Abstract
Secondary structures of the G-rich strand of human telomere DNA fragments G3(TTAG3)n, n = 1–16, have been studied by means of circular dichroism spectroscopy and PAGE, in solutions of physiological potassium cation concentrations. It has been found that folding of these fragments into tetraplexes as well as tetraplex thermostabilities and enthalpy values depend on the number of TTAG3 repeats. The suggested topologies include, e.g. antiparallel and parallel bimolecular tetraplexes, an intramolecular antiparallel tetraplex, a tetraplex consisting of three parallel chains and one antiparallel chain, a poorly stable parallel intramolecular tetraplex, and both parallel and antiparallel tetramolecular tetraplexes. G3(TTAG3)3 folds into a single, stable and very compact intramolecular antiparallel tetraplex. With an increasing repeat number, the fragment tetraplexes surprisingly are ever less thermostable and their migration and enthalpy decrease indicate increasing irregularities or domain splitting in their arrangements. Reduced stability and different topology of lengthy telomeric tails could contribute to the stepwise telomere shortening process.
INTRODUCTION
Eukaryotic chromosomes are linear and have telomeres at their ends (1). A ribonucleoprotein complex, telomerase, ensures replication of the telomere (2). The telomere DNA is composed of the (TTAGGG)n/(CCCTAA)n repeats in humans and the G-rich strand forms a single-stranded overhang on the 3′ end of the molecule (3). The overhang folds into a hairpin containing guanine–guanine base pairs (4–6), or a DNA guanine tetraplex (7) under suitable conditions. The tetraplexes were detected in vivo (8–10), they are recognized by some proteins (11–14) and are probable to play an important role in telomerase regulation (15,16). The telomerase is inhibited by guanine tetraplex structures (17). Telomeres are highly conserved in human chromosomes (18); their length, however, is heterogeneous (19). The length of the telomeric DNA is related to aging and cancer (20,21). For example, telomere shortening triggers a p53-dependent cell cycle arrest via accumulation of G-rich single-stranded DNA fragments (22).
Guanine tendency to association has been known for many years (23–26). X-ray crystallographic (27,28) and solution NMR (29,30) studies showed that oligonucleotides related to telomere repeats associated into various types of tetraplex structures. Short oligonucleotides such as TTAG3, TG3T or TTG4 associate into tetramolecular parallel-stranded tetraplexes (31–33). Longer fragments generate hairpins associating into bimolecular tetraplexes in which the two hairpin strands have an antiparallel alignment (32). The human telomeric DNA sequence AG3(TTAG3)3 was shown (29) to form an intramolecular tetraplex in which each strand had one parallel and one antiparallel neighbor. Quite a different tetraplex type was observed (34) in crystals of human telomere DNA fragments TAG3TTAG3T and AG3(TTAG3)3. Their bimolecular and intramolecular tetraplexes contained parallel strands with three linking trinucleotide loops positioned on the sides of the quadruplex core, in a propeller-like arrangement (34).
Telomeric G-rich DNA fragments were studied by a number of methods (35–38) including circular dichroism (CD) spectroscopy (39–46). Parallel tetraplexes provide diagnostic CD spectra dominated by a strong positive band at 260 nm whereas antiparallel tetraplexes have the positive band at 290 nm (39,47). However, we have frequently observed CD spectra characteristic of parallel tetraplexes even with intramolecular and bimolecular tetraplexes that apparently should be antiparallel. For a long time, we had been wrestling with the question of how it is possible that CD spectroscopy, which is such a sensitive and reliable indicator of DNA secondary structure, fails in this case. The solution was the same as in previous cases. CD spectroscopy provided correct information. The explanation was found thanks to the crystal structures showing (34) that loops of bimolecular as well as intramolecular tetraplexes can be located on the sides of the tetraplex core, permitting to preserve the parallel orientation of the strands. Here we take advantage of this knowledge to study guanine tetraplex topology of human telomere DNA fragments differing by the number of (TTAG3) repeats.
MATERIALS AND METHODS
The oligonucleotides were purchased from VBC Genomics Bioscience Research (Vienna). The lyophilized oligonucleotides were dissolved in 1 mM sodium phosphate and 0.3 mM EDTA, pH 7, to give a stock solution concentration of ∼100 OD/ml. The precise sample concentrations were determined from their absorption measured at 90°C in the above buffer using molar extinction coefficients calculated according to Gray et al. (48). The UV absorption spectra were measured on a UNICAM 5625 UV/VIS spectrometer. Before starting the experiments, the samples of the G-rich oligonucleotides were denatured (10′ at 90°C) in the above low salt solution to remove aggregates. The sample was then left to cool to room temperature. The salt was added to the denatured sample at 0°C or at room temperature and the formation of tetraplexes was followed. In this way the CD spectral changes were reproducible starting from the same initial conditions.
CD spectra were measured using a Jobin-Yvon Mark VI dichrograph in 0.1 and 1 cm pathlength Hellma cells, placed in a thermostatted holder. The DNA concentration was chosen to give an absorption of ∼0.6–0.8 at the absorption maximum, which gives an optimum signal-to-noise ratio. CD was expressed as the difference in the molar absorption of the right-handed and left-handed circularly polarized light, Δɛ, in units of M−1 cm−1. To compare the oligonucleotides of different lengths and different primary structures, the molarity (M) was related to the number of guanine residues in the DNA samples.
The pH values were adjusted using either HCl or NaOH and checked directly in CD cells using a Sentron Red-Line electrode and a Sentron Titan pH meter. The buffer was sodium or potassium phosphate. KCl or NaCl were also added directly to the cells. The salt and DNA concentrations were then corrected for the increase of the sample volume.
