Abstract
We report the characterization of a novel factor, Rsa4p (Ycr072cp), which is essential for the synthesis of 60S ribosomal subunits. Rsa4p is a conserved WD-repeat protein that seems to localize in the nucleolus. In vivo depletion of Rsa4p results in a deficit of 60S ribosomal subunits and the appearance of half-mer polysomes. Northern hybridization and primer extension analyses of pre-rRNA and mature rRNAs show that depletion of Rsa4p leads to the accumulation of the 27S, 25.5S and 7S pre-rRNAs, resulting in a reduction of the mature 25S and 5.8S rRNAs. Pulse–chase analyses of pre-rRNA processing reveal that, at least, this is due to a strong delay in the maturation of 27S pre-rRNA intermediates to mature 25S rRNA. Furthermore, depletion of Rsa4p inhibited the release of the pre-60S ribosomal particles from the nucleolus to the nucleoplasm, as judged by the predominantly nucleolar accumulation of the large subunit Rpl25-eGFP reporter construct. We propose that Rsa4p associates early with pre-60S ribosomal particles and provides a platform of interaction for correct processing of rRNA precursors and nucleolar release of 60S ribosomal subunits.
INTRODUCTION
Ribosome biogenesis is a fundamental multistep process that, in eukaryotes, takes place largely within the nucleolus (1). Late steps in both 40S and 60S ribosomal subunit (r-subunit) synthesis occurs in the nucleoplasm and after export of precursor particles to the cytoplasm (2). Most of our understanding of eukaryotic ribosome biogenesis has been obtained from the model yeast, Saccharomyces cerevisiae (3,4). In the yeast nucleolus, three of the four rRNAs (18S, 5.8S and 25S) are transcribed as a single large primary transcript by RNA polymerase I and processed to the first detected rRNA precursor (pre-rRNA), the so-called 35S pre-rRNA. The fourth rRNA (5S) is independently transcribed as a pre-rRNA (pre-5S) by RNA polymerase III. In the 35S pre-rRNA, the mature rRNA sequences are separated by two internal transcribed spacers (ITS1 and ITS2) and flanked by two external transcribed spacers (5′ ETS and 3′ ETS), which must be precisely and efficiently processed for mature rRNA formation (see Figure 1).
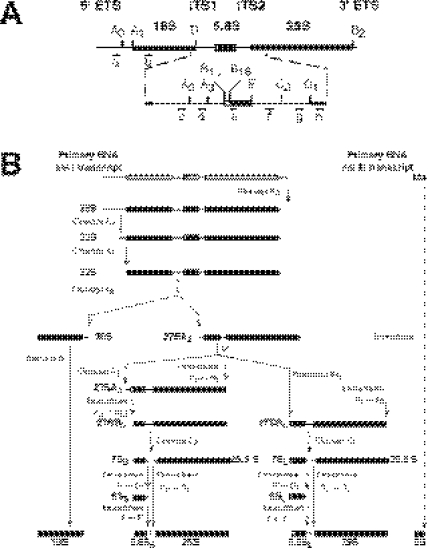
Figure 1.
Pre-rRNA processing in S.cerevisiae. (A) Structure and processing sites of the 35S pre-rRNA. This precursor contains the sequences for the mature 18S, 5.8S and 25S rRNAs that are separated by two internal transcribed spacer sequences, ITS1 and ITS2, and flanked by two external transcribed spacer sequences, 5′ ETS and 3′ ETS. The mature rRNA species are shown as bars and the transcribed spacer sequences as lines. The processing sites and their locations as well as the various probes used are indicated. (B) Pre-rRNA processing pathway. The primary RNA pol I transcript undergoes covalent modifications (data not shown), and it is cleaved at its 3′ end to yield the 35S pre-rRNA, which is the longest detectable precursor. The 35S pre-rRNA is cleaved at site A0 to generate the 33S pre-rRNA. This molecule is subsequently processed at sites A1 and A2, resulting in the separation of the pre-rRNAs destined for the small and large ribosomal subunits. The final maturation of the 20S precursor takes place in the cytoplasm, where cleavage at site D yields the mature 18S rRNA. The 27SA2 precursor is processed by two alternative pathways that both lead to the formation of mature 5.8S and 25S rRNAs. In the major pathway, the 27SA2 precursor is first cleaved at site A3 and then, the 27SA3 precursor is exonucleolytically digested 5′→3′ up to site B1S to yield the 27SBS precursor. A minor pathway processes the 27SA2 or the 27SA3 molecule at site B1L by an as yet unknown mechanism, producing the 27SBL pre-rRNA. While processing at sites B1S and B1L is being completed, the 3′ end of the mature 25S rRNA is generated by 3′→5′ trimming to site B2. The subsequent processing of both 27SB species appears to be identical. Cleavage at site C2 generates the 25.5S and 7S pre-rRNAs. The 7S pre-rRNA is 3′→5′ trimmed to the 3′ end of the mature 5.8S rRNA. The 25.5S species is 5′→3′ digested to the mature 25S rRNA. The primary RNA pol III transcript is trimmed to the 3′ end of the mature 5S rRNA. The data presented in this study suggest that Rsa4p is required for efficient processing of all steps, leading from 27SA2 pre-rRNA to mature 25S and 5.8S rRNAs. For reviews on pre-rRNA processing and the known processing enzymes (4,5).
Maturation of rRNAs is a well-defined pathway (see Figure 1) and involves numerous trans-acting factors that are required for the processing and covalent rRNA modification reactions, such as small nucleolar RNA–protein (snoRNP) complexes, endonucleases and exonucleases, and different base methylases (4,5). Concomitantly to rRNA maturation, the pre-rRNAs assemble in an ordered manner with the r-proteins and a large number of trans-acting factors that are generally referred to as r-subunit assembly factors (for examples of trans-acting factors see http://www.medecine.unige.ch/~linder/proteins.html).
The process of r-subunit assembly is still poorly understood (2,6,7). An outline of the process was provided by the gradient analysis in the 1970s, which identified 90S, 66S and 43S pre-ribosomal particles (8). Recent advances in the proteomic field have now allowed the prediction of several distinct pre-ribosomal particles and the draw of a new model for the maturation of both 40S and 60S r-subunits (see Figure 2 and its legend). From this, a large number of non-ribosomal proteins have been identified in the pre-ribosomal particles (9–14). Many of these factors remain uncharacterized so far, therefore, are without an assigned function in RNA metabolism (2,3).
