Abstract
Hybridization of fluorescent molecular beacons provides real-time detection of RNA secondary structure with high specificity. We used molecular beacons to measure folding and unfolding rates of the Tetrahymena group I ribozyme under native conditions. A molecular beacon targeted against 15 nt in the 5′ strand of the P3 helix specifically hybridized with misfolded forms of the ribozyme, without invading the native tertiary structure. The beacon associated with the misfolded ribozyme 300 times more slowly than with an unstructured oligonucleotide containing the same target sequence, suggesting that the misfolded ribozyme core remains structured in the absence of Mg2+. The rate of beacon hybridization under native conditions revealed a linear relationship between the free energy of unfolding and Mg2+ concentration. A small fraction of the RNA population unfolded very rapidly, suggesting parallel unfolding in one step or through misfolded intermediates.
INTRODUCTION
RNA secondary structure rearrangements are important for genetic regulation and for folding and assembly of RNA–protein complexes. For example, metabolite-sensing riboswitches control mRNA transcription and translation by switching between alternative secondary structures (1). During pre-mRNA splicing, a cascade of helix exchange reactions controls spliceosome assembly and the recycling of snRNP complexes (2). Among ribozymes, competition between alternative secondary structures dictates the extent of self-cleavage (3,4,5) and even the catalytic function of the RNA (6).
RNA secondary structures can be probed by selective hybridization of complementary oligonucleotides, which are easily targeted to specific sequences (7–10). Zarrinkar and Williamson (11) used DNA oligomers and RNase H to investigate the kinetic folding pathway of the Tetrahymena ribozyme. Hybridization of the oligonucleotide can also be monitored by FRET or by a change in the fluorescence of a single dye conjugated to an oligonucleotide (12).
We wished to determine whether fluorescently tagged ‘molecular beacons’ can measure changes in RNA secondary structure in real time, without requiring site-specific labeling of the RNA target. Molecular beacons are hairpin oligonucleotide probes that were originally described by Tyagi and Kramer (13) for use in real-time PCR. The beacons are conjugated to a fluorophore on one end and a quencher on the other. In the absence of target, self-complementary arms base pair and bring the fluorophore in close proximity to the quencher, resulting in little fluorescence. Hybridization of the loop with the target sequence forces the stem of the beacon apart, separating the two dyes and resulting in an increase in fluorescence.
An advantage of molecular beacons over single dye probes is that the presence of target is signaled by an increase in fluorescence over a low baseline, which is readily detected even over a large concentration of unhybridized beacon (13). The greater signal-to-noise ratio allows the beacons to be used in situations where conventional fluorescent probes may not be useful, such as the detection of mRNA in live cells (14–16), detection of a specific mRNA sequence amongst many others (17) and detection of DNA binding proteins (18).
Molecular beacons are often more specific than linear probes (19). However, they typically require a ∼15 nt target sequence (13). They also hybridize more slowly with their targets than linear probes, because the hairpin must open in order to separate the fluorophore and quencher (20). Therefore, to be useful in RNA folding, the stability of the beacon hairpin should be optimized to minimize the baseline fluorescence while allowing rapid hybridization.
The utility of molecular beacons for studying RNA folding was tested by targeting a misfolded intermediate in the well-characterized folding pathway of the Tetrahymena L-21 ribozyme (21). Because the intensity of the fluorescent signal and the hybridization rate of the beacon depend on the concentration and the accessibility of the target, the relative levels of individual folding intermediates can be quantified. Here, we show that unfolding of the ribozyme can be measured under native conditions using molecular beacons. A small fraction of the ribozyme binds the beacon very rapidly, suggesting that the ribozyme unfolds by more than one pathway.
MATERIALS AND METHODS
Preparation of RNA
The L-21 ribozyme was prepared by in vitro transcription as described previously (22). The oligoribonucleotide 5′-rGCAAGACCGUCAAAU was purchased from Dharmacon, deprotected according to the manufacturer's protocol and used without further purification.
Molecular beacons
Fluorescently labeled molecular beacon MB-P3: 5′(6-FAM)-dGGGCTATTTGACGGTCTTGCAGCCC-3′(C6-NH-DABCYL) was purchased from Trilink Biotechnologies. The linear probe L-P3: 5′-dATTTGACGGTCTTGC-3′ fluorescein was purchased from Sigma Genosys. Concentrations were determined from the absorption at 260 nm; MB-P3: ɛ260 = 267 OD/µmol; L-P3: ɛ260 = 130 OD/µmol.
Fluorescence measurements
The fluorescence experiments were performed using a PerkinElmer LS-50B Luminescence Spectrometer, using an excitation wavelength of 496 nm and a 515 nm emission wavelength. All experiments were carried out in splicing buffer [50 mM HEPES, 100 nM (NH4)2SO4 and 1 mM EDTA] at 37°C. Where stated, the RNA was incubated in 0.8 M urea in splicing buffer before the addition of the beacon. The background intensity of 100 nM beacon was recorded before 300 nM L-21 was added to the cuvette. The fluorescence intensity was then measured over time. The relative change in fluorescence was obtained from ΔF = [F(t) − Fmin]/(Fmax−Fmin), where F(t) is the fluorescence intensity at each time, Fmin is the baseline fluorescence in the absence of target and Fmax is the maximum fluorescence at equilibrium, respectively.