Non-denaturing PAGE was performed in a thermostatted submersible apparatus (SE-600; Hoefer Scientific, San Francisco). Gels (16%, 29:1 monomer/bis ratio), 14 cm × 16 cm × 0.1 cm in size, were run for 20 h at 70 V (∼5 V/cm) and 0°C. DNA (2–3 µg) (∼10 µl of 0.7 mM DNA) was loaded on gels. The electrophoreses were run in the Robinson-Britton buffer pH 7.2 (26 mM mixture of boric, phosphoric and acetic acids plus 69 mM NaOH). Heteroduplexes of the studied oligonucleotides with their complementary C-rich strands were used as length markers. They were formed by mixing equimolar concentrations of both strands in low salt (1 mM Na phosphate plus 0.3 mM EDTA, pH 7) solution, heating to 90°C, followed by slow annealing to room temperature. The gels were stained with Stains-All (Sigma). Densitometry was performed using a Personal Densitometer SI, Model 375-A (Molecular Dynamics, Sunnyvale, CA).
The enthalpy of the individual tetraplexes was determined from the melting curves generated by monitoring a 290 nm CD band as a function of temperature according to (49). The transition enthalpy values were estimated only for fragments that melt in a two-state manner. The melting was measured in 10 mM potassium phosphate plus 0.15 M KCl, pH 7.
RESULTS
The results of this study follow from comparative CD spectroscopy, gel migration and UV absorption experiments on a set of 20 oligonucleotides containing pieces of human telomere DNA of various lengths and starting and terminal nucleotides. Here we mostly deal with the ‘blunt-ended’ fragments, as the TTA residues on the 5′ or 3′ molecule ends influence DNA tetraplex formation. The influence is, however, distinct in short and long molecules. Major conclusions of the paper are not influenced by the TTA tails. The study takes advantage of our experience with CD spectroscopy, which sensitively and reliably discriminates among various DNA secondary structures including variants of guanine tetraplexes (47). We have also used gel migration reflecting the number of associated molecules in the tetraplexes and compactness of their folding and UV absorption that reflects guanine tetraplex formation as well (50).
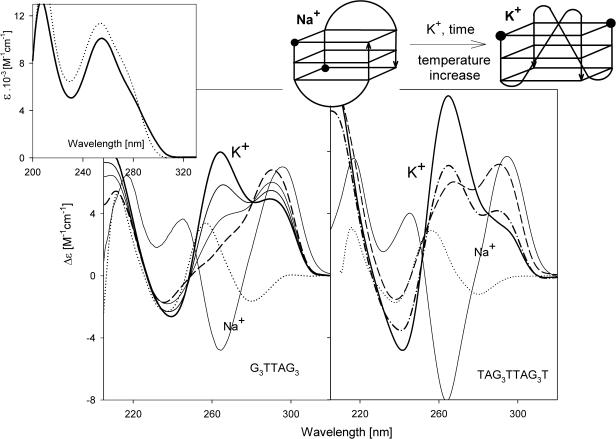
The first oligonucleotide studied is G3TTAG3. This 9mer provided a CD spectrum at low salt concentration (1 mM Na phosphate, 0.3 mM EDTA and pH 7, room temperature) corresponding to a denatured single strand (Figure 1, left). Sodium as well as potassium cations generated a dominant positive CD band at 290 nm reflecting the presence of an antiparallel-stranded guanine tetraplex (Figure 1, left). Changes in the UV absorption spectrum, i.e. hypochromism of the absorption maximum and mainly the increase of absorption at 295 nm, as compared with the denatured sample, also hint (50) at guanine tetraplex formation (Figure 1, inset). The antiparallel tetraplex, however, is unstable at 150 mM KCl because it isomerizes, with very slow kinetics (days), into a structure providing a dominant CD band at 260 nm, diagnostic (39,40) of a parallel guanine tetraplex. This isomerization is facilitated by increasing temperature. However, the isomerization was not finished at any conditions tested so that the oligonucleotide always coexisted in the antiparallel and parallel guanine tetraplexes in K+-containing solutions. In line with this conclusion following from the CD spectroscopy studies, gel migration revealed a coexistence of two bimolecular species of G3TTAG3 (Figure 2). One migrated faster than the heteroduplex of G3TTAG3 with the complementary strand C3TAAC3 and the other migrated more slowly. The faster species probably corresponds to the antiparallel tetraplex, and the slower one—whose population increases with time and temperature—to the parallel tetraplex. No such isomerization towards the parallel tetraplex was observed with G3TTAG3 in the presence of Na+ ions. In the presence of K+, a similar oligonucleotide TAG3TTAG3T crystallized (34) as a peculiar bimolecular tetraplex with a parallel orientation of strands whose loops were located on the sides of the tetraplex core (see the sketch in Figure 1). This oligonucleotide, however, adopted not only this parallel but also an antiparallel tetraplex in aqueous solution containing K+ ions (38). Our CD (Figure 1, right) and gel migration (Figure 2) data consistently demonstrated that TAG3TTAG3T coexisted in two bimolecular tetraplexes. Exposure to higher temperatures shifted the equilibrium to the side of the parallel tetraplex. Simultaneously, the population of the slower electrophoretic band increased. Both of these related oligonucleotides thus form both antiparallel and parallel bimolecular tetraplexes in K+-containing solutions. The 9mer G3TTAG3 isomerizes between the two conformers by means of a two-state process.
Figure 1.
CD spectra of G3TTAG3 (left) and TAG3TTAG3T (right) measured at 25°C (dots) in 1 mM Na phosphate plus 0.3 mM EDTA, pH 7 immediately after thermal denaturation. Measurements in 10 mM potassium phosphate plus 0.15 M KCl were carried out (left): immediately (dashes) and 1, 3 and 10 days (from the thinnest to the thickest line) after K+ addition. The samples were kept and measured at 25°C; (right): after two days keeping at 0°C and measured at 0°C (dashes) and 25°C (dash–dots) and after three days keeping at 25°C and measured at 25°C (thick line). The thin full line spectra in both panels correspond to the samples kept for two days in 10 mM Na phosphate plus 0.15 M NaCl at 0°C and measured at 0°C. Inset: UV absorption spectra of G3TTAG3 in 10 mM K phosphate plus 0.15 M KCl at 91°C (dots) and 25°C (solid line). The sketch shows the antiparallel and parallel bimolecular tetraplexes of TAG3TTAG3T. The balls in the sketch refer to the oligonucleotide 5′ ends.