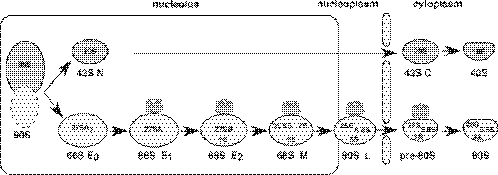
Figure 2.
Current model for the formation, maturation and export of 66S pre-ribosomal particles in S.cerevisiae. A series of distinct particles are predicted to be intermediates during the synthesis of 60S r-subunits. These are termed, according to their position in the pathway, early 0 (E0), early 1 (E1), early 2 (E2) and middle (M) 66S pre-ribosomal particles and late (L) and cytoplasmic pre-60S ribosomal particles. All these particles are defined by the purification of complexes associated with TAP-tagged version of selected ribosomal subunit assembly factors (9,12,13,37,61). The pre-rRNAs abundantly associated with the different particles are indicated. Note that although in this figure the 5S rRNA assembles late in the pathway, its exact binding position is not clear. The data presented in this study and the literature suggest that Rsa4p associates with early pre-60S r-subunits and releases into the cytoplasm. Rsa4, grey barrel; nucleolus, rectangle; pre-ribosomal particles and mature r-subunits, light balloons; and nuclear envelope, rods. For further description of the ribosomal subunit assembly pathway (2,3,62).
In this paper, we described the functional analysis of Ycr072cp, one of those as yet unknown non-ribosomal proteins, which we named Rsa4p (for ribosome assembly 4). Rsa4p is an essential and evolutionarily conserved member of the superfamily of WD-repeat proteins and is mainly composed of a repetitive arrangement of eight WD domains. WD-repeat proteins are critically important for many cellular processes, including pre-rRNA processing and r-subunit assembly (15). Rsa4p has been identified as a component of most stable nucleolar, nucleoplasmic and cytoplasmic pre-60S r-particles that have been affinity purified (see Figure 2 and references therein). Our results are consistent with these data and demonstrate that Rsa4p localizes predominantly in the nucleolus and is required for different pre-rRNA processing steps during the maturation of pre-60S r-subunits. In addition, Rsa4p seems to be required for optimal intra-nuclear transport of pre-60S r-particles. We hypothesized that Rsa4p associates with early pre-60S r-particles and provides a stable platform for the interaction of several other r-subunit assembly and export factors through the complete pathway of 60S r-subunit biogenesis.
MATERIALS AND METHODS
Strains, media and microbiological methods
The strains used in this study are derived from the strain W303 (MATa/MATα ura3-1/ura3-1 trp1-1/trp1-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 ade2-1/ade2-1 can1-100/can1-100) (16). The strain WDG72 was constructed by sequential transformation of W303-1A (MATa) with plasmid p3872 and afterwards with the 3.4 kb EcoRI–HindIII fragment from the pD72C plasmid. Transformants showing Trp and Ura prototrophy were selected and disruption of the RSA4 locus was confirmed by Southern blot. This strain allows growth in media containing galactose as a carbon source but impedes growth in media containing glucose.
Yeast strains were grown at 30°C either in rich YPD medium or in YPGal medium. When indicated, cells were also grown in a synthetic medium containing either 2% galactose (SGal) or glucose (SD) as a carbon source and supplemented with specific nutrients required for the auxotrophic deficiencies. Yeast media are described precisely (17,18).
Yeast genetic methods were carried out by following standard protocols (17). Transformation of yeast cells was carried out using the lithium acetate procedure (19).
The Escherichia coli strain DH5α (20) was used for subcloning and amplification of plasmid DNA. Luria–Bertani (LB) supplemented with appropriate antibiotics was used as routine growth medium for these cells (21).
Plasmids
BSD is a Bluescript KS+ derived plasmid that contains the 4.3 kb EcoRI genomic fragment where RSA4 is located. It was obtained after digestion of the original library clone distributed for chromosome III sequencing with EcoRI (22). Plasmid BSD was digested with PstI and SacI and the 2.36 kb fragment containing the open reading frame (ORF) and 400 bp from both the 5′ and the 3′ ends was cloned into pFL36 (23), digested with the same enzymes, yielding plasmid pFL72.
Plasmid p3872 expresses Rsa4p under the control of the GAL1 promoter. Two new restriction sites were created by PCR with mutagenic oligonucleotides, using plasmid BSD as a template. The 26mer 5′-CGATAAACACACTCGAGCACAATATACAGA (changes in the sequence are underlined) creates a new XhoI restriction site 17 nt upstream of the initial ATG. The second 26mer 5′-GGAGTAAGTTGCTGCAGGTAAACGGGC introduces a new PstI site 222 nt downstream of the stop codon. Centromeric plasmid pFL39 (with TRP1 as a marker) (23) was digested with EcoRI and PstI, and then the 750 bp EcoRI–XhoI fragment from plasmid pCG542, containing the GAL1–GAL10 promoter (24), along with the amplified fragment, previously digested with XhoI and PstI, was inserted. The resulting plasmid was labelled p3872.
Plasmid pD72C was constructed for gene disruption in three steps. First, a 1 kb HindIII–DraI fragment from the 5′ upstream region of RSA4 was cloned between the HindIII–HincII sites of pUC19 (25). Second, the URA3 marker from pFL38 (23) was cloned as a 1.1 kb BglII fragment in the BamHI site of pUC19. Finally, the 1.3 kb HincII–EcoRI fragment from BSD (3′-untranslated region of RSA4) was inserted between the SmaI–EcoRI sites of pUC19.
Plasmid YCplac111-RSA4-eGFP was constructed as follows: a ∼3 kb XhoI–NdeI fragment from pFL72 was blunt-ended and cloned into SmaI–restricted YCplac111-yeGFP/TCYC1 (a gift from M. Hall). One candidate in the appropriate orientation, YCplac111-7eGFP, was selected. Then, a PCR was performed using pFL72 as a template and using the oligonucleotides 5′-1551-YCR072C (5′-TGGGTTCTCTGCGTTTC-3′, placed 59 bp upstream of the sole BamHI site present in RSA4 ORF) and 3′-XBAI-YCR072C (5′-GCTCTAGAATGCGTCCACAATCTTAC-3′, complementary to the end of the RSA4 ORF but lacks the stop codon; an XbaI site is underlined). The 1 kb PCR product was digested with BamHI and XbaI and cloned into YCplac111-7eGFP, which was also digested with BamHI and XbaI, yielding the plasmid YCplac111-RSA4-eGFP.
