Abstract
Directly upstream of the Halobacterium salinarum transducer genes basT and htpIV we identified two open reading frames (orfs) with significant homologies to genes encoding binding proteins for amino acids and compatible solutes, respectively. Behavioral testing of deletion mutants indicates that halobacterial chemotaxis towards branched-chain amino acids as well as compatible osmolytes of the betaine family requires both a binding and a transducer protein. We therefore named the binding/transducer proteins BasB/BasT for branched-chain and sulfur-containing amino acids and CosB/CosT for compatible solutes. Our data support a signaling mechanism with the binding proteins functioning as lipid-anchored receptors interacting with the extracellular domain of their cognate transducers. Inspection of the halobacterial genome suggests that BasB and CosB exclusively mediate chemotaxis responses without any additional role in transport, which is in contrast to bacterial binding proteins, which are always part of ABC transport systems. The CosB/CosT system is the first instance of a chemotaxis signaling pathway for organic osmolytes in the living world and natural abundance 13C-NMR analysis of cytoplasmic extracts suggests that H.salinarum utilizes these solutes for osmotic adaptation.
Keywords: compatible solute/glycine betaine/lipoprotein/methyl-accepting chemotaxis/osmotic stress
Introduction
Chemotaxis in archaea, much as in bacteria, is based on monitoring levels of extracellular attractants or repellents and transmitting this information via the generation of intracellular two-component phosphorylation cascades to the flagellar motor (Rudolph and Oesterhelt, 1995). As a result, the cell modulates the direction of flagellar rotation and therefore migrates towards favorable surroundings. In eubacteria, a methyl-accepting chemotaxis protein (MCP) or a periplasmically localized substrate-binding protein commonly acts as the primary receptor for the attractant or repellent molecules (Stock and Surette, 1996). In the first case, a chemotactic signal is elicited by direct interaction of the ligand and the periplasmic domain of the integral membrane MCP. If the receptor is a binding protein, the MCP functions as a transducer, transmitting the signal from the binding protein to the two-component system made up of the histidine kinase CheA and the response regulator CheY. Mediating chemotactic responses is, however, not the sole functional repertoire of binding proteins. Their predominant role is, rather, mediating solute accumulation as part of ABC transport systems (Ehrmann et al., 1998; Detmers et al., 2001). Here, they initiate uptake of specific substrates, like sugars or peptides, by binding and passing the solutes to the integral membrane components of their cognate ABC transporters. The genes encoding binding proteins are generally localized in the ABC transport clusters on the chromosomes, even if they function in both transport and chemotaxis (Boos and Lucht, 1996). The classical example of a dual functioning binding protein is the maltose-binding protein (MBP) in Escherichia coli, which interacts either with the ABC transporter complex for maltose uptake or with the chemotaxis transducer Tar for sensing the extracellular concentration of maltose (Stock and Surette, 1996). In the archaeon Halobacterium salinarum, so far more than a dozen genes encoding halobacterial transducer proteins (Htps) have been identified, which are homologs of the bacterial MCPs (Rudolph et al., 1996; Zhang et al., 1996a). Recent sequencing data indicate that the halobacterial genome encodes some 18 members of the Htp family (Ng et al., 2000). None of them is known to involve binding proteins for transmitting tactic responses. Here, we present the first evidence of binding protein-dependent chemotaxis in archaea: BasB (branched chain and sulfur-containing amino acid binding protein) mediates responses to amino acids, and CosB (compatible solute binding protein) mediates chemotaxis to organic osmolytes.
Living cells throughout the kingdoms have evolved the capacity to accumulate small organic compounds in order to adapt to environments of high osmotic strength (Kempf and Bremer, 1998; Martin et al., 1999). These molecules, which are accumulated by uptake or de novo synthesis, include carbohydrates, polyols, amino acids and derivatives thereof, e.g. glycine betaine (Martin et al., 1999). The intracellular amassing of such solutes compensates for the external hypertonicity that otherwise would negatively affect the cell’s physiology by loss of turgor and dehydration of the cytoplasm. These osmolytes are generally referred to as ‘compatible solutes’ because they do not impair vital cellular functions even if amassed at molar concentrations, i.e. they are compatible with the physiology of the cell (Brown, 1976). Because they protect against osmotic stress, compatible solutes are often also termed osmoprotectants. Another route followed to maintain positive turgor in hypertonic surroundings is intracellular accumulation of inorganic ions (Galinski, 1995; Kempf and Bremer, 1998). Uptake of potassium ions has been demonstrated as an early response on extracellular osmotic upshifts in certain eubacteria, including E.coli and Bacillus subtilis (Kempf and Bremer, 1998; McLaggan et al., 1994). However, long-term adjustment to hypertonic conditions requires accumulation of organic osmolytes in these organisms. Generally, it appears that synthesis or uptake of organic osmolytes is essential for non-halophilic species to grow at elevated salinity (Csonka and Epstein, 1996; Martin et al., 1999). Truly halophilic organisms, in contrast, are thought to survive the extremely low water activity of their natural habitats predominantly by inorganic ion accumulation (Brown, 1976; Galinski, 1995). Consistently, intracellular potassium concentrations of >4 M have been detected for extreme halophilic archaea, namely the Halobacterium species, which thrive at saturated (>4 M) NaCl concentrations (Christian and Waltho, 1962). For these species, amassing of organic solutes for osmoadaptative reasons has not been considered so far. Here we provide the first evidence that H.salinarum has not only (i) evolved a specific chemotactic detection system for trimethylammonium compounds, but also (ii) amasses these compatible solutes intracellularly.
Results
Identification of two putative binding-protein orthologs in the halobacterial genome
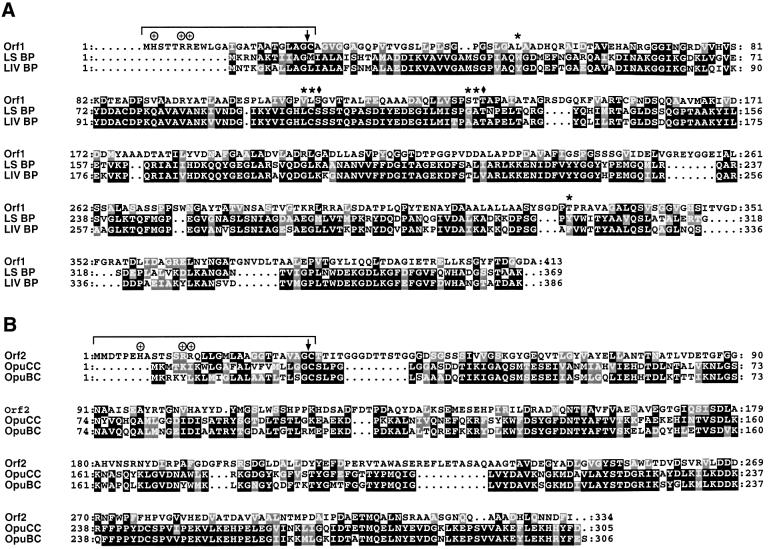
In the course of the H.salinarum genome sequencing project (D.Oesterhelt, unpublished) we inspected the chromosomal loci of several htp genes. Directly upstream of basT, which encodes a chemotaxis transducer for amino acids (Kokoeva and Oesterhelt, 2000), and htpIV, which is of unknown function (Rudolph et al., 1996), we identified two open reading frames (orfs) (Figure 1A, orf1 and orf2). The sequence information has been deposited in DDBJ/EMBL/GenBank (accession numbers AJ438168 and AJ438169). The existence of both orfs has been confirmed by others in a closely related strain, Halobacterium sp. NRC-1 (Ng et al., 2000). A BLAST search (Altschul et al., 1997) of the DDBJ/EMBL/GenBank databases with the predicted translation products of orf1 (413 residues) and orf2 (334 residues) revealed significant sequence similarities to binding proteins for branched-chain amino acids (Orf1) and quaternary amines (Orf2). Figure 2A shows an alignment of Orf1 with the precursors of the E.coli binding proteins LS-BP and LIV-BP. LS-BP specifically binds leucine, whereas LIV-BP exhibits a broader substrate range including leucine, isoleucine and valine. Both binding proteins have been shown to feed into the same integral membrane ABC transport complex for leucine uptake (Adams et al., 1990). Among the conserved residues in Orf1 are Ser79 and Thr102 of LIV-BP (marked with diamonds in Figure 2A), which are assumed to be crucial for substrate binding. According to the crystal structure of LIV-BP with bound leucine, the substrate is held in place primarily by hydrogen bonding of its α-ammonium and α-carboxylate groups to Ser79 and Thr102 (Sack et al., 1989). Also, the hydrophobic character of the binding pocket, which buries the substrate’s side chain, appears to be conserved in Orf1 (indicated by asterisks).
