism(s) by which synaptic activation induces the translation of specific mRNAs is not known, some evidence suggests that it could be similar to that which occurs in early development (Wu et al., 1998). In Xenopus oocytes, for example, several dormant mRNAs are recruited for translation only when they undergo cyto plasmic polyadenylation (Richter, 2000). Polyadenylation, in turn, is controlled by the CPE (general structure of UUUUUAU), a sequence in the 3′-untranslated region (UTR) of responding mRNAs, and its binding protein cytoplasmic polyadenylation element-binding protein (CPEB). The recent identification of CPEB in the mammalian central nervous system (CNS) suggested that polyadenylation-induced translation might occur in this tissue as well as in oocytes (Wu et al., 1998). Indeed, experience-dependent activation of synapses results in the polyadenylation and translation of the CPE-containing α-Ca2+/calmodulin-dependent protein kinase II (αCaMKII) mRNA, but not of the CPE-lacking neurofilament (NF) mRNA (Wu et al., 1998). Moreover, the increased αCaMKII synthesis in synaptoneurosomes isolated from the visual cortex of dark-reared rats exposed to light (i.e. visual experience) is inhibited if the animals are injected with 3-(2-carboxypiperazin-4-yl) propyl-1-phosphonic acid [CPP; an antagonist of N-methyl-d-aspartate (NMDA)] or cordycepin (3′-deoxyadenosine, a blocker of polyadenylation) prior to light exposure (Wells et al., 2001). These observations suggest that αCaMKII synthesis may involve the NMDA receptor (which mediates visual experience) signaling and a polyadenylation event. However, where polyadenylation takes place and how it can be regulated in response to synaptic stimulation are unknown.
To begin to investigate not only the subcellular location at which experience-dependent polyadenylation occurs (i.e. synapse, soma or nucleus), but also the signaling events that may be involved, it is enormously useful to consider the interplay of factors that mediate cytoplasmic polyadenylation in early development. While the CPEs of dormant Xenopus oocyte mRNAs are associated with CPEB, the second element essential for polyadenylation, AAUAAA, is unoccupied. The induction of polyadenylation requires the Aurora kinase (also known as Eg2 or IAK1 in other nomenclature) (Andresson and Ruderman, 1998; Nigg, 2001), which phosphorylates CPEB Ser174 (Mendez et al., 2000a). This event induces CPEB to recruit the multisubunit cleavage and polyadenylation specificity factor (CPSF) to the AAUAAA sequence (Mendez et al., 2000b), which in turn probably attracts active poly(A) polymerase (PAP) to the end of the mRNA. The mechanism by which polyadenylation induces translation involves maskin, a CPEB-associated protein that also interacts with eIF4E, the cap-binding protein (Stebbins-Boaz et al., 1999). The binding of maskin to eIF4E precludes the binding of eIF4G to eIF4E, an interaction that is necessary for the correct positioning of the 40S ribosomal subunit on the end of the mRNA (Gingras et al., 1999). At a time that is coincident with and caused by polyadenylation, maskin dissociates from eIF4E (Cao and Richter, 2002), thereby allowing eIF4G to bind eIF4E and properly assemble the translation initiation complex (Stebbins-Boaz et al., 1999; reviewed in Mendez and Richter, 2001; Q.P.Cao and J.D.Richter, submitted).
In this study, we show that the multiple components involved in cytoplasmic polyadenylation in Xenopus oocytes are also present at synapses of hippocampal neurons where they support NMDA receptor activity-dependent polyadenylation. We also demonstrate that NMDA receptor stimulation results in the activation of Aurora, which phosphorylates CPEB on a key residue that is essential for polyadenylation. Thus, these data not only show that there is an extraordinary conservation of a key translational control mechanism in early development and in the CNS, but they also point to one molecular basis of synaptic plasticity.
Results
Localization of polyadenylation/translation factors at synapses
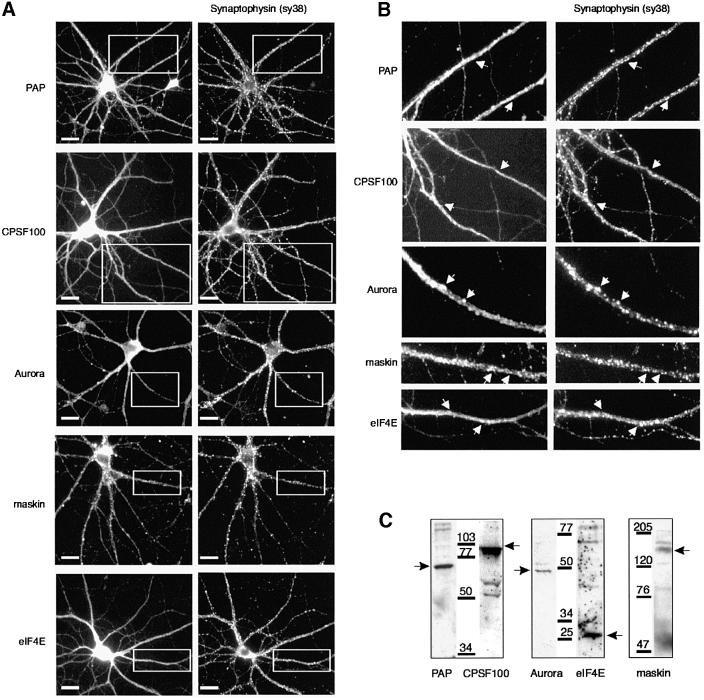
Cultures of rat primary hippocampal neurons were probed with antibodies directed against PAP, the 100 kDa subunit of CPSF (CPSF100), Aurora, maskin and eIF4E (all of the antibodies have been used previously for immunolocalization; Hodgman et al., 2001), as well as those against synaptophysin and αCaMKII, which serve as synaptic markers. Figure 1A shows that immunoreactivity for all these polyadenylation/translation factors was evident in the cell body, and in some cases was distributed as puncta along the dendrites, which is similar to that observed for CPEB (Wu et al., 1998). Immunoreactivity for PAP and CPSF100 was also observed in nuclei, an expected result because these proteins are primarily nuclear. Some of the immunoreactivity for all the factors was also co-localized with synaptophysin (arrows in Figure 1B), suggesting that the polyadenylation machinery is located near or at synapses. The specificity of the antibodies used for the immunolocalization was assessed in a series of western blots shown in Figure 1C. Antibodies directed against PAP, CPSF, Aurora, eIF4E and maskin each primarily detected bands corresponding to the appropriate molecular weights of the proteins.
Fig. 1. Immunolocalization of polyadenylation/translation factors in dendrites of cultured hippocampal neurons. (A) Fourteen day rat primary hippocampal cultures were immunostained with antibodies directed against poly(A) polymerase (PAP), the 100 kDa subunit of CPSF (CPSF100), Aurora, maskin and eIF-4E, and co-stained with monoclonal antibody for synaptophysin (sy38) which locates synapses. (B) A higher magnification of dendrites taken from selected areas (white rectangles) in (A) demonstrates that these factors occasionally are co-localized with synapses (arrows). (C) The specificity of the anti-PAP, CPSF, Aurora, eIF-4E and maskin antibodies was evaluated by immunoblotting using the mouse brain extract. Each of these antibody preparations recognized one major band of the expected molecular weight (arrows), PAP, 70 kDa; CPSF, 100 kDa; Aurora, 47 kDa; eIF4E, 25 kDa; maskin, 150 kDa. Scale bars, 10 µm.
