Abstract
RNA interference (RNAi) is a process of post-transcriptional gene silencing initiated by double-stranded RNAs, including short interfering RNA (siRNA). Silencing is sequence-specific and RNAi has rapidly become central to the study of gene function. RNAi also carries promise for selective silencing of viral and endogenous genes causal for disease. To detect the very low levels of siRNA effective for RNAi we modified the 3′ end of the sense strand of siRNA with a nuclease-resistant DNA hairpin. We show that the modified siRNA-DNA construct (termed ‘crook’ siRNA) functions as a primer for the PCR and describe a novel, yet simple PCR protocol for its quantification (amolar levels/cell). When transfected into mammalian cells, crook siRNA induces selective mRNA knock-down equivalent to its unmodified siRNA counterpart. This new bifunctional siRNA construct will enable future in vivo studies on the uptake, distribution and pharmacokinetics of siRNA, and is particularly important for the development of siRNA-based therapeutics. More generally, PCR-based detection of siRNA carries wide-ranging applications for RNAi reverse genetics.
INTRODUCTION
Synthetic short interfering RNAs (siRNAs) are widely employed to study eukaryotic gene function by RNA interference (RNAi), and siRNA also holds promise for the development of novel therapeutics that circumvent the potential dangers of gene therapy (1–4). The induction of RNAi by siRNA is highly efficient and there is a need for sensitive quantification of siRNA delivery and distribution within multicellular systems. The aim of this work was to construct an siRNA derivative that (i) enables its detection by PCR-based technology and (ii) retains RNAi activity for selective knock-down of targeted mRNA in mammalian cells. Our approach was to modify the sense strand of the siRNA duplex to enable its use as a primer for the PCR. Accordingly the 3′ end of the sense strand of siRNA (21 nt) was extended by a 19 nt DNA sequence to allow the siRNA to function as a PCR primer. The end of the DNA primer sequence, d(GCGAAGC), forms a stable hairpin structure and confers nuclease resistance (5–9). This siRNA–DNA structure is envisaged as a shepherd's crook and is termed crook siRNA. As biological model for testing the ability of crook siRNA to induce RNAi, we used a line of human cervical cancer cells (SiHa) positive for human papilloma virus type 16 (HPV16) (10). These cells express viral E6 and E7 genes and we used E7 siRNA (11) as foundation for the construction of E7 crook siRNA (Figure 1a). First we tested the ability of E7 crook siRNA to function as a primer for PCR amplification of DNA. For this purpose, we developed a simple protocol which is applicable for use with different siRNA sequences linked with a common DNA extension to form the crook siRNA.
Figure 1.
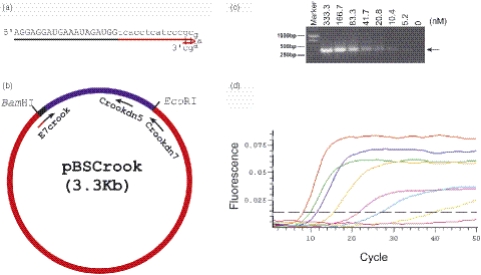
Design and application of E7 crook siRNA for use in quantitative PCR. (a) Sequence and schematic representation of E7 crook siRNA. (b) Design of the crook template and positions of upstream (E7 crook siRNA) and downstream primers (crook dn7 and crook dn5) for PCR amplification. The crook template (312 bp) is derived from the CMV promoter and is cloned into pBlueScript (pBS) to give pBSCrook (Materials and Methods). (c) Visualization of crook template DNA after PCR amplification using E7 crook siRNA and crook dn 7 as primers and agarose gel electrophoresis. E7 crook siRNA was added in a limiting dilution series, as indicated; crook template and crook dn7 primer were present in excess for the reaction (see text). (d) Read-out from quantitative PCR showing effects of E7 crook siRNA dilution series (333.3 to 5.2 nM) on product amplification and plateau levels (see text).
