Abstract
We present a new support vector machine (SVM)-based approach to predict the substrate specificity of subtypes of a given protein sequence family. We demonstrate the usefulness of this method on the example of aryl acid-activating and amino acid-activating adenylation domains (A domains) of nonribosomal peptide synthetases (NRPS). The residues of gramicidin synthetase A that are 8 Å around the substrate amino acid and corresponding positions of other adenylation domain sequences with 397 known and unknown specificities were extracted and used to encode this physico-chemical fingerprint into normalized real-valued feature vectors based on the physico-chemical properties of the amino acids. The SVM software package SVMlight was used for training and classification, with transductive SVMs to take advantage of the information inherent in unlabeled data. Specificities for very similar substrates that frequently show cross-specificities were pooled to the so-called composite specificities and predictive models were built for them. The reliability of the models was confirmed in cross-validations and in comparison with a currently used sequence-comparison-based method. When comparing the predictions for 1230 NRPS A domains that are currently detectable in UniProt, the new method was able to give a specificity prediction in an additional 18% of the cases compared with the old method. For 70% of the sequences both methods agreed, for <6% they did not, mainly on low-confidence predictions by the existing method. None of the predictive methods could infer any specificity for 2.4% of the sequences, suggesting completely new types of specificity.
INTRODUCTION
Many pharmacologically important peptides in bacteria, fungi and some plants are synthesized nonribosomally by multimodular peptide synthetases (NRPS) (1,2). Prominent examples of such peptides are antibiotics, such as actinomycin, bacitracin, cephalosporins, penicillins and vancomycin, the antitumor peptide bleomycin and the immunosuppressant cyclosporin A. NRPS belong to the family of megasynthetases, which are among the largest known enzymes, with molecular weights of up to ∼2.3 MDa (∼21 000 residues) (3). They possess several modules, each of which contains a set of enzymatic domains that, in their specificity, number and organization, determine the primary structure of the corresponding peptide products (2) [see Figure 1; for a recent review on NRPS see Sieber and Marahiel (1) and Lautru and Challis (4)]. The adenylation domain (A domain), which is the subject of this study, specifically recognizes and activates one amino acid (or hydroxy acid) that will subsequently be appended to the nascent peptide chain by other NRPS domains. Based on the crystal structure of the phenylalanine activating A domain of the NRPS gramicidin synthetase A (GrsA), Conti et al. (5) determined 10 residue positions that are crucial for substrate binding and catalysis. These residues are within a radius of ∼5.5 Å around the phenylalanine bound in the active site. The predictive method described by Stachelhaus et al. (2) and Challis et al. (6) is based on the high structural conservation of the binding pocket, with a root mean square deviation (RMSD) of the Cα atoms of <1 Å (7), reflected by a relatively high mutual sequence similarity of 26–56% (8) of NRPS A domains. Therefore, Stachelhaus et al. (2) and Challis et al. (6) concluded that the 10 decisive residues of GrsA will line up with the corresponding positions of other A domains in a multiple sequence alignment, and can be extracted to form a ‘specificity-confering code’. The specificity of uncharacterized A domains can then be inferred based on the ‘code’ of domains with known specificity (6) or based on consensus sequences for each specificity (2). In this paper, we present a new method for predicting the specificity of A domains by machine learning using the physico-chemical fingerprint of the residues lining the active site of the enzymatic domain (8 Å around the bound substrate). The generality of the approach makes it applicable to the prediction of functional subspecificities of other classes of enzymes that share a conserved structure but catalyze different substrates (see Discussion). We use a state-of-the-art encoding of residues into feature vectors for machine learning based on the physico-chemical properties of the amino acids and utilize an up-to-date training dataset of A domains with known specificity that we have compiled from the literature.
Figure 1.
Module and domain structure of NRPS. Above (in the middle): one complete NRPS consisting of three modules. Below: enzymatic domains that are contained in a complete module: Cond: condensation domain, Adenyl: adenylation domain (A domain), N-Meth: N-methylation domain (optional, does not appear in all NRPS), PCP: thiolation domain (Peptidyl Carrier Protein domain), Epi: epimerization domain (optional). Other optional domains are heterocyclation, oxidation, reduction and formylation domains. The substrate specificity of the adenylation domain is the subject of this study.
MATERIALS AND METHODS
Acquisition of a collection of A domains with known specificity
The HMMER package (9) and self-written Perl scripts were used to search for NRPS in the protein databases UniProt/TrEMBL/Swiss-Prot (10,11), requiring the occurrence of a complete NRPS module with at least one condensation domain, one A domain (AMP-binding) and one peptidyl carrier domain [Pfam (12) accession numbers PF00668, PF00501 and PF00550]. The same software was also used to extract the AMP-domains from NRPS sequences to generate profile hidden Markov models (HMMs) of parts of domains and to extract certain positions of subdomains that were aligned against HMMER profiles. The programs ClustalW (13), T-Coffee (14) and MUSCLE (15,16) were used for generating multiple sequence alignments that were further manually curated for good alignment of core sequences, structural ‘anchors’ and putative constituents of binding pockets. Specificity annotations of extracted A domains were either obtained directly from the literature or by following references (PubMed links, gene name, organism, authors, etc.) given in database entries of proteins.
Extraction of homologous positions of A domains
The ‘Biochemical Algorithms Library’ [BALL, (17)] and a simple Python script were used to extract residues that have at least one atom at a given distance from the bound phenylalanine in the GrsA-Phe crystal structure [PDB ID 1amu, (5)]. In a multiple alignment of different A domains with the protein sequence of GrsA-Phe the residue positions that lined up with certain residues in GrsA-Phe were extracted; we ensured that all extracted residues lie in conserved gap-free segments, to allow for a reliable inference of their structural and functional relevance (see Figure 2 for illustration).
Figure 2.
Phenylalanine bound to gramicidin synthetase A activation domain. (a) The 10 residues (green) that are in direct contact with the substrate phenylalanine (ball and stick representation) are shown. These 10 residues are the basis for the specificity prediction method by Stachelhaus et al. (2). (b) Same as in (a) but the residues are in the space filling representation. (c) The residues in green (at a distance of up to 5.5 Å from phenylalanine) are surrounded by all the 34 residues (purple) at a distance of up to 8 Å from phenylalanine. The predictive method described here bases on these 34 amino acids and encodes them by their physico-chemical properties. Representations were created using BALLView (17, www.ballview.org).