To calculate the equilibrium binding constant (Kd) the fluorescence intensity of 2 nM MB-P3 was recorded in splicing buffer at 37°C, with 0–250 nM 15mer oligonucleotide. The beacon and target were equilibrated 5 min after each addition. Titrations performed with 50 nM MB-P3 and 0–2 µM L-21 ribozyme were equilibrated 20 min per addition. The data were fit to a single-site binding isotherm ΔF = v = [RNA]/(Kd + [RNA]), in which v is the fraction of bound beacon and the concentration of free target [RNA] = [RNA]0 − v[B]0was calculated iteratively. [RNA]0 and [B]0 are the total concentrations of RNA and beacon, respectively.
Stopped-flow fluorescence spectroscopy
The hybridization rates of 50 nM MB-P3 or L-P3 with a complementary 15mer RNA were measured in splicing buffer at 37°C using an Applied Photophysics Stopped Flow (SX 18MV). The fluorescent probe and 0–1 µM 15mer (final) were mixed in a 1:1 ratio with a dead time of 1.8 ms. The sample was excited at 496 nm and the emission intensity was measured using a 515 nm cut-off filter. At least three separate experiments were performed at each oligonucleotide concentration.
The normalized change in fluorescence was fit to ΔF = A1(1 − e−λ1t) + A2(1 − e−λ2t), where A1 and A2 are the amplitudes and λ1 and λ2 are the observed rate constants of each phase. The observed rate constants of the fast phase obtained from separate trials were averaged. The slow phase typically represented <20% of the total change in fluorescence and was ignored. The slow phase may reflect unfolding of the beacon, as the rate constant (0.05 s−1) was independent of the target concentration but sensitive to the stability of the stem (data not shown). The hybridization kinetics of the molecular beacon with the 15mer were assumed to follow a two-state mechanism, [RNA] + [B] ↔ [B · RNA], for which kobs = kon [RNA]0 + koff when the total concentration of target [RNA]0 is greater than the concentration of beacon.
Ribozyme unfolding
L-21 ribozyme RNA (1.05 µM) was incubated in 10 µl splicing buffer and 0–6 mM MgCl2 at 50°C for 20 min, then 37°C for an additional 30 min. This was to ensure that the majority of the RNA bypassed any folding traps and formed the native state whenever possible (23,24). The pre-folded RNA was diluted into the same buffer containing 250 nM MB-P3. Final conditions were 30 nM RNA and 250 nM MB-P3 in 350 µl buffer. The solution was mixed by inverting the cuvette, and the change in fluorescence recorded over 120 min at 37°C. The baseline fluorescence of MB-P3 was recorded at 37°C before the RNA was added. The change in fluorescence was fit to the double exponential equation above. Unfolding data in 5 and 6 mM MgCl2 were fit to a semi-logarithmic expression to find the rate constant of the slow kinetic phase, because of the small amplitude of the unfolding reaction. Reported rate constants are the average of 3–4 trials.
The unfolding kinetics was analyzed according to the mechanism in Scheme 1. First, we assumed that RNA closing is faster than hybridization of the beacon, and
| 1 |
Binding and dissociation rate constants of the beacon with the unfolded ribozyme (kon and koff) were taken from reactions with the 15mer RNA in 0 and 6 mM MgCl2. These values were linearly interpolated to obtain kon at intermediate Mg2+ concentrations. In addition, the opening and closing rates of the RNA, kop and kcl, were obtained by solving expressions for the observed rate constants λ1, λ2 and amplitude A:
| 2 |
| 3 |
The amplitude was taken from the maximum change in fluorescence ΔF relative to ΔF for the same concentration of 15mer RNA.
Scheme 1.
Mechanism of beacon hybridization.
RESULTS
Beacon design
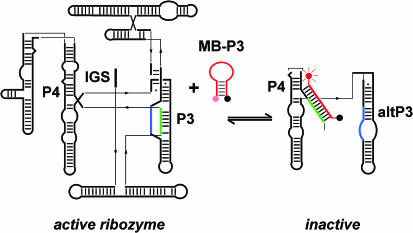
To test whether molecular beacons could be used to monitor RNA folding reactions, we chose the well-studied Tetrahymena thermophila ribozyme as a model system. While <5% of the ribozyme folds directly to its native structure, the majority of the population passes through misfolded intermediates in which the 5′ strand of the paired region P3 is displaced by the alternative pairing AltP3 (21). The beacon MB-P3 was targeted against the displaced strand of P3, such that the beacon should only bind the misfolded or unfolded ribozyme RNA (Figure 1). Oligonucleotides complementary to this sequence were previously used to characterize folding intermediates containing AltP3 (21).
Figure 1.
Molecular beacon targeted against the ribozyme core. Hybridization of the molecular beacon MB-P3 (red) with its target separates the dye (red sphere) and DABCYL quencher (black sphere), resulting in increased fluorescence. MB-P3 is complementary to nt 92–106 in the Tetrahymena ribozyme, which corresponds to the 5′ strand of helix P3 (green), part of the central triple helix and the base of P4. The 5′ strand of P3 is released in misfolded intermediates containing the alternative pairing altP3 between J8/7 (blue) and the 3′ half of P3.