MATERIALS AND METHODS
Construction of crook DNA template
Forward primer 1 that contained crook sequence at the 5′ end (5′-tcacctcatcccgcgaagcccatatatggagttcc-3′) and reverse primer 1 (5′-agcgatgactaatacgtagatgtac-3′) were used to amplify the primary crook template from the CMV promoter sequence. The final crook DNA template was amplified with forward primer 2 (5′-cgcggatcctcacctcatcccgcga-3′) and reverse primer 2 (5′-gcggaattcaagtaggaaagtcccataaggt-3′) and cloned into pBS at BamHI/EcoRI, as pBSCrook (Figure 1b). The PCRs were performed using Qiagen Hotstart PCR kit and the cycle as follows: 95°C for 15 min, 30 cycles of 94°C for 45 s, 58°C for 45 s, 72°C for 1 min, then 72°C for 5 min. In pBSCrook template, there are two downstream primers usable for E7 crook detection, designated crook dn5 (5′-gtcaatagggggcgtacttg-3′) and crook dn7 (5′-gtaatacgactcactatagggcgaattggg-3′), which give single products of 250 and 400 nt, respectively.
Cell culture
Human HPV16 positive cell line SiHa was cultured with MEM plus 10% FCS (Sigma), 1.0 mM sodium pyruvate and 0.1 mM non-essential amino acids. Human colorectal carcinoma HCT116 cells were cultured in DMEM with 10% FCS. HeLa cells containing stably integrated HIV-1 Tat gene under the Tet-Off promoter, firefly luciferase gene under HIV-1 LTR and Renilla luciferase gene under CMV promoter as described previously (12) were cultured as exponentially growing subconfluent monolayers on plates in DMEM supplemented with 10% Tet System Approved FBS (Clontech). All the cell lines were maintained with penicillin 100 U/ml and streptomycin 100 µg/ml at 37°C in a 5% CO2 incubator.
Total RNA preparation
The transfected cells were washed three times with PBS, and harvested by trypsinization. The cells were counted and washed with PBS, after centrifugation at 200 g for 5 min to pellet. The cell pellets were lysed in Lysis buffer A (140 mM NaCl, 10 mM Tris and 0.5% NP-40, pH 8.0) on ice for 10 min. The lysis solutions were extracted with 1:1 vol of phenol once and 1:1 vol of chloroform/isoamyl alcohol (24:1) once. The total RNA were precipitated by adding 1/10 vol of 3 M NaOAc (pH 5.2), 1 µl glycoblue (Ambion) and 2.5 vol of ice-cold 100% ethanol at −20°C for at least 1 h. After centrifugation at 12000 g for 30 min at 4°C, the RNA pellets were washed once with 75% ice-cold ethanol, dried and resuspended in 100 µl RNase/DNase-free ddH2O.
Detection of E7 crook siRNA by real-time PCR
The real-time PCRs were performed using DNA Engine Opticon 2 System (MJ Research, USA) and QuantiTect SYBR Green PCR kit (Qiagen, UK). In each reaction, the final concentration of pBSCrook template and downstream primer Crook dn7 are 200 pg/µl and 0.33 µM, respectively. For the standard curve, the input of E7 crook was diluted from 333.3 to 5.2 nM (Figure 1c). The cycle was performed as follows: 94°C for 15 min, 50 cycles of 94°C for 45 s, 60°C for 45 s and 72°C for 45 s. For the detection of E7 crook in transfected cells, 500 ng of total RNA was used in each reaction. For E7 crook dose-dependent detection, 1 µg total RNA was used for each reaction. All quantification statistics were calculated using Sigma plot 8.0. The standard curves were plotted as shown in Figure 2b. The equation for quantification of E7 crook in transfected cells is Yi = Y1 + (Xi − X1) × (Y2 − Y1)/(X2 − X1) where X represents C(T) cycle value and Y represents E7 crook amount value.
Figure 2.