Processing of the collection of A domains for machine learning
Starting with the current set of A domain sequences with known specificity (as described above), the 34 residues at a distance of up to 8 Å from the bound phenylalanine in GrsA were extracted and duplicate sequences were removed; sequences with similar specificities (see Results and Discussion) were clustered to composite specificities. Clusters comprising fewer than five sequences were discarded.
Support vector machines (SVMs)
Theory
SVMs are playing an increasingly important role in the field of computational biology. For an in-depth overview of the current research and applications to computational biology see Schölkopf et al. (18). We will give a short introduction here. Assume that we have a series of examples, each associated with a number d of features (either numerical or binary values), then we can treat each example simply as a d-dimensional vector in a d-dimensional space ℒ. If we want to construct a binary classification of the examples, i.e. label each d-dimensional data point as ‘positive’ or ‘negative’, a simple and intuitive way would be to construct a separating (hyper)plane, which separates the positive and negative data points. If the data are linearly separable, a separating hyperplane can be found, which maximizes the distance between the data points that are closest to the hyperplane, the so-called support vectors (SVs), and the hyperplane itself. Once we have determined these points that ‘support the plane’ we can write down a decision function that will assign a label to any new data point (+ or −).
The classical SVMs are ‘inductive’ SVMs. Here the training data that are used to build the model should ideally cover the whole problem space; the model is then used to predict the labeling of new data points. In most biological datasets the number of labeled data points is rather small, but a large number of unlabeled data points (e.g. unannotated proteins) is available. To take advantage of these additional unlabeled data, the so-called ‘transductive’ SVMs (TSVMs) have been developed (19,20). To address the problem of learning with unlabeled data (often called ‘semi-supervised’ or ‘transductive learning problem’), TSVMs assume that the missing labels of the unlabeled data points are consistent with their positions in the hyperspace in two aspects: (i) nearby points and (ii) points on the same structure (typically referred to as a cluster or a manifold) are likely to share the same label (21).
Performance estimates
To assess the accuracy of any classifying algorithm there exist several statistics on the number of true positive, false positive, true negative and false negative predictions [TP, FP, TN and FN; see Baldi et al. (22) for a review]:
More precisely, the error rate gives the fraction of data points that is classified incorrectly, the recall gives the fraction of really positive data points that is contained in the predicted positives, the precision specifies the fraction of TPs in all as positive predicted data points. MCC uses all four numbers (TP, TN, FP and FN), is symmetric with respect to FP and FN, and may often provide a much more balanced evaluation of the prediction than the above statistics (22).
Two similar tests are widely used for determining the above parameters: leave-one-out (loo) tests and x-fold cross-validations (with x typically 3, 5 or 10). In a loo test the predictive model is trained on a dataset that has been reduced by one data point. The generated model is then used to give a prediction for the removed data point. The whole procedure is repeated for each single data point of the set. In a, say, 5-fold cross-validation the dataset is divided randomly into five parts; one fifth of the dataset is removed, the model is trained on the rest, the so-called training data, and the prediction is made for the fifth, the so-called test data; the procedure is repeated for all the remaining one-fifth of the dataset. Initially, we evaluated our models for the different composite specificities using both x-fold cross-validation (3-, 5- and 10-fold, each three times repeated with randomized splits) and loo. Since both tests yielded extremely similar results, here we report only on the results of the loo, the most fine-grained form of cross-validation. A more thorough evaluation of the accuracy would require two levels of cross-validation (i.e. nested cross-validation) (23). However, as the models considered here are relatively simple and do not allow for strong fitting of the data, use of a a straight loo test is sufficient for our purposes.
Feature representation based on physico-chemical properties of amino acids
From each A domain we extracted a signature of 34 amino acids. This consisted of all residues with at least one atom ≤8 Å from the bound substrate. Residues of the A10 core motif (8) were not included because they are extremely highly conserved and do not vary between different specificities (NGK, K=Lys517). We encoded each amino acid by normalized real values representing their physico-chemical properties. We used amino acid indices from AAindex (24–26) and Neumaier et al. (27, http://www.mat.univie.ac.at/~neum/software/protein/aminoacids.html) to describe
the number of hydrogen bond donors (28),
volume (32),
secondary structure preferences for beta-turns, beta-sheets and alpha-helices (33),
hydrophobicity with a three-dimensional vector (27) and
the isoelectric point (29).
We standardized the values in such a way that the interval of ±1 SD (calculated from the value distribution of each AAindex file) is projected onto the interval of ±1. StandardizedValue = (IndexValue − MeanIndexValue)/StandardDeviation, and thus obtained a vector of 408 features for each A domain. The choice of these properties is discussed in Results and Discussion.
SVM implementation
In this study, we used the program package SVMlight (34, svmlight.joachims.org) for training SVM models on data and classification of data. This program also implements algorithms for training large TSVMs. The algorithm proceeds by solving a sequence of optimization problems lower-bounding the solution using a form of local search. For details see Joachims (20). SVMlight can efficiently compute loo testing; loo provides ‘almost unbiased’ estimates for error rate, recall (= sensitivity Sn) and precision (= specificity Sp) (svmlight.joachims.org).
Choice of optimal kernel function and parameters
SVMlight provides linear, polynomial, radial basis (RBF) and sigmoid kernel functions. Two parameters, C and j, are common to all kernel functions. The parameter C is the penalty that is assigned to erroneous training points that cannot be classified correctly. If the features are normalized as described above one can put C = 1 as a starting point for a grid search around this value (in this study C ∈ {1/32, 1/16, …, 1, 2, …, 32}). The cost-factor j determines how training errors on positives examples outweigh errors on negative examples. The usual initial estimation j0 [see Morik et al. (35)] of the cost-factors by the fraction of negative over positive training examples was also used in this study, with values of j0 in the order of 10, depending on the ratio of the dataset. To determine the optimal value for j, a grid search was applied as well with j ∈ {1/32 j0, 1/16 j0, …, 1j0, 2j0, …, 32j0}. The nonlinear kernel functions have additional parameters. The RBF kernel function has an additional parameter σ, with σ2 ≈ mean(‖xi − xj‖2), that is approximately the mean of the squared Euclidian distances of all pairs of data points. To be precise SVMlight uses a parameter γ for the RBF kernel, with γ = 1/2σ2. The approximation given above can then be used as a starting point for a grid search to find the best value for σ2. In this study the same factors as for the optimization of C and j were used, multiplied by the initial approximation of γ.