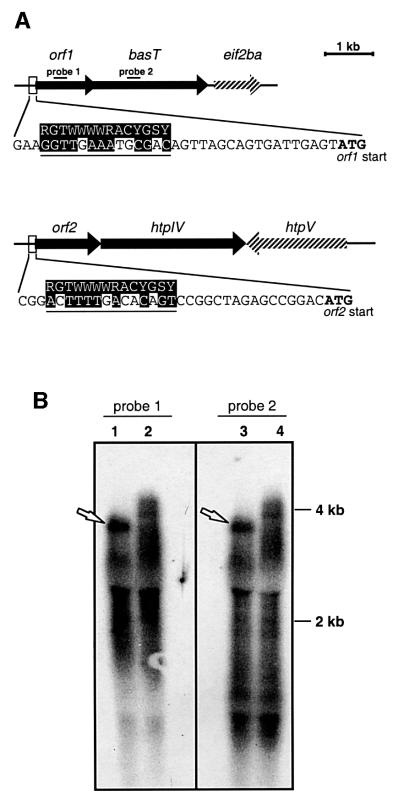
Fig. 1. (A) Genomic organization of the basT and htpIV loci. Nucleotides matching the consensus sequence for a strong halobacterial promoter (indicated on top) are shaded (R = A or G; Y = C or T; W = A or T; S = G or C; N = A, C, G or T). eif2ba, putative translation initiation factor (Ng et al., 2000); htpV encodes a halobacterial transducer protein of unknown function (Rudolph et al., 1996); (B) Northern blot analysis of the BasT locus. Lanes 1 and 3, wild type; lanes 2 and 4, strain Mev-BasT (Kokoeva and Oesterhelt, 2000). Represented are the X-ray films after hybridization with radiolabeled probes. Lanes 1 and 2 were hybridized with a probe to orf1 (probe 1), and lanes 3 and 4 with a probe to basT (probe 2). Sequence positions of the probes are indicated in (A). Each lane contains 5 µg of total RNA.
Fig. 2. (A) Alignment of Orf1 with the precursors of LS-BP and LIV-BP from E.coli. Residues interacting with the α-carboxyl and α-amino groups of the ligand (leucine) are marked by diamonds and residues forming the hydrophobic binding pocket are indicated by asterisks. (B) Alignment of Orf2 with OpuCC and OpuBC of B.subtilis. Putative signal peptides are indicated and the peptidase cleavage sites are marked by vertical arrows. Conserved and similar residues are boxed in black and grey, respectively.
Orf2 exhibits strong sequence similarity to the binding proteins OpuCC and OpuBC, which mediate uptake of compatible solutes in B.subtilis as part of the ABC trans port systems OpuC and OpuB, respectively (Figure 2B). OpuBC specifically mediates the accumulation of choline, whereas OpuCC has a broader substrate range including the trimethylammonium compounds carnitine, choline and glycine betaine (Kappes et al., 1999).
Due to the characteristics of their cell envelopes (Kandler and König, 1993), archaea as well as Gram-positive bacteria lack a periplasmic compartment that is typical of Gram-negative bacteria (Beveridge, 1999). Therefore, binding proteins of archaea and Gram-positive bacteria are presumably all attached to the membrane by lipid anchors or transmembrane segments (Ehrmann et al., 1998). Consistent with lipid anchoring, the N-termini of both Orf1 and Orf2 show the diagnostic features of signal peptides of lipoprotein precursors (Sutcliffe and Russell, 1995): (i) a positively charged N-terminal region, (ii) a hydrophobic central region, (iii) an average signal peptide length of 20–30 amino acids and (iv) conservation of the consensus sequence for a signal peptidase cleavage site (Leu/Val/Ile-Ala/Ser-Gly/Ala-Cys) at positions –3 to +1 including the invariable cysteine, which covalently links the lipids. These features are also evident in OpuCC and OpuBC of the Gram-positive bacterium B.subtilis (Figure 2B) as well as in halocyanin, an archaeal protein shown to be modified by ether lipids (Mattar et al., 1994; see Discussion).
Chromosomal organization of the orf1 and orf2 loci
orf1 shares four nucleotides with basT, which encodes the membrane-bound chemotaxis transducer for branched chain (Leu, Ile, Val) and sulfur-containing (Met, Cys) amino acids (Kokoeva and Oesterhelt, 2000). Four nucleotides following the stop of orf2 starts htpIV, whose predicted protein topology resembles that of BasT with two transmembrane segments flanking an extracellular domain (Rudolph et al., 1996). Both orf1 and orf2 are also preceded by sequences with significant similarity to the consensus sequence of a strong halobacterial promoter (Danner and Soppa, 1996), whereas no promoter-like structure could be identified directly upstream of basT or htpIV (Figure 1A). Based on these findings we hypothesized that orf1 and orf2 form transcriptional units with their cognate transducers. Northern blot analysis of the orf1/basT locus indicates that this is indeed the case (Figure 1B). Both a probe to orf1 and a probe to basT resulted in identical hybridization patterns (probes indicated in Figure 1A). In spite of a high background signal, a relatively distinct band at ∼4 kb can be identified for both probes (Figure 1B, lanes 1 and 3; arrows), which agrees with the length of the putative orf1/basT transcriptional unit. A bas mutant strain, Mev-BasT, serves as control (Kokoeva and Oesterhelt, 2000; Supplementary table I available at The EMBO Journal Online). This strain carries a 6.8 kb insertion in the basT gene (lanes 2 and 4). In this case, the signal extends to higher molecular weights, indicating a larger transcript most likely resulting from the insertional modification.