Sucrose gradient centrifugation, protein and RNA analyses
Polyribosome preparations, polysome analyses, and r-subunit preparations were performed as described previously (26). Gradient analysis was performed with an ISCO UA-6 system, with continuous monitoring at A254. Fraction analyses of proteins were performed as described previously (26).
Total yeast protein extracts were prepared and analysed by western blotting, according to the standard procedures (17,21). Rabbit polyclonal antibodies were generated against the partially purified recombinant Rsa4p protein (E. Sanz-Martínez, and M. Remacha unpublished data).
RNA extraction, northern hybridization and the primer extension analysis were carried out according to the standard procedures (27). In all experiments, RNA was extracted from samples corresponding to 10 OD600 units of exponentially grown cells and 5 or 2.5 µg were loaded in gels or used to primer extension reactions. Sequences of oligonucleotides used for RNA hybridization and primer extension analyses have been described previously (28). Pulse–chase labeling of pre-rRNA was performed as described previously (26), using 250 µCi [methyl-3H]methionine (70–85 Ci/mmol; Amersham) per 40 OD600 of yeast cells.
Fluorescence microscopy
The strain WDG72 was first transformed with YCplac111-RSA4-eGFP and then subjected to p3872 plasmid segregation on SD-Leu plates. The plasmid YCplac111-RSA4-eGFP complemented the rsa4 allele to almost the wild-type extent (data not shown). For localization, strain WDG72 expressing the Rsa4-eGFPp fusion protein from YCplac111-RSA4-eGFP was further transformed with pRS314-DsRedNOP1 (a gift from J. Bassler). Then, several transformants were grown to the mid-log phase in SD-Leu-Trp liquid medium, washed and resuspended in water. Acquisition was carried out in a Leica DMR microscope equipped with a DC camera, according to the manufacturer's instructions. Digital images were processed with Adobe Photoshop 7.0.
To test pre-ribosomal particle export, strains W303-1A and WDG72 were transformed with either pRS315-RPL25-eGFP, which allows the localization of Rpl25-eGFP (a gift from O. Gadal) or pRS315-Rps2-eGFP, which allows the localization of Rps2-eGFP (a gift from O. Gadal). In an independent experiment, strains W303-1A and WDG72 were transformed with both pASZ311-Rpl25-eGFP and pUN100-DsRedNOP1 (gifts from O. Gadal) and transformants were selected in SGal-Leu-Ade liquid medium. Localization was studied from cells grown in SGal-Leu-Ade liquid medium or at various time points after transfer from liquid SGal-Leu-Ade to liquid SD-Leu-Ade.
RESULTS
Rsa4p is an essential evolutionarily conserved protein
The RSA4 gene is essential for viability (E. Sanz-Martínez and M. Remacha, unpublished data), as also reported in a systematic deletion analysis (29). RSA4 encodes a protein of 515 amino acids with a calculated molecular mass of 57 kDa and pI 9.3. Sequence analysis and database comparison shows that Rsa4p belongs to the superfamily of WD-repeat containing proteins (15). Precisely, the WD motif (30) is present in up to seven copies at the middle and carboxyl-end domain of the protein, five of which fully match the consensus sequence as found in Prosite (PDOC00574). The repeated unit comprises, on the average, 42 amino acids, with a 12-residue core region that is highly conserved (Figure 3). A distinctive feature in this protein is the presence between repeats IV and V of an 82 amino acids insertion, which resembles, in fact, two less conserved WD repeats. Within this insertion, there is a highly hydrophilic region of 29 amino acids with a predicted alpha helix secondary structure.
Figure 3.
RSA4 encodes a member of the WD-repeat protein family. Protein sequence is aligned to enhance the presence of the seven WD motifs. Those in bold face completely match the motif as it is found in Prosite (PDOC00574). The most highly conserved residues among the seven repeats are underlined. The hydrophilic region between domains IV and V is double underlined. In the N-terminal end, the putative nuclear location site is also underlined.
We identified clear sequence counterparts in other organisms, including other yeasts (i.e. Kluyveromyces lactis and Schizosaccharomyces pombe, Oryza sativa, Caenorhabditis elegans, Drosophila melanogaster, Xenopus laevis and Homo sapiens) (Supplementary Table 1). Sequence comparison between the individual WD-repeats of these proteins shows the highest conservation in the most C-terminus repeats, the lowest similarity being in the region between domains IV and V (Supplementary Table 1). Nevertheless, as no significant homology to any other members of the superfamily was found, this region is exclusive for this group of WD-repeat containing proteins. Remarkably, the homology for the WD-repeats I and II of Rsa4p is restricted to its yeast orthologues, having a much lower homology with the remaining five, which is similar to the homology detected with non-related WD-repeat containing proteins.
In addition to the WD-repeats, the encoded Rsa4p polypeptide has a 130 amino acid long N-terminal domain, which contains one putative nuclear localization signal (NLS, from residues 6 to 14) (Figure 3). Comparison of this N-end sequence extension with the orthologue proteins mentioned above (Supplementary Table 1) reveals that significant homology is essentially restricted to the yeast proteins.
Conditional expression of Rsa4p
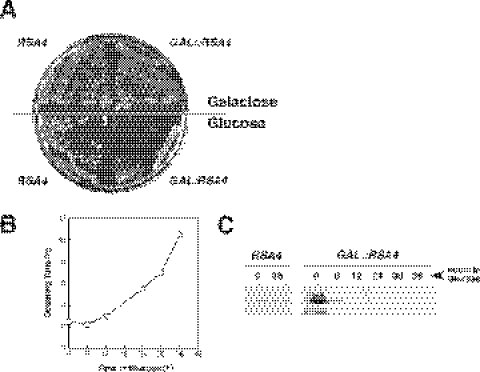
Given the fact that Rsa4p is essential, a conditional system for the phenotypic analysis was established. The strain W303-1A was sequentially transformed first with the plasmid p3872, containing RSA4 under the GAL1 promoter, and afterwards with the EcoRI–HindIII linearized pD72C plasmid, containing an RSA4::URA3 construct, to disrupt the genomic copy (see Materials and Methods). Transformants Trp+ and Ura+ were selected and disruption of the RSA4 locus was confirmed by Southern blot (data not shown). One of these transformants, named WDG72, was selected for further analysis. Growth of WDG72 was identical to that of the isogenic wild-type strain on YPGal plates, showing the GAL::RSA4 construct to be functional (Figure 4A). However, this construct resulted in a strong slow-growth phenotype on YPD plates (Figure 4A). After WDG72 was shifted from liquid YPGal medium to liquid YPD medium, the growth rate remained similar to that of the wild-type control strain for the first 6 h but then progressively decreased to a doubling time of >10 h after 36 h in YPD medium (Figure 4B). The growth rate of the isogenic wild-type strain was almost unaffected when shifted from liquid YPGal to YPD (data not shown). Concomitant with the decrease in the growth rate, WDG72 cells were depleted of Rsa4p, as detected by the western blot analysis (Figure 4C). The growth defect of WDG72 in glucose-based medium is directly related to the presence or absence of Rsa4p in the cell since when WDG72 is transformed with the centromeric plasmid pFL72, carrying an intact copy of RSA4, the transformants grow normally both in glucose and galactose-based media (data not shown).