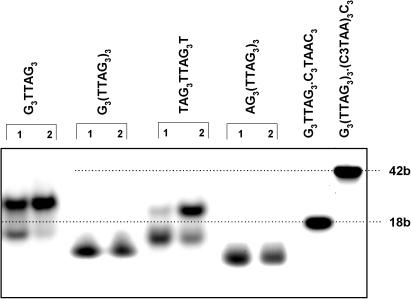
Figure 2.
Electrophoresis run in the Robinson–Britton buffer plus 0.15 M KCl, pH 7.2, at 2°C. The samples were incubated one day in the electrophoretic buffer at 0°C (lanes 1) and at 37°C (lanes 2). The sample of G3TTAG3 was loaded immediately (lane 1) on the gel and after two days incubation (lane 2) at room temperature. The heteroduplexes G3TTAG3•C3TAAC3 and G3(TTAG3)3•(C3TAA)3C3 were used as markers for mobility of 18 and 42 base long fragments, respectively.
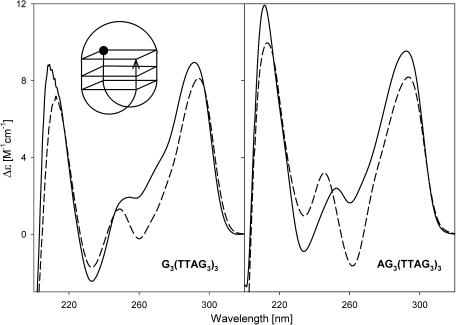
The intramolecular tetraplex of AG3(TTAG3)3 was also parallel-stranded with the loops on the sides in the crystal (34). However, this oligonucleotide generated an antiparallel tetraplex in aqueous solution, both in the presence of NaCl or KCl as indicated by CD (Figure 3). The same was true with G3(TTAG3)3. This is by far not the first example of DNA adopting qualitatively different conformers in aqueous solution and the crystalline state (51). The antiparallel tetraplexes of G3(TTAG3)3 and AG3(TTAG)3 were formed easily without any observable kinetics and any annealing. We observed no CD changes indicating isomerization towards parallel arrangement starting from low to 300 mM concentrations of KCl, after exposure of the sample to heating to various temperatures or pH changes. Both AG3(TTAG3)3 and G3(TTAG3)3 always provided only a single electrophoretic band (Figure 2) irrespective of the various solution conditions and irrespective of various sample treatments. Both oligonucleotides migrated significantly faster in the gel than would be expected for a single-stranded molecule of the same length (Figure 2). Hence the antiparallel tetraplexes of AG3(TTAG)3 and G3(TTAG3)3 are strictly defined, very compact monomolecular structures. Both tetraplexes melted cooperatively with increasing temperature and the melting was a two-state process, consistent with the above notion of only a single tetraplex at the beginning of melting. An antiparallel tetraplex of AG3(TTAG3)3 was observed (29) by NMR in solution in the presence of sodium cations (Figure 3, sketch).
Figure 3.
CD spectra of G3(TTAG3)3 and AG3(TTAG3)3 in 1 mM Na phosphate plus 0.3 mM EDTA, pH 7, measured at 0°C (dashes) and in 10 mM K phosphate plus 0.15 M KCl (solid lines). The same spectra were obtained at 0°C and at room temperature. The sketch shows the tetraplex structure of G3(TTAG3)3 or AG3(TTAG3)3. The ball in the sketch refers to the oligonucleotide 5′ end.
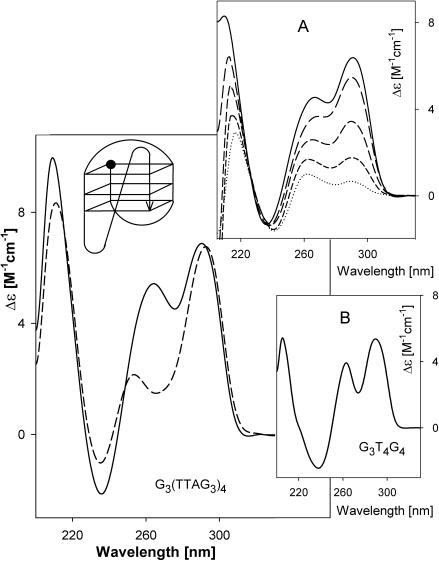
The CD spectrum of G3(TTAG3)4 in K+ solutions contains a decreased positive band at 290 nm compared with G3(TTAG)3 and an additional small positive band at 260 nm (Figure 4). Similar features were observed in the CD spectrum of G3TTAG3 (Figure 1). This means that the fourth TTAG3 repeat, containing the fifth G3 block, caused an increase in conformational features characteristic of a parallel-stranded tetraplex. In this case, however, there is not a mixture of two different structures, but a single strict arrangement. No exchange of 290 and 260 nm bands takes place, in contrast to the behavior described in Figure 1. The CD spectra neither change with time nor with temperature within the 0–50°C interval. Melting of the G3(TTAG3)4 tetraplex is a two-state process (isoelliptic points at 227 and 238 nm; Figure 4, inset A), which again implies a single melting structure. Electrophoresis also demonstrates a single intramolecular species of G3(TTAG3)4 (Figure 5). Hence the data may suggest that the extra TTAG3 repeat was used to generate a side loop, so that the resulting tetraplex would contain three parallel and one antiparallel strands (sketch in Figure 4). Such a structure consisting of three parallel and one antiparallel strands was reported (52) to be formed by a bimolecular tetraplex of G3T4G4. Its CD spectrum (Figure 4, Inset B) is essentially the same as the CD spectrum of G3(TTAG3)4.