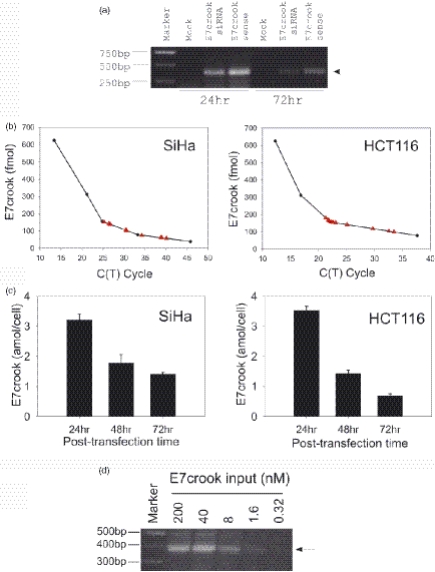
Recovery and quantification of E7 crook siRNA after transfection into human cells in culture. (a) Recovery of E7 crook siRNA duplex and also of single-stranded E7 crook siRNA sense strand at 24 and 72 h after transfection in to SiHa cells visualized indirectly by its ability to prime the amplification of the crook template. Parallel cell cultures were transfected with either duplex E7 crook siRNA or with single-stranded E7 crook sense RNA, as indicated. (b) Quantification of E7 crook siRNA (red triangles) using standard curves derived from a dilution series of E7 crook siRNA (Materials and Methods; internal standard dilution curves were performed for each experiment). E7 crook siRNA was recovered with total RNA 24 h after transfection into either SiHa or HCT116 cells, as indicated. (c) Recovery levels of E7 crook siRNA at 24, 48 and 72 h post-transfection into SiHa and HCT116 cells as indicated. (d) Detection of E7 crook siRNA from SiHa cells transfected with different input doses of the crook siRNA, as indicated.
RNAi as evidenced by mRNA knock-down and cellular phenotype
siRNAs [Figure 1; (11)] were synthesized and HPLC purified (Dharmacon). For annealing of the siRNAs, 20 µM complementary RNAs were incubated in annealing buffer (20 mM Tris–HCl, pH 7.5, 10 mM MgCl and 50 mM NaCl) for 1 min at 90°C followed by 1 h at 37°C. For transfection the SiHa and HCT116 cells were trypsinized and subbed into 6-well plates (10 cm2) without antibiotics, 1.5 × 105 cells per well. After 24 h the cells were transfected with siRNA formulated into liposomes (Oligofectamine; Life Technologies) according to the manufacturer's instructions. Routine siRNA concentration was 0.58 µg per 1.5 × 105 cells per well (equivalent to 200 nM). In addition, an siRNA dilution series from 200 nM, 40 nM, 8 µM, 1.6 nM to 0.32 nM was performed. The final volume of culture medium was 1.5 ml per well. Cells were harvested for analysis at various times thereafter as indicated in Results. Each experiment was carried out four or more times. Transfection efficiencies were established by transfecting with liposomes containing FITC-dextran (FD-150; Sigma). For E7 mRNA amplification by real-time PCR the primers were 5′-CGGAATTCATGCATGGAGATACACCTACAT-3′ and 5′-CGGGAAGCTTATGGTTTCTGAGAACAGATGG-3′, and the thermal cycle was 47°C for 30 min; 94°C for 2 min; then 30 cycles of 94°C for 45 s, 58°C for 45 s and 72°C for 2 min; followed by 72°C for 5 min. For lamin A/C mRNA amplification the primers 5′-AAGCAGCGTGAGTTTGAGAGC-3′ and 5′-AGGGTGAACTTTGGTGGGAAC-3′ were used in the thermal cycle: 50°C for 30 min, 94°C for 15 min, then 30 cycles of 94°C for 45 s, 58°C for 45 s, 72°C for 1 min, followed by 80°C for 15 s. For cell cycle analysis the cells were harvested, washed with PBS and fixed in 90% ethanol overnight at −20°C. The fixed cells were pelleted, washed in PBS and resuspended in PBS containing 0.1 µg/ml propidium iodide with 200 U/ml RNase A and analysed by using FACS. Apoptotic cells were identified using annexin-V-Fluos (Boehringer) following the manufacturer's protocol.