The 15 nt loop of the beacon was complementary to nt 92–106 in the Tetrahymena L-21 ribozyme. The sequence of the stem was designed using MFOLD (25), such that it would form only the desired hairpin and no additional base pairs within the loop. The most promising beacon sequences were assayed by native gel mobility shift, to determine whether they bound the unfolded RNA but not the native ribozyme in 6 mM MgCl2 (data not shown). The beacon chosen for further study (MB-P3) had a 5 bp stem with a predicted ΔG of −2 to −3 kcal/mol. We found that this stability provided an optimal balance between low baseline fluorescence and rapid hybridization with the target, in agreement with recent findings (26) The beacon was derivatized on the 5′ end with 6-FAM and on the 3′ end with the quencher DABCYL.
MB-P3 selectively binds non-native ribozyme
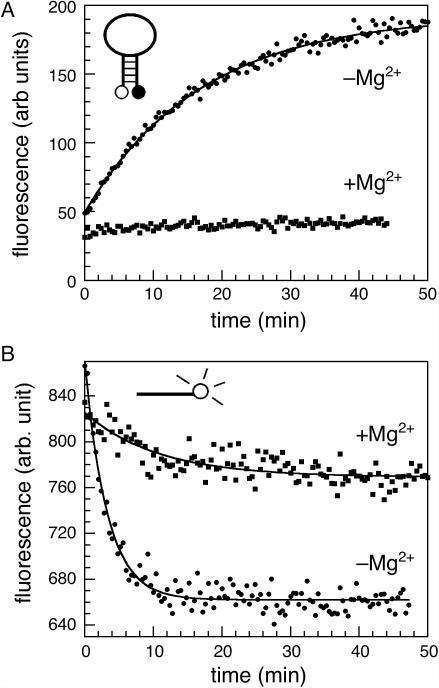
In the absence of magnesium, the Tetrahymena L-21 ribozyme predominantly forms structures containing the non-native Alt P3 helix (21). Thus, the target site of the beacon MB-P3 should be unpaired and available for hybridization with the probe. When 100 nM MB-P3 was added to 300 nM L-21 ribozyme RNA in splicing buffer without magnesium ion, the fluorescence increased by a factor of 4.7 (Figure 2A). The fluorescence intensity did not change, however, when the L-21 ribozyme was folded for 30 min in 6 mM MgCl2 before the beacon was added. These results showed that the beacon can distinguish between the folded and misfolded states of the ribozyme, and that it does not disrupt the native RNA.
Figure 2.
Molecular beacon only binds misfolded RNA. (A) Fluorescence of molecular beacon MB-P3 (100 nM) in the presence of 300 nM L-21 ribozyme RNA at 37°C. The beacon bound ribozyme RNA preincubated in splicing buffer with no MgCl2 (−Mg2+) but not RNA folded in 6 mM MgCl2 (+Mg2+) prior to addition of the beacon. (B) Fluorescence of 100 nM linear probe L-P3 in the presence of folded or unfolded L-21 ribozyme, as in (A). Curves represent best fit to for MB-P3 and for L-P3. MB-P3, kobs = 0.059 min−1; L-P3, kobs = 0.31 min−1 (−Mg2+).
We also created a linear probe (L-P3) by derivatizing a 15mer of the same sequence as the MB-P3 loop with 3′ fluorescein only. L-P3 fluorescence is quenched upon hybridization with RNA, due to stacking between the fluorescein dye and a G in the adjacent duplex (12,27,28). L-P3 also preferentially hybridized with misfolded RNA (Figure 2B), but the decrease in fluorescence intensity was only 20%. In general, we found that L-P3 was less sensitive than the beacon MB-P3, especially when the probe was in molar excess of the target. In contrast, fluorescence of the molecular beacon increases from a low baseline in the absence of target and provides a useful signal-to-noise ratio over a range of target and Mg2+ concentrations (see below).
Ribozyme structure slows MB-P3 hybridization kinetics
Although the MB-P3 beacon selectively binds unfolded or misfolded ribozyme RNA, the observed hybridization rate was much slower than expected for association of the beacon with an unstructured target. The beacon bound 300 nM ribozyme at a rate of 0.067 ± 0.008 min−1 in the absence of magnesium (Figure 2), about five times slower than the linear probe L-P3 (0.33 ± 0.03 min−1). Thus, slow hybridization of the beacon with the ribozyme might be explained by the need for the beacon hairpin to unpair (20).
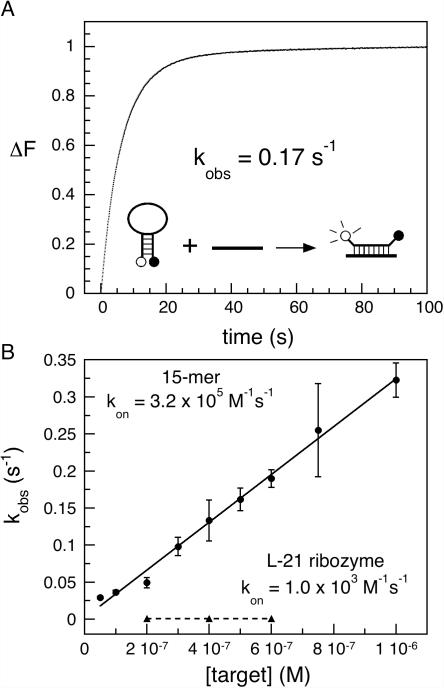
To determine the maximum hybridization rate of the beacon, the rate constant for the association of 50 nM MB-P3 with a complementary 15 nt oligoribonucleotide was measured by stopped-flow fluorescence spectroscopy (Figure 3A). MB-P3 bound the oligomer 300-fold faster than the unfolded L-21 ribozyme (kobs = 5.8 min−1 versus 0.067 min−1 in 300 nM RNA). For the 15mer RNA, the second order rate constant kon was 3.2 × 105 M−1 s−1 (Figure 3B), again 5-fold slower than kon for the linear probe (1.5 × 106 M−1 s−1). For the L-21 ribozyme, however, kon was ∼1.0 × 103 M−1 s−1 (Figure 3B). Thus, the structure of the RNA target inhibited the hybridization kinetics much more than the hairpin in the molecular beacon.