Multiclass problem
After having trained the SVM models for each composite specificity it is necessary to combine the predictions of all models to one single prediction for the ‘large’ and ‘small’ clusters. The most widely used method [according to Vert and co-workers (18)] is to combine the scores (= distance of the classified point to the hyperplane) by a max rule: the SVM that outputs the largest score is used to assign the specificity to the unknown sequence. If all single SVMs return ‘negative’, then no final prediction will be possible. This does not necessarily mean that the unknown sequence has a very ‘exotic’ specificity, but possibly that the single model of the actual specificity might give a false negative answer. Because the quality of the single models differs, we decided to multiply the scores by the squared MCC value of the model. As the MCC is a quality measure close to 1 for very good models, and decreases with the reliability of the models, this allows for a reasonable scaling of the scores. In the relatively rare case of several ‘positive’ answers, the one with the highest scaled score will be used in the evaluation of the overall predictive error of the combined model. However the predictive program (NRPS predictor) will list all the models that return a positive value.
RESULTS AND DISCUSSION
A current set of annotated specificities
Because the large majority of NRPS sequences deposited in public sequence databases are poorly annotated and the annotation quality and syntax differs from author to author, keyword-based search strategies in an automated manner are infeasible. Therefore we first manually collected all 160 A domain sequences used by Stachelhaus et al. (2). We then scanned the UniProt/TrEMBL/Swiss-Prot protein database (10,11) with profile HMMs for complete NRPS modules. We required modules to be complete (one condensation-, one A- and one pp-binding domain), as we hereby avoided extracting very similar enzymes, such as acyl-CoA ligases. For the 245 detected sequences we followed the PubMed (www.pubmed.gov) literature references in the UniProt entry or tried to find the associated articles via PubMed (searching for gene name, organism, authors, etc.). Thus we were able to find 227 additional A domain sequences. We joined this dataset with the sequences of J. Ravel's NRPS BLAST server (6) and finally obtained a set of 397 A domains with known specificity (fully listed in the Supplementary Data). We required that the specificity annotation was based on experimental evidence, by an ATP-PPi-exchange reaction (36) or, when the specificity was inferred by the co-linearity rule based on the ordered composition of the peptide product, that the inference was confirmed by an unambiguous match with a known ‘specificity-conferring code’ (2,6) of another A domain. The number of occurrences of the different specificities in these 397 A domain sequences are depicted in Table 1.
Table 1.
Distribution of the 397 adenylation domains with known specificity on their substrates
| Specificity | Occurrence | Specificity | Occurrence | Specificity | Occurrence |
|---|---|---|---|---|---|
| 3-me-Glu | 1 | Dhb | 15 | Phe | 11 |
| 4pPro | 1 | Dhpg | 8 | Phg | 1 |
| Aad | 10 | Dht | 4 | Pip | 5 |
| Abu | 2 | D-lyserg | 1 | Pro | 16 |
| Aeo | 1 | Gln | 8 | Sal | 2 |
| Ala | 34 | Glu | 12 | Ser | 22 |
| Ala-b | 3 | Gly | 12 | Ser-Thr | 2 |
| Ala-d | 1 | His | 1 | Tcl | 1 |
| Alaninol | 1 | Hpg | 19 | Thr | 24 |
| Arg | 5 | Hyv-d | 1 | Trp | 3 |
| Asn | 14 | Ile | 11 | Tyr | 14 |
| Asp | 12 | Iva | 7 | Val | 27 |
| Bht | 7 | Leu | 31 | Valhyphaa | 1 |
| Bmt | 1 | Lys | 5 | Vol | 1 |
| Cys | 23 | Lys-b | 2 | ||
| Dab | 4 | Orn | 10 |
Besides the proteinogenic amino acid in three letter code there are the following known rare specificities: 3-me-Glu, 3-methyl-glutamate; 4pPro, 4-propyl-proline; Aad, 2-amino-adipic acid; Abu, 2-amino-butyric acid; Aeo, 2-amino-9,10-epoxy-8-oxodecanoic acid; Ala-b, β-alanine; Ala-d, d-alanine; Alaninol; Bht, beta-hydroxy-tyrosine; Bmt, (4R)-4[(E)-2-butenyl]-4-methyl-l-threonine; Dab, 2,4-diamino-butyric acid; Dhb, 2,3-dihydroxy-benzoic acid; Dhpg = Dpg, 3,5-dihydroxy-phenyl-glycine; Dht, dehydro-threonine = Dhbu = 2,3-dehydroaminobutyric acid; D-lyserg, d-lysergic acid; Hpg, 4-hydoxy-phenyl-glycine; Hyv-d, 2-hydroxy-valeric acid; Iva, isovaline; Lys-b, β-lysine; Orn, ornitine; Phg, phenyl-glycine; Pip, pipecolic acid; Sal, salicylic acid; Tcl, (4S)-5,5,5-trichloro-leucine; Valhyphaa, valine or hydrophobic amino acid; Vol, valinol.
Inferring functional and structural relevance of residues in a structurally conserved context
When comparing firefly luciferase (another AMP-binding enzyme that activates luciferin) and GrsA, a structure-based alignment reveals that 67% of the alpha-carbon positions are conserved to within 3 Å. The RMSD is 2.6 Å, although both enzymes share only 16% sequence similarity (2,5,7). Furthermore, the RMSD calculated over the Cα atoms enclosed in a sphere of radius 9 Å, centered at the GrsA residue Asp-235 in the active site, is 0.95 Å (7). Owing to the much higher similarity between GrsA and other NRPS A domains [between 30 and 80%; (37)] the conformation of their mainchains is likely to be even more similar, particularly around the substrate-binding pocket. Therefore, in a multiple sequence alignment of other NRPS A domains with GrsA those residues that align with the residues that line the active site can be expected to be involved in the specific substrate recognition and binding of the homologue A domain. To make sure that we included all residue positions that might have an interaction with the substrate, or might be influenced by, or adapted to, the residues that interact directly with the substrate, we decided to extract all residues up to a distance of 8 Å from the substrate in GrsA. A steric cell of 8 Å was similarly used by Lilien et al. (38) for an energy simulation of the GrsA active site. In Figure 2 we illustrate the residues at a distance up to 5.5 Å and 8 Å in immediate and mediate contact with the substrate phenylalanine, respectively.