RNAi as evidenced by the luciferase assay
siRNAs comprising the following sense strands were used (5′–3′): GL3-FFL siRNA (13), CUUACGCUGAGUACUUCGAtt; GL3-FFL crook siRNA, CUUACGCUGAGUACUUCGAtcacctcatcccgcgaagc; GL3-FFL mism siRNA, CGUACGCGGAAUACUUCGAtt; and as a negative siRNA control, non-specific control VI (Dharmacon). To assess RNAi a dual luciferase reporter assay was used. In each experiment, two identical 96-well plates were prepared with 104 HeLa Tet-Off/Tat/luc-f/luc-R cells per well and incubated at 37°C for 24 h. One of the plates was used for the luciferase assay and the other for the cytotoxicity assay. After annealing in Dharmacon siRNA buffer, siRNAs were prepared at a concentration of 500 nM in Opti-MEM (Invitrogen) serum-free medium and formed a complex with a 1:200 vol of Lipofectamine 2000 (Invitrogen). After 20 min at room temperature, subsequent dilutions were prepared from siRNA/Lipofectamine 2000 mixture. Cells were incubated with the mixtures for 3 h, washed with PBS and left for additional incubation in DMEM/10% FBS for 24 h. Cell lysates were prepared and analysed using the Dual Luciferase Reporter Assay System (Promega) and relative light units for both firefly and Renilla luciferase read sequentially using a Berthold Detection Systems Orion Microplate luminometer. Toxicity was determined by measurement of the proportion of live cells colorimetrically using CellTiter 96 AQueous One Solution Assay (Promega). The absorbance at 490 nm was read using a Molecular Devices Emax microplate reader. Each data point was averaged over two replicates of three separate experiments.
RESULTS
Concept for PCR-based quantification of E7 crook siRNA
The PCR is routinely employed to detect and to quantify a DNA template (or RNA template via reverse transcription) by cyclical amplification in vitro. The sample containing the DNA template is first heated to separate the complementary strands of DNA, including those of the DNA template of interest. Two opposing DNA primers are included, which are designed to complement two defined sequences within the DNA template and separated by a known distance. These upstream and downstream primers serve to initiate synthesis of a DNA product of predicted length, synthesis being dependent upon the presence of the template. Importantly in standard PCR, the two PCR primers are added in excess for the reaction.
In the present study, we have altered the standard PCR protocol to allow for a limiting quantity of the E7 crook siRNA (upstream) primer. Thus the E7 crook siRNA primer is limiting for the reaction, whereas the second (downstream) primer and the DNA template are added in excess for the reaction. Under these conditions it was predicted that the initial rate of template amplification would correlate with the initial concentration of the E7 crook siRNA primer. It was also anticipated that, during the repeated cycles of the PCR process, the E7 crook siRNA primer would become diluted to the point where it could no longer support the priming of new strand DNA synthesis, even though the downstream primer and the DNA template remain in excess: at this point template amplification should plateau.
Design of E7 crook siRNA and PCR template
The sequence of the E7 crook siRNA (sense strand) and the design of the crook DNA template is shown schematically (Figure 1a and b). The crook DNA template is derived from the cytomegalovirus (CMV) promoter sequence and, for ease of production and general accessibility, is cloned into pBS to give the pBSCrook plasmid (3.3 kb; Materials and Methods). In the pBSCrook plasmid we identified two alternative downstream sites as targets for reverse PCR primers (designated crook dn5 and crook dn7; see Materials and Methods and Figure 1b). When used in conjunction with E7 crook siRNA (upstream primer) the crook dn5 and crook dn7 primers are predicted to give single products of 250 and 400 nt, respectively.
PCR-based detection and quantification of E7 crook siRNA
The ability of E7 crook siRNA to support PCR was demonstrated using crook dn5 or crook dn7 as downstream primer. Upon amplification single PCR products of 250 and 400 nt, respectively, were observed (see Figure 1c for crook dn7 primer). To test the sensitivity of the reaction, we employed a dilution series of E7 crook siRNA ranging from 333.3 down to 5.2 nM. An excess of downstream DNA primer (crook dn7) and crook template was used throughout (Materials and Methods). A single melting point spanning 81.5–83°C was obtained (data not shown) and, as predicted, the product yield tended to plateau at lower levels with increasing dilution of the E7 crook siRNA primer (Figure 1d). A single PCR product (400 nt) was detectable (Figure 1c and d).
Similar sensitivity has been reported by Overhoff et al. (14) using a complex, indirect method for the detection of siRNA by liquid hybridization with a 32P-labelled probe. Hybridization is followed by a nuclease protection step and the resultant radiolabelled samples are analysed by PAGE and blotting onto nylon membrane. A phosphorimager is used to detect and to quantify the radioactive signal on the blots. In addition to being indirect, the hybridization selectivity and sensitivity of the 32P-labelled probe will vary depending upon siRNA mismatch tolerance and also upon competing total cellular RNA sequences.