Figure 3.
Binding kinetics of beacon MB-P3 by stopped-flow fluorescence. (A) Sample trace from stopped-flow spectrometer of reaction between 50 nM MB-P3 and 500 nM 15mer oligomer target in splicing buffer. The change in fluorescence, ΔF, was fit (gray line) to two exponential phases with rate constants λ1 = 0.17 s−1 (86%) and λ2 = 0.049 s−1 (14%). The rate constant λ2 may reflect opening of the beacon hairpin. (B) Observed rate constants for the change in fluorescence (kobs = λ1) versus the total concentration of target RNA. Line is the best fit to kobs = kon[RNA]0 + koff. Closed circle: 15mer RNA, kon = 3.23 × 105 M−1s−1, koff = 1.7 × 10−3 s−1; closed square: L-21 ribozyme, kon = 1.0 × 103 M−1s−1, koff ≈ 5.7 × 10−4 s−1. Error bars represent the standard deviation of three trials. In 6 mM MgCl2, kon = 7.1 × 105 M−1s−1 for the 15mer.
To test whether structure in the ribozyme RNA inhibits beacon association, 0.8 M urea was added to the binding reaction. This concentration of urea destabilizes the misfolded intermediates without unfolding the native ribozyme (29). The hybridization rate of MB-P3 increased ∼2-fold in 0.8 M urea, to 0.124 ± 0.005 min−1. A similar acceleration was observed on the hybridization kinetics of L-P3, indicating that this effect was not simply due to destabilization of the beacon hairpin.
From the slow hybridization of the molecular beacon and L-P3 with ribozyme RNA, we concluded that the core of the L-21 ribozyme is partially structured in the absence of magnesium, preventing access of the beacon to its target. This result is consistent with native gel electrophoresis (30) and hydroxyl radical footprinting data (31) showing that monovalent salts present in splicing buffer, which contains 100 mM ammonium sulfate, are sufficient to stabilize folding intermediates of the ribozyme. Hybridization of the beacon could be inhibited by base pairs in P4, which overlap the target site, the non-native helix altP1 which base pairs with the 5′ strand of P3 (21), and tertiary interactions between peripheral stem–loops P2.1 and P9.1 (30,32,33).
Binding free energy of molecular beacon
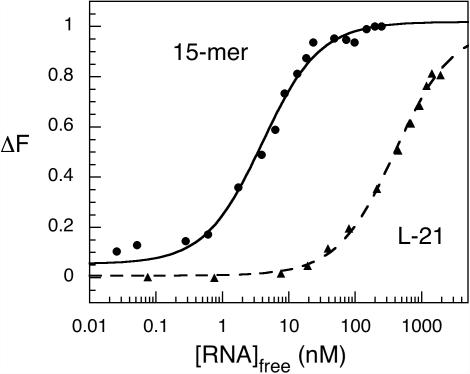
The affinity of the molecular beacon MB-P3 for target RNA was determined from titrations with either a 15mer RNA target or the L-21 ribozyme. The dissociation constants in splicing buffer without Mg2+ were 4.0 nM (Kd,oligo) and ∼400 nM (Kd,RNzyme), respectively (Figure 4). The binding reaction with the L-21 ribozyme was not saturated, so its Kd was estimated from the slope of the titration curve. As the base pairs between the target and the molecular beacon are the same in both complexes, the 100-fold difference between the dissociation constants of the 15mer and the L-21 ribozyme must represent the additional free energy needed to unfold the ribozyme in order for the molecular beacon to bind (Equation 4).
| 4 |
From the difference in the binding constants at 37°C, the free energy of unfolding was about 2.8 kcal/mol in the absence of MgCl2. This is equivalent to opening 1–3 bp (25).
Figure 4.
Beacon binds an oligomer more tightly than misfolded ribozyme. Equilibrium titrations of MB-P3 with complementary target RNA in splicing buffer without MgCl2 at 37°C. RNA oligomer target (closed circle), 15 nt oligomer plus 2 nM MB-P3, Kd = 4.0 ± 0.4 nM; (closed square), L-21 ribozyme plus 50 nM MB-P3, Kd ≈ 400 nM. The fraction of bound probe is proportional to the change in fluorescence, ΔF. For ribozyme titration, the maximum fluorescence was adjusted so that the fitted curve saturated near 1.
Measuring ribozyme unfolding under native conditions
Since MB-P3 forms a stable complex with its complement, it can trap unfolded RNA molecules. The rate of MB-P3 hybridization, which is sensitive to the accessibility of its target, was used to measure unfolding of the ribozyme core under native conditions. As the Mg2+ concentration increases, tertiary interactions in the ribozyme become more stable, and fluctuations that permit the P3 helix to open become less frequent. Therefore, the hybridization kinetics of the beacon should reflect the stability of the ribozyme tertiary structure and become slower as the magnesium concentration is raised.
The observed rate of beacon association with the ribozyme (kobs) depends on the intrinsic rate of hybridization with an unstructured target (kon) and the probability of the ribozyme unfolding, with kop and kcl the rates of unfolding and refolding the target as illustrated in Scheme 1.