Figure 4.
CD spectra of G3(TTAG3)4 in 1 mM Na phosphate plus 0.3 mM EDTA, pH 7 (dashes) and in 10 mM K phosphate plus 0.15 M KCl (solid line) measured at 0°C. Inset A: CD spectra of G3(TTAG3)4 in 10 mM K phosphate plus 0.15 M KCl measured at 45, 61, 68, 73 and 79°C (from the solid to the dotted line). Inset B: CD spectrum of G3T4G4 tetraplex measured in the same solution at 0°C. The sketch shows a suggested structure of G3(TTAG3)4 tetraplex. The ball refers to the oligonucleotide 5′ end.
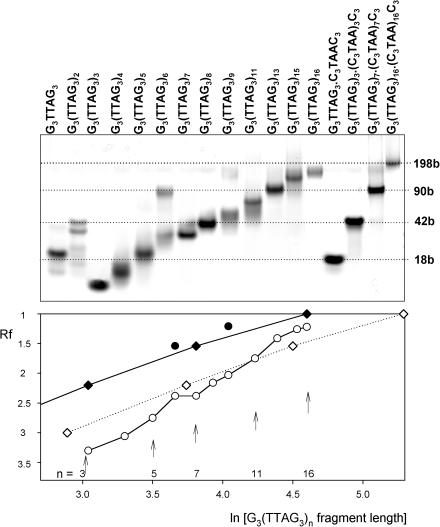
Figure 5.
Upper panel: electrophoresis of the telomere fragments (primary structures given above the lanes) run in the Robinson-Britton buffer plus 0.15 M KCl, pH 7.2, at 2°C. The samples were incubated one day in the electrophoretic buffer before loading on the gel. The heteroduplexes with complementary C-rich strands serve as markers for 18, 42, 90 and 198 base long fragments. Bottom panel: Rf—mobility related to that of G3(TTAG3)16•(C3TAA)16C3 heteroduplex as a function of natural logarithm of fragment lengths: heteroduplexes serving as markers (diamonds); bimolecular tetraplexes (solid circles); intramolecular tetraplexes (open circles). The dotted line with open diamonds simulates mobility of single-stranded molecules.
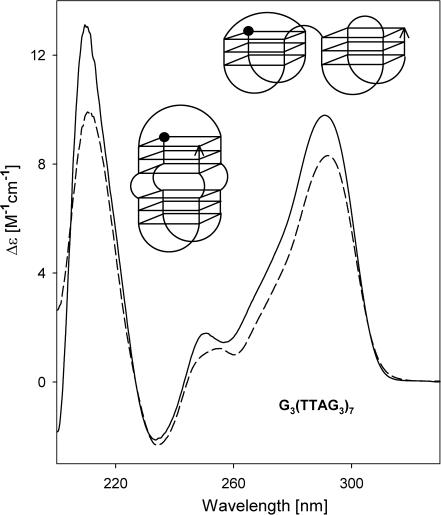
The CD spectrum of G3(TTAG3)7 is very similar to that of G3(TTAG3)3 (Figure 6). G3(TTAG3)7 migrated slightly faster than the heteroduplex of G3(TTAG3)3•(C3TAA)3C3 (Figure 5) indicating that G3(TTAG3)7 folded into an intramolecular tetraplex. The intramolecular antiparallel tetraplex melted through a two-state process (data not shown) and this oligonucleotide showed no sign of isomerization into an alternative structure. The thermal stability of the tetraplex of G3(TTAG3)7 was, however, lower than that of G3(TTAG3)3 (see below) and its migration was not so anomalous (Figure 5). The folding of G3(TTAG3)7 was therefore not as compact as that of G3(TTAG3)3.
Figure 6.
CD spectra of G3(TTAG3)7 in 1 mM Na phosphate plus 0.3 mM EDTA, pH 7 (dashed line) and 10 mM K phosphate plus 0.15 M KCl (solid line), both at 0°C. The sketches show two alternative arrangements of the G3(TTAG3)7 tetraplex fitting the results obtained. The ball refers to the oligonucleotide 5′ end.
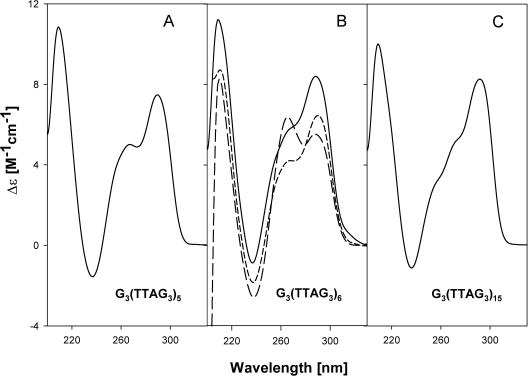
The CD spectrum of G3(TTAG3)5 displayed an increase in the 290 nm band and a decrease in that at 260 nm compared with G3(TTAG3)4 (Figure 7A). The tetraplex of G3(TTAG3)5 was intramolecular again (Figure 5). We expected G3(TTAG3)6 to have a suitable primary structure to generate a parallel tetraplex containing four TTAG3 repeats in the tetraplex core and three additional repeats in the side loops observed in the crystal (34). However, this was not the case. The CD band at 260 nm (characteristic of the parallel tetraplex) further decreased and that at 290 nm increased (Figure 7B) as compared with G3(TTAG3)4. It follows from electrophoretic measurements (Figure 5) that the oligonucleotide formed not only intramolecular but also bimolecular tetraplexes. However, increasing temperature enhanced, with long kinetics, the CD band at 260 nm with a simultaneous compensatory decrease of the CD band at 290 nm. Simultaneously the intramolecular tetraplex dominated in this sample as indicated by electrophoresis run at room temperature (data not shown). Hence increasing temperature induced a slow isomerization of the antiparallel tetraplex into a parallel intramolecular tetraplex, which, however, melted at a relatively low temperature (Figure 8). Thus, the intramolecular tetraplex, having three TTAG3 repeats in the loops, was rather unstable (Figure 8). The electrophoretic bands of G3(TTAG3)6 were diffused (Figure 5), thus indicating conformational exchanges.