In contrast, crook siRNA quantification is both direct and represents a constant for different siRNAs since detection is reliant upon the DNA component of the crook siRNA construct (Figure 1). The siRNA component of crook siRNA can be varied according to the chosen gene target. Moreover our non-radioactive approach exploits the simple principles of PCR. Thus, crook siRNA may be employed to explore aspects of RNAi that have hitherto proved problematic for study owing to the very low levels of siRNA involved. With this objective in mind we next asked if crook siRNA can be recovered from cells and quantified in the presence of cellular RNA.
Recovery and quantification of crook siRNA from transfected mammalian cells
Previously we have shown that routine transfection of E7 siRNA into SiHa cervical cancer cells induces growth arrest and apoptosis (11). In contrast, the growth and viability of non-virally infected cells, such as normal human diploid fibroblasts and HCT116 human colorectal cancer cells, are unaffected by E7 siRNA (11). Using SiHa and HCT116 cells, we now asked if E7 crook siRNA can be recovered from cells after liposomal transfection (the percentage cells transfected was between 70 and 80% for both SiHa and HCT116 cells). After transfection, the cells were harvested at various times up to 72 h and total RNA was extracted as described in Materials and Methods. Care was taken to wash the cells extensively before lysis, thus removing unincorporated liposomal/crook siRNA complexes before cellular RNA extraction. A single product of the expected size was obtained by quantitative PCR (see Figure 2a for SiHa samples). Parallel controls show that the single-stranded E7 crook siRNA (sense strand) is also recoverable up to at least 72 h after transfection into SiHa cells and amplifies to give a single PCR product (Figure 2a). This is important since it demonstrates that the crook siRNA sense strand is stable in cells. This stability is probably attributable to the DNA hairpin structure d(GCGAAGC), which is known to be nuclease resistant and thermostable (5–9). In the context of RNAi the detection of crook sense siRNA is likely to reflect crook siRNA duplexes plus crook sense siRNA that has been unwound and dissociated from the antisense strand during RISC complex formation.
We conclude that E7 crook siRNA can be recovered after transfection into mammalian cells and can be detected by PCR. Single-stranded E7 crook sense strand routinely gave higher levels of recovery compared with double-stranded E7 crook siRNA (see Figure 2a). One possible explanation is that duplex E7 crook siRNA enters into protein complexes associated with RNAi (see below) and that such complexes impair the recovery and/or ability of crook siRNA to function as a primer for crook template PCR amplification.
The amount of E7 crook siRNA recovered from transfected cells was calculated using the standard curve derived from quantitative PCR of known dilutions of E7 crook siRNA (ranging from 333.3 to 5.2 nM; Figure 2b). Samples were analysed in triplicates and the recovery of E7 crook siRNA from both the SiHa and the HCT116 cells was remarkably reproducible (Figure 2b). At 24 h post-transfection, similar amounts of E7 crook siRNA were recovered from the SiHa and the HCT116 cells (Figure 2c). Subsequently, the two cell lines differed as follows. The recovery level of crook siRNA from HCT116 cells fell by approximately half during each 24 h period (Figure 2c; 48 and 72 h time points). This corresponds with the doubling time of this cell line and is likely to reflect dilution of the transfected E7 crook siRNA with each cell division. In contrast, E7 crook siRNA levels in the SiHa cells showed little decline after 48 h (Figure 2c). This is consistent with the transient growth arrest normally induced by E7 siRNA 48–72 h post-transfection and before apoptosis of the SiHa cervical cancer cells (11). It also confirms stability of the crook siRNA construct under the experimental conditions. Recovery of crook siRNA from cells transfected with either 200 nM, 40 nM, 8 µM, 1.6 nM or 0.32 nM was also tested. Crook siRNA was readily detectable down to 8 nM input dose (Figure 2d).