Figure 4.
Conditional system for the phenotypic analysis. (A) Growth comparison of the strains WDG72 (GAL::RSA4) and W303-1A (RSA4). The cells were streaked on YPGal (Galactose) and YPD (Glucose) plates and incubated at 30°C for 3 days. (B) Growth curve of WDG72 at 30°C, after transferring exponential cells from YPGal to YPD medium, for up to 36 h. (C) Depletion of the Rsa4p protein; whole-cell extracts were prepared from W303-1A and WDG72, harvested at the indicated times after the shift to YPD medium. Equal amounts of protein were separated by 12% SDS–PAGE and Rsa4p was detected by western blotting using polyclonal rabbit antibodies.
In vivo depletion of Rsa4p leads to a deficiency in 60S ribosomal subunits
To assess the function of Rsa4p in ribosome biogenesis, we first assayed steady-state levels of polysomes in the GAL::RSA4 (WDG72) and RSA4 (W303-1A) strains in YPGal or after shifting to YPD. Both, the GAL::RSA4 and RSA4 strains showed normal polysome profiles when grown in YPGal, including normal levels of free 40S and 60S r-subunits, monosomes (and 80S free couples) and polysomes (Figure 5A and data not shown). However, when shifted for 24 h to YPD, WDG72 cells showed a strong decrease in the levels of free 60S r-subunits, an increase in the levels of free 40S r-subunits and an overall decrease in the 80S peak and in polysomes (Figure 5B). In addition, there was an accumulation of half-mer polysomes (Figure 5B). Wild-type cells showed no alteration in the polysome profile when transferred to YPD (data not shown). These results indicate that depletion of Rsa4p leads to a deficit in 60S r-subunits relative to 40S r-subunits.
Figure 5.
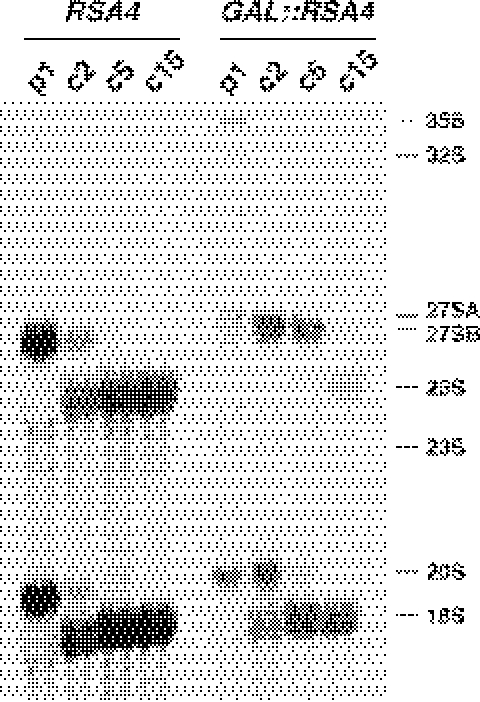
Depletion of Rsa4p results in deficit in free 60S r-subunits and accumulation of half-mer polysomes. WDG72 (GAL::RSA4) was grown in YPGal (A) and then shifted to YPD for 24 h (B). Cells were harvested at an OD600 of ∼0.8 and equal amounts of cells extracts (10 A260 units) were resolved in 7–50% (w/v) sucrose gradients. The A254 was read continuously. Sedimentation is from left to right. The peaks of free 40S and 60S ribosomal subunits, 80S free couples/monosomes and polysomes are indicated. Half-mers are indicated by arrows.
Rsa4p is required for normal pre-rRNA processing
To study in more detail the role of Rsa4p in 60S r-subunit metabolism, we analysed the effects of Rsa4p depletion on the processing of 35S pre-rRNA. Total RNA was isolated from RSA4 and GAL::RSA4 strains at various time points after transfer from liquid YPGal to liquid YPD, and steady-state levels of premature and mature rRNA species were subjected to northern and primer extension analyses. Different oligonucleotides hybridizing to defined regions of the 35S pre-rRNA transcript were used to monitor specific processing intermediates (see Figure 1). As shown in Figure 6A, depletion of Rsa4p caused a marked decrease in 25S rRNA but only a slight decrease in 18S rRNA steady-state levels, which is in agreement with the polysome profile results. Upon depletion, the GAL::RSA4 strain accumulated the 35S pre-rRNA; clear accumulation was observed 24 h after transfer to YPD and it increased with time. The levels of 20S pre-rRNA were reduced in the Rsa4p-depleted strain, commencing also 24 h after transfer to YPD medium (Figure 6A). This reduction is concomitant to the appearance of an aberrant 23S RNA, which extends from the 5′ ETS to site A3, and to the slight decrease in mature 18S rRNA (Figure 6A). Moreover, the 27SB pre-rRNA was strongly accumulated at 24 h and at later time points. Interestingly, the 27SA2 pre-rRNA seems also to accumulate slightly 24 h after the transfer to glucose medium, but its levels are reduced at later time points.
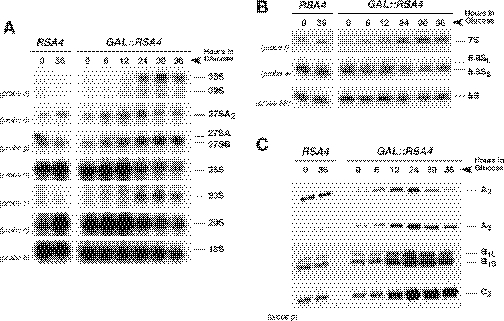
Figure 6.