Figure 7.
CD spectra of (A) G3(TTAG3)5, (B) G3(TTAG3)6 and (C) G3(TTAG3)15 in 10 mM potassium phosphate plus 0.15 M KCl measured at 0°C. In addition, the (B) contains CD spectra of G3(TTAG3)6 in the same solvent but measured at 37°C immediately after increasing temperature (short dashes), and after one day incubation at 37°C (long dashes).
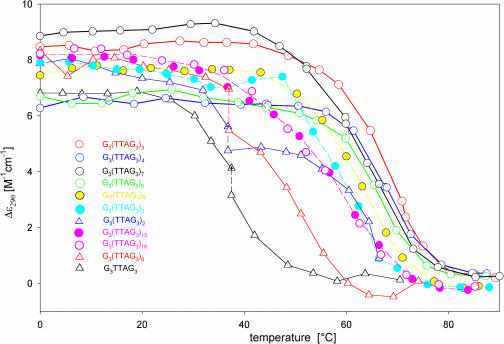
Figure 8.
Temperature dependences of the telomere DNA fragments in 10 mM potassium phosphate plus 0.15 M KCl, pH 7, monitored by changes in Δɛ290. All points, with the exception of those marked with a cross, correspond to equilibrium values. The equilibrium was reached within 110 min with G3TTAG3 and 20 h with G3(TTAG3)2 and G3(TTAG3)6. Solid lines and open circles: G3(TTAG3)3 (red), G3(TTAG3)4 (blue), G3(TTAG3)5 (green), and G3(TTAG3)7 (black); dashed lines and solid circles: G3(TTAG3)8 (yellow), G3(TTAG3)9 (cyan); G3(TTAG3)15 (violet solid circles) and G3(TTAG3)16 (violet open circles); thin solid lines and triangles: G3(TTAG3)2 (blue), G3(TTAG3)6 (red), and G3TTAG3(black).
G3(TTAG3)2 behaved in a similar way. A bimolecular tetraplex, whose core was formed by two repeats of each of the two participating molecules, had one repeat in the loop, or an overhang on its 5′ or 3′ end. It was unstable again and the oligonucleotide preferred to form tetramolecular tetraplexes (Figure 5). Increasing temperature led to time-dependent changes in the CD spectrum (Figure 8), reflecting an isomerization into a parallel tetraplex. Electrophoresis revealed formation of three kinds of bimolecular tetraplexes, the population of which increased with temperature (data not shown), and two kinds of tetramolecular tetraplexes. The latter tetraplexes probably differ by strand orientation. Thus even tetramolecular tetraplexes may be both parallel-stranded and antiparallel-stranded. The G3(TTAG3)2 conformation was relatively thermostable (Figure 8), for which the dominating tetramolecular G3(TTAG3)2 tetraplexes are probably responsible.
G3(TTAG3)8 only generated an intramolecular tetraplex like G3(TTAG3)4. G3(TTAG3)9 provided a diffuse band corresponding to an intramolecular tetraplex and a low population of a bimolecular tetraplex. We observed no isomerization with these oligonucleotides. Their tetraplex structures probably are too long to permit formation of side loops. The CD spectrum remained of the same type, with the dominant band at 290 nm, corresponding to an antiparallel tetraplex but with a shoulder from the shorter wavelength side (Figure 7C). This held for all longer fragments including G3(TTAG3)11 and G3(TTAG3)15 with the integral multiples of the four G3 blocks. The CD spectrum no longer corresponded to a regular intramolecular antiparallel tetraplex. Longer fragments obviously cannot generate a strict intramolecular tetraplex, maybe also owing to the possibility of the sequence to associate in both antiparallel and parallel way. Irregularities probably occur in the tetraplex structures of the long G3(TTAG3)n fragments, which are consistent with the observed decrease in thermostability with the increasing repeat number (Table 1).
Table 1.
Melting temperatures (Tm) and enthalpy (ΔH) values for the oligonucleotides G3(TTAG3)n
| G3(TTAG3)n | n = 3 | 4 | 5 | 6 | 7 | 8 | 9 | 11 | 13 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tm (°C) | 67 | 67.8 | 65.4 | 50.3 | 63 | 62.2 | 59.5 | 57.1 | 58.6 | 56.6 | 56.3 |
| ΔH (kJ mol−1) | −202 | −201 | −193 | — | −140 | −160 | — | −133 | −122 | −105 | −106 |
Table 1 also includes ΔH values calculated from the melting curves of telomeric fragments which adopt only a single type of arrangement. Table 1 shows that G3(TTAG3)3 displays the highest value of ΔH. The value corresponds to the published literature data (40,50). The enthalpies then decrease with the increasing repeat number. The close enthalpy values of the tetraplexes of G3(TTAG3)3, G3(TTAG3)4 and G3(TTAG3)5 indicate that their tetraplex cores are in principle similar, being formed by one unit of three guanine tetrads, though their detailed tetraplex structures are different. The sharp enthalpy decrease of the G3(TTAG3)7 tetraplex suggests that its structure is divided into more cooperative units which melt independently. This may be a consequence of TTA loops between two triads of guanine tetrads (the lower sketch in Figure 6), or, more probably, the triads may be positioned side by side like beads connected by the TTA sequence (the upper sketch in Figure 6). This alternative arrangement is supported by the enhanced electrophoretic mobility of G3(TTAG3)7 tetraplex. A further substantial decrease of ΔH observed with longer fragments indicates that their tetraplex structures are split into more units and/or contain irregularities. This is consistent with the decreased thermostabilities of the tetraplexes of the longer fragments. Simultaneously the gel mobility is decreased, indicating less compact arrangements of the tetraplexes.