Construction and expression analysis of mutants in the orf1/basT locus
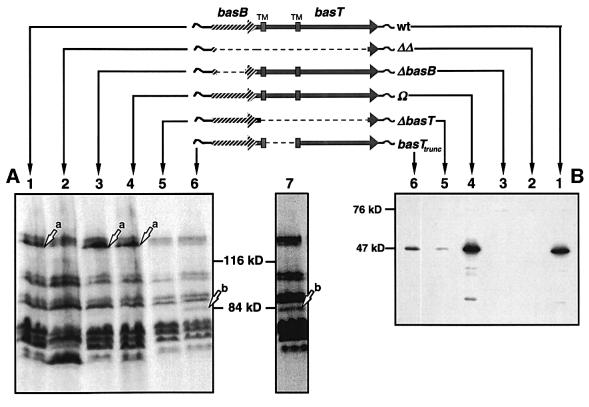
Given the binding protein homologies of Orf1, the chromosomal colocalization of orf1 and basT, and the northern blot results, we reasoned that Orf1 might play an essential role in the chemotactic response towards the BasT-specific attractants. Therefore we gave Orf1 the conceptual name BasB, for branched-chain and sulfur-containing amino acid binding protein. To test our assumption, we generated specific bas locus mutants by using a two-step strategy and examined them behaviorally (see also Supplementary data). First, the entire bas locus including orf1 and basT was deleted from the halobacterial chromosome, resulting in strain ΔΔ. Then, strain ΔΔ was transformed with suicide vectors carrying the bas operon in its native form or modified by in-frame deletion of basB or basT, which resulted in strains Ω, ΔbasB and ΔbasT, respectively. Another mutant strain, basTtrunc, was also generated using this technique. This strain carries the intact basB followed by a truncated basT gene lacking the sequence stretch that encodes the predicted extracellular domain of the transducer. For a schematic representation of the mutant strains, whose genotypes were confirmed by Southern analysis (not shown), see Figure 3.
Fig. 3. (A) Patterns of [3H]methyl-labeled proteins of the bas mutant strains in comparison with the wild type. Protein gels were exposed to X-ray films for 1 (lanes 1–6) or 2 (lane 7) weeks. Lanes: 1, wild type; 2, ΔΔ; 3, ΔbasB; 4, Ω; 5, ΔbasT; 6 and 7, basTtrunc. Arrows indicate the positions of BasT (a) and BasTtrunc (b). (B) Immunochemical detection of BasB in total protein extracts using anti-BasB antibodies. Lanes: 1, wild type; 2, ΔΔ; 3, ΔbasB; 4, Ω; 5, ΔbasT; 6 and 7, basTtrunc.
The bas mutants were analyzed for BasT expression by taking advantage of BasT’s ability to accept methyl groups (Kokoeva and Oesterhelt, 2000). First, halobacterial cells were radiolabeled with l-[methyl-3H]methionine, then their proteins were extracted and separated by SDS–PAGE. The corresponding fluorograph (Figure 3A) demonstrates that strains ΔΔ (lane 2) and ΔbasT (lane 5) do not express BasT, as they lack the BasT-specific band (Kokoeva and Oesterhelt, 2000). Conversely, this band is present in the wild-type, ΔbasB and Ω strains (arrow a, lanes 1, 3 and 4; for a better resolution compare Figure 5, arrow b). Note that a deviation of apparent (>116 kDa) from calculated (84.8 kDa) molecular weight as observed for BasT holds true for many halophilic proteins and is presumably due to their acidic nature (Storch et al., 1999). The basTtrunc strain also lacks the band corresponding to BasT (Figure 3A, lane 6). However, in this strain a new band appears that is absent in the wild type as well as in all bas mutants (arrow b, lanes 6 and 7). As it migrates at much lower molecular weights, this band very likely reflects the truncated BasT protein, indicating that it is still capable of covalent methyl group modification. Furthermore, this band is restricted to the membrane fraction of the protein extracts suggesting that it is membrane bound like the native BasT protein (not shown).
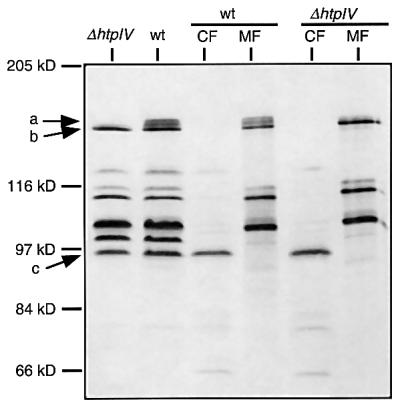
Fig. 5. Autofluorogram of [3H]methylated protein extracts of strains ΔhtpIV and S9 wild type (wt). CF and MF, cytoplasmic and membrane fractions, respectively. Arrows indicate the positions of HtpIV (a), BasT (b) and the soluble transducer protein Car (c).
The expression of BasB in the mutant strains was analyzed by western blotting using a polyclonal antiserum raised against a peptide of BasB. The antiserum detects a single band migrating at 47 kDa in the wild type (Figure 3B, lane 1), which is also present in strain Ω (lane 4) but absent in ΔΔ andΔbasB (lanes 2 and 3). In contrast to BasT, the apparent molecular weight of BasB closely matches the calculated molecular weight of the mature protein (41 kDa). Western analysis also revealed that strains ΔbasT and basTtrunc exhibit much less BasB protein than the wild type (Figure 3B, lanes 5 and 6), which is surprising given that both mutant strains carry an intact basB gene in its native 5′ chromosomal context. It could be speculated that the stability of the corresponding transcripts is impaired in those mutant strains. Alternatively, it is possible that the membrane association of BasB is strongly stabilized, i.e. protected against proteolytic degradation, by BasT, presumably via its extracellular domain, which is missing in basTtrunc. Note that expression of the halophilic phototaxis receptor sensory rhodopsin I (SRI) also appears to depend on the presence of its cognate transducer protein HtrI (Perazzona et al., 1996; see Discussion).