If beacon binding is slower than reclosing of the RNA (kon[B]<<kcl), then the observed rate constant for beacon association kobs ≈ Kop(kon[B]), with Kop = kop/kcl and [B] as the concentration of unbound beacon. Thus, the equilibrium constant for RNA unfolding, Kop, can be estimated by comparing the rate of hybridization with the ribozyme and the 15mer RNA, where the latter is taken to represent an unstructured target (kon). Because this method relies on the rate of beacon hybridization, it can be used under conditions that favor the folded RNA. This is similar to the EX2 hydrogen exchange conditions used to describe protein folding kinetics under native conditions (34).
Mg2+-dependence of unfolding
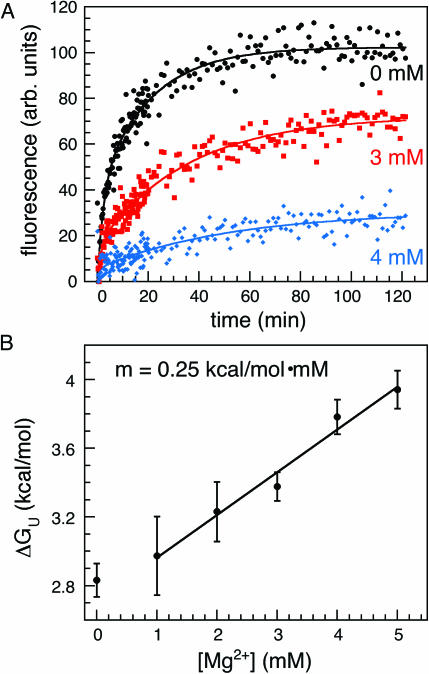
As expected, the extent and the rate of MB-P3 hybridization depended on Mg2+ concentration (Figure 5A). Using the EX2 approximation as described above, the free energy of unfolding at 37°C was obtained from hybridization of MB-P3 (Table 1). As shown in Figure 5B, ΔGU varied linearly with the Mg2+ concentration, over almost the entire range of concentrations tested. Thus, hybridization of the beacon correlates with the relative stability of the ribozyme. The linear free energy relationship ΔGU = ΔGU(0) + m · [Mg2+] resulted in an m-value of 0.25 kcal mol−1 mM−1. This is smaller than the m-values of 0.5 kcal mol−1 mM−1 and 1.2 kcal mol−1 mM−1 reported for folding and unfolding of the pre-rRNA at 30°C, as measured by native gel electrophoresis (35). The gel assay for folding primarily measures the tertiary structure of the entire RNA. In contrast, hybridization of MB-P3 reflects the availability of the 5′ side of P3 for base pairing, which may require opening of secondary (and tertiary) structure in the misfolded intermediates. This could explain why ΔGU appears less sensitive to Mg2+ by this method.
Figure 5.
Unfolding of the ribozyme assayed by beacon hybridization. The L-21 ribozyme (30 nM) was allowed to fold in splicing buffer plus 0–6 mM MgCl2, then mixed with 250 nM MB-P3 to trap unfolded RNA (see Materials and Methods). The rate of hybridization was measured from the change in fluorescence. (A) Sample traces of the increase in fluorescence as MB-P3 hybridizes to the unfolded ribozyme. (Closed circle) Black, 0 mM MgCl2; (closed square) red, 3 mM MgCl2; (closed diamond) blue, 4 mM MgCl2. The results were fit to a biphasic rate equation (see Materials and Methods). The rate of the burst phase was ∼0.3–0.9 min−1. (B) A linear fit to the free energy of unfolding ΔGU versus magnesium concentration yielded m = 0.25 kcal mol−1 mM−1. Free energies were calculated from the kinetics of beacon association at each Mg2+ concentration as described in the text and Table 1. Error bars are from the standard deviation of three trials.
Table 1.
Unfolding of the Tetrahymena ribozyme at 37°C
| [Mg2+] (mM) | kobs (min−1)a | ΔFrelb | kon[B] (min−1)c | kop (min−1) | ΔGUd (kcal/mol) |
|---|---|---|---|---|---|
| 0 | 0.049 ± 0.008 | 0.88 | 4.8 | 0.61 | 2.8 ± 0.1 |
| 1 | 0.047 ± 0.017 | 0.83 | 5.8 | 0.58 | 3.0 ± 0.2 |
| 2 | 0.036 ± 0.01 | 0.77 | 6.8 | 0.45 | 3.2 ± 0.2 |
| 3 | 0.032 ± 0.004 | 0.65 | 7.7 | 0.40 | 3.4 ± 0.1 |
| 4 | 0.019 ± 0.003 | 0.59 | 8.7 | 0.23 | 3.8 ± 0.1 |
| 5 | 0.016 ± 0.003 | 0.41 | 9.7 | 0.20 | 3.9 ± 0.1 |
| 6 | 0.023 ± 0.001 | 0.37 | 10.6 | 0.28 | 3.8 ± 0.1 |
aObserved rate constant for association of 250 nM MB-P3 plus 30 nM L-21 ribozyme in splicing buffer plus 0–6 mM MgCl2. Rate constants are for the major slow phase of hybridization (see Materials and Methods).
bAmplitude of change in fluorescence, ΔF, in ribozyme relative to ΔF in 15mer RNA under the same conditions. Uncertainty is ±20%.
cAssociation rate of the molecular beacon with 15mer RNA oligomer. Values in 2–5 mM MgCl2 were interpolated from measurements in 0 and 6 mM MgCl2.
dFree energy of unfolding from Equation 1, where Kop = kop/kcl and ΔGU = −RT ln Kop at 37°C, assuming kcl = 1 s−1. Dissociation of the beacon koff was 1.7 × 10−3 s−1. Error in ΔGU was propagated from the uncertainties in kobs.