DISCUSSION
The guanine tetraplex is a biologically relevant alternative of the Watson and Crick duplex of DNA. For example, it occurs at the ends of chromosomes where it may regulate telomere metabolism (6,7). Many dozens of papers were written about telomeres and some of them describe folding of the G-rich telomere strand into different types of guanine tetraplexes (27–38). However, many questions still remain open. For example, how the tetraplex folding depends on the number of DNA repeats in the telomere? Solution of this problem could help to understand the problem of telomere shortening, which accompanies important biological phenomena, such as aging or cancer (20).
In this paper, we study a DNA set of 20 human telomere fragments including G3(TTAG3)n, where n = 1–16. The study was performed at physiological concentrations of potassium cations using mostly CD spectroscopy and gel electrophoresis. CD spectroscopy is a very sensitive and reliable indicator of DNA secondary structure. It is a unique method in its ability to discriminate between non-cooperative conformational changes within a single structure and conformational isomerizations between distinct structures. The solution conditions inducing conformational changes can easily be changed directly in the CD cells, which enables mapping of the whole conformational space of the studied DNA molecule (47).
We show in this paper that tetraplex conformation, and the tetraplex topology in particular, depends on the number of (TTAG3) repeats in the human telomere. The shortest fragment G3TTAG3 coexists in the antiparallel and parallel tetraplexes (Figures 1 and 2). The parallel tetraplex was observed in the crystal of TAG3TTAG3T (34). Temperature influences the conformational equilibrium in solution while higher temperatures stabilize the parallel tetraplex. Coexistence of antiparallel and parallel tetraplexes was also observed with TAG3TTAG3T in solution by NMR (38).
The 21mer G3(TTAG3)3 contains four G3 blocks and folds into an intramolecular antiparallel tetraplex at all conditions tested in this work (Figures 2 and 3). We observed no indication of its isomerization into the parallel tetraplex adopted by a related AG3(TTAG3)3 in the crystalline state (34). In addition, we show here that this 22mer does not generate the parallel tetraplex in solution, either (Figures 2 and 3).
Surprisingly, extension of G3(TTAG3)3 by one TTAG3 repeat retains the tetraplex intramolecular (Figure 5) but the CD spectrum indicates an increased population of parallel tetraplex features (Figure 4). The oligonucleotide does not, however, adopt a mixture of tetraplex conformers but a single discrete conformation. We suggest that the extra TTAG3 repeat might be used to generate the side loop observed in the crystal (34), so that the tetraplex of G3(TTAG3)4 would contain three parallel strands and one antiparallel strand (Figure 4). Such a tetraplex was observed by NMR with G3T4G4 (52). Its CD spectrum is principally the same as in the case of G3(TTAG3)4 (Figure 4, inset B). The suggested 3 + 1 model fits our observations. However, it is to be noted that CD spectroscopy does not directly reflect strand orientation. We have numerous indications (M. Vorlíčková, unpublished data) that the population of syn/anti geometries of guanosine glycosidic torsion angles is what determines the CD spectral shape of the guanine tetraplexes. The syn/anti geometry is related to strand orientation but not absolutely.
Similar to G3(TTAG3)3, the 45mer G3(TTAG3)7 generates only a single stable conformer, i.e. an intramolecular antiparallel tetraplex (Figures 5 and 6). The CD spectra of G3(TTAG3)3 and G3(TTAG3)7 are almost identical. However, G3(TTAG3)7 is surprisingly less thermostable (Figure 8), and has a distinctly lower enthalpy (Table 1). It also does not migrate as fast as G3(TTAG3)3, even though it migrates faster than it would correspond to a monotonous dependence of Rf as a function of logarithm of fragment lengths (Figure 5, lower panel). This suggests that the G3(TTAG3)3 tetraplex has a very compact globular shape, whereas the tetraplex of G3(TTAG3)7 lacks this exceptionality.
We expected that the 39mer G3(TTAG3)6 would be the proper candidate to generate a stable parallel intramolecular tetraplex with three TTAG3 repeats in the side loops. This, however, was not the case. Apart from a monomolecular tetraplex, the 39mer dimerized into a bimolecular tetraplex (Figure 5). The CD spectrum of the tetraplexes displayed still increased ellipticity at 290 nm as compared with those of G3(TTAG3)4 and G3(TTAG3)5, and a compensatory decrease at 260 nm, which hints at an elevated amount of structural features characteristic of antiparallel tetraplexes (Figure 7B). Increasing temperature, however, induced a slow increase of the 260 nm CD band, indicating a conversion of the oligonucleotide conformation towards a parallel tetraplex. A simultaneous increase in the population of the monomolecular fraction in this sample (data not shown) indicated that the expected intramolecular parallel tetraplex was formed. The arising tetraplex, however, was rather unstable because the oligonucleotide melted at relatively very low temperatures (Figure 8). The 15mer G3(TTAG3)2, containing one redundant repeat, behaved in a similar way. Increasing temperature also induced a time-dependent isomerization towards a parallel tetraplex arrangement. The oligonucleotide formed not only bimolecular but also tetramolecular tetraplexes (Figure 5). The bimolecular tetraplex yielded three weak electrophoretic bands corresponding to three possible positions of the repeat that was not included in the tetraplex core, i.e. in the middle, or on the 3′ or 5′ ends. The population of bimolecular tetraplexes slightly increased with increasing temperature. G3(TTAG3)2, however, predominantly formed tetramolecular tetraplexes, which probably were responsible for a significantly higher thermostability of the oligonucleotide than was observed with G3(TTAG3)6 (Figure 8). It is interesting that two types of tetramolecular tetraplexes were formed by G3(TTAG3)2 (Figure 5). Obviously, even tetramolecular tetraplexes may associate not only in parallel but also in antiparallel orientation.