Clustering of sequences with similar specificities
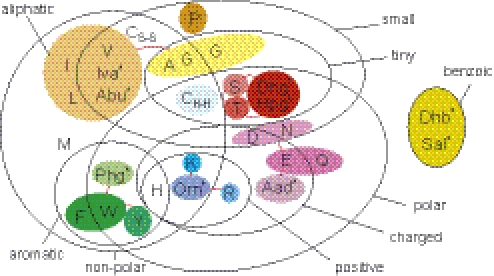
For a reliable prediction of specificities, one ideally desires to have a training set of sequences for each distinct specificity. In reality we often find A domains with considerably high side specificities that lead to alternate peptide products that differ at the corresponding position, such as in the case of tyrocidine: in the tyrocidine biosynthesis operon [Bacillus brevis, TYCB_BREPA) (39) the A domain TycB_m3 activates l-tryptophan with 100% relative activity [in an ATP-PPi-exchange reaction (36)], and l-phenylalanine with 48%, but is annotated as l-phenylalanine-activating because (d-)phenylalanine was found in the product. It is also possible that in biochemical specificity tests [ATP-PPi-exchange reaction (36) with recombinant A domains] a considerable side specificity might be detected but the alternative substrate is not incorporated in vivo, for example, because of sterical reasons in the further processing of the nascent peptide: the A domain of BarD of the barbamide (bar) biosynthetic gene cluster has 100% specificity for leucine and valine, and 80% for trichloroleucine but the in vivo incorporation of valine was experimentally excluded (40). Because there might exist considerably high side specificity, we addressed this problem by clustering specificities for amino acids with very similar physico-chemical properties. For this clustering we also took observations of Challis et al. (6) into account. Challis et al. (6) analyzed the predicted binding pockets of most A domains known to date. Based on the ‘code’ of eight amino acids closest to the substrate they pointed out that specificities for physico-chemically similar substrates often only differ in single residues. An experimentally verified example is the directed mutagenesis of Ala322Gly in GrsA, increasing its specificity to Trp (2). We decided to consider two different kinds of clusterings: grouping specificities into few large clusters and into more small clusters. Forming larger clusters, i.e. putting together more different but similar specificities into one composite specificity, has the advantages of (i) obtaining larger positive datasets for SVM training (yielding more reliable models), (ii) covering a larger spectrum of sequence variations, (iii) covering a larger subspace in the hyperspace, (iv) lowering the risk of over-fitting and, finally, (v) allowing for recognition of new substrates that are very similar to the substrate specificities in the cluster. However, forming smaller clusters by clustering similar specificities only where they are necessary (e.g. Phe/Trp, see above) has the advantage of allowing for more concrete/precise predictions, but at a higher risk of over-fitting owing to a reduced number of positive training data. Table 2 and Figure 3 illustrate the specificities that have been clustered.
Table 2.
Clustering of amino acids with similar physico-chemical properties and/or similar substrate binding pockets (6) into composite specificities
| Large clusters | Small clusters | ||
|---|---|---|---|
| Gly (12), Ala (20), Val (22), Leu (22), Ile (7), Abu (2), Iva (7) | Apolar, aliphatic side chains | Gly (12), Ala (20) | Tiny size, hydrophilic, transition to aliphatic |
| Val (22), Leu (22), Ile (7), Abu (2), Iva (7) | Aliphatic, branched hydrophobic side chain | ||
| Ser (13), Thr (16), Ser/Thr (1), Dhpg (7), Hpg (13) | Aliphatic chain or phenyl group with -OH | Ser (13) | Serine-specific |
| Thr (16) | Threonine-specific | ||
| Dhpg (7), Hpg (13) | Polar, uncharged (hydroxy-phenyl) | ||
| Phe (11), Trp (3), Phg (1), Tyr (12), Bht (6) | Aromatic side chain | Phe (11), Trp (3) | Unpolar aromatic ring |
| Tyr (12), Bht (6) | Polar aromatic ring | ||
| Asp (8), Asn (13), Glu (9), Gln (6), Aad (7) | Aliphatic chain ending with H-bond donor | Asp (8), Asn (13) | Asp-Asn-hydrogen bond acceptor |
| Glu (9), Gln (6) | Glu-Gln-hydrogen bond acceptor | ||
| Aad (7) | 2-Amino-adipic acid | ||
| Cys (17) | Polar, uncharged (aliphatic chain with -SH group at the end) | – | – |
| Orn (8), Lys (3), Arg (5) | Long positively charged side chain (aliphatic chain with -NH2 group at the end) | Orn (8) | Orn and hydroxy- Orn specific |
| Arg (5) | Arg-specific | ||
| Pro (16), Pip (4) | Cyclic aliphatic chain with polar group | Pro (16) | Pro-specific |
| Dhb (9), Sal (2) | Hydroxy-benzoic acid derivates (no amino group) | No small cluster, no separation possible | – |
The numbers in parentheses denote the counts of domains with unique 8 Å sequence. Please note that the division of large into small clusters was not always possible owing to the small amount of available training data. Also see Figure 3.
Figure 3.
Venn diagram grouping amino acids by common physico-chemical properties according to Taylor (49). The colored sets show how similar amino acids have been clustered to composite specificities of A domains. To get larger clusters several smaller clusters were joined, as indicated by red lines connecting colored sets. This clustering is based on conclusions by Challis et al. (6) on cross-specificities of A domains and own groupings according to physical–chemical properties. An asterisk indicates rare non-proteinogenic amino acids, for abbreviations see Table 1.