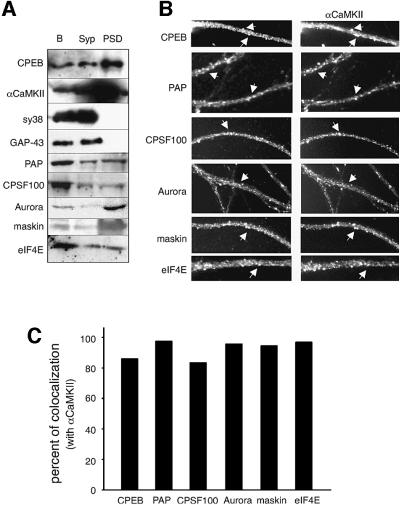
To determine whether the polyadenylation/translation factors are at least partially postsynaptic, the mouse brain was fractionated into synaptosome (Syp) and postsynaptic density (PSD) compartments (Figure 2A). Western blots of these fractions revealed the presence of CPEB and αCaMKII in the PSD, which are positive markers for the fractionation procedure (Wu et al., 1998); the absence of synaptophysin (sy38) and growth-associated protein-43 (GAP-43) in the PSD served as negative controls. All the other polyadenylation/translation factors were detected in the PSD, although only Aurora and maskin were enriched in that fraction (Figure 2A). Because PAP, CPSF100 and eIF4E are known to function in other regions of neurons, their lack of enrichment in the PSD was an expected result. These observations suggest that polyadenylation-induced translation could take place in the synapto-dendritic compartment.
Fig. 2. Identification of polyadenylation/translation factors at the postsynaptic density (PSD). (A) Immunoblots were prepared from total adult mouse brain homogenate (B), synaptosome (Syp) and detergent-extracted PSD fractions, and probed with the antibodies noted above. The blots were also probed with antibodies directed against CPEB, αCaMKII, synaptophysin (sy38) and GAP-43. (B) Cultured rat primary hippocampal neurons were treated with Triton X-100 prior to fixation and then immunostained with CPEB antibody, as well as the antibodies noted above and a monoclonal antibody directed against αCaMKII. The arrows denote regions of apparent co-localization. (C) The degree of co-localization of polyadenylation/translation factors with αCaMKII in dendrites is quantified.
In cultured hippocampal neurons, αCaMKII is particularly concentrated at excitatory synapses within dendritic spines where it mediates signal transduction in response to calcium influx (Kennedy, 1998; Allison et al., 2000). Because synaptic activity triggers αCaMKII protein synthesis (Ouyang et al., 1997, 1999) as well as αCaMKII mRNA polyadenylation (Wu et al., 1998), it seemed plausible that the polyadenylation/translation factors might be clustered at the synapses that contain this kinase. Accordingly, we immunostained hippocampal neurons with antibodies for the factors noted above as well as antibody for αCaMKII. In this case, however, the cells were treated with detergent prior to fixation (Allison et al., 2000), which should emphasize synaptic staining by decreasing the overall (i.e. soluble and non-synaptic) intensity of immunoreactivity in dendrites (Figure 2B). Indeed, this regimen revealed that the detergent-insoluble polyadenylation/translation factors were often coincident with αCaMKII, and thus located at synapses. While it is important to bear in mind that there is a significant amount of detergent-soluble polyadenylation/translation factors in dendrites, the amount that is detergent insoluble is >80% coincident with the αCaMKII synaptic marker (Figure 2C). These data indicate that the polyadenylation/translation machinery is most prevalent at excitatory synapses.
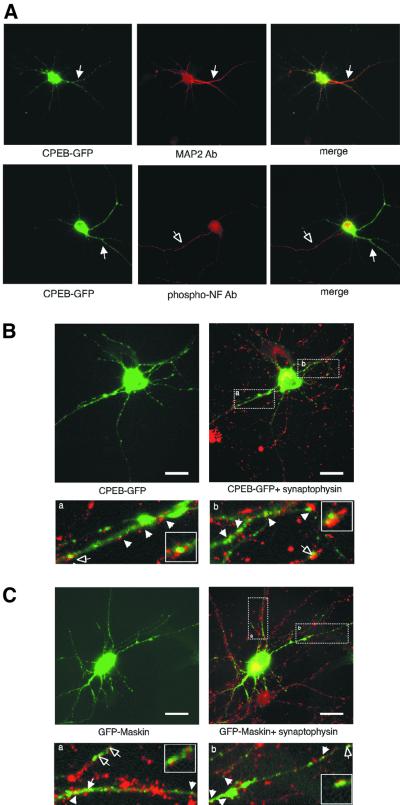
One caveat of any immunocytochemical analysis, including those described above, is the possibility of antibody cross-reactivity with other factors, irrespective of whether western blots show immunoreactivity with a single protein species. Therefore, to confirm the specificity of two of our immunostaining results, we infected hippocampal neurons with Sindbis virus expressing CPEB–green fluorescent protein (GFP) or GFP–maskin fusion proteins, and assessed whether these exogenous chimeric proteins were transported to dendrites and took up residence at or near synapses. First, Figure 3A confirms that CPEB–GFP is present in dendrites but not axons [co-localizes with microtubule-associated protein 2 (MAP2) but not phospho-NF]. This staining pattern also confirms that these cultures contain neurons with differentiated axons and dendrites (Ulfig et al., 1998). Both CPEB–GFP and GFP–maskin were expressed in the cytoplasm, formed particles and were transported to dendrites (Figure 3B and C, left). Some of the CPEB–GFP or GFP–maskin-containing particles in dendrites (Figure 3A and B, arrows) were co-localized with synaptophysin immunostaining (red), as indicated by a yellow (merge) color. In control experiments, non-fusion GFP distributed homogeneously in the whole neuron and did not form puncta in dendrites (data not shown). Also, because relatively young (7-day-old) neurons were used for virus infection, there were not as many synapses formed as in the 2-week-old neurons we used for immunostaining. In addition, because some synapses were formed on non-infected cells, the degree of apparent co-localization of CPEB–GFP or GFP–maskin with synaptophysin was much less than that observed by antibody staining. Moreover, many of the CPEB–GFP and GFP–maskin particles contain RNA and are trafficked to distal regions of dendrites, and thus would not be expected to be localized predominantly to synapses (Y.-S.Huang, E.Barbarese, J.Carson, Q.Cao and J.D.Richter, in preparation). Nonetheless, using an assay that does not rely upon antibody staining, we show that CPEB and maskin can localize to synaptic regions
Fig. 3. CPEB–GFP and GFP–maskin in dendrites. (A) CPEB–GFP co-localizes with the dendritic marker MAP2 (solid arrow), but not with the axonal marker phospho-neurofilament (open arrow) in cultured hippocampal neurons. The distribution of CPEB–GFP (B) and GFP–maskin (C) in cultured neurons immunostained with an antibody against the synaptic marker synaptophysin (red, right) is also shown. The magnified images in (A) and (B) show regions of co-localization between CPEB or maskin with synaptophysin (yellow, arrow). The selected synaptic area denoted by the hollow arrow is shown as a high-power image in the inset. Scale bars, 10 µm.