Effects of Rsa4p depletion on steady-state levels of pre-rRNAs and mature rRNAs. The strains W303-1A (RSA4) and WDG72 (GAL::RSA4) were grown in YPGal medium and then shifted to YPD medium. Cells were harvested at indicated times and total RNAs were extracted. (A) Equal amounts of total RNA (5 µg) were resolved on a 1.2% agarose–formaldehyde gel and transferred onto a nylon membrane. The same membrane was hybridized with different probes. (B) Equal amounts of total RNA (2.5 µg) were resolved on a 7% polyacrylamide–urea gel, transferred onto a nylon membrane and hybridized consecutively with different probes. (C) Equal amounts of total RNA (5 µg) were used for primer extension analysis. Probe g was labelled and used for the reactions. Note that this probe allows detection of 27SA2 (as the stop at site A2), 27SA3 (as the stop at site A3), and both 27SB (as stops at sites B1L and B1S) and 25.5S (as the stop at site C2). Probe names are indicated between parentheses on the left (except for probe 5S, see Figure 1A for their location in the 35S pre-rRNA).
The analysis of low-molecular-weight rRNA species revealed a mild decrease in the steady-state levels of the 5.8S and 5S rRNAs but an accumulation of the 7S pre-rRNAs, 24 h after the transfer to glucose medium (Figure 6B).
To further characterize pre-rRNA processing in the GAL::RSA4 strain, we assessed the levels of the 27S and 25.5S pre-rRNAs by primer extension analyses. As shown in Figure 6C, the primer extension stops at sites C2, B1L and B1S, and increased at 12 h after transfer to YPD medium, indicating that 25.5S, 27SBL and 27SBS pre-rRNAs were accumulated after depletion of Rsa4p. The level of the primer extension at site A2, the 5′ end of the 27SA2 pre-rRNA increased 12 h after transfer to YPD medium but decreased at 30 and 36 h time points. A similar observation was made for site A3, the 5′ end of the 27SA3 pre-rRNA (Figure 6C). The level of the 27SA3 pre-rRNA was, however, reduced to a lesser extent. All these results are fully consistent with the results obtained by northern hybridization. However, since primer extension is a more sensitive technique, the accumulation of these pre-rRNAs is readily detected earlier at 12 h after transfer to glucose medium.
To study the kinetics of rRNA production, we analysed the effects of Rsa4p depletion on the synthesis and processing of pre-rRNA by pulse–chase labelling with [methyl-3H]methionine. Both, the RSA4 and GAL::RSA4 strains were grown in liquid YPGal medium and then shifted to liquid minimal glucose medium lacking methionine (SD-Met) for 24 h. At this time point, the GAL::RSA4 strain was doubling every 6.5 h compared with 2.5 h for the RSA4 strain. The cells were pulse-labelled for 1 min, then chased for 2, 5 and 15 min with an excess of ice-cold methionine and total RNA was extracted and analysed. In the wild-type RSA4 strain, the 35S precursor was converted rapidly into 32S pre-rRNA and then into 27S and 20S species, which were further processed into 25S and 18S mature rRNAs, respectively (Figure 7). After 5 min of chase, most of the labelled species were mature rRNAs. Consistent with the northern blot and primer extension data, depletion of Rsa4p resulted in a mild delay of 35S pre-rRNA processing, as shown by the 35S pre-rRNA abundance and persistence compared with the wild-type in early chase time points (Figure 7). This was combined with some delay in the formation of the 27S and 20S pre-rRNAs and the appearance of traces of the aberrant 23S species (Figure 7). The processing pathway leading from 20S pre-rRNA to the 18S rRNA was not significantly affected. In contrast, the 27S precursors persisted until the end of the chase. As a consequence, almost no labelled mature 25S rRNA was detected (Figure 7).
Figure 7.
Depletion of Rsa4p inhibits pre-rRNA processing of 27S precursors. The strains W303-1A (RSA4) and WDG72 (GAL::RSA4) were grown in YPGal medium and then shifted for 24 h to SD-Met medium. Cells were pulse-labelled with [methyl-3H]methionine for 1 min, and then chased with a large excess of ice-cold methionine for 2, 5 and 15 min. Total RNA was extracted and 20 000 c.p.m. was loaded and separated on a 1.2% agarose–formaldehyde gel, transferred onto a nylon membrane and visualized by fluorography. The positions of the different pre-rRNAs and mature rRNAs are indicated.
Taken all together, these data demonstrate that all pre-rRNA processing steps leading from 27SA2 to 25S and 5.8S rRNAs are inhibited in the Rsa4p-depleted strain. Moreover, depletion of Rsa4p mildly delays processing at sites A0, A1 and A2, which is frequently seen in most strains defective in 60S r-subunit biogenesis and probably owing to indirect effects (4).
Depletion of Rsa4p impairs the intra-nuclear transport of pre-60S r-particles
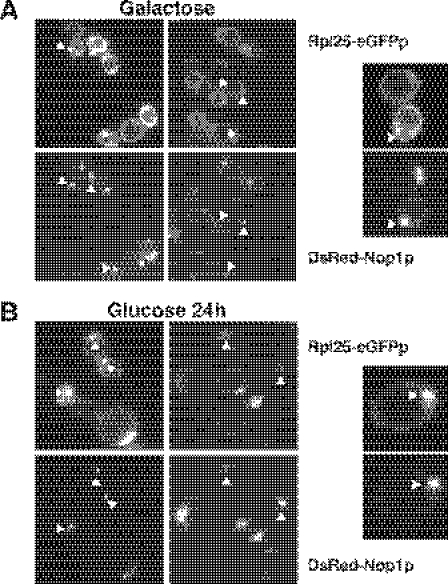
To test whether Rsa4p is required for r-subunit export, the RSA4 and GAL::RSA4 strains were transformed with a plasmid expressing a GFP-tagged form of the 60S r-protein Rpl25p. The localization of the Rpl25-eGFPp was determined by fluorescence microscopy after growth in liquid minimal galactose medium (SGal-Ade-Leu) and at a different time after transfer to liquid minimal glucose medium (SD-Ade-Leu), as described previously (31,32). Rpl25-eGFPp was predominately cytoplasmic in wild-type cells grown in galactose medium and glucose medium, indicating that nascent pre-60S r-particles are processed and rapidly exported to the cytoplasm (data not shown). As shown in Figure 8A, Rpl25-eGFPp was also detected primarily in the cytoplasm of GAL::RSA4 cells grown in galactose medium. In contrast, Rpl25-eGFP was found to accumulate in the nucleus of GAL::RSA4 cells 24 h after a shift to glucose medium (Figure 8B). The GFP-decorated region mainly coincides with the nucleolus, identified by the red fluorescently DsRed-Nop1p marker (see below) (Figure 8B). A similar assay for nuclear export of pre-40S r-particles was performed by the fluorescence microscopy analysis of the RSA4 and GAL::RSA4 strains transformed with a plasmid expressing an eGFP-tagged form of the 40S r-protein Rps2p (33). Both strains showed comparable cytoplasmic localization of Rps2-eGFPp in liquid minimal galactose medium or 24 h after transfer to liquid minimal glucose medium (data not shown).