SVMs: particularities
Feature representation based on physico-chemical properties of amino acids
From each A domain we extracted the signature of 34 amino acids at ≤8 Å from the bound substrate (see Materials and Methods). Each amino acid was encoded by 12 different values representing its physico-chemical properties, obtaining a vector of 408 features for each sequence. Chemical properties chosen were the number of hydrogen bond donors, the polarity and the hydrophobicity of the residues, and the isoelectric point; physical properties were volume and the preferences to appear in different secondary structures. We chose these properties because they are the key factors in the decision on how well a given substrate might bind to the defined set of residues and thus make sense, biologically, chemically, and physically. If the positions of the active site residues are given and fixed in space (as we conclude they are here) then these properties describe well the inside of the active site keyhole. If for any reason (e.g. a very large/different substrate) the binding pocket structure is altered, then we expect to see residues that have a different secondary structure preference at the positions that we extract from the profile alignment. Therefore, it makes sense also to encode the secondary structure preferences.
SVM implementation
In this study, we based our predictions on SVMs that implement the intuitive idea of separating two data ‘clouds’ by a geometric plane (see Materials and Methods for details), as implemented in SVMlight. We used an innovative variant of SVMs, the so-called TSVMs, that not only take into account the labeled training data but also integrate unlabeled data, in our case sequences with unknown specificity. We tried different kernel functions in our experiments, including linear, polynomial, radial basis and sigmoid functions. In a grid search we determined the optimal kernel parameters using SVMlights built-in loo test functionality. For linear and radial kernel functions (RBFs) we got the best results (for error rate, specificity, sensitivity and MCC), varying from case to case. When the linear kernel was equally good or better, then we preferred it over the RBF kernel for simplicity of the models, else we chose the RBF kernel. After the determination of the optimal kernel function and parameters, we gathered 646 uncharacterized A domain sequences from UniProt (10), as described in Materials and Methods.
For each cluster of composite specificity, we prepared a feature file with the sequences belonging to this specificity labeled +, all other sequences with different but known specificity labeled −, and the uncharacterized sequences labeled 0 (i.e. unlabeled). We used SVMlight in transductive mode to build models. With a self-written Perl script we ran loo cross-validation to check error rate, recall (sensitivity), precision (specificity) and MCC. For each cluster we trained a TSVM, as described above, to obtain a model for each composite specificity.
SVM quality assessment
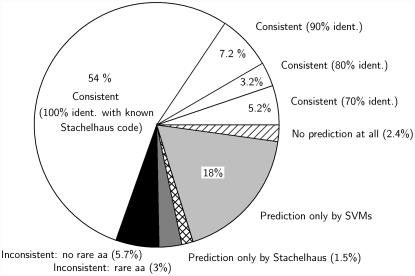
The number of available positive training data points varied between the different clusters and was sometimes quite small. Although the quality of models in machine learning depends on the amount of training data available, previous findings show that, besides the highly conserved overall structure of the binding pocket common to all A domains, the composition of residues lining the active site of sequences with the same specificity are even more conserved (2), which should allow one to obtain relatively good models. In a rigorous quality assessment of the generated models (Table 3), we could show that most SVM models yield good to very good results (MCC = 0.85–1). Some yield very poor results, such as the SVM with the composite specificity for Phe=Trp=Phg=Tyr=Bht (MCC = 0.85) or for Gly=Ala (MCC = 0.84). An explanation for the low performance of the model for very large aromatic amino acids could be that there exist a few, but spatially very different configurations of the binding pocket, for which it is impossible to generate one discriminative model. The problems with the glycine/alanine model could lie in the small size of the substrates; as Challis et al. (6) already suggested, there might be many degenerate solutions to activate these substrates. Similarly, the quality of the model for proline specificity is poor. As Lautru et al. (1) pointed out, only 4–5 residues at the top of the selectivity pocket are likely to be in direct contact with proline's relatively compact side chain based on homology modelings of the binding pockets. We obtained bad performance for models that aim at distinguishing between phenylalanine/tyrosine and all other amino acids, because there are sequences known, such as tyrocidine synthetase TycC_M3, with specificities for both Tyr and Trp, and others that have a specificity for Phe and Trp. A tabular overview of all the results of the quality assessment of the models is shown in Table 3. As the predictive quality of the models was estimated by loo tests on the set of sequences with known specificity (training data) one needs to check whether the test data (sequences with unknown specificities) are drawn from the same distribution. To check this we compared the mean pairwise distance of the training data with the mean pairwise distance between the training and test data. The mean of the Euclidian distance within the training data was 18.9 (SD 2.9) and the mean distance between the training and test data was 18.8 (SD 2.5). Because both distributions are very similar it is safe to assume that the performance of our models on the test data will be similarly good. To finally obtain one model for all ‘large’ and all ‘small’ clusters we score the results of different models using the returned distance of the data point to the hyperplane multiplied by the square of the MCC. This scaling makes sense because the MCC reflects the reliability of each model (see Materials and Methods). Our ‘large’ clusters cover 282 of the 300 specificities, the ‘small’ clusters cover 273 sequences. We ran an loo test on both multi-class models. The ‘large’ cluster model gave 260 correct predictions, 30 incorrect predictions and 10 times it gave no prediction, corresponding to a total error rate of 13%, or 7.8% on the sequences that the models were trained for. The ‘small’ cluster model gave 231 correct predictions, 44 incorrect predictions and did not decide for 25 sequences, corresponding to an error rate of 23%, or 15%, respectively. Given the set of 300 unique 8 Å signature sequences, we also evaluated the performance of a sequence-based model that uses the 34 amino acids signature. To first