Tetraplexes of human telomere fragments longer than G3(TTAG3)7 all provided the same CD spectra with a dominant band at 290 nm and a shoulder at 260 nm (Figure 7). In no case, including G3(TTAG3)11 and G3(TTAG3)15, did the CD spectra correspond to the pure antiparallel tetraplexes adopted by G3(TTAG3)3 and G3(TTAG3)7. In addition to the intramolecular tetraplex, the 57mer G3(TTAG3)9 also formed bimolecular tetraplexes (Figure 5). No isomerization, however, was detected with increasing temperature with this oligonucleotide. All other longer oligonucleotides exclusively formed intramolecular tetraplexes (Figure 5). The increasing number of (TTAG3) repeats surprisingly caused thermal destabilization of their tetraplexes (Figure 8 and Table 1), a decrease in their ΔH values and a slower migration in the gel (Figure 5). A comparison of the migration of telomere fragments containing 4, 5, 6 and 7 G3 blocks, as well as of fragments containing 8, 9 and 10 blocks, suggests that the repeats not included in the tetraplex core slow down the migration. However, ΔH values of G3(TTAG3)3, G3(TTAG3)4 and G3(TTAG3)5 tetraplexes are nearly the same. This indicates that the fragments generate a single similar tetraplex unit, consisting of three guanine tetrads, though detailed tetraplex arrangements are different. The large decrease of ΔH values with the tetraplex of G3(TTAG3)7 indicates tetraplex splitting into more cooperative units, perhaps two beads shown in Figure 6. The value of ΔH further decreases with longer telomeric fragments, which may reflect an increasing number of cooperative units in the structure. The relatively faster migration of fragments containing 4, and to a lesser extent 8 and 12 G3 blocks, may support the possibility of their folding into beads, like in the case of nucleosomes. We will further study this possibility. In general, longer fragments migrate more slowly, the longer they are. It seems that increasing amounts of the (TTAG3) repeats in the fragment increase the number of available conformations. The presence of various loops on the tetraplex sides then probably leads to irregularities in tetraplex arrangements and, consequently, to the structure destabilization.
The most stable tetraplex is formed by G3(TTAG3)3. Hence the four G3 blocks in the human telomere repeats, and not the longer segments, are the most promising candidates for tetraplex formation in vivo. Reduced stability and different topology of lengthy telomeric tails could contribute to the stepwise telomere shortening process.
Acknowledgments
The authors thank Mgr Daniel Renciuk for undertaking preliminary DMS experiments. The study was supported by grant A 4004201 from the Grant Agency of the Academy of Sciences of the Czech Republic, grant NM/7634-3 from the Ministry of Health of the Czech Republic, and by the institutional grant A50040507. Funding to pay the Open Access publication charges for this article was provided by the above A4004201 and NM/7634-3 grants.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn E.H., Greider C.W. Telomeres. Plainview, NY, USA: Cold Spring Harbor Laboratory; 1995. [Google Scholar]
- 2.Cech T.R., Nakamura T.M., Lingner J. Telomerase is a true reverse transcriptase. A review. Biochemistry (Mosc) 1997;62:1202–1205. [PubMed] [Google Scholar]
- 3.Londono-Vallejo A., DerSarkissian H., Cazes L., Thomas G. Differences in telomere length between homologous chromosomes in humans. Nucleic Acids Res. 2001;29:3164–3171. doi: 10.1093/nar/29.15.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson E., Hardin C.C., Walk S.K., Tinoco I.J., Blackburn E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine–guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 5.Choi K.-H., Choi B.-S. Formation of a hairpin structure by telomere 3′ overhang. Biochim. Biophys. Acta. 1994;1217:341–344. doi: 10.1016/0167-4781(94)90298-4. [DOI] [PubMed] [Google Scholar]
- 6.Sundquist W.I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 7.Murchie A.I.H., Lilley D.M.J. Tetraplex folding of telomere sequences and the inclusion of adenine bases. EMBO J. 1994;13:993–1001. doi: 10.1002/j.1460-2075.1994.tb06344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao E.H., Sun X.G., Zhang X.Y., Li J.W., Bai C.L. Fold-back tetraplex DNA species in DNase I-resistant DNA isolated from HeLa cells. J. Biomol. Struct. Dyn. 2000;17:871–878. doi: 10.1080/07391102.2000.10506575. [DOI] [PubMed] [Google Scholar]
- 9.Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H., Plückthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lew A., Rutter W.J., Kennedy G.C. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc. Natl Acad. Sci. USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang G., Cech T.R. The β subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 12.Laporte L., Thomas G.J., Jr Structural basis of DNA recognition and mechanism of quadruplex formation by the β subunit of the Oxytricha telomere binding protein. Biochemistry. 1998;37:1327–1335. doi: 10.1021/bi972400d. [DOI] [PubMed] [Google Scholar]
- 13.Frantz J.D., Gilbert W. A yeast gene product, G4p2, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem. 1995;270:9413–9419. doi: 10.1074/jbc.270.16.9413. [DOI] [PubMed] [Google Scholar]
- 14.Muniyappa K., Anuradha S., Byers B. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol. Cell. Biol. 2000;20:3648–3658. doi: 10.1128/mcb.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher T.M., Sun D., Salazar M., Hurley L.H. Effect of DNA secondary structure on human telomerase activity. Biochemistry. 1998;37:5536–5541. doi: 10.1021/bi972681p. [DOI] [PubMed] [Google Scholar]