Functional analysis of the bas mutants
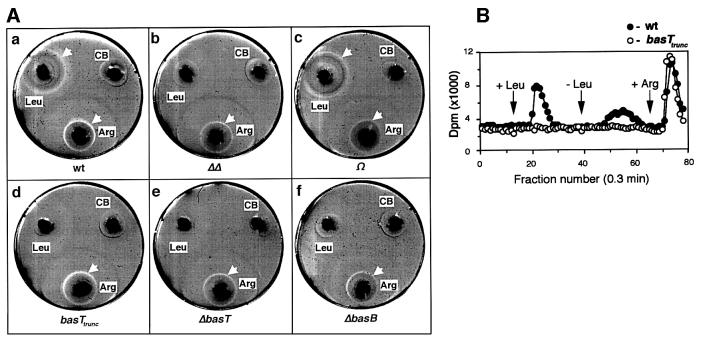
The chemotactic ability of the mutant strains was assessed by the chemical-in-plug (CIP) method, which has recently been adapted for the behavioral testing of halobacterial cells (Storch et al., 1999). For the assay, hard agar plugs containing the test compound were pushed into soft agar, which included the motile cells (Figure 4A). The formation of bright rings around the plugs indicates a chemotactic response (see arrows in Figure 4A). The rings represent zones depleted from cells that migrate up the attractant gradient towards the plug. Arginine, a chemoattractant specifically sensed by the soluble transducer protein Car (Storch et al., 1999) served as a positive control. Figure 4A reveals that cells lacking either the binding protein homolog (Figure 4A,f), the transducer (Figure 4A,e) or both (Figure 4A,b) have lost the capacity to respond to leucine, while they retained normal taxis to arginine. The same behavior is also seen for cells carrying a truncated transducer (Figure 4A,d). On the other hand, complementation with the native operon (strain Ω, Figure 4A,c) completely restored wild-type behavior (Figure 4A,a). Identical results were obtained with the other BasT-specific attractants, valine, isoleucine, methionine and cysteine (not shown). These findings strongly indicate that the chemotactic response to all those amino acid attractants requires both the transducer protein BasT and the binding protein BasB. However, considering the low levels of BasB in ΔbasT and basTtrunc as detected by immunoblotting, one can not decide whether the lack of the entire transducer protein (ΔbasT) or just the deletion of its extracellular domain (basTtrunc) is sufficient for the abolition of the plug plate response. Given that the chemotactic ring formation presumably requires an intact wild-type (BasT and BasB) detection unit, the plug plate assay might be inappropriate to assess any residual chemotactic activity based on the few BasB/BasTtrunc detection units that could still assemble in strain basTtrunc. Thus, an alternative chemotaxis test was employed, the so-called flow assay, which simply monitors changes in the turnover rate of volatile methyl groups (Storch et al., 1999). These rate changes are supposed to reflect the methylation/demethylation of specific transducer proteins. Figure 4B demonstrates that stimulation with 5 mM leucine does not affect the methyl group turnover in basTtrunc although the truncated transducer appears to be expressed and methylatable in those cells (Figure 3A). This result points to a detection mode with the extracellular loop, which is missing in basTtrunc, playing a crucial part in signal transmission and/or signal complex assembly (see Discussion).
Fig. 4. Behavioral analysis of the bas mutants. (A) CIP assay of the wild-type (a), ΔΔ (b), Ω (c), basTtrunc (d), ΔbasT (e) and ΔbasB (f) strains. Chemotactic depletion rings are indicated (white arrows). Hard agar plugs filled with arginine (Arg) or with chemotaxis buffer alone (CB) served as positive and negative controls, respectively. (B) Amino acid-induced (5 mM, arrows) changes in the rate of [3H]methyl group release of the H.salinarum strains wild type (closed circles) and basTtrunc (open circles). Subsequent arginine stimulations at 5 mM served as a control.
HtpIV is a membrane-bound and methylatable protein
Considering that orf2, located upstream of htpIV, most likely encodes an ortholog of binding proteins for trimethylammonium compounds and that both genes are presumably cotranscribed, we revisited the unpublished strain ΔhtpIV, which carries a deletion in the htpIV gene. In this strain, ∼1 kb of the htpIV coding sequence is replaced by a cassette conferring mevinolin resistance (see Supplementary data). The replaced sequence stretch (Arg374 to Ser694) of the HtpIV protein includes the signaling domain, which relays the chemotactic signal to the histidine kinase CheA. The successful deletion was confirmed by Southern analysis (not shown).
HtpIV expression was revealed by radiolabeling of the cells and electrophoretic separation of the protein extracts as described above (Figure 3A). The corresponding fluorograph of Figure 5 shows a band doublet in the S9 wild-type extracts (arrow a), which migrates slightly more slowly than the band corresponding to BasT (arrow b). This band doublet is absent in ΔhtpIV. HtpIV exhibits two sites (Glu567 and Glu767), whose neighboring sequences perfectly match the consensus sequence for chemotaxis methylation sites (Le Moual and Koshland, 1996). Thus, the appearance of a band doublet could be due to differentially methylated forms of HtpIV, since it is well known of enteric transducer proteins that their electrophoretic mobility correlates with the extent of covalent methyl modification. Higher methylation levels, for instance, were shown to increase the migration rate in polyacrylamide gels (Nowlin et al., 1988). Like BasT and in contrast to the cytoplasmic transducer Car (arrow c), HtpIV is found exclusively in the membrane fraction of halobacterial cells (Figure 5, lanes ‘CF’ and ‘MF’). This is in good agreement with the results of the hydropathy analysis predicting two transmembrane segments for HtpIV (Rudolph et al., 1996). Together, the analysis of [3H]methyl-labeled proteins provided clear evidence that HtpIV is a membrane-bound member of the MCP family that is absent in strain ΔhtpIV.
Functional analysis of HtpIV
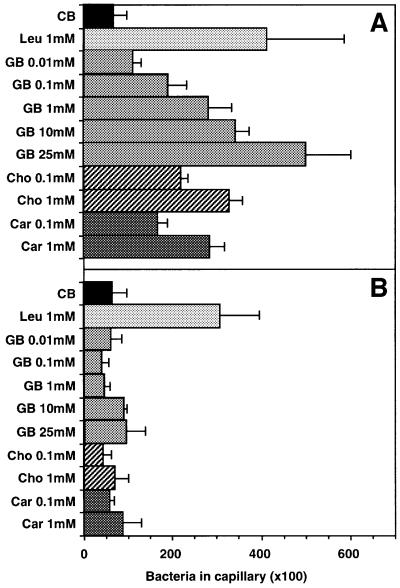
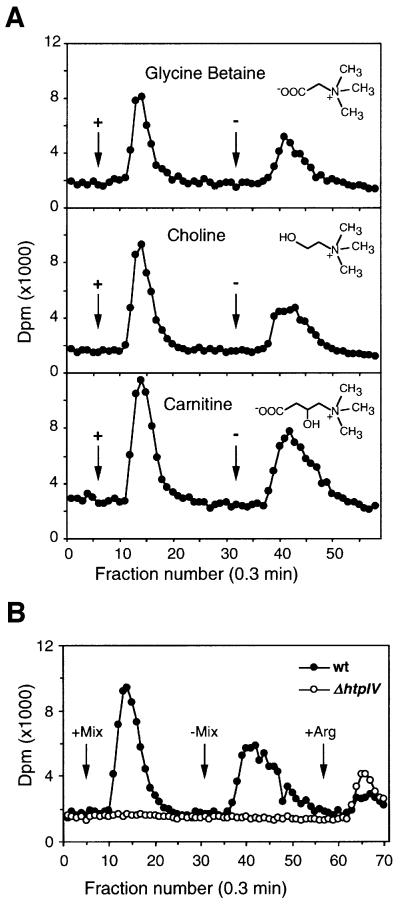
Orf2 shows significant homology to two proteins of B.subtilis: OpuBC, which binds choline, and OpuCC, which binds a broad range of trimethylammonium compounds including carnitine, glycine betaine and choline. We therefore chose these three compounds for the behavioral tests. Figure 6 shows the results of the capillary assay. This test measures the amount of cells migrating into micro-capillaries containing chemical solutions, which reflects the chemotaxis potency of the given compound (Storch et al., 1999). Clearly, cells deleted in htpIV have lost the capacity to respond to the quaternary amines tested while they showed normal response to the BasB/BasT-mediated attractant leucine. In contrast, the wild-type cells migrated at a concentration of 1 mM equally well into capillaries filled with any one of the quaternary amines. Glycine betaine was tested over a range of different concentrations indicating a threshold response <0.01 mM and a peak response at ≥25 mM. The response pattern at 0.1 and 1 mM for carnitine and choline is close to that observed for glycine betaine, pointing to a comparable chemotactic potency of all three quaternary amines in H.salinarum. We also carried out CIP assays. However, the cells did not form any chemotactic rings in response to the three quaternary amines, suggesting that these attractants are metabolized poorly or not at all by the cells (see Discussion). The capillary assay demonstrates that halobacterial cells respond strongly to compatible solutes of the betaine family and that these chemotactic responses are specifically mediated by the halobacterial transducer protein HtpIV. As HtpIV is methylatable we expected that stimulation with these chemotaxis attractants causes specific changes in methyl group turnover in halobacterial cells as already demonstrated for the BasT attractants (see above). The flow assay analysis (Figure 7A) shows that the onset (+) and offset (–) of stimulation with 5 mM choline, carnitine or glycine betaine causes a burst in methanol release, with the ‘on’ signal slightly larger than the ‘off’ signal. This response pattern resembles that for the halobacterial amino acid attractants (Figure 4B; Kokoeva and Oesterhelt, 2000). In contrast to wild-type cells, simultaneous stimulation with all three quaternary amines did not affect the methyl group turnover of ΔhtpIV cells (Figure 7B). These results confirm that HtpIV is the specific mediator of the chemotaxis response towards trimethylammonium compounds. In correspondence with the BasB/BasT system we therefore defined HtpIV as CosT, compatible solute transducer protein, and Orf2 as CosB, compatible solute binding protein.