get a overview of the clustering of the 34 amino acid sequences, we built a phylogenetic tree [using a maximum-likelihood method (IQPNNI) (41,42), visualized with SplitsTree (43) (www.splitstree.org), see Supplementary Data]. When we analyze the tree, we see—overall—a clustering of similar specificities. Looking at details, we detect some ‘incompatible’ specificities in some subtrees, for example, in one where most A domains of the fungus Trichoderma virens TEX1 gene (Uniprot accession no. Q8NJX1) cluster despite their different specificities. The reason might be that, by increasing the number of amino acid positions from 8 or 10 to 34, we also capture more of the species phylogenetic signal. We also tested the performance of a BLAST (44) search using as database the 300 sequences with known specificity. Using the closest BLAST match to infer the specificity, 233 sequences would have been annotated correctly, corresponding to an error rate of 22.3%. This indicates that BLAST could be helpful especially for rare specificities and, therefore, we plan to integrate it in a future version of the NRPSpredictor. However, the BLAST strategy is inferior to the SVM strategy because it cannot build a generalizing SVM model for a specificity, but only finds the closest sequence(s). To assess the accuracy of the predictions on ‘new’ sequences, that are not very similar to the others with known specificity, we re-trained models only with sequences with a certain minimum distance and still got acceptable results (see Supplementary Data). To further examine the reliability and usefulness of our new method, we applied our prediction program to all 1230 adenylation domains in the June 2005 version of UniProt (10) (the proteins were extracted from the database as described in Materials and Methods). We compared the consistency of our predictions with the predictions based on the ‘specificity-conferring code’. (To automate this method by Stachelhaus et al. (2) we automatically extracted the 10 amino acid code and scored it against the collection of 10 amino acid codes of known specificities, requiring at least the identity of 7 of the 10 positions for a ‘match’.) For 70% of the sequences both predictors gave consistent predictions, which underlines the usability of our approach. The new SVM-based method could predict the specificities for 18% of the sequences, where the sequence-based method by Stachelhaus et al. (2) cannot. Thus, there are 2.4% for which neither method gives a prediction. For 1.5% only the traditional method could give a prediction. About 8.8% of the sequences are inconsistently classified by the old and the new method. Of them 3% are rare specificities that the SVMs were not trained for. An illustration of these comparisons are shown in Figure 4. If we accept only ≥80% matches for a positive ‘Stachelhaus’-prediction, we observe that the number of sequences for which no predictor can predict anything increases by 2.5% and the specificities that can only be predicted by the TSVMs increase by 8%. We also observe that the number of inconsistent predictions drops by 6.5%, the number of sequences only predicted by the Stachelhaus method drops by 1% and the number of consistent predictions decreases by 4%. We interpret this observation such that the Stachelhaus predictions at 70% are less reliable and give rise to more inconsistent predictions.
Table 3.
Results of cross-validating the different SVMs by loo
| Specificity of SVM | Positive training points | Kernel type | Leave-one-out cross-validation | Quality of SVM | |||
|---|---|---|---|---|---|---|---|
| Error | Sn | Sp | MCC | ||||
| Large clusters | 282 Labeled and 664 unlabeled data points (18 + 646) | ||||||
| Dhb=Sal | 11 | l | 0.4 | 100 | 92 | 96 | ++ |
| Asp=Asn=Glu=Gln=Aad | 43 | r | 1.4 | 100 | 91 | 95 | ++ |
| Pro=Pip | 20 | r | 0.7 | 90 | 100 | 95 | ++ |
| Cys | 17 | r | 0.7 | 100 | 89 | 94 | ++ |
| Ser=Thr=Dhpg=Dpg=Hpg | 50 | r | 2.5 | 96 | 91 | 92 | ++ |
| Gly=Ala=Val=Leu=Ile=Abu=Iva | 92 | r | 4.3 | 95 | 93 | 90 | + |
| Orn=Lys=Arg | 16 | l | 0.7 | 88 | 88 | 87 | + |
| Phe=Trp=Phg=Tyr=Bht | 33 | r | 3.2 | 88 | 85 | 85 | 0 |
| Small clusters | 273 Labeled and 673 unlabeled data points (27 + 646) | ||||||
| Dhb=Sal | 11 | l | 0 | 100 | 100 | 100 | ++ |
| Aad | 7 | l | 0 | 100 | 100 | 100 | ++ |
| Glu=Gln | 15 | l | 0 | 100 | 100 | 100 | ++ |
| Dhpg=Dpg=Hpg | 20 | l | 0.4 | 100 | 95 | 97 | ++ |
| Ser | 13 | l | 0.4 | 92 | 100 | 96 | ++ |
| Cys | 17 | l | 0.7 | 100 | 89 | 94 | ++ |
| Thr | 16 | l | 0.7 | 94 | 94 | 93 | ++ |
| Pro | 16 | r | 0.7 | 94 | 94 | 93 | ++ |
| Asp=Asn | 21 | l | 1.1 | 90 | 95 | 92 | ++ |
| Val=Leu=Ile=Abu=Iva | 60 | l | 2.9 | 92 | 95 | 91 | + |
| Orn | 8 | l | 0.7 | 88 | 88 | 87 | + |
| Gly=Ala | 32 | l | 3.3 | 81 | 90 | 84 | 0 |
| Tyr | 18 | r | 2.2 | 94 | 77 | 84 | 0 |
| Arg | 5 | l | 0.7 | 80 | 80 | 80 | 0 |
| Phe=Trp | 14 | l | 3.7 | 57 | 67 | 60 | 0 |
The more training data that are available the more reliable the trained predictive models are. The ‘quality of SVM’ in the last column, therefore, is a qualitative measure for the MCC. Kernel type l stands for linear kernel and r stands for radial basis function kernel. Error rate, sensitivity (Sn), specificity (Sp) and Mathews correlation coefficient (MCC) are given in percentage.
Figure 4.
Results of a comparison of the new SVM-based method with the sequence-based prediction method based on the ‘specificity-confering code’ by Stachelhaus et al. (2) and Challis et al. (6) (For simplicity we refer to the latter as the ‘Stachelhaus method’): of the 1230 adenylation domains (with HMMER automatically extracted from the June 2005 version of UniProt) 70% or 858 obtained consistent predictions by both predictors (white sectors). For most of these consistent predictions (54% of the total or 666) the Stachelhaus method was based on an exact match with a known ‘specificity-conferring code’, the others had at least an 70% match. To 2.4% or 29 sequences none of the predictors can assign any specificity (no match ≥70%, diagonal hatches). An 18% or 217 sequences could be classified only by the SVMs and not by the Stachelhaus method (light gray sector), and 18 A domains (1.5%) could not be classified by the SVMs but by the Stachelhaus method (cross-hatched), two of them are rare specificities. The Stachelhaus predictions for the rest are mainly based on 70% matches to known specificity ‘codes’. For 108 sequences (8.8%) the predictions were inconsistent but 38 of them (3% of the total, gray sector) had matches to rare amino acids that were not used for training the SVMs. The remaining 70 incompatible predictions were mainly based on ≤80% identity matches with known ‘specificity-conferring codes’ (black sector).