Polyadenylation at synapses
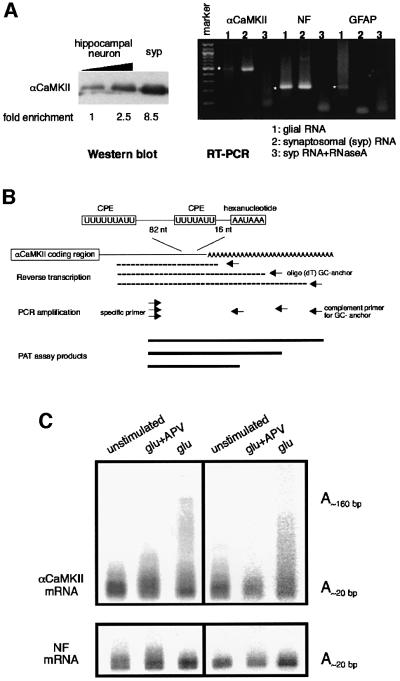
Although αCaMKII mRNA undergoes poly(A) elongation in the visual cortex of dark-reared rats exposed to light (Wu et al., 1998), the subcellular location where this process occurs (i.e. nucleus, soma or synapse) is not known. To assess whether polyadenylation occurs in the synapto-dendritic compartment in response to synaptic activity, as might be inferred from the data in Figures 1 and 2, we performed a series of experiments with synaptosomes isolated from cultured hippocampal neurons and from the hippocampus. These synaptosomes, which are isolated by iso-osmotic Percoll gradient centrifugation, have been shown by electron microscopy to contain intact presynaptic sacs attached to postsynaptic sealed vesicles (Kiebler et al., 1999; Bagni et al., 2000), and have been demonstrated to support protein synthesis (Bagni et al., 2000). The presence of dendritic mRNAs and polyribosomes in the postsynaptic vesicle, and the observation that αCaMKII protein is synthesized de novo in synaptosomes, demonstrates a translational competence in the postsynaptic compartment (Bagni et al., 2000). Thus, isolated synaptosomes seemed ideally suitable for studying synaptic mRNA polyadenylation.
Synaptosomes prepared from cultured hippocampal neurons were highly enriched for the synaptic marker αCaMKII, as shown by western blotting, and for αCaMKII and NF mRNAs as analyzed by RT–PCR (Figure 4A). However, RT–PCR failed to reveal the presence of GFAP mRNA, demonstrating that they lacked astrocyte contamination. The synaptosomes were exposed to glutamate, which was followed by RNA extraction and poly(A) tail analysis by the PCR-based poly(A) test (PAT) assay (Salles and Strickland, 1999). Figure 4B shows the positions of the two CPEs within the 3′-UTR of αCaMKII mRNA as well as a schematic of the assay procedure. Figure 4C shows duplicate experiments where glutamate stimulated polyadenylation of αCaMKII mRNA, but not of NF mRNA, which served as the CPE-lacking negative control. Synaptosome αCaMKII mRNA contained a poly(A) tail of ∼20 nucleotides, which was elongated up to ∼160 nucleotides following glutamate stimulation. Because 2-amino-5-phosphono-valerate (APV), a specific antagonist of the NMDA activity, prevented the glutamate-induced polyadenylation of αCaMKII mRNA, we infer that the signaling pathway leading to polyadenylation is mediated by the NMDA receptor.
Fig. 4. αCaMKII mRNA polyadenylation in synaptosomes is stimulated by glutamate. (A) Immunoblot for αCaMKII showing enrichment of synaptosomes (lanes 1 and 2, 1 and 2.5 µg of protein from cultured neurons; lane 3, 1 µg of protein from synaptosome). The intensity of the bands was analyzed by scanning densitometry. The purity of the synaptosomal RNAs was measured by RT–PCR of αCaMKII, neurofilament (NF) and glial fibrilary acid protein (GFAP) mRNAs (amplified bands are denoted by the dots) using total RNA from cultured glial cells (lane 1), synaptosomes (lane 2) and synaptosomes treated with RNase A (lane 3). Marker, 100 bp marker. (B) Schematic diagram of the PCR-based poly(A) tail assay. The synaptosomal RNA is annealed with oligo(dT) fused to a GC-rich anchor and reverse transcribed, which is followed by PCR amplification using a primer specific for either αCaMKII or NF mRNA. Because the oligo(dT) can anneal anywhere along the length of a poly(A) tail, mRNAs with long tails will yield amplification products of diverse size, the largest of which will approximates the largest size of the tail. Conversely, mRNAs with short tails will yield amplification products that are correspondingly short and discrete in size. (C) Synaptosomes were prepared from cultures of 14-day-old hippocampal neurons by differential centrifugation in Percoll–sucrose. The synaptosmes were then mock treated, or treated with glutamate plus APV or with glutamate only. Total RNA was then extracted and the degree of polyadenylation of αCaMKII and NF mRNAs was determined by a PAT assay. Results of PAT assays from two different synaptosome preparations are shown. The approximate lengths of the poly(A) tails are indicated.
Phosphorylation at synapses
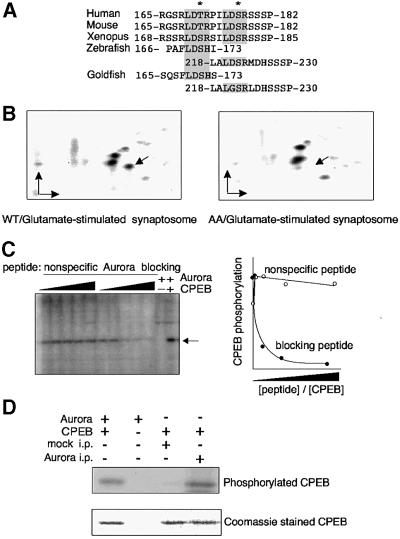
The observation that Aurora kinase is present at synapses suggests that it might be activated by synaptic stimulation and phosphorylate CPEB on the serine (in the frog) or threonine (in the mouse) that resides within the LDS/TR motif that is present in all vertebrate CPEB proteins (Mendez and Richter, 2001; Figure 5A). This phosphorylation event is necessary for polyadenylation (Mendez et al., 2000a). To assess this possibility, we prepared synaptosomes from the mouse hippocampus and stimulated them with glutamate. An extract was then prepared and primed with [γ-32P]ATP and purified recombinant wild-type CPEB, or a CPEB with alanine for serine substitutions in the two LDSR motifs (note that the double mutation was used because of the possibility that the second LDSR might be phosphorylated in a compensatory fashion if only the first was mutated). Following incubation, CPEB was isolated, digested with trypsin and subjected to two-dimensional phosphopeptide mapping (Mendez et al., 2000a). We assessed the phosphorylation of exogenous CPEB because there is insufficient endogenous protein to perform two-dimensional phosphopeptide mapping.