Fig. 6. Capillary assay of S9 wild-type (A) and ΔhtpIV (B) strains. Cells were incubated with capillaries filled with the test compound in chemotaxis buffer (CB) for 2 h at 37°C in the dark. The amount of cells migrating into the capillaries was quantified by plating. Error bars indicate the SEM of four independent experiments. GB, glycine betaine; Cho, choline; Car, carnitine.
Fig. 7. Volatile [3H]methyl group release of S9 wild-type and ΔhtpIV cells stimulated with quarternary amines. Assay conditions were as in Figure 4B. (A) Stimulation of wild-type cells with either glycine betaine, choline or carnitine at 5 mM. (B) Cells were stimulated with a mix of the three quaternary amines at 5 mM each.
Accumulation of compatible solutes
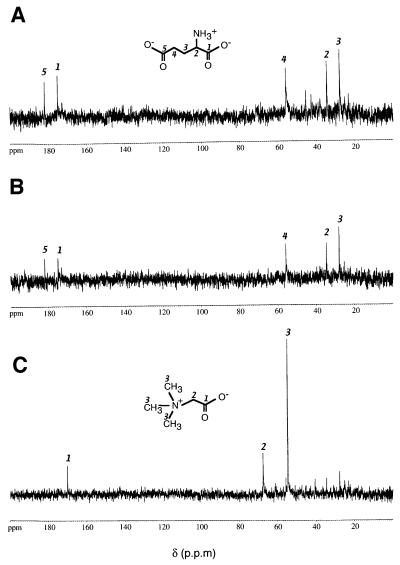
Having identified quaternary amines as chemotaxis attractants in H.salinarum, we asked what role these compounds play in the context of halobacterial physiology. Considering that these solutes are used by numerous organisms for osmoadaptive reasons, we carried out natural abundance 13C-NMR on ethanolic extracts from halobacterial cells grown at 4 M NaCl. Figure 8A shows the 13C-NMR spectrum of an extract of cells grown in complex medium. Five major resonances could be detected (181.8, 175.0, 55.7, 34.4 and 27.6 p.p.m.), which are consistent with the chemical shifts of the five carbons of l-glutamate (Lai et al., 1995). This amino acid is a well known osmoregulator observed throughout the kingdoms (Martin et al., 1999). The glutamate resonances also clearly dominate the spectrum when cells are grown in synthetic medium (Figure 8B). As this medium does not contain glutamate (or glutamine), this result demonstrates that H.salinarum is able to amass this amino acid by de novo synthesis. The 13C-NMR analysis also reveals that halobacterial cells accumulate high amounts of glycine betaine when it is provided in the medium. Growth in synthetic medium supplemented with 10 mM glycine betaine results in the spectrum shown in Figure 8C. The three strong resonances (170.1, 67.6 and 54.7 p.p.m.) identify glycine betaine as the major small organic molecule in the cytoplasm, whereas the signals of the glutamate carbons are clearly reduced as compared with unsupplemented medium (Figure 8B). Note that the NMR analyses were carried out under pulsing conditions where the CHn moieties are not saturated. Therefore, integrated intensities can be used to estimate roughly the relative concentrations of solutes (see Materials and methods). Accordingly, the relative reduction of the glutamate peaks of Figure 8C compared with Figure 8B indicates that accumulation of glycine betaine downregulates the net accumulation of glutamate.
Fig. 8. Natural abundance 13C-NMR spectra of ethanolic cell extracts. H.salinarum strain S9 was grown at 4 M NaCl in complex medium (A), synthetic medium (B) or synthetic medium supplemented with 10 mM glycine betaine (C). Resonances for l-glutamate (A and B) and glycine betaine (C) are indicated by carbon numbers as assigned in the structural formulae.
Discussion
In this study, we present the first experimental evidence for binding protein-dependent chemotaxis in archaea. Our findings suggest that binding proteins function as primary receptors in mediating responses to amino acids and compatible osmolytes in H.salinarum: (i) we identified two orfs, basB and cosB, which encode orthologs of eubacterial binding proteins and both of which are located directly upstream of chemotaxis transducer genes; (ii) northern analysis of the bas locus indicates that basB and its cognate transducer gene basT form a transcriptional unit; (iii) the results of the behavioral analysis of bas mutant strains is strong evidence that both basT and basB are required for halobacterial responses to branched-chain and sulfur-containing amino acids.
BasB and CosB presumably exert their functions as membrane-attached proteins. Their N-termini exhibit features that are diagnostic for signal peptides of microbial lipoproteins. These features are also evident in halo cyanin, a blue copper protein of unknown function in Natronobacter pharaonis, an alkalohalophilic archaeon closely related to H.salinarum (Mattar et al., 1994). Analysis by mass spectroscopy indicated that the N-terminal cysteine of halocyanin is covalently modified by C20 diphytanyl diether lipids, which commonly occur in archaeal organisms. Bacterial lipoproteins, on the contrary, are generally anchored by fatty acids (Wu, 1996). Given the N-terminal similarities to halocyanin, it is conceivable that BasB and CosB are also anchored by such ether lipids to the halobacterial membrane. Interestingly, sequence analysis of the completed halobacterial genome (Ng et al., 2000) based on this lipoprotein signature indicated that probably all binding protein orthologs are lipoproteins in H.salinarum (M.V.Kokoeva, K.-F.Storch and D.Oesterhelt, unpublished). However, lipid anchoring is apparently not the only mode of membrane attachment in archaea. Recent work in Sulfolubus sulfataricus indicates that in this archaeon binding proteins are tethered to the cell membrane via transmembrane segments (Elferink et al., 2001).