Figure 8.
Depletion of Rsa4p leads to accumulation of Rpl25p-eGFP in the nucleolus. WDG72 (GAL::RSA4) was transformed with Rpl25p-eGFP and DsRed-Nop1p plasmids. Selected candidates were grown in liquid SGal-Leu-Ade medium (galactose) (A) and then shifted to liquid SD-Leu-Ade medium (glucose) for 24 h (B). Cells were harvested, washed and resuspended in water and inspected in the fluorescence microscope. Selected cells are shown in a magnified picture. Triangles point to nucleolar fluorescence.
We conclude that upon depletion of Rsa4p, intra-nuclear transport of pre-60S r-particles is blocked at an early stage, most probably before the release of preribosomes from the nucleolus.
Rsa4p localizes to the nucleolus
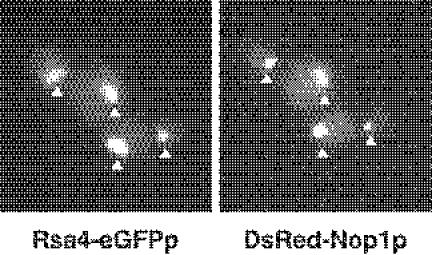
To establish the subcellular localization of Rsa4p, we constructed a strain in which the wild-type RSA4 gene was replaced by a RSA4-eGFP fusion allele (see Materials and Methods). This allele is not fully functional but supports almost wild-type growth at 30°C (data not shown). Examination by fluorescence microscopy of the cells expressing Rsa4-eGFPp revealed a bright, nuclear signal (Figure 9). To allow visualization of the nucleolus, the RSA4-eGFP strain was additionally transformed with a plasmid expressing DsRed-tagged Nop1p (31). This decorated the crescent-shaped region of the nucleus that is characteristic of the yeast nucleolus (31). As shown in Figure 9, the nucleolar signal of DsRed-Nop1p colocalizes with the signal of Rsa4-eGFPp, indicating that Rsa4-eGFPp is localized predominantly in the nucleolus. Similar results have been independently obtained by a systematic protein localization study (34). Interestingly, the putative human orthologue of Rsa4p (human BAA91621) is also a nucleolar protein (35). The predominant localization of the Rsa4p–eGFPp fusion protein in the nucleolus suggest that Rsa4p is a nucleolar protein. This is in agreement with a specific role of Rsa4p in ribosome biogenesis.
Figure 9.
Rsa4p localized predominantly to the nucleolus. In vivo localization of Rsa4p-eGFP and DsRed-Nop1p was determined by fluorescence microscopy. Triangles point to nucleolar fluorescence.
DISCUSSION
Rsa4p is predicted to be involved in 60S r-subunit biogenesis. It has been shown that Rsa4p is a component of a set of stable pre-60S r-particles that were purified with different TAP-tagged version of trans-acting factors. These particles have substantially different pre-rRNA and protein compositions (12,36–38). In agreement, we have found that most Rsa4p sediments in sucrose gradients to positions expected for pre-60S r-particles (data not shown). Based on all these data, we and others have deduced that Rsa4p might associate with early nucleolar pre-60S r-particles and dissociate in the cytoplasm after the final structural rearrangements of cytoplasmic 60S pre-ribosomal subunits (3,12).
In this study, we present several lines of evidences that Rsa4p is indeed involved in 60S r-subunit biogenesis. First, depletion of Rsa4p leads to a deficit of 60S r-subunits and the appearance of half-mer polysomes. Second, when pre-rRNA processing was examined, we found that a number of steps in the processing of 27S pre-rRNA to mature 25S rRNA were inhibited in the Rsa4p-depleted strain (see below). Moreover, the transport of pre-60S r-particles from the nucleolus to the nucleoplasm is likely impaired upon depletion of Rsa4p. Finally, Rsa4p has an NLS and seems to be enriched in the nucleolus.
A striking feature is the pre-rRNA processing phenotype observed upon Rsa4p depletion. This resulted in the reduction of both 25S and 5.8S rRNAs. We found a mild delay of early pre-rRNA cleavages at sites A0, A1 and A2. This delay is an unspecific phenotype observed in many other mutations that interfere with the synthesis of mature rRNA from the large r-subunit. It has been proposed that these pre-rRNA processing defects arise from inefficient recycling of trans-acting factors that improperly disassemble from defective 60S pre-ribosomal particles (32). Most importantly, we observed the accumulation of all pre-rRNA species from early 27SA2 to 7S and 25.5S precursors. This involves Rsa4p in almost all different steps of 60S r-subunit maturation, therefore, Rsa4p is not likely a bona fide pre-rRNA processing factor, such as a nuclease, but it could instead be involved in r-subunit assembly. Inhibition of 27S pre-rRNA processing steps is accompanied by nucleolar retention of pre-60S r-particles. This defect is apparently specific as export of small r-subunits was unaffected. Depletion of or mutation in a set of other protein trans-acting factors has specific export defects of pre-60S r-particles. Most of them lead to a nuclear (nucleoplasmic) rather than a nucleolar accumulation. These proteins include among others, Efl1p, Ipi1p, Ipi3p, Nog2p, Noc2p, Noc3p, Nug1p, Nug2p, Rea1p, Rix1p, Rrp12p and Tif6p (32,37–41). In addition, depletion of or mutations in Noc1p, Nog1p, Nop7p or Rix7p seem to cause a nucleolar retention of pre-60S r-particles (32,42–44). However, when studied, depletion of or mutation in any of the above factors did not result in pre-rRNA processing defects identical to those described for the Rsa4p depletion. Some resulted in a strong reduction of all precursors from the 27SB pre-rRNA as mutants in Noc2p and Noc3p (32). The phenotype observed upon depletion of Nog2p was the accumulation of the pre-rRNA intermediates 27SBS and 7SS (37). Mutation in Nop7p, Rix7p or Tif6p showed some accumulation of 27SB precursors and depletion of 7S pre-rRNAs (42,44,45). Next, reduction in 27SB precursors and a mild to slight accumulation of 7S pre-rRNA have been detected for mutants in Nug1p and Nug2p (38). Conditional strains for Ipi1p, Ipi3p and Rea1p showed only inhibition of 7S pre-rRNA processing, without apparent accumulation of this precursor (39). Finally, mutation in Rix1p had no effect on pre-rRNA processing (38). Based on these data, we conclude that Rsa4p carries out a specific function in pre-rRNA processing, assembly and export that are distinct from that performed for many other trans-acting factors required for nuclear export of pre-60S r-particles from the nucleolus. Since Rsa4p is conserved during evolution, orthologues found in higher eukaryotes are expected to perform similar functions.