- 16.Sun D., Lopez-Guajardo C., Quada J., Hurley L., Von Hoff D. Regulation of catalytic activity and processivity of human telomerase. Biochemistry. 1999;38:4037–4044. doi: 10.1021/bi982249n. [DOI] [PubMed] [Google Scholar]
- 17.Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 18.Moyzis R.K., Buckingham J.M., Cram L.S., Dani M., Deaven L.L., Jones M.D., Meyne J., Ratliff R.L., Wu J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lansdorp P.M., Verwoerd N.P., van de Rijke F.M., Dragowska V., Little M.T., Dirks R.W., Raap A.K., Tanke H.J. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 20.Hastie N., Dempster M., Dunlop M., Thompson A., Green D., Allshire R. Telomere reduction in human colorectal-carcinoma and with aging. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 21.Blackburn E.H. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 22.Saretzki G., Sitte N., Merkel U., Wurm R., von Zglinicki T. Telomere shortening triggers a p53-dependent cell cycle arrest via accumulation of G-rich single stranded DNA fragments. Oncogene. 1999;18:5148–5158. doi: 10.1038/sj.onc.1202898. [DOI] [PubMed] [Google Scholar]
- 23.Ralph R.K., Connors W.J., Khorana H.G. Secondary structure and aggregation in deoxyguanosine oligonucleotides. J. Am. Chem. Soc. 1962;84:2265–2266. [Google Scholar]
- 24.Gellert M., Lipsett M., Davies D. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2019. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray D.M., Bollum F.J. A circular dichroism study of poly dG, poly dC and poly dG:dC. Biopolymers. 1974;13:2087–2102. doi: 10.1002/bip.1974.360131011. [DOI] [PubMed] [Google Scholar]
- 26.Guschlbauer W., Chantot J.-F., Thiele D. Four-stranded nucleic acid structures 25 years later: from guanosine gels to telomer DNA. J. Biomol. Struct. Dyn. 1990;8:491–511. doi: 10.1080/07391102.1990.10507825. [DOI] [PubMed] [Google Scholar]
- 27.Laughlan G., Murchie A.I.H., Norman D.G., Moore M.H., Moody P.C.E., Lilley D.M.J., Luisi B. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science. 1994;265:520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- 28.Cáceres C., Wright G., Gouyette C., Parkinson G., Subirana J.A. A thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+/Na+ ions. Nucleic Acids Res. 2004;32:1097–1102. doi: 10.1093/nar/gkh269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Patel D.J. Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol. 1993;234:1171–1183. doi: 10.1006/jmbi.1993.1668. [DOI] [PubMed] [Google Scholar]
- 31.Aboul-ela F., Murchie A.I.H., Lilley D.M.J. NMR study of parallel-stranded tetraplex formation by the hexadeoxynucleotide d(TG4T) Nature. 1992;360:280–282. doi: 10.1038/360280a0. [DOI] [PubMed] [Google Scholar]
- 32.Jin R.Z., Gaffney B.L., Wang C., Jones R.A., Breslauer K.J. Thermodynamics and structure of a DNA tetraplex—a spectroscopic and calorimetric study of the tetramolecular complexes of d(TG3T) and d(TG3T2G3T) Proc. Natl Acad. Sci. USA. 1992;89:8832–8836. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Patel D.J. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplex with anti glycosidic torsion angles in solution. Biochemistry. 1992;31:8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- 34.Parkinson G.N., Lee M.P., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 35.Miura T., Thomas G.J. Structural polymorphism of telomere DNA: interquadruplex and duplex–quadruplex conversions probed by Raman spectroscopy. Biochemistry. 1994;33:7848–7856. doi: 10.1021/bi00191a012. [DOI] [PubMed] [Google Scholar]
- 36.Phan T.P., Mergny J.L. Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Risitano A., Fox K. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 38.Phan A.T., Patel D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balagurumoorthy P., Brahmachari S.K., Mohanty D., Bansal M., Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balagurumoorthy P., Brahmachari S.K. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 41.Giraldo R., Suzuki M., Chapman L., Rhodes D. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: A circular dichroism study. Proc. Natl Acad. Sci. USA. 1994;91:7658–7662. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W., Wu P., Ohmichi T., Sugimoto N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
- 43.Li W., Miyoshi D., Nakano S., Sugimoto N. Structural competition involving G-quadruplex DNA and its complement. Biochemistry. 2003;42:11736–11744. doi: 10.1021/bi034168j. [DOI] [PubMed] [Google Scholar]
- 44.Dapic V., Abdomerovic V., Marrington R., Peberdy J., Rodger A., Trent J.O., Bates P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hazel P., Huppert J., Balasubramanian S., Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 46.Rujan I.N., Meleney C., Bolton P.H. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vorlickova M., Kypr J., Sklenar V. In: Encyclopedia of Analytical Science. 2nd edn. Worsfold P.J., Townshend A., Poole C.F., editors. Vol. 6. Oxford: Elsevier; 2005. pp. 391–399. [Google Scholar]
- 48.Gray D.M., Hung S.-H., Johnson K.H. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 49.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 50.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 51.Kypr J., Chladkova J., Zimulova M., Vorlickova M. Aqueous trifluorethanol solutions simulate the environment of DNA in the crystalline state. Nucleic Acids Res. 1999;27:3466–3473. doi: 10.1093/nar/27.17.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sket P., Crnugelj M., Plavec J. d(G3T4G4) forms unusual dimeric G-quadruplex structure with the same general fold in the presence of K+, Na+ or NH4+ ions. Bioorg. Med. Chem. Lett. 2004;12:5735–5744. doi: 10.1016/j.bmc.2004.08.009. [DOI] [PubMed] [Google Scholar]