Colocalization of receptor and transducer genes: a general principle in halobacteria?
In both bacteria and archaea the bas/cos systems represent the first case in which substrate-binding and transducer proteins are organized in operon-like structures on the chromosome. Generally, genes encoding substrate-binding proteins are either part of ABC transport operons or located in their close vicinity, even if the binding proteins additionally function as receptors in chemotaxis (Boos and Lucht, 1996). Intimate clustering of cognate receptor and transducer genes, however, appears not to be restricted to the bas/cos systems in halophilic archaea. Also, the retinal proteins SRI and SRII, which mediate halobacterial phototaxis, are presumably cotranscribed with their respective transducer proteins, HtrI and HtrII (Yao and Spudich, 1992; Zhang et al., 1996b). There is evidence that SRI and SRII associate in signaling complexes with their cognate transducers (Schmies et al., 2001) and that photostimulation causes structural changes in these complexes, which are then relayed into the cytoplasmic signaling cascade via the histidine kinase CheA (Sasaki and Spudich, 2000).
Moreover, analysis of the halobacterial genome indicates that colocalization of binding protein and transducer genes is not restricted to the BasB/T and CosB/T units in this organism. A further dozen orfs have been annotated as binding protein orthologs in Halobacterium (Ng et al., 2000), most of which are organized in ABC transport clusters. However, two of them, yufN and potD (DDBJ/EMBL/GenBank accession numbers AAG19258 and AAG19271), overlap with htr6 and htr18, respectively, which represent new members of the halobacterial transducer family of unknown function (Ng et al., 2000). At the protein level, PotD shows significant sequence similarity to spermidine/putrescine-binding proteins, suggesting that the PotD/Htr18 system mediates chemotaxis toward this compound class. Yet BLAST searches as of 5 November 2001 did not point to any specific substrate for YufN. Interestingly, potD follows downstream of htr18 on the chromosome, whereas yufN is located upstream of htr6 as for the bas and cos units.
Given also that the integral membrane modules of ABC transporters are usually conserved among species, we examined the halobacterial genome for such modules, which are specific for substrates of BasB, CosB or PotD. However, no respective homolog was found, suggesting that ABC transport systems are not involved in the accumulation of these substrates. Thus, BasB, CosB and PotD might exclusively mediate chemotaxis responses without any additional function in transport, which is in contrast to the bacterial binding proteins involved in chemotaxis. Based on these findings it could be speculated that halophilic archaea evolved such chromosomal colocalization due to their need for a precisely regulated stoichiometry of the transducer and receptor modules. Immunochemical analysis of BasB expression (Figure 3B) indicates that the expression of the binding protein depends on the presence of its cognate transducer protein, plausibly via stabilizing protein–protein interactions. It would therefore be economically reasonable to implement the required stoichiometry already at the transcript level.
With respect to the mechanism of signal detection, it appears that the bas/cos systems deviate substantially from their photosensory counterparts. Interchanging domains of HtrI and HtrII revealed that only their transmembrane domains define the specificity of interaction with their cognate photoreceptors (Zhang et al., 1999a), whereas omitting the extracellular domain of HtrII does not abro gate SRII-mediated phototaxis. The behavior of basTtrunc, however, indicates that BasT requires its extracellular domain to mediate tactic responses. Therefore, the following mechanism of signaling for the Bas and Cos systems could be proposed: substrate binding of the lipid-anchored BasB (CosB) induces conformational changes, which are relayed to the cognate transducer via its extracellular domain. This results in an altered signaling state of the transducer protein, which subsequently affects the activity of the histidine kinase CheA. A similar mode of signal transduction is well known for enterobacterial chemotaxis to maltose, where a maltose-loaded MBP interacts with a specific periplasmic portion of the transducer protein Tar (Zhang et al., 1999b). To elucidate whether the extracellular domains of CosT and BasT are sufficient for response specificity, in analogy to the transmembrane segments of HtrI/HtrII, equivalent chimeric studies are needed with the extracellular domains swapped between BasT and CosT.
Chemotactic detection and accumulation of compatible solutes: what for in extreme halophiles?
The presence of a dedicated chemotaxis detection system for trimethylammonium compounds in H.salinarum gives rise to two basic questions: (i) why has this archaeon evolved such a system and (ii) what role do these molecules play in this organism? Trimethylammonium compounds function as osmotic regulators in a variety of species throughout the kingdoms. In this function, they are also referred to as compatible solutes. The majority of these solutes have at least three properties in common: they have no (net) charge under physiological conditions, they are very soluble and they are preferentially excluded from the immediate hydration sphere of proteins (Galinski, 1995; Kempf and Bremer, 1998). This latter property particularly is accounted for the protein-stabilizing effect of compatible solutes (Timasheff, 1992). Glycine betaine and other quaternary amines have already been shown to function as osmoprotectants not only in eubacteria and eukarya but also in a variety of methanogenic archaea (Martin et al., 1999). Our results indicate that these compounds might be involved in osmoadaptive processes in H.salinarum as well. First, natural abundance 13C-NMR of ethanolic extracts reveals that halobacterial cells accumulate glycine betaine when growing in medium supplemented with this quaternary amine. The NMR spectra indicate that glycine betaine is amassed at concentrations dramatically higher than for any other small organic molecule except for glutamate (see below). Secondly, the lack of chemotactic responses in the plug plate assay indirectly indicates that those quaternary amines are not metabolized by the cells. As has been discussed previously, formation of depletion rings apparently relies, at least in part, on the metabolic conversion of the attractant compound (Storch et al., 1999). These results meet two basic requirements for compatible solute functioning in osmoregulation: accumulation in the cytoplasm at high concentrations and compatibility with the cell’s physiology.
At first glance, it appears surprising that organic osmolytes could play a role in this archaeon. Extreme halophilic organisms like the Halobacterium species were generally considered to adapt to their high-osmolar habitats by accumulating inorganic ions, primarily K+ (Galinski, 1995; Martin et al., 1999). Indeed, cytoplasmic K+ concentrations of up to 4.6 M have been reported for H.salinarum when grown at 4 M NaCl (Christian and Waltho, 1962). Although intracellular amounts of Cl– have never been quantified, it was concluded that accumulation of KCl is key to osmoadaptation in these extreme halophiles (Galinski, 1995). This notion is corroborated by the presence of specific KCl uptake systems in H.salinarum, including a Δψ-driven K+ uniporter and the unique light-driven chloride pump halorhodopsin (Wagner et al., 1978; Kolbe et al., 2000; Ng et al., 2000).
Our 13C-NMR data demonstrate that glutamate is the sole small organic molecule that is highly accumulated by de novo synthesis in halobacteria. It is conceivable, therefore, that glutamate, in addition to chloride, might function as a counterion for K+ in halobacterial cells, balancing the electrical state of the cytoplasm. An intimate regulatory relationship between K+ and glutamate accumulation in response to osmotic stress is well known for enteric bacteria (McLaggan et al., 1994). Furthermore, amassing of glutamate has also been demonstrated by 13C-NMR for several methanogenic archaea (Martin et al., 1999) and in the haloalkalophilic Natronococcus occultus, which is, like H.salinarum, a member of the Halobacteriacaea family (Desmarais et al., 1997).