CONCLUSION
During the past 5 years SVM-based machine learning has been extensively applied within the field of bioinformatics, such as to the classification of genes and proteins, prediction along the DNA or protein strand, microarray gene expression and to other problems [for a recent review see Noble (45)]. Here we describe a new application of SVMs to functional subtyping of the substrate specificities of a class of enzymes based on the physico-chemical fingerprint of the residues that form the substrate-binding pocket. To take advantage of the abundant amount of unannotated data, we use an implementation (20) of TSVMs, introduced by Vapnik (19) in 1998. TSVMs have been shown to be superior to inductive SVMs in a similar application, the prediction of receptor binding compounds based on three-dimensional properties of the molecule (46,47), where also a large number of unlabeled data were available. Our results prove a high reliability of the predictions, even though the currently available amount of training data is relatively low, leaving room for further improvement with a growing number of annotated A domains. When applying our method and the sequence-based method (2,6) to a set of over 1000 adenylation domains currently detectable in UniProt (10), in summary, the new method can predict the specificities for 18% more sequences than the old one, while being consistent within the 70% that both methods predict. For 2.4% of the sequences, none of the method can give any prediction. Moreover, the inconsistent predictions, where both methods disagree have a large amount of ‘Stachelhaus’ predictions at 70% identity. This illustrated that there is still a large amount of sequences for which a prediction is very uncertain or impossible. Interestingly, we can observe that in those ‘difficult’ sequences the ratio of eukaryotic sequences is more than two times higher than it is on average, indicating that the eukaryotic A domains might have developed alternative substrate-binding patterns. In cases where both methods give consistent predictions, the method by Stachelhaus et al. (2) gives a more concrete prediction, since it decides for one specificity, whereas our method decides for one composite specificity that usually stands for more than one substrate. Nevertheless, we would like to emphasize that the combination of the ‘old’ and our method gives a new powerful prediction tool that can be directly used by the scientists working in the field. Our results confirm the applicability of the SVM-based strategy for substrate specificity prediction and that it should be considered also for the prediction of the subtypes of other enzymes, e.g. nucleotidyl cyclases, protein kinases, lactate/malate dehydrogenases and trypsin-like serine proteases, a selection used by Hannenhalli and Russell (48).
AVAILABILITY OF THE PROGRAM
An implementation of the described method, called the NRPSpredictor, is freely available to the NRPS community via our server reachable at http://www-ab.informatik.uni-tuebingen.de/software. The web-interface allows one to upload or paste-in the (multi-)fasta file(s) of the protein sequence(s) to be analyzed. The adenylation domains are automatically extracted, as well as the residues of the ‘specificity-confering code’ and the residues 8 Å around the substrate. The predictions of each model for each cluster of composite specificity are given as well as the best matches of the ‘specificity-confering code’ to known specificities. The results are presented as an HTML output as well as a short text-based report and a tabular output that can be viewed with a spreadsheet program.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors would like to thank Andreas Wietzorrek (Department for Microbiology/Biotechnology, University of Tübingen, Germany) for help with the collection of annotated A domains, Jacques Ravel (TIGR, Rockville, MD) for kindly providing us with the sequences of the NRPS BLAST server, Timothy Davison (Max Planck Institute for Biological Cybernetics, Tübingen, Germany) for the valuable discussion, Andreas Biegert (University of Tübingen, Germany) who implemented the toolbox webfrontend, where the NRPSpredictor is integrated, as well as Esther Rheinbay (University of Tübingen, Germany) for the useful hints during correction of the manuscript. Author TW was supported by a grant of the German Ministry for Education and Research (BMBF-Fkz: 0313105). Author CR's and DHH's funding as well as the payment of the Open Access publication charges for this article was provided by the Deutsche Forschungsgemeinschaft (funding of the ZBIT, BIZ 1/1-2 and BIZ 1/1-3).
Conflict of interest statement. None declared.
REFERENCES
- 1.Sieber S.A., Marahiel M.A. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 2.Stachelhaus T., Mootz H.D., Marahiel M.A. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 3.Wiest A., Grzegorski D., Xu B.-W., Goulard C., Rebuffat S., Ebbole D.J., Bodo B., Kenerley C. Identification of peptaibols from Trichoderma virens. J. Biol. Chem. 277:20862–20868. doi: 10.1074/jbc.M201654200. [DOI] [PubMed] [Google Scholar]
- 4.Lautru S., Challis G.L. Substrate recognition by nonribosomal peptide synthetase multi-enzymes. Microbiology. 2004;150:1629–1636. doi: 10.1099/mic.0.26837-0. [DOI] [PubMed] [Google Scholar]
- 5.Conti E., Stachelhaus T., Marahiel M.A., Brick P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidins. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Challis G.L., Ravel J., Townsend C.A. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 7.Di Vincenzo L., Grgurina I., Pascarella S. In silico analysis of the adenylation domains of the freestanding enzymes belonging to the eucaryotic nonribosomal peptide synthetase-like family. FEBS J. 2005;272:929–941. doi: 10.1111/j.1742-4658.2004.04522.x. [DOI] [PubMed] [Google Scholar]
- 8.Marahiel M.A., Stachelhaus T., Mootz H.D. Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem. Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 9.Durbin R., Eddy S.R., Krogh A., Mitchison G. Biological Sequence Analysis. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- 10.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson J., Higgins D., Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 15.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohlbacher O., Lenhof H. BALL—rapid software prototyping in computational molecular biology. Biochemicals Algorithms Library. Bioinformatics. 2000;16:815–824. doi: 10.1093/bioinformatics/16.9.815. [DOI] [PubMed] [Google Scholar]
- 18.Schölkopf B., Tsuda K., Vert J., editors. Kernel Methods in Computational Biology. Cambridge, MA: MIT Press; 2004. [Google Scholar]
- 19.Vapnik V.N. The Nature of Statistical Learning Theory. New York, NY: Wiley-Interscience; 1998. [Google Scholar]
- 20.Joachims T. Transductive inference for text classification using support vector machines. In: Bratko I., Dzeroski S., editors. Proceedings of the Sixteenth International Conference on Machine Learning (ICML'99); Bled, Slovenia. San Francisco, CA, USA: Morgan Kaufmann Publishers; 1999. pp. 200–209. [Google Scholar]
- 21.Zhou D., Bousquet O., Lal T., Weston J., Schölkopf B. Learning with Local and Global Consistency. Adv. Neural Inform. Process. Syst. 2004;16:321–328. [Google Scholar]
- 22.Baldi P., Brunak S., Chauvin Y., Andersen C.A.F., Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 23.Markowetz F., Spang R. Molecular diagnosis. Classification, model selection and performance evaluation. Methods Inf. Med. 2005;44:438–443. [PubMed] [Google Scholar]
- 24.Kawashima S., Kanehisa M. Aaindex: amino acid index database. Nucleic Acids Res. 2000;28:374. doi: 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomii K., Kanehisa M. Analysis of amino acid indices and mutation matrices for sequence comparison and structure prediction of proteins. Protein Eng. Des. Sel. 1996;9:27–36. doi: 10.1093/protein/9.1.27. [DOI] [PubMed] [Google Scholar]
- 26.Nakai K., Kidera A., Kanehisa M. Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Eng. Des. Sel. 1988;2:93–100. doi: 10.1093/protein/2.2.93. [DOI] [PubMed] [Google Scholar]
- 27.Neumaier A., Huyer W., Bornberg-Bauer E. Hydrophobicity analysis of amino acids. 1999.