Ribozyme opening and closing kinetics
The intrinsic association and dissociation rate constants were obtained from hybridization with the 15 nt RNA target (Figure 3B). The rate of binding to the L-21 ribozyme was measured in 0–6 mM MgCl2, by diluting pre-folded RNA into buffer containing 250 nM MB-P3 at 37°C. As the Mg2+ concentration was raised, the maximum fluorescence of the sample decreased, indicating less total beacon hybridization (Figure 5A). A small burst of hybridization was followed by slower association of the beacon with the ribozyme. The hybridization kinetics was fit to a biphasic exponential rate equation, yielding kobs = 0.049 min−1 in no MgCl2 for the major slow phase of the reaction (Table 1). Assuming that kon[B] = 4.8 min−1 is smaller than kcl and koff = 0.10 min−1 is negligible, we obtained Kop = 0.010 and ΔGU = 2.8 kcal/mol in splicing buffer with no Mg2+. This is the same within error as the estimate for ΔGU obtained from titration with the beacon (Figure 3).
The opening rate of the MB-P3 target sequence can be estimated from the refolding rate of the RNA (kcl). If the ribozyme refolds directly to the native state (U→N) with a rate of 1 s−1 (30,36), then kop is ∼0.4 min−1 in 3 mM MgCl2. This agrees with estimates of the unfolding rate from native gel electrophoresis [0.45 min−1 (35)]. Similar estimates would apply if the compact folding intermediates of the ribozyme are also inaccessible to the beacon, as these intermediates also form in 1–3 s (37). On the other hand, if the ribozyme must refold through metastable intermediates (I→N) in order to close the beacon target site, the assumption that kon[B] ≪ kcl is no longer valid because refolding [∼1 min−1 (30,38)] is slower than hybridization.
Therefore, we also calculated values of kcl, kop and Kop by fitting the amplitudes and observed rate constants for beacon hybridization to the rate equation for the mechanism in Scheme 1 (see Materials and Methods). The rates of ribozyme folding and unfolding were 0.51 min−1 and 0.022 min−1, respectively, in 3 mM MgCl2. The unfolding free energies obtained in this manner were smaller than those above, ranging from 1.1 kcal/mol in no MgCl2 to 2.6 kcal/mol in 6 mM MgCl2. However, they yielded a similar m-value of 0.27 kcal mol−1 mM−1 (data not shown). We believe that this method of obtaining Kop is less reliable because it is sensitive to uncertainty in the amplitude of the fluorescence change.
Comparison of beacon with global unfolding by native PAGE
To visualize the unfolding process and hybridization of the beacon, the ribozyme–beacon complex was monitored by native PAGE under the same conditions as the fluorescence assays (Figure 6). The ribozyme RNA was equilibrated in splicing buffer plus 2 mM MgCl2, and then mixed with 250 nM MB-P3. Aliquots from the reaction were loaded directly onto a non-denaturing polyacrylamide gel as described previously (35). Duplicate reactions were prepared with either radiolabeled ribozyme or radiolabeled molecular beacon.
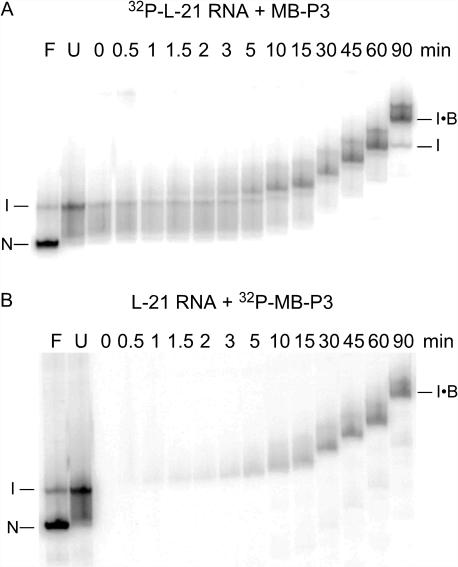
Figure 6.
Ribozyme unfolding and beacon hybridization by native gel electrophoresis. L-21 ribozyme (30 nM) was allowed to fold in splicing buffer plus 2 mM MgCl2, then mixed with 250 nM MB-P3 beacon at 37°C. Aliquots were loaded directly on a native 4% polyacrylamide gel containing 3 mM MgCl2 (35) at the times shown above the lanes. Duplicate reactions were carried out with (A) 32P-labeled RNA and unlabeled MB-P3 or (B) unlabeled RNA and 32P-labeled MB-P3. Lanes U, unfolded 32P-ribozyme in no Mg2+; F, folded 32P-ribozyme in 6 mM MgCl2. N, native ribozyme; I, misfolded intermediate; I•B, ribozyme–beacon complex. The gels were run continuously during the experiment.