Crook siRNA induces selective mRNA knock-down
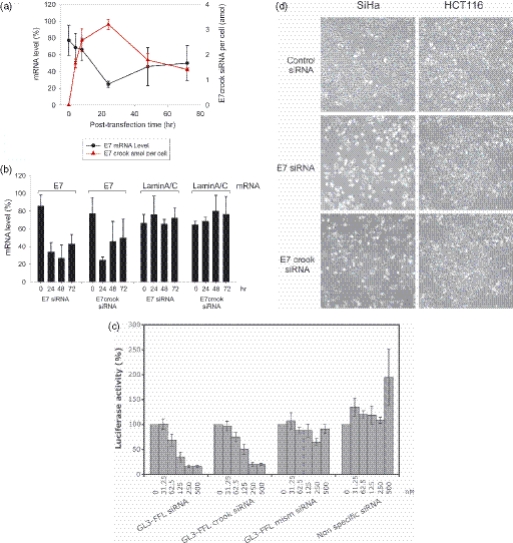
To ascertain if E7 crook siRNA is competent for the induction of RNAi, we tested for selective knock-down of viral E7 mRNA in the SiHa cells (Figure 3a and Materials and Methods). E7 mRNA knock-down after transfection with E7 crook siRNA was clearly evident at 24 h post-transfection (Figure 3a and b). To determine if knock-down was selective for E7 mRNA we also measured lamin A/C mRNA levels. The results show that lamin A/C mRNA is unaffected by E7 crook siRNA at all time points tested (0, 4, 8, 24, 48 and 72 h; see Figure 3b for 24, 48 and 72 h time points). We, therefore, conclude that E7 mRNA knock-down reflects selective targeting by E7 crook siRNA, rather than general non-selective mRNA degradation. Quantification of E7 crook siRNA in these same samples reveals that maximal uptake occurs within 24 h and shows a reciprocal correlation between cellular crook E7 siRNA and E7 mRNA levels in the cells (Figure 3a).
Figure 3.
RNAi induced by crook siRNA. (a) Cellular levels of E7 crook siRNA (red dashed line) and E7 mRNA (solid black line) in SiHa cervical cancer cells at different times following a single transfection of the E7 crook siRNA. Cells were harvested for analysis at 0, 4, 8, 24, 48 and 72 h post-transfection. (b) Comparison of E7 mRNA knock-down by E7 crook siRNA and by unmodified E7 siRNA at 24, 48 and 72 h post-transfection into SiHa cells. Also shown are laminA/C mRNA levels, included as non-target negative control for E7 siRNA selectivity. (c) Comparison of firefly luciferase activity for GL3-FFL siRNA and GL3-FFL crook siRNA, with controls of GL3-FFL mismatched siRNA and non-specific siRNA (normalized to cell count). (d) Appearance of SiHa cervical cancer cells (expressing HPV E7 mRNA) and HCT116 colorectal cancer cells (negative for HPV E7 mRNA) 72 h post-transfection with E7 crook siRNA or with unmodified E7 siRNA, as indicated. Apoptosis of SiHa cells transfected with E7 siRNA was confirmed by annexin V staining and FACS analysis (Materials and Methods).
We were also interested in comparing the effects of E7 crook siRNA with unmodified E7 siRNA. Parallel cultures of SiHa cells were transfected with either E7 crook siRNA or unmodified E7 siRNA. The cells were harvested for analysis at 24, 48 and 72 h post-transfection. The results demonstrate selective E7 mRNA knock-down and were essentially the same for both E7 crook siRNA and the unmodified E7 siRNA (Figure 3b). Thus, we conclude that crook siRNA (i) retains the ability to induce RNAi and (ii) appears to be functionally comparable with unmodified siRNA.
RNAi competency of crook siRNA was confirmed using a second, independent assay in which reporter plasmids are used to detect post-transcriptional gene silencing indirectly. Briefly, the assay employed HeLa cells encoding a stably integrated exogenous expression vector for GF3-firefly luciferase (GL3-FFL) under control of the HIV-1 promoter (13,14). When transfected with GL3-FFL siRNA (19 nt) (15), a dose-dependent reduction in luciferase activity is observed (Figure 3c), but no reduction was seen for a triple mismatched siRNA control or non-specific siRNA control. We also show that GF3-FFL crook siRNA (Materials and Methods) gives a similar dose–response to that obtained with the unmodified GF3-FFL siRNA for the knock-down of luciferase activity (Figure 3c and Materials and Methods). A second specificity control employed Renilla luciferase constitutively expressed under the CMV promoter (12,13,15). Again as expected, no reduction in Renilla luciferase activity was observed after transfection with either GL3-FFL crook siRNA or the unmodified GL3-FFL siRNA (data not shown). Further, the GL3-FFL crook siRNA showed dose-dependent PCR amplification analogous to that shown in Figure 1c (data not shown). Thus, we demonstrate equivalent functionality for crook siRNA and unmodified siRNA in two independent experimental models for the induction of RNAi in mammalian cells.