- 28.Fauchere J.L., Charton M., Kier L.B., Verloop A., Pliska V. Amino acid side chain parameters for correlation studies in biology and pharmacology. Int. J. Pept. Protein Res. 1988;32:269–278. doi: 10.1111/j.1399-3011.1988.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerman J.M., Eliezer N., Simha R. The characterization of amino acid sequences in proteins by statistical methods. J. Theoret. Biol. 1968;21:170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]
- 30.Radzicka A., Wolfenden R. Comparing the polarities of the amino acids: side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral aqueous solution. J. Biochem. 1988;27:1664–1670. [Google Scholar]
- 31.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 32.Tsai J., Taylor R., Chothia C., Gerstein M. The packing density in proteins: standard radii and volumes. J. Mol. Biol. 1999;290:253–266. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
- 33.Chou P.Y., Fasman G.D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 34.Joachims T. Making large-scale SVM learning practical. In: Schölkopf B., Burges C., Smola A., editors. Advances in Kernel Methods. Cambridge, MA, USA: MIT-Press; 1999. pp. 169–184. [Google Scholar]
- 35.Morik K., Brockhausen P., Joachims T. Combining statistical learning with a knowledge-based approach—a case study in intensive care monitoring. Proceedings of the Sixteenth International Conference on Machine Learning (ICML'99); San Francisco, CA, USA. 1999. pp. 268–277. [Google Scholar]
- 36.Miller G., Lipman M. Release of infectious Epstein–Barr virus by transformed marmoset leukocytes. Proc. Natl Acad. Sci. USA. 1973;70:190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turgay K., Krause M., Marahiel M.A. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol. Microbiol. 1992;6:529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. 2743–2744. [DOI] [PubMed] [Google Scholar]
- 38.Lilien R.H., Stevens B.W., Anderson A.C., Donald B.R. A novel ensemble-based scoring and search algorithm for protein redesign, and its application to modify the substrate specificity of the gramicidin synthetase A phenylalanine adenylation enzyme. In: Gusfield D., Bourne P., Istrail S., Pevzner P., Watermann M., editors. Proceedings of the Eighth Annual International Conference on Research in Computational Molecular Biology (RECOMB 2004); San Diego, CA, USA. New York, NY, USA: ACM; 2004. pp. 46–57. [DOI] [PubMed] [Google Scholar]
- 39.Mootz H.D., Marahiel M.A. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang Z., Flatt P., Gerwick W.H., Nguyen V.A., Willis C.L., Sherman D.H. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (pks)-non-ribosomal peptide synthetase (nrps) origin involving an unusual trichloroleucyl starter unit. Gene. 2002;296:235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 41.Vinh L.S., von Haeseler A. IQPNNI: moving fast through tree space and stopping in time. Mol. Biol. Evol. 2004;21:1565–1571. doi: 10.1093/molbev/msh176. [DOI] [PubMed] [Google Scholar]
- 42.Minh B.Q., Vinh L.S., von Haeseler A., Schmidt H.A. pIQPNNI—parallel reconstruction of large maximum likelihood phylogenies. Bioinformatics. 2005 doi: 10.1093/bioinformatics/bti594. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 43.Huson D., Bryant D. Application of phylogenetic networks in evolutionary studies. MBE. 2005 doi: 10.1093/molbev/msj030. (in press) [DOI] [PubMed] [Google Scholar]
- 44.Altschul S., Madden T., Schffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noble S.W. Support Vector Machine Applications In Computational Biology. Cambridge, MA: MIT Press; 2004. pp. 71–92. [Google Scholar]
- 46.Schölkopf B., Guyon I., Weston J. Statistical Learning and Kernel Methods In Bioinformatics. The Netherlands: IOS Press Amsterdam; 2003. pp. 1–21. [Google Scholar]
- 47.Weston J., Pérez-Cruz F., Bousquet O., Chapelle O., Elisseeff A., Schölkopf B. Feature selection and transduction for prediction of molecular bioactivity for drug design. Bioinformatics. 2003;19:764–771. doi: 10.1093/bioinformatics/btg054. [DOI] [PubMed] [Google Scholar]
- 48.Hannenhalli S., Russell R. Analysis and prediction of functional sub-types from protein sequence alignments. J. Mol. Biol. 2000;303:61–76. doi: 10.1006/jmbi.2000.4036. [DOI] [PubMed] [Google Scholar]
- 49.Taylor W. The classification of amino acid conservation. J. Theoret. Biol. 1986;119:205–218. doi: 10.1016/s0022-5193(86)80075-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.