When native 32P-labeled ribozyme in 6 mM Mg2+ is shifted to 2 mM Mg2+, it unfolds within 15 s to intermediates migrating above the native ribozyme (Figure 6A) (30). Complexes containing 32P-labeled beacon, however, appeared more slowly. The rate of hybridization measured by gel shift was 0.026 min−1 in 2 mM MgCl2, similar to that measured by fluorescence (0.036 min−1; Table 1). The ribozyme–beacon complexes migrated more slowly than the ribozyme folding intermediates alone, suggesting that most of the tertiary structure of the ribozyme must unfold in order for the beacon to bind. This is consistent with the weak affinity of the beacon for the ribozyme RNA, even in the absence of Mg2+ (Figure 4).
DISCUSSION
Fluorescent probe for RNA unfolding
Our results show that molecular beacons can be used as probes for folding and unfolding of the Tetrahymena ribozyme. The beacon MB-P3 reliably discriminated between the folded and misfolded or unfolded forms of the RNA, and the hybridization rate of the beacon decreased as the stability of the ribozyme structure at 37°C increased. Because the molecular beacon forms a stable complex with the ribozyme, it can trap the unfolded RNA, allowing the reaction dynamics to be studied even under conditions that favor the native state. This is analogous to the use of hydrogen exchange to monitor protein unfolding under native conditions (34). For ‘EX2’ conditions, the concentration of the beacon should be low so that its association with the RNA is slower than folding. Alternatively, if binding is much faster than the conformational change to be studied (‘EX1’), the extent of hybridization can be used to monitor the progress of the RNA folding reaction (11).
Structured unfolding intermediates
The hybridization kinetics of molecular beacon MB-P3 revealed that its complementary target in the 5′ strand of the P3 pseudoknot remains partially structured in splicing buffer, even in the absence of Mg2+. By comparing the hybridization equilibrium and kinetics of the ribozyme with an unstructured oligonucleotide, we estimate that an additional ΔGU of 2.8 kcal/mol is required to expose the MB-P3 target in splicing buffer. These results were initially unexpected, as P3 is mispaired in most misfolded intermediates. Native gel electrophoresis, however, showed that MB-P3 only binds the least compact forms of the ribozyme (Figure 6). Thus, the slow hybridization kinetics of the beacon can be explained by the extensive unfolding required to expose sequences in the ribozyme core to an oligonucleotide probe.
Ribozyme unfolding pathway
Both ensemble and single molecular experiments (29,36) have shown that a small fraction of the Tetrahymena ribozyme folds directly to the native state, while the majority folds through an ensemble of metastable intermediates. Although the ribozyme population is expected to unfold along the same parallel pathways, this has not been demonstrated biochemically. A mixture of intermediates with different electrophoretic mobilities is typically produced within the 15–30 s dead time of our native PAGE assay (29). However, the intermediates might be formed by partial unfolding from the native state to the intermediate (N→I), or by complete unfolding (N→U) and rapid collapse back to I.
That we still observe compact (fast migrating) intermediates at short times in the presence of the beacon suggests that at least some of the ribozyme unfolds directly to misfolded intermediates (N→I), as the completely unfolded RNA would have been trapped by the beacon. The burst of beacon hybridization (see Figure 5A) may include signal from a small percentage of the RNA that unfolds directly from N→U. We still observed this burst when the ribozyme RNA was pre-equilibrated with MB-P3 beacon in 6 mM MgCl2, before unfolding was triggered by the addition of EDTA (data not shown). These experiments support the possibility that the core of the ribozyme can reorganize within a compact state, as suggested by footprinting and small angle X-ray scattering results (32,37,39).
CONCLUSIONS
Molecular beacons are advantageous for monitoring RNA conformation, because they do not require modification of the RNA, they are relatively easy to design and they can be used in vivo or in cell-free extracts (14,15). A disadvantage of molecular beacons is that they require a relatively large complementary target sequence to ensure that the complex is stable. Hybridization of probes to the 15 nt target sequence used in these studies appears to disrupt most tertiary interactions in the Tetrahymena ribozyme. Thus, molecular beacons are likely to be most useful for detecting the rearrangement of RNA secondary structure during RNA folding, RNP assembly or RNA-dependent regulation.
Because molecular beacons can be designed to pair with single-stranded regions of assembly intermediates, they can select particular structures within a heterogeneous RNA population. With their low background signal, molecular beacons can detect small populations of intermediates that are not be observable with other probes. This feature allowed us to probe unfolding of the Tetrahymena ribozyme under conditions in which the native RNA is stable. Importantly, we find that the core of the misfolded ribozyme in 200 mM requires ∼2.8 kcal/mol to unfold, and that the RNA likely unfolds through different parallel pathways.