Moreover, it has been shown for E.coli that after the early K-glutamate response upon osmotic upshock, the cells accumulate glycine betaine and proline as compatible solutes if they are present in the medium (Csonka and Epstein, 1996). The accumulation of these neutral solutes is in turn accompanied by depletion of the cytoplasmic K+ and glutamate pools (Cayley et al., 1992). This depletion was interpreted as an osmoadaptive strategy of E.coli cells, as high intracellular K+ levels are believed to interfere with physiological functions in non-halophilic bacteria (Csonka and Epstein, 1996). Halobacteria, however, are supposed to be true halophilic organisms, which do seem to require high intracellular potassium concentration for optimal growth and thus for their cellular functions. None the less, we find halobacterial cells accumulating glycine betaine and at the same time reducing their glutamate levels as evidenced by 13C-NMR. In the light of the E.coli data, it could be speculated that halobacteria also concomitantly drain their K+ pools under these conditions. Interestingly, in vitro studies have clearly demonstrated that high concentrations of betaine or sorbitol keep halophilic enzymes native, even in the complete absence of salts (Cadenas and Engel, 1994; Vuillard et al., 1995).
In conclusion, the evolution of dedicated sensing and presumably uptake systems is strong evidence that amassing of osmolytes of the betaine family provides a selective advantage for H.salinarum in its natural habitats. However, preliminary growth tests of halobacterial cells in the presence of glycine betaine, carnitine or choline do not indicate any major effect of these compounds on cell propagation (data not shown). Hence, detailed growth analyses and cell survival tests might be required to reveal the physiological relevance of compatible solutes for H.salinarum.
Materials and methods
Strains and culture conditions
A highly motile single colony isolate of H.salinarum strain S9 was used throughout (Storch et al., 1999). Halobacterial cells were grown under standard conditions in complex medium or in synthetic medium as described in Oesterhelt and Krippahl (1983) with the following modifications: dl-serine and dl-phenylalanine were substituted by l-serine and l-phenylalanine, respectively.
Northern blotting analysis
RNA extraction and northern blot analysis was carried out essentially as described (Sambrook et al., 1989). Probes to basB (probe 1) and basT (probe 2) were generated by amplification from halobacterial genomic DNA using the primer pairs 5′-ACAGCTCCTCGTCAGCCC-3′/ 5′-ACCAGCTCGTCGATGACG-3′ and 5′-TCGTGGTGGCACTCGTGG-3′/5′-TCGAGGTCCGTGATGAGC-3, respectively. The PCR products were radiolabeled by random priming using the Prime-It-Kit (Amersham) according to the manufacturer’s protocol. X-ray films were exposed to the membrane for 24 h.
Western blotting analysis and behavioral assays
Total protein from halobacterial cells was obtained by acetone extraction as described (Storch et al., 1999). Antibodies raised in rabbits against a BasB-specific peptide (AKIVDDDMYAAAD) conjugated to keyhole limpet hemocyanin were obtained from a commercial source (Davids Biotechnologie, Germany). For immunoblotting, the antibodies were used at a dilution of 1:5000 and BasB was detected using the ECL western blotting kit (AP Biotech). CIP and capillary assays were performed as described (Storch et al., 1999; Kokoeva and Oesterhelt, 2000).
In vivo radiolabeling, detection of halobacterial transducer proteins and volatile methyl group production experiments
Radiolabeling of halobacterial cells and visualization of tranducer proteins was carried out as described in Storch et al. (1999). Analysis of volatile methyl group production (flow assay) was performed as described (Kokoeva and Oesterhelt, 2000).
Extraction of intracellular solutes and 13C-NMR spectroscopy
S9 cells were grown in 200 ml of either complex or synthetic medium supplemented with or without 10 mM glycine betaine using 500 ml Erlenmeyer flasks on a shaker (220 r.p.m, 37°C). The NaCl concentration in either medium was 4 M. When a density of 0.6 × 108 cells/ml was reached, cells were harvested by centrifugation (6000 g, 45 min). Cell pellets were suspended in 1.5 ml 70% (v/v) ethanol, sonicated for 5 min in a bath sonicator and centrifuged (11 000 g, 10 min, 4°C). The extraction step was repeated twice, the supernatants were pooled and most of the solvent was removed by rotary evaporation. Final sample drying was achieved by lyophilization. For NMR analysis, samples were redissolved in 0.5 ml D2O and filtered through a Pasteur pipette filled with glass wool. 1H WALTZ-decoupled 13C-NMR spectra were recorded with a Bruker AMX 400 spectrometer at 100.6 MHz with a 5 mm broadband probe head. Acquisition parameters included a 26 kHz sweep width, 32k datapoints, 35° pulse angle and 30 000 transients. Given that the CHn moieties in the organic solutes investigated have nuclear Overhauser effects (NOEs) of ∼3, the CHn are not saturated under those acquisition conditions and thus integrated intensities can be used to estimate the relative abundance of the compounds (Lai et al., 1995). Chemical shifts were measured relative to dioxane (66.3 p.p.m.).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Isolde Sonnenbichler for performing the NMR recordings, Nadja Patenge for technical help with the northern blots and Benjamin A.Horwitz for critical reading of the manuscript.
References
- Adams M.D., Wagner,L.M., Graddis,T.J., Landick,R., Antonucci,T.K., Gibson,A.L. and Oxender,D.L. (1990) Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J. Biol. Chem., 265, 11436–11443. [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaeffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Beveridge T.J. (1999) Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol., 181, 4725–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos W. and Lucht,J.M. (1996) Periplasmic binding protein-dependent ABC transporters. In Neidhardt,R.C.I., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington DC, pp. 1175–1209.
- Brown A.D. (1976) Microbial water stress. Bacteriol. Rev., 40, 803–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas Q. and Engel,P.C. (1994) Activity staining of halophilic enzymes: Substitution of salt with a zwitterion in non-denaturing electrophoresis. Biochem. Mol. Biol. Int., 33, 785–792. [PubMed] [Google Scholar]
- Cayley S., Lewis,B.A. and Record,M.T. (1992) Origins of the osmoprotective properties of betaine and proline in Escherichia coli K-12. J. Bacteriol., 174, 1586–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian J.H.B. and Waltho,J.A. (1962) Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim. Biophys. Acta, 65, 506–508. [DOI] [PubMed] [Google Scholar]
- Csonka L.N. and Epstein,W. (1996) Osmoregulation. In Neidhardt,R.C.I., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff, W.S., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington DC, Vol. 2, pp. 1210–1223.