Fig. 5. Glutamate stimulation of CPEB phosphorylation in synaptosomes. (A) Alignment of vertebrate CPEB proteins denoting the two conserved LDS/TR Aurora phosphorylation motifs. (B) Phosphopeptide maps of CPEB. Purified E.coli-expressed CPEB (containing the two Aurora phosphorylation motifs noted in A), or a mutant CPEB (right) containing alanine for serine substitutions in these motifs (AA), was phosphorylated by an extract prepared from glutamate-stimulated synaptosomes in the presence of [γ-32P]ATP. In each case, CPEB was then resolved by SDS–PAGE, electroblotted onto a PVDF membrane, digested with trypsin and subjected to two-dimensional phosphopeptide mapping. The arrow denotes the LDS/TR-containing phosphopeptide. Arrows also indicate the origin and the direction of the TLE (horizontal) and ascending chromatography. (C) Inhibition of CPEB phosphorylation by an Aurora-blocking peptide. CPEB, as well as increasing amounts of the Aurora-blocking peptide (RGSRLDTRP ILDSRSSL), or a non-specific peptide (WHWLQLKPGQPMY) (Mendez et al., 2000a) were incubated in a synaptosome extract together with [γ-32P]ATP. Phospho-CPEB (arrow) was then resolved by one-dimensional SDS–PAGE and the signal was quantified by a phosphoimager and plotted against the molar ratio of non-specific or blocking peptide versus CPEB. The electrophoretic migration of CPEB phosphorylated in vitro by recombinant Aurora is also presented. (D) Immunoprecipitation of Aurora from synaptosomes. Aurora was immunoprecipitated from extracts prepared from synatoptosomes derived from cultured hippocampal neurons with Aurora-specific antibody or, as a control, mock precipitated with pre-immune serum. Kinase buffer, CPEB and [γ-32P]ATP were then added to the immunoprecipitate and CPEB phosphorylation was analyzed as described above. The figure also shows that equal amounts of CPEB, as detected by Coomassie Blue staining, were present in the appropriate reactions.
Figure 5B demonstrates that while wild-type CPEB yielded three prominent phosphopeptides, the mutant CPEB containing LDAR in place of the LDSR motif yielded only two (arrows). When unstimulated synapotosomes were used as the kinase source and wild-type CPEB was used as the substrate, the same three phosphopeptides were detected, and were present in the same relative amounts as those observed with glutamate-stimulated synaptosomes (data now shown). Consequently, these data demonstrate that a synaptosome kinase, whose catalytic activity is indistinguishable from that of Aurora (Mendez et al., 2000a), phosphorylates CPEB at a critical residue that is necessary for cytoplasmic polyadenylation.
To identify this kinase, we first employed an LDS/TR motif-containing peptide that we have shown previously is a specific competitor of Aurora activity (Mendez et al., 2000a). When added to a hippocampal neuron synaptosome extract, this peptide inhibited CPEB phosphorylation, whereas an unrelated peptide had no effect (Figure 5C). We have confirmed the activity of Aurora in synaptosomes by immunoprecipitating it and assessing whether it had the capacity to phosphorylate wild-type CPEB (Figure 5D). While the pellet from a mock immunoprecipitation (with pre-immune serum) had no kinase activity, the Aurora-containing immunoprecipitate was indeed capable of phosphorylating CPEB. Con sequently, these data show that Aurora resides in the synapto-dendritic compartment where it can phosphorylate CPEB.
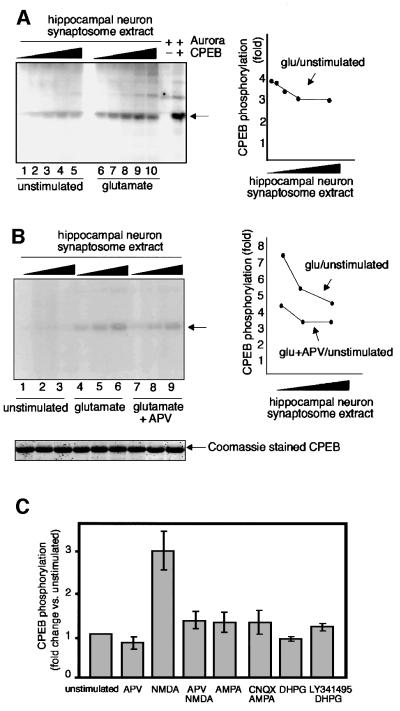
Next, we determined whether synaptosomal Aurora activity could be stimulated by glutamate treatment. However, because two-dimensional phosphopeptide mapping is not particularly suitable for a quantitative analysis, we employed one-dimensional PAGE to resolve phospho-CPEB. Figure 6A shows that glutamate treatment enhanced, by 4-fold, the phosphorylation of CPEB. The decreasing ratio in the phosphorylation of CPEB (glutamate stimulated to unstimulated) as a function of increasing amounts of extract is a reflection of substrate saturation in the glutamate-stimulated extract. In another experiment, glutamate stimulated the phosphorylation of CPEB by up to 7-fold (Figure 6B), which indicates the variability in the response of hippocampal synaptosomes to the neurotransmitter. Because glutamate treatment of synaptosomes proportionately enhanced the phosphorylation of all three phosphopeptides (cf. Figure 5B), including LDSR (data not shown), these results accurately reflect the stimulation of Aurora kinase activity by glutamate.
Fig. 6. Glutamate stimulates CPEB phosphorylation through NMDA receptor signaling. (A) Synaptosomes prepared from cultured hippocampal neurons were either mock treated or treated with 0.3 mM glutamate. Extracts were then prepared and supplemented with E.coli-expressed CPEB as well as [γ-32P]ATP. Following a 10 min incubation, phospho-CPEB (arrow) was resolved by SDS–PAGE and the signal quantified by a phosphoimager. The change in phosphorylation by glutamate was expressed as fold change relative to that of the unstimulated extract and plotted against the increasing amount of synatosome extract used (lanes 1 and 6, 0.125 µl; lanes 2 and 7, 0.25 µl; lanes 3 and 8, 0.5 µl; lanes 4 and 9, 1 µl; lanes 5 and 10, 2 µl) (B) Synaptosomes from cultured hippocampal neurons were treated with glutamate as described above, as well as with glutamate plus APV. CPEB phosphorylation (arrow) was monitored and plotted as described above (lanes 1, 4 and 7, 0.5 µl; lanes 2, 5 and 8, 1 µl; lanes 3, 6 and 9, 2 µl). The lower part of the figure shows that equal amounts of CPEB, as detected by Coomassie Blue staining, was present in all reactions. (C) Synaptosomes prepared from hippocampal tissues were treated with 120 µM APV, 50 µM NMDA, 10 µM AMPA, 20 µM CNQX, 50 µM DHPG, 10 µM LY341495 or combinations thereof. The synaptosomal extracts were prepared and used for CPEB phosphorylation assays. The changes in the degree of CPEB phosphorylation from different extracts were normalized to that which was unstimulated, and expressed as a fold change. The bar graph is the summary of three independent experiments. All error bars represent the SEM. The degree of CPEB phosphorylation from NMDA-treated extracts is significantly different from the unstimulated extract (P < 0.05, by Student’s t-test). The phosphorylation of CPEB by the other agonist/antagonist-treated extract was not statistically different from the unstimulated extract.
To determine whether the NMDA receptor mediates the glutamate-enhanced activity of Aurora, we treated synaptosomes with APV as well as with glutamate, and measured the resulting amount of CPEB phosphorylation (Figure 6B). Depending on the amount of extract used, this stimulation was inhibited by APV by as much as 50%, which indicates that the NMDA receptor is involved in Aurora activation.