Acknowledgments
The authors thank Priya Ramaswamy for initial characterization of L-P3. This work was supported by a grant from the NIH (GM46686). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mandal M., Breaker R.R. Gene regulation by riboswitches. Nature Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 2.Madhani H.D., Guthrie C. Dynamic RNA–RNA interactions in the spliceosome. Annu. Rev. Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 3.Woodson S.A., Cech T.R. Alternative secondary structures in the 5′ exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991;30:2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- 4.Fedor M.J., Uhlenbeck O.C. Substrate sequence effects on ‘hammerhead’ RNA catalytic efficiency. Proc. Natl Acad. Sci. USA. 1990;87:1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrotta A.T., Been M.D. A toggle duplex in hepatitis delta virus self-cleaving RNA that stabilizes an inactive and a salt-dependent pro-active ribozyme conformation. J. Mol. Biol. 1998;279:361–373. doi: 10.1006/jmbi.1998.1798. [DOI] [PubMed] [Google Scholar]
- 6.Schultes E.A., Bartel D.P. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science. 2000;289:448–452. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- 7.Uhlenbeck O.C., Chirikjian J.G., Fresco J.R. Oligonucleotide binding to the native and denatured conformers of yeast transfer RNA(Leu)3. J. Mol. Biol. 1974;89:495–504. doi: 10.1016/0022-2836(74)90478-1. [DOI] [PubMed] [Google Scholar]
- 8.LeCuyer K.A., Crothers D.M. The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry. 1993;32:5301–5311. doi: 10.1021/bi00071a004. [DOI] [PubMed] [Google Scholar]
- 9.Sohail M., Akhtar S., Southern E.M. The folding of large RNAs studied by hybridization to arrays of complementary oligonucleotides. RNA. 1999;5:646–655. doi: 10.1017/s1355838299982195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Disney M.D., Childs J.L., Turner D.H. New approaches to targeting RNA with oligonucleotides: inhibition of group I intron self-splicing. Biopolymers. 2004;73:151–161. doi: 10.1002/bip.10520. [DOI] [PubMed] [Google Scholar]
- 11.Zarrinkar P.P., Williamson J.R. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 12.Cardullo R.A., Agrawal S., Flores C., Zamecnik P.C., Wolf D.E. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 1988;85:8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 14.Sokol D.L., Zhang X., Lu P., Gewirtz A.M. Real time detection of DNA. RNA hybridization in living cells. Proc. Natl Acad. Sci. USA. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bratu D.P., Cha B.J., Mhlanga M.M., Kramer F.R., Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santangelo P.J., Nix B., Tsourkas A., Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vet J.A., Majithia A.R., Marras S.A., Tyagi S., Dube S., Poiesz B.J., Kramer F.R. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl Acad. Sci. USA. 1999;96:6394–6399. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyduk T., Heyduk E. Molecular beacons for detecting DNA binding proteins. Nat. Biotechnol. 2002;20:171–176. doi: 10.1038/nbt0202-171. [DOI] [PubMed] [Google Scholar]
- 19.Bonnet G., Tyagi S., Libchaber A., Kramer F.R. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsourkas A., Behlke M.A., Rose S.D., Bao G. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 2003;31:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan J., Woodson S.A. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J. Mol. Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 22.Zaug A.J., Grosshans C.A., Cech T.R. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme–substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 23.Walstrum S.A., Uhlenbeck O.C. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990;29:10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- 24.Herschlag D., Cech T.R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 25.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 26.Bockisch B., Grunwald T., Spillner E., Bredehorst R. Immobilized stem–loop structured probes as conformational switches for enzymatic detection of microbial 16S rRNA. Nucleic Acids Res. 2005;33:e101. doi: 10.1093/nar/gni101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin P.Z., Pyle A.M. Stopped-flow fluorescence spectroscopy of a group II intron ribozyme reveals that domain 1 is an independent folding unit with a requirement for specific Mg2+ ions in the tertiary structure. Biochemistry. 1997;36:4718–4730. doi: 10.1021/bi962665c. [DOI] [PubMed] [Google Scholar]
- 28.Walter N.G., Burke J.M. Real-time monitoring of hairpin ribozyme kinetics through base-specific quenching of fluorescein-labeled substrates. RNA. 1997;3:392–404. [PMC free article] [PubMed] [Google Scholar]
- 29.Pan J., Thirumalai D., Woodson S.A. Folding of RNA involves parallel pathways. J. Mol. Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 30.Pan J., Deras M.L., Woodson S.A. Fast folding of a ribozyme by stabilizing core interactions: evidence for multiple folding pathways in RNA. J. Mol. Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 31.Takamoto K., He Q., Morris S., Chance M.R., Brenowitz M. Monovalent cations mediate formation of native tertiary structure of the Tetrahymena thermophila ribozyme. Nature Struct. Biol. 2002;9:928–933. doi: 10.1038/nsb871. [DOI] [PubMed] [Google Scholar]
- 32.Russell R., Zhuang X., Babcock H.P., Millett I.S., Doniach S., Chu S., Herschlag D. Exploring the folding landscape of a structured RNA. Proc. Natl Acad. Sci. USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shcherbakova I., Gupta S., Chance M.R., Brenowitz M. Monovalent ion-mediated folding of the Tetrahymena thermophila ribozyme. J. Mol. Biol. 2004;342:1431–1442. doi: 10.1016/j.jmb.2004.07.092. [DOI] [PubMed] [Google Scholar]
- 34.Bai Y., Sosnick T.R., Mayne L., Englander S.W. Protein folding intermediates: native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan J., Thirumalai D., Woodson S.A. Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics, and kinetics. Proc. Natl Acad. Sci. USA. 1999;96:6149–6154. doi: 10.1073/pnas.96.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuang X., Bartley L.E., Babcock H.P., Russell R., Ha T., Herschlag D., Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 37.Sclavi B., Sullivan M., Chance M.R., Brenowitz M., Woodson S.A. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 38.Rook M.S., Treiber D.K., Williamson J.R. An optimal Mg(2+) concentration for kinetic folding of the tetrahymena ribozyme. Proc. Natl Acad. Sci. USA. 1999;96:12471–124716. doi: 10.1073/pnas.96.22.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das R., Kwok L.W., Millett I.S., Bai Y., Mills T.T., Jacob J., Maskel G.S., Seifert S., Mochrie S.G., Thiyagarajan P., et al. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J. Mol. Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]