- Danner S. and Soppa,J. (1996) Characterization of the distal promoter element of Halobacteria in vivo using saturation mutagenesis and selection. Mol. Microbiol., 19, 1265–1276. [DOI] [PubMed] [Google Scholar]
- Desmarais D., Jablonski,P.E., Fedarko,N.S. and Roberts,M.F. (1997) 2-Sulfotrehalose, a novel osmolyte in haloalkaliphilic archaea. J. Bacteriol., 179, 3146–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmers F.J.M., Lanfermeijer,F.C. and Poolman,B. (2001) Peptides and ATP binding cassette peptide transporters. Res. Microbiol., 152, 245–258. [DOI] [PubMed] [Google Scholar]
- Ehrmann M., Ehrle,R., Hofmann,E., Boos,W. and Schloesser,A. (1998) The ABC maltose transporter. Mol. Microbiol., 29, 685–694. [DOI] [PubMed] [Google Scholar]
- Elferink M.G.L., Albers,S.V., Konings,W.N. and Driessen,A.J.M. (2001) Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol., 39, 1494–1503. [DOI] [PubMed] [Google Scholar]
- Galinski E.A. (1995) Osmoadaptation in bacteria. Adv. Microb. Physiol., 37, 272–328. [PubMed] [Google Scholar]
- Kandler O. and König,H. (1993) Cell envelopes of archaea: Structure and chemistry. In Kates,M., Kushner,D.J. and Matheson,A.T. (eds), The Biochemistry of Archaea (Archaebacteria). Elsevier Science Publishers, Amsterdam, pp. 223–259.
- Kappes R.M., Kempf,B., Kneip,S., Boch,J., Gade,J., Meier-Wagner,J. and Bremer,E. (1999) Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol., 32, 203–216. [DOI] [PubMed] [Google Scholar]
- Kempf B. and Bremer,E. (1998) Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol., 170, 319–330. [DOI] [PubMed] [Google Scholar]
- Kokoeva M.V. and Oesterhelt,D. (2000) BasT, a membrane-bound transducer protein for amino acid detection in Halobacterium salinarum. Mol. Microbiol., 35, 647–656. [DOI] [PubMed] [Google Scholar]
- Kolbe M., Besir,H., Essen,L.-O. and Oesterhelt,D. (2000) Structure of the light-driven chloride pump halorhodopsin at 1.8 Å resolution. Science, 288, 1390–1396. [DOI] [PubMed] [Google Scholar]
- Lai M.C., Ciulla,R., Roberts,M.F., Sowers,K.R. and Gunsalus,R.P. (1995) Extraction and detection of compatible intracellular solutes. In Robb,F.T., Place,A.R., Sowers,K.R., Schreier,H.J., DaSarma,S. and Fleischmann,E.M. (eds), Archaea: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 349–368.
- Le Moual H. and Koshland,D.E. (1996) Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol., 261, 568–585. [DOI] [PubMed] [Google Scholar]
- Martin D.D., Ciulla,R.A. and Roberts,M.F. (1999) Osmoadaptation in archaea. Appl. Environ. Microbiol., 65, 1815–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar S., Scharf,B., Kent,S.B.H., Rodewald,K., Oesterhelt,D. and Engelhard,M. (1994) The primary structure of halocyanin, an archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J. Biol. Chem., 269, 14939–14945. [PubMed] [Google Scholar]
- McLaggan D., Naprstek,J., Buurman,E.T. and Epstein,W. (1994) Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J. Biol. Chem., 269, 1911–1917. [PubMed] [Google Scholar]
- Ng W.V. et al. (2000) Genome sequence of Halobacterium species NRC-1. Proc. Natl Acad. Sci. USA, 97, 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlin D.M., Bollinger,J. and Hazelbauer,G.L. (1988) Site-directed mutations altering methyl-accepting residues of a sensory transducer protein. Proteins, 3, 102–112. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D. and Krippahl,G. (1983) Phototrophic growth of halobacteria and its use for isolation of photosynthetically deficient mutants. Ann. Microbiol., 134B, 137–150. [DOI] [PubMed] [Google Scholar]
- Perazzona B., Spudich,E.N. and Spudich,J.L. (1996) Deletion mapping of the sites on the HtrI transducer for sensory rhodopsin I interaction. J. Bacteriol., 178, 6475–6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J. and Oesterhelt,D. (1995) Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium.EMBO J., 14, 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J., Nordmann,B., Storch,K.F., Gruenberg,H., Rodewald,K. and Oesterhelt,D. (1996) A family of halobacterial transducer proteins. FEMS Microbiol. Lett., 139, 161–168. [DOI] [PubMed] [Google Scholar]
- Sack J.S., Saper,M.A. and Quiocho,F.A. (1989) Periplasmic binding protein structure and function refined X-ray structures of the leucine-isoleucine-valine-binding protein and its complex with leucine. J. Mol. Biol., 206, 171–192. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Habor, NY.
- Sasaki J. and Spudich,J.L. (2000) Proton transport by sensory rhodopsins and its modulation by transducer-binding. Biochim. Biophys. Acta, 1460, 230–239. [DOI] [PubMed] [Google Scholar]
- Schmies G., Engelhard,M., Wood,P.G., Nagel,G. and Bamberg,E. (2001) Electrophysiological characterization of specific interactions between bacterial sensory rhodopsins and their transducers. Proc. Natl Acad. Sci. USA, 98, 1555–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J.B. and Surette,M.G. (1996) Chemotaxis. In Neidhardt,R.C.I., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, Vol. 1, pp. 551–573.
- Storch K.F., Rudolph,J. and Oesterhelt,D. (1999) Car: a cytoplasmic sensor responsible for arginine chemotaxis in the archaeon Halobacterium salinarum. EMBO J., 18, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe I.C. and Russell,R.R.B. (1995) Lipoproteins of Gram-positive bacteria. J. Bacteriol., 177, 1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timasheff S.N. (1992) Water as ligand: Preferential binding and exclusion of denaturants in protein unfolding. Biochemistry, 31, 9857–9864. [DOI] [PubMed] [Google Scholar]
- Vuillard L., Madern,D., Franzetti,B. and Rabilloud,T. (1995) Halophilic protein stabilization by the mild solubilizing agents nondetergent sulfobetaines. Anal. Biochem., 230, 290–294. [DOI] [PubMed] [Google Scholar]
- Wagner G., Hartmann,R. and Oesterhelt,D. (1978) Potassium uniport and ATP synthesis in Halobacterium halobium. Eur. J. Biochem., 89, 169–179. [DOI] [PubMed] [Google Scholar]
- Wu H.C. (1996) Biosynthesis of lipoproteins. In Neidhardt,R.C.I., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington DC, pp. 1005–1014.
- Yao V.J. and Spudich,J.L. (1992) Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc. Natl Acad. Sci. USA, 89, 11915–11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Brooun,A., McCandless,J., Banda,P. and Alam,M. (1996a) Signal transduction in the Archaeon Halobacterium salinarium is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc. Natl Acad. Sci. USA, 93, 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Brooun,A., Mueller,M.M. and Alam,M. (1996b) The primary structures of the Archaeon Halobacterium salinarium blue light receptor sensory rhodopsin II and its transducer, a methyl-accepting protein. Proc. Natl Acad. Sci. USA, 93, 8230–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.N., Zhu,J.Y. and Spudich,J.L. (1999a) The specificity of interaction of archaeal transducers with their cognate sensory rhodopsins is determined by their transmembrane helices. Proc. Natl Acad. Sci. USA, 96, 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gardina Paul,J., Kuebler Ann,S., Kang Hui,S., Christopher Jon,A. and Manson Michael,D. (1999b) Model of maltose-binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc. Natl Acad. Sci. USA, 96, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]