A plausible explanation for our results takes into account that Rsa4p belongs to the family of WD-repeat proteins. Typically, members of this family adopt a propeller-like structure (15,46). Residues on the top and bottom of the propeller are involved in binding of specific proteins (47,48). In processes such as mRNA splicing or pre-mRNA 3′-end formation, which involve the formation of large multicomponent complexes, WD-repeat proteins are thought to function as scaffolding molecules to hold together the components of these complexes (49–52). In other processes, such as signalling transduction and translation, WD-repeat proteins serve as adaptors to present a protein substrate to other different proteins. For instance, yeast WD-repeat protein Asc1p (and its mammalian orthologue RACK1) is a 40S r-protein that recruits at the ribosome a variety of proteins, including RNA-binding proteins, translation factors and proteins involved in signal transduction. The connection of the ribosome with these different proteins provides a mechanism to regulate translation activity (48,53). We favour a model in which Rsa4p acts as a platform of interaction for several other trans-acting factors that are used for different structural rearrangements of the pre-60S r-particles during their maturation. These rearrangements control the transport from the nucleolus to the nucleoplasm and from there to the cytoplasm. Indeed, 60S r-subunit export require several AAA-ATPases and GTPases (37–39,42,54). In the absence of Rsa4p, the lack or delay in the interaction of the Rsa4p partners with the pre-60S r-particles might inhibit or strongly delay pre-rRNA processing at different steps, impede the proper recycling of trans-acting factors associated with pre-60S r-particles and block the release of these particles from the nucleolus.
Rsa4p is about the 20th WD-repeat protein that has been described as having a role during ribosome synthesis in S.cerevisiae. Many of them (Pwp2p, Dip2p, Utp13, Utp18 and Utp21) form a stably 25–30S complex required for the binding of U3 snoRNP and the Mpp10/Imp3/Imp4 complex to the 35S pre-rRNA during formation of 90S pre-ribosomal particles (55,56). WD-repeat proteins, such as Erb1p (57), Mak11p (36) and Ytm1p (13), are required for optimal 60S r-subunit biogenesis. Mak11p and Ytm1p have not been well characterized so far. Depletion of Erb1p results in a strong reduction of the different 27S pre-rRNAs species and 7S pre-rRNA (57). Unfortunately, these pre-rRNA phenotypes do not provide any clear insight into the role of Erb1p during 60S r-subunit assembly. Finally, two other WD-proteins Rrb1p and Sqt1p seem to serve as chaperones for assembly onto pre-60S r-particles of the 60S r-proteins Rpl3p and Rpl10p, respectively (58–60).
In conclusion, the most significant effect of Rsa4p depletion in yeast cells is the block of the synthesis of 60S r-subunits at all steps of 27S pre-rRNA processing. This is accompanied by the nucleolar retention of pre-60S r-particles. Taking into account the activity of WD-repeat proteins and the fact that Rsa4p remain associated with most pre-60S r-particles, it is reasonable to speculate that Rsa4p acts as an adaptor for recruitment of trans-acting factors required for pre-rRNA processing, assembly and export within different pre-60S r-particles (see Figure 2). Different approaches (i.e. synthetic lethal or two-hybrid studies) are now required to determine those trans-acting factors that work as Rsa4p interacting partners.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors are indebted to J. Bassler, O. Gadal, M. N. Hall and D. Kressler for their gift of the material used in this study. The authors are grateful to M. J. Quintero for technical assistance and S. Chávez and J. P. G. Ballesta for encouragement and helpful discussions. The authors thank D. Haun for style supervision. This work was supported by grants from the Spanish Ministry of Education and Science and FEDER (BFU2004-00252 to J.d.l.C and BFU2004-03079 to M.R.). The authors also thank the Fundación Ramón Areces (Madrid) for an institutional grant to the Centro de Biología Molecular ‘Severo Ochoa’ and the Andalusian Government (CVI271) for support. Funding to pay the Open Access publication charges for this article was provided by aforementioned grants from the Spanish Ministry of Education and Science and FEDER.
Conflict of interest statement. None declared.