During the course of our studies, we noticed that there was little change in CPEB phosphorylation or αCaMKII mRNA polyadenylation in response to glutamate if Aurora kinase activity was already high and αCaMKII mRNA already contained a long poly(A) tail in unstimulated synaptosomes. This occurred more often in synaptosomes prepared from hippocampal tissue than from cultures, quite possibly because the basal activity of the NMDA receptor was already quite high in brain tissue. These observations, plus the results from Figures 4 and 6 indicating that Aurora kinase activity and polyadenylation could be inhibited by APV, suggested that we may be able to prepare ‘quiescent’ synaptosomes from the hippocampal tissues by adding APV to the initial homogenization buffer to silence NMDA receptor signaling. Synaptosomes prepared in this manner were treated subsequently with APV, NMDA or these agents in combination. From three independent experiments, we found that NMDA receptor activation increased the Aurora activity, as measured by CPEB phosphorylation, by 3-fold (Figure 6C). The NMDA treatment of synaptosomes, like glutamate, proportionately enhanced the phosphorylation of all three CPEB phosphopeptides (cf. Figure 5B), including LDSR (data not shown).
In similar experiments, we employed 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) or LY341495, which antagonize the α-amino-3-hydroxy-5-methylisoxazole-4-proprionate (AMPA) receptors or metabotropic glutamate receptors (mGluRs), respectively, during synaptosome preparation. The synaptosomes were subsequently treated with AMPA, CNQX, DHPG, LY341495 or combinations thereof, and then assayed for CPEB phosphorylation. From three experiments, we found that modulation of AMPA receptor and mGluR had no effect on the Aurora activity (Figure 6C).
NMDA receptor signaling activates Aurora and stimulates polyadenylation
If the activation of Aurora through NMDA receptor signaling leads to endogenous CPEB phosphorylation, the polyadenylation of a CPE-containing RNA, i.e. αCaMKII mRNA, should be commensurate with the change of the kinase activity. Accordingly, we determined Aurora kinase activity and performed PAT assays on the same preparation of synaptosomes that were either untreated or treated with NMDA. Figure 7A shows that not only did NMDA enhance CPEB phosphorylation by 3-fold, it also induced the polyadenylation of αCaMKII mRNA, but not of NF mRNA (Figure 7B). These results, together with those presented in Figures 4 and 6, show that NMDA receptor signaling results in Aurora activation, CPEB Ser174 phosphorylation and αCaMKII mRNA polyadenylation in the synapto-dendritic compartment of hippocampal neurons and tissues.
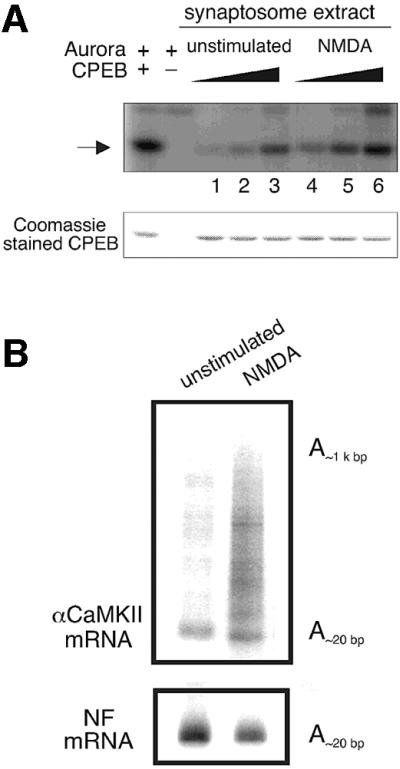
Fig. 7. Activation of NMDA receptor in synaptosomes results in CPEB-phosphorylation and αCaMKII polyadenylation. Unstimulated synaptosomes, or those treated with NMDA as described in Figure 6C, were divided in half; one portion was used for a CPEB phosphorylation experiment (A) while the other portion was used for PAT assays (B). The CPEB phosphorylation (arrow) was measured by using variable amounts of synaptosomal extracts (lanes 1 and 4, 0.1 µl; lanes 2 and 5, 0.5 µl; lanes 3 and 6, 2.5 µl). The Coomassie Blue staining of CPEB shows that equal amounts of substrates were present in all reactions. For the PAT assays, the poly(A) lengths of αCaMKII and neurofilament (NF) mRNA were determined.
Discussion
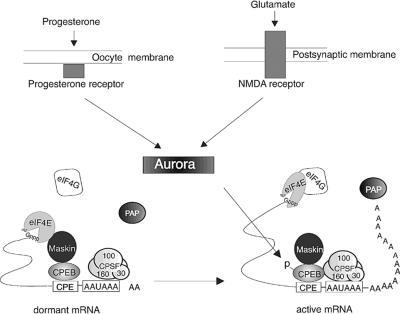
The results presented in this study demonstrate that all the factors known to control polyadenylation-induced translation in oocytes are also present at synapses of hippocampal neurons (Figure 8). Glutamate-stimulated synaptosomes promote both polyadenylation of the CPE-containing αCaMKII mRNA and the Aurora-catalyzed phosphorylation of CPEB on the key residue that mediates polyadenylation. Because polyadenylation is abrogated by APV and stimulated by NMDA, the signaling pathway that culminates in this mRNA modification is mediated by the NMDA receptor. Taken together, these observations define both a signaling pathway and a process that could underlie translation-dependent forms of synaptic plasticity.
Fig. 8. Conserved mechanism of polyadenylation-induced translation in oocytes and neurons. When dormant, CPE-containing mRNAs have short poly(A) tails and are associated with CPEB. CPEB is bound by maskin, which in turn is bound by the cap-binding protein eIF4E. The binding of maskin to eIF4E precludes an interaction of eIF4G with eIF4E, thereby preventing the association of the 40S ribosomal subunit with the mRNA. Polyadenylation-induced translational recruitment is initiated by progesterone binding to a possible surface-associated receptor in oocytes, or by glutamate binding to the integral membrane NMDA receptor in neurons. In both cases, a signal is transduced and, in an unknown number of steps, Aurora is activated. This kinase phosphorylates CPEB, which causes it (CPEB) to associate with and recruit CPSF to the AAUAAA polyadenylation hexanucleotide. CPSF brings poly(A) polymerase (PAP) to the end of the mRNA, where it catalyzes poly(A) addition. Coincident with, and possibly caused by polyadenylation, maskin and eIF4E dissociate, which allows eIF4G to bind eIF4E and correctly position the ribosomal subunit on the mRNA. 160, 100 and 30 refer to the molecular sizes of the CPSF subunits, and 7mGppp refers to the mRNA cap.
Other studies have shown that synapto-dendritic αCaMKII protein synthesis is stimulated upon synaptic activation (Ouyang et al., 1997; Scheetz et al., 1997; Wu et al., 1998; Bagni et al., 2000; Aakalu et al., 2001). This present study, as well as the initial investigation by Wu et al. (1998), strongly argues that this specific protein synthesis event is controlled by cytoplasmic polyadenylation at synapses. This assertion is also buttressed by other recent results showing that the translational activation of a reporter transcript by glutamate in hippocampal neurons requires a CPE in the 3′-UTR (Wells et al., 2001). More importantly, the experience-dependent αCaMKII protein synthesis in the visual cortex (Wu et al., 1998) is sensitive to cordycepin (3′-deoxyadenosine), an inhibitor of poly(A) elongation (Wells et al., 2001). Thus, cytoplasmic polyadenylation mediates translation in the CNS as well as in oocytes.