REFERENCES
- 1.Olson M.O., Dundr M., Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- 2.Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 3.de la Cruz J., Kressler D., Linder P. Ribosomal subunit assembly. In: Olson M.O.J., editor. Nucleolus. Georgetown: Kluwer academic. Landes Bioscience/eurekah.com; 2004. pp. 258–285. [Google Scholar]
- 4.Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 5.Raué H.A. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine. In: Olson M.O.J., editor. Nucleolus. Georgetown: Kluwer academic. Landes Biosciences/Eurekah.com; 2004. pp. 199–222. [Google Scholar]
- 6.Fatica A., Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 7.Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 8.Trapman J., Retèl J., Planta R.J. Ribosomal precursor particles from yeast. Exp. Cell Res. 1975;90:95–104. doi: 10.1016/0014-4827(75)90361-4. [DOI] [PubMed] [Google Scholar]
- 9.Dez C., Froment C., Noaillac-Depeyre J., Monsarrat B., Caizergues-Ferrer M., Henry Y. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Biol. Cell. 2004;24:6324–6337. doi: 10.1128/MCB.24.14.6324-6337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 12.Nissan T.A., Bassler J., Petfalski E., Tollervey D., Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C.J., et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 14.Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2002;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith T.F., Gaitatzes C., Saxena K., Neer E.J. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 16.Thomas B.J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 17.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. Vol. 2. New York, NY: John Wiley & Sons, Inc.; 1994. pp. 13.10.11–13.14.17. [Google Scholar]
- 18.Kaiser C., Michaelis S., Mitchell A. Methods In Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 19.Gietz D., St Jean A., Woods R.A., Schiestl R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Oliver S.G., van der Aart Q.J., Agostoni-Carbone M.L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J.P., Benit P., et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 23.Bonneaud N., Ozier-Kalogeropoulos O., Li G.Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 24.Cid A., Perona R., Serrano R. Replacement of the promoter of the yeast plasma membrane ATPase gene by a galactose-dependent promoter and its physiological consequences. Curr. Genet. 1987;12:105–110. doi: 10.1007/BF00434664. [DOI] [PubMed] [Google Scholar]
- 25.Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 26.de la Cruz J., Kressler D., Rojo M., Tollervey D., Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 1998;4:1268–1281. doi: 10.1017/s1355838298981158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venema J., Planta R.J., Raué H.A. In vivo mutational analysis of ribosomal RNA in Saccharomyces cerevisiae. In: Martin R., editor. Protein Synthesis: Methods and Protocols. Vol. 77. Totowa, NJ: Humana Press; 1998. pp. 257–270. [DOI] [PubMed] [Google Scholar]
- 28.Rosado I.V., de la Cruz J. Npa1p is an essential trans-acting factor required for an early step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 2004;10:1073–1083. doi: 10.1261/rna.7340404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 30.van der Voorn L., Ploegh H.L. The WD-40 repeat. FEBS Lett. 1992;307:131–134. doi: 10.1016/0014-5793(92)80751-2. [DOI] [PubMed] [Google Scholar]
- 31.Gadal O., Strauß D., Kessl J., Trumpower B., Tollervey D., Hurt E. Nuclear export of 60S ribosomal subunit depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milkereit P., Gadal O., Podtelejnikov A., Trumtel S., Gas N., Petfalski E., Tollervey D., Mann M., Hurt E., Tschochner H. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell. 2001;105:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 33.Milkereit P., Strauss D., Bassler J., Gadal O., Kuhn H., Schutz S., Gas N., Lechner J., Hurt E., Tschochner H. A Noc-complex specifically involved in the formation and nuclear export of ribosomal 40S subunits. J. Biol. Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- 34.Habeler G., Natter K., Thallinger G.G., Crawford M.E., Kohlwein S.D., Trajanoski Z. YPL.db: the Yeast Protein Localization database. Nucleic Acids Res. 2002;30:80–83. doi: 10.1093/nar/30.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scherl A., Couté Y., Déon C., Callé A., Kindbeiter K., Sánchez J.C., Greco A., Hochstrasser D., Díaz J.J. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saveanu C., Namane A., Gleizes P.E., Lebreton A., Rousselle J.C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saveanu C., Bienvenu D., Namane A., Gleizes P.E., Gas N., Jacquier A., Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baßler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 39.Galani K., Nissan T.A., Petfalski E., Tollervey D., Hurt E. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J. Biol. Chem. 2004;279:55411–55418. doi: 10.1074/jbc.M406876200. [DOI] [PubMed] [Google Scholar]
- 40.Senger B., Lafontaine D.L., Graindorge J.S., Gadal O., Camasses A., Sanni A., Garnier J.M., Breitenbach M., Hurt E., Fasiolo F. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 41.Oeffinger M., Dlakic M., Tollervey D. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 2004;18:196–209. doi: 10.1101/gad.285604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gadal O., Strauss D., Braspenning J., Hoepfner D., Petfalski E., Philippsen P., Tollervey D., Hurt E. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 2001;20:3695–3704. doi: 10.1093/emboj/20.14.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karl T., Önder K., Kodzius R., Pichová A., Wimmer H., Thür A., Hundsberger H., Löffler M., Klade T., Beyer A., et al. GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr. Genet. 1999;34:419–429. doi: 10.1007/s002940050416. [DOI] [PubMed] [Google Scholar]
- 44.Oeffinger M., Leung A., Lamond A., Tollervey D., Lueng A. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA. 2002;8:626–636. doi: 10.1017/s1355838202020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basu U., Si K., Warner J.R., Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 2001;75:1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Nocker S., Ludwig P. The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics. 2003;4:50. doi: 10.1186/1471-2164-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson D.K., Cerna D., Chew E. The 1.1 Å structure of the spindle checkpoint protein Bub3p reveals functional regions. J. Biol. Chem. 2005;280:13944–13951. doi: 10.1074/jbc.M412919200. [DOI] [PubMed] [Google Scholar]
- 48.Baum S., Bittins M., Frey S., Seedorf M. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem. J. 2004;380:823–830. doi: 10.1042/BJ20031962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohnacker M., Barabino S.M., Preker P.J., Keller W. The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′ end-processing complex. EMBO J. 2000;19:37–47. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayadi L., Miller M., Banroques J. Mutations within the yeast U4/U6 snRNP protein Prp4 affect a late stage of spliceosome assembly. RNA. 1997;3:197–209. [PMC free article] [PubMed] [Google Scholar]
- 52.Albers M., Diment A., Muraru M., Russell C.S., Beggs J.D. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA. 2003;9:138–150. doi: 10.1261/rna.2119903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerbasi V.R., Weaver C.M., Hill S., Friedman D.B., Link A.J. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 2004;24:8276–8287. doi: 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kallstrom G., Hedges J., Johnson A. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dosil M., Bustelo X.R. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- 56.Krogan N.J., Peng W.T., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 57.Pestov D.G., Stockelman M.G., Strezoska Z., Lau L.F. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 2001;29:3621–3630. doi: 10.1093/nar/29.17.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schaper S., Fromont-Racine M., Linder P., de la Cruz J., Namade A., Yaniv M. A yeast homolog of chromatin assembly factor 1 is involved in early ribosome assembly. Curr. Biol. 2001;11:1885–1890. doi: 10.1016/s0960-9822(01)00584-x. [DOI] [PubMed] [Google Scholar]
- 59.Eisinger D.P., Dick F.A., Denke E., Trumpower B.L. SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant-negative mutations of the ribosomal protein gene QSR1. Mol. Cell. Biol. 1997;17:5146–5155. doi: 10.1128/mcb.17.9.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iouk T.L., Aitchison J.D., Maguire S., Wozniak R.W. Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome synthesis. Mol. Cell. Biol. 2001;21:1260–1271. doi: 10.1128/MCB.21.4.1260-1271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fatica A., Cronshaw A.D., Dlakic M., Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 62.Dez C., Tollervey D. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.