In this study, we demonstrate that the degree of the polyadenylation correlates with the degree of the Aurora activation. In some synaptosome preparations, αCaMKII mRNA contained a long poly(A) tail when the Aurora activity was high in the unstimulated control, leading to no significant changes upon glutamate stimulation. If we inhibited the NMDA receptor with APV during synaptosome preparation and reduced the basal Aurora activity in the unstimulated control, we consistently obtained the NMDA receptor-mediated Aurora activation and polyadenylation of αCaMKII mRNA in synaptosomes isolated from both hippocampal neurons and tissues. Because only the αCaMKII but not NF mRNA is polyadenylated, it is unlikely that activation of the NMDA receptor enhances the overall PAP activity.
Polyadenylation may be one of several mechanisms that regulate translation in neurons. Scheetz et al. (2000) have shown that stimulation of the NMDA receptor in synaptoneurosomes enhances the rate of αCaMKII peptide chain elongation, an event that appears to be controlled by eEF2 phosphorylation. Our results and those of Scheetz et al. (2000) are not mutually exclusive; indeed, the stimulation of initiation (by polyadenylation) and elongation (by eEF2) would seem to ensure that polypeptides are synthesized to a maximum extent. Another mechanism that controls protein synthesis in neurons, at least in Aplysia, is dependent on the kinase FRAP/mTOR (Yanow et al., 1998; Casadio et al., 1999). In this case, rapamycin, which inactivates FRAP/mTOR signaling, inhibits 5-hydroxytryptamine-induced protein synthesis in sensory cell neurites (Casadio et al., 1999). While FRAP/mTOR regulates many growth-related phenomena (Schmelzle and Hall, 2000), its effects on translation are mediated through p70S6 kinase (Jefferies et al., 1997) and a group of eIF4E-binding proteins known collectively as the eIF4EBPs (Gingras et al., 1999). Recently, rapamycin has been shown to inhibit BDNF-induced synaptic potentiation in hippocampal slices (Tang et al., 2002), suggesting that the FRAP/mTOR pathway, as well as the Aurora pathway, is likely to influence translation at synapses in mammals.
Recently, Aakalu et al. (2001) introduced a GFP reporter flanked by the 5′- and 3′-UTRs of αCaMKII mRNA into cultured hippocampal neurons and observed GFP synthesis in response to BDNF stimulation that was clustered at some hot spots in the vicinity of synapses in ‘optically’ isolated intact dendrites (Aakalu et al., 2001). Although one might infer that BDNF induces Aurora activity (because both lead to αCaMKII synthesis), this is not necessarily the case. BDNF acts both pre- and postsynaptically to facilitate synaptic transmission. Presynaptically, BDNF appears to induce glutamate release (Li et al., 1998), which in turn would activate the NMDA receptor in the postsynaptic membrane subsequently to trigger Aurora-dependent polyadenylation and translation. Alternatively, BDNF could bind and activate postsynaptic TrkB receptors and stimulate the local translation machinery, possibly through the FRAP/mTOR pathway (Yanow et al., 1998; Casadio et al., 1999). Whether the CPEB-mediated polyadenylation of αCaMKII mRNA is an event partially involved in this BDNF-stimulated local translation is unknown.
An important series of experiments involving CPEB in the control of neural activity has been reported in Aplysia. This species contains two CPEB-like proteins (for a discussion of CPEB proteins in vertebrates and invertebrates see Mendez and Richter, 2001; Mendez et al., 2002), at least one of which is phosphorylated (Liu et al., 2001) and translationally induced (Si et al., 2001) in sensory neurons in response to serotonin-induced long- term facilitation (LTF). The activation of CPEB probably results in the polyadenylation of neuronal actin mRNA in the synaptosome (Liu et al., 2001), which might account for the synthesis of cytoskeletal elements necessary for new synaptic connection. Most importantly, the blocking of CPEB activity in neurons abrogates the maintenance of LTF (Si et al., 2001), strongly suggesting that CPEB plays an import role in regulating synaptic plasticity. Whether Aurora phosphorylates CPEB in Aplysia neurons in response to serotonin treatment will be interesting to determine.
Finally, our experiments point to an interesting connection between factors involved in malignant transformation and neuronal plasticity. Aurora, which we show is a key regulatory kinase at synapses, is found on centrosomes in many somatic cells, is overexpressed in some tumors and is often associated with aneuploidy (Bischoff et al., 1998; Nigg, 2001). Deleted in colorectal cancer (DCC), a possible tumor suppressor, is also a component of the netrin receptor in neurons, and hence modulates axon guidance (Kolodziej, 1997). Thus, depending on the incoming signal, the activity of these molecules influences such divergent cellular responses as cell division and neuronal plasticity.
Materials and methods
Cell culture and immunostaining
Cultures of rat hippocampal neurons were prepared using previously described methods (Banker and Goslin, 1988). Briefly, the hippocampus was dissected from 18 or 19 d.p.c. rat embryos and dissociated by trypsin and trituration through a Pasteur pipet. The neurons were plated on poly-l-lysine-coated coverslips at a density of 6000/cm2, and co-cultured with a layer of glial cells in minimal essential medium (MEM) with N2 supplement (Gibco). The cells were then fixed with 4% formaldehyde in phosphate-buffered saline (PBS) at 37°C for 15 min, permeabilized with 0.1% Triton X-100 for 5 min, and blocked with 10% horse serum and 1% goat serum in PBS for 1 h. For some experiments, the neurons were treated with 0.02% Triton X-100 for 2 min at room temperature before fixation. The cells were treated with primary antibodies directed against synaptophysin (sy38), αCaMKII (6G9) (both from Chemicon), eIF4E (gift of N.Sonenberg, McGill University), CPSF100 (gift of J.Manley, Columbia University), Aurora (gift of J.Ruderman, Harvard University), maskin (Stebbins-Boaz et al., 1999) and PAP (Gebauer and Richter, 1995) and secondary goat α-rabbit IgG (Alexa-594 labeled) or goat α-mouse IgG (Alexa-488 labeled). Fluorescent images of the neurons were obtained on a Zeiss microscope with 63×, 1.4 NA lens and processed using Iplab, NIH Image and AdobePhotoshop softwares. Where noted, the intensity of 100 puncta containing each of the polyadenylation/translation factors was measured using the NIH Image software. The intensity of the αCaMKII signal in the puncta as set is at least 40% of that of the polyadenylation factors to be considered co-localized.
Sindbis virus construction and infection
The pSinRep5 CPEB–GFP plasmid was published previously (Groisman et al., 2000). The XhoI–BamHI fragment containing full-length maskin was cloned into the pEGFP-C3 plasmid (Clontech), resulting in pEGFP-maskin. The GFP–maskin fragment was excised from the pEGFP-maskin by digestion with NheI and HincII and ligated to XbaI–StuI-digested pSinRep5 plasmid (Invitrogen). The preparation of the Sindbis virus followed the manufacturer’s protocol (Invitrogen). Seven-day-old hippocampal neurons (plated at a density of 12 000/cm2) were infected with the virus expressing either CPEB–GFP or GFP–maskin for 1 h. The coverslips with the virus-infected neurons were then placed back onto the dishes containing a layer of glial cells and incubated for an additional 4 h before immunostaining with MAP2 (AP20; Chemicon), phospho-NF (SMI 312; Sternberg’er Monoclonals Incorporated) or sy38 antibodies as described above.
Postsynaptic density preparation and western blotting
PSD fractions were prepared from 3-week-old mouse brain as described by Carlin et al. (1980), and the protein in them was determined by the BCA assay (Pierce). About 30 µg of total protein from the initial homogenate, synaptosomes and PSD fractions were separated by SDS–PAGE, blotted and probed with antibodies for CPEB, PAP, CPSF100, eIF4E, maskin, Aurora (IAK1, Transduction Laboratory), synaptophysin (sy38), αCaMKII (6G9) and GAP-43 (91E12), which was followed by secondary antibody conjugated to horseradish peroxidase and developed with the chemiluminescence reaction.
Synaptosome preparation and PAT assay
Approximately 14 × 106 14-day-old cultured hippocampal neurons were harvested in 7 ml of homogenization buffer [0.32 M sucrose, 0.1 mM EDTA, 0. 25 mM dithiothreitol (DTT), 2 mM HEPES pH 7.4], disrupted by homogenization, and nuclei and cell debris were pelleted. The supernatant was centrifuged at 14 000 g for 10 min to pellet mitochondria and synaptosomes. The pellet was resuspended in 4 ml of gradient buffer (homogenization buffer containing 0.25 M sucrose but lacking DTT) and loaded onto a sucrose–Percoll discontinuous gradient as described (Dunkley et al., 1986). The synaptosome-containing fraction at the 15–23% Percoll interface was collected and washed with 3 vols of PBS, followed by centrifugation at 12 000 g for 5 min. The synaptosome pellet was resuspended in 200 µl of buffer (10 mM Tris pH 7.5, 2.2 mM CaCl2, 0.5 mM Na2HPO4, 0.4 mM KH2PO4, 4 mM NaHCO3 and 80 mM NaCl) (Bagni et al., 2000), and ∼40 µl of this synaptosome fraction were incubated with 0.3 mM glutamate and 10 µM glycine, or 0.3 mM glutamate plus glycine and 0.1 mM APV for 10 min at 37°C. The RNA was extracted from the synaptosome fraction and used for a PAT assay with the message-specific primers and procedure as described by Wu et al. (1998). In the present study, however, the PCR contained 5 µCi of [α-32P]dATP. The purity of the synaptosomal RNA was analyzed by RT–PCR using three sets of specific primers: NF sense, 5′-GAGATGTATTACGCAAAGTAC and antisense, 5′-CCAGTATGACCTTTATTGAGC; GFAP sense, 5′-GTTGTGTTCAAGCAGCCTGG and antisense 5′-CCAGTGAGTAAAGGTGACAG; the primers for αCaMKII were described previously (Wu et al., 1998).
CPEB phosphorylation
Synaptosomes, prepared from cultured hippocampal neurons and stimulated with glutamate essentially as described above, were suspended in H1 kinase solution (80 mM β-glycerophosphate, 20 mM EGTA, 15 mM MgCl2, 50 mM Na3VO4, 0.2% Triton X-100, 10 µg/ml each of leupeptin, pepstatin and chymostatin, and 0.5 µM okadaic acid). To a serial dilution of this mixture (0.125, 0.25, 0.5, 1 and 2 µl) was added 1 µl (2 µg) of purified Escherichia coli-expressed histidine-tagged CPEB ΔC (encompassing residues 1–290), 1 µl (10 µCi) of [γ-32P]ATP and buffer (20 mM Tris pH 7.7, 10 mM MgCl2, 50 mM KCl, 1 mM DTT) to a final volume of 30 µl. The mixture was incubated for 15 min at 30°C and the proteins then resolved by SDS–PAGE.
For two-dimensional phosphopeptide mapping, synaptosome-phosphorylated CPEB (wild-type or LDSR to LDAR mutant) was resolved by SDS–PAGE, electroblotted onto PVDF membrane, located by autoradiography, digested with trypsin and subjected to two-dimensional phosphopeptide mapping (Boyle et al., 1991; Mendez et al., 2000a).
In other experiments, synaptosome extracts were primed with [γ-32P]ATP and CPEB (0.25 µg) as well as a peptide that specifically blocks Aurora activity (RGSRLDTRPILDSRSSC, 1, 5, 15 or 40 µg) or a non-specific peptide (WHWLQLKPGQPMY, 1, 5, 15 or 40 µg). Following incubation, CPEB was resolved by SDS–PAGE. For all experiments, radioactive products were visualized and quantified with a phosphoimager.
Synaptosome preparation from the rat hippocampal tissue
Synaptosomes from rat hippocampal tissue were prepared in a similar way to that described above, except that the hippocampus was homogenized in buffer containing a glutamate receptor antagonist, 120 µM APV, 20 µM CNQX or 10 µM LY341495, to minimize the NMDA, AMPA or mGluR receptor activity, respectively. After Percoll gradient centrifugation and washing in PBS to free the synaptosomes of antagonist (APV, CNQX or LY341495), it was treated with 120 µM APV, 50 µM NMDA, 10 µM AMPA, 20 µM CNQX, 50 µM DHPG, 10 µM LY341495 or combinations thereof, for 5 min at 37°C, followed by the CPEB phosphorylation or PAT assay as described above.
Acknowledgments
Acknowledgements
We thank David Wells, Steve Lambert and Raul Mendez for aid and advice, and Nahum Sonenberg, Jim Manley and Joan Ruderman for antibodies. We are grateful to Mark Bear, Justin Fallon, Steve Lambert, Lori Lorenz, David Wells and members of the Richter laboratory for helpful discussions, essential advice and encouragement. This work was supported by a Program Project Grant from the NIH (NS 39321). Y.-S.H. was supported by the Charles A.King Trust fellowship from the Medical Foundation.
References
- Aakalu G., Smith,W., Nguyen,N., Jiang,C. and Schuman,E.M. (2001) Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron, 30, 489–502 [DOI] [PubMed] [Google Scholar]
- Allison D.W., Chervin,A.S., Gelfand,V.I. and Craig,A.M. (2000) Postsynaptic scaffolds of excitatory and inhibitory synapses in hippocampal neurons: maintenance of core components independent of actin filaments and microtubules. J. Neurosci., 20, 4545–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresson T. and Ruderman,J. (1998) The kinase Eg2 is a component of the Xenopus oocyte progesterone-activated signaling pathway. EMBO J., 17, 5627–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C., Mannucci,L., Dotti,C. and Amaldi,F. (2000) Chemical stimulation of synaptosomes modulates α-Ca2+/calmodulin-dependent protein kinase II mRNA association to polysomes. J. Neurosci., 20, RC76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G. and Goslin,K. (1988) Developments in neuronal cell culture. Nature, 336, 185–186. [DOI] [PubMed] [Google Scholar]
- Bischoff J.R. et al. (1998) A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J., 17, 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle W.J., van der Geer,P. and Hunter,T. (1991) Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol., 201, 110–149. [DOI] [PubMed] [Google Scholar]
- Carlin R.K., Grab,D.J., Cohen,R.S. and Siekevitz,P. (1980) Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J. Cell Biol., 86, 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio A., Martin,K.C., Giustetto,M., Zhu,H., Chen,M., Bartsch,D., Bailey,C.H. and Kandel,E.R. (1999) A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell, 99, 221–237. [DOI] [PubMed] [Google Scholar]
- Crino P.B. and Eberwine,J. (1996) Molecular characterization of the dendritic growth cone: regulated mRNA transport and local protein synthesis. Neuron, 17, 1173–1187. [DOI] [PubMed] [Google Scholar]
- Dunkley P.R., Jarvie,P.E., Heath,J.W., Kidd,G.J. and Rostas,J.A. (1986) A rapid method for isolation of synaptosomes on Percoll gradients. Brain Res., 372, 115–129. [DOI] [PubMed] [Google Scholar]
- Gebauer F. and Richter,J.D. (1995) Cloning and characterization of a Xenopus poly(A) polymerase. Mol. Cell. Biol., 15, 1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught,B. and Sonenberg,N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Groisman I., Huang,Y.S., Mendez,R., Cao,Q., Theurkauf,W. and Richter,J.D. (2000) CPEB, maskin and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell, 103, 435–447 [DOI] [PubMed] [Google Scholar]
- Hodgman R., Tay,J., Mendez,R. and Richter,J.D. (2001) CPEB phosphorylation and cytoplasmic polyadenylation are catalyzed by the kinase IAK1/Eg2 in maturing mouse oocytes. Development, 128, 2815–2822. [DOI] [PubMed] [Google Scholar]
- Huber K.M., Kayser,M.S. and Bear,M.F. (2000) Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science, 288, 1254–1257. [DOI] [PubMed] [Google Scholar]
- Jefferies H.B., Fumagalli,S., Dennis,P.B., Reinhard,C., Pearson,R.B. and Thomas,G. (1997) Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J., 16, 3693–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacharmina J.E., Job,C., Crino,P. and Eberwine,J. (2000) Stimulation of glutamate receptor protein synthesis and membrane insertion within isolated neuronal dendrites. Proc. Natl Acad. Sci. USA, 97, 11545–11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. and Schuman,E.M. (1996) A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science, 273, 1402–1406. [DOI] [PubMed] [Google Scholar]
- Kennedy M.B. (1998) Signal transduction molecules at the glutamatergic postsynaptic membrane. Brain Res. Brain Res. Rev., 26, 243–257. [DOI] [PubMed] [Google Scholar]
- Kiebler M.A., Lopez-Garcia,J.C. and Leopold,P.L. (1999) Purification and characterization of rat hippocampal CA3-dendrititc spines associated with mossy fiber terminals. FEBS Lett., 445, 80–86 [DOI] [PubMed] [Google Scholar]
- Kolodziej P.A. (1997) DCC’s function takes shape in the nervous system. Curr. Opin. Genet. Dev., 7, 87–92. [DOI] [PubMed] [Google Scholar]
- Li Y.X., Zhang,Y., Lester,H.A., Schuman,E.M. and Davidson,N. (1998) Enhancement of excitatory neurotransmitter release induced by BDNF in cultured hippocampal neurons. J. Neurosci., 18, 10231–10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chen,D.B. and Schwartz,J.H. (2001) Polyadenylation during long-term facilitation (LTF) of Aplysia sensory neuron synapses. Soc. Neurosci. Abstr., 594.13. [Google Scholar]
- Martin K.C., Barad,M. and Kandel,E.R. (2000) Local protein synthesis and its role in synapse-specific plasticity. Curr. Opin. Neurobiol., 10, 587–592. [DOI] [PubMed] [Google Scholar]
- Mendez R. and Richter,J.D. (2001) Translational control by CPEB: a means to the end. Nature Rev. Mol. Cell. Biol., 2, 521–529. [DOI] [PubMed] [Google Scholar]
- Mendez R., Barnard,D. and Richter,J.D. (2002) Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J., 21, 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R., Hake,L.E., Andresson,T., Littlepage,L.E., Ruderman,J.V. and Richter,J.D. (2000a) Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature, 404, 302–307. [DOI] [PubMed] [Google Scholar]
- Mendez R., Murthy,K.G., Ryan,K., Manley,J.L. and Richter,J.D. (2000b) Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell, 6, 253–259. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. (2001) Mitotic kinases as regulators of cell division and its checkpoints. Nature Rev. Mol. Cell. Biol., 2, 21–32. [DOI] [PubMed] [Google Scholar]
- Ouyang Y., Kantor,D., Harris,K.M., Schuman,E.M. and Kennedy,M.B. (1997) Visualization of the distribution of autophosphorylated calcium/calmodulin-dependent protein kinase II after tetanic stimulation in the CA1 area of the hippocampus. J. Neurosci., 17, 5416–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y., Rosenstein,A., Kreiman,G., Schuman,E.M. and Kennedy,M.B. (1999) Tetanic stimulation leads to increased accumulation of Ca(2+)/calmodulin-dependent protein kinase II via dendritic protein synthesis in hippocampal neurons. J. Neurosci., 19, 7823–7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J.D. (2000) Influence of polyadenylation-induced translation on metazoan development and neuronal synaptic function. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 785–805.
- Richter J.D. and Lorenz,L.J. (2002) Selective translation of mRNAs at synapses. Curr. Opin. Neurobiol., in press. [DOI] [PubMed] [Google Scholar]
- Salles F.J. and Strickland,S. (1999) Analysis of poly(A) tail lengths by PCR: the PAT assay. Methods Mol. Biol., 118, 441–448. [DOI] [PubMed] [Google Scholar]
- Scheetz A.J., Nairn,A.C. and Constantine-Paton,M. (1997) N-methyl-d-aspartate receptor activation and visual activity induce elongation factor-2 phosphorylation in amphibian tecta: a role for N-methyl-d-aspartate receptors in controlling protein synthesis. Proc. Natl Acad. Sci. USA, 94, 14770–14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz A.J., Nairn,A.C. and Constantine-Paton,M. (2000) NMDA receptor-mediated control of protein synthesis at developing synapses. Nature Neurosci., 3, 211–216. [DOI] [PubMed] [Google Scholar]
- Schmelzle T. and Hall,M.N. (2000) TOR, a central controller of cell growth. Cell, 103, 253–262. [DOI] [PubMed] [Google Scholar]
- Si K., Giustetto,M., Hsu,R. and Kandel,E.R. (2001) CPEB, a regulator of local protein synthesis, is also a candidate for a protein synthesis dependent mark in synapse-specific long-term facilitation in Aplysia. Soc. Neurosci. Abstr., 682.3. [Google Scholar]
- Stebbins-Boaz B., Cao,Q., de Moor,C.H., Mendez,R. and Richter,J.D. (1999) Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell, 4, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Steward O. and Schuman,E.M. (2001) Protein synthesis at synaptic sites of dendrites. Annu. Rev. Neurosci., 24, 299–325 [DOI] [PubMed] [Google Scholar]
- Tang S.J., Reis,G., Kang,H., Gingras,A.C., Sonenberg,N. and Schuman,E.M. (2002) A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc. Natl Acad. Sci. USA, 99, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfig N., Nickel,J. and Bohl,J. (1998) Monoclonal antibodies SMI 311 and SMI 312 as tools to investigate the maturation of nerve cells and axonal patterns in human fetal brain. Cell Tissue Res., 291, 433–443. [DOI] [PubMed] [Google Scholar]
- Wells D.G., Dong,X., Quinlan,E.M., Huang,Y.S., Bear,M.F., Richter,J.D. and Fallon,J.R. (2001) A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J. Neurosci., 21, 9541–9548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. et al. (1998) CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of αCaMKII mRNA at synapses. Neuron, 21, 1129–1139. [DOI] [PubMed] [Google Scholar]
- Yanow S.K., Manseau,F., Hislop,J., Castellucci,V.F. and Sossin,W.S. (1998) Biochemical pathways by which serotonin regulates translation in the nervous system of Aplysia.J. Neurochem., 70, 572–583. [DOI] [PubMed] [Google Scholar]