E7 crook siRNA is non-cytotoxic but induces a cellular phenotype determined by the siRNA component of the hybrid molecule
Although SiHa cells express exogenous viral genes, this nonetheless reflects natural gene expression and HPV infection, and viral gene expressions are causal for cancerous transformation of cervical epithelial cells in vivo. Previously we have identified HPV E7 as survival factor for HPV16-positive cervical cancer cells (11) and have shown that a single dose of E7 siRNA induces apoptosis in SiHa cervical cancer cells, peaking at 72 h post-transfection. We now show that a single dose of E7 crook siRNA similarly induces apoptosis in the SiHa cells (Figure 3d; left-hand panels). We observed induction of SiHa cell apoptosis down to 8 nM (data not shown) and were able to recover and quantify the crook siRNA at these low levels of input (see above; Figure 2a). Lower input levels were also detectable (data not shown) but were ineffective for the induction of apoptosis.
In contrast to the SiHa cells, apoptosis was not observed in parallel cultures of HCT116 cells (which lack HPV E7 mRNA) (Figure 3d; right-hand panels), and cell growth and proliferation were unaffected by E7 crook siRNA up to 200 nM. Thus we substantiate the phenotypic consequences of E7 knock-down by RNAi (SiHa cells), and also demonstrate that the crook siRNA construct is not intrinsically cytotoxic for human epithelial cells (HCT116).
DISCUSSION
Since crook siRNA is designed to be bifunctional, with RNAi initiated by the siRNA component of the molecule and PCR priming conferred by the crook DNA extension (see above), an important aspect of this novel technology is its ease of adaptation for use with siRNAs in general. Given the similarities between crook siRNA and unmodified siRNA for the induction of RNAi (this study; Figure 3b and c), we anticipate that simple PCR-based detection and quantification of crook siRNA will prove valuable in the development of RNAi therapeutics and informative on siRNA cellular uptake, intracellular localization, tissue distribution and pharmacokinetics. However, the applications of this new technology are not restricted to therapeutic development, or even to mammalian systems. We anticipate that PCR-based detection of siRNAs (such as crook siRNA) will prove a powerful adjunct for RNAi-based studies in general.
Acknowledgments
This work was funded by core funding from Yorkshire Cancer Research (to J.M.) and the Medical Research Council (to M.J.G.). Funding to pay the Open Access publication charges for this article was provided by Yorkshire Cancer Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Hutvagner G., Zamore P.D. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 3.Tomari Y., Zamore P.D. Perspectives: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 4.Hannon G.J., Rossi J.J. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 5.Hirao I., Nishimura Y., Tagawa Y., Watanabe K., Miura K. Extraordinarily stable mini-hairpins: electrophoretical and thermal properties of the various sequence variants of d(GCGAAAGC) and their effect on DNA sequencing. Nucleic Acids Res. 1992;20:3891–3896. doi: 10.1093/nar/20.15.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan I.M., Coulson J.M. A novel method to stabilise antisense oligonucleotides against exonuclease degradation. Nucleic Acids Res. 1993;21:2957–2958. doi: 10.1093/nar/21.12.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshizawa S., Ueda T., Ishido Y., Miura K., Watanabe K., Hirao I. Nuclease resistance of an extraordinarily thermostable mini-hairpin DNA fragment, d(GCGAAGC) and its application to in vitro protein synthesis. Nucleic Acids Res. 1994;22:2217–2221. doi: 10.1093/nar/22.12.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Réfrégiers M., Laigle A., Jollès B., Chinsky L. Fluorescence resonance energy transfer analysis of the degradation of an oligonucleotide protected by a very stable hairpin. J. Biomol. Struct. Dyn. 1996;14:365–371. doi: 10.1080/07391102.1996.10508131. [DOI] [PubMed] [Google Scholar]
- 9.Jollès B., Réfrégiers M., Laigle A. Opening of the extraordinarily stable mini-hairpin d(GCGAAGC) Nucleic Acids Res. 1997;25:4608–4613. doi: 10.1093/nar/25.22.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yee C., Krishnan-Hewlett I., Baker C.C., Schlegel R., Howley P.M. Presence and expression of human papillomavirus sequence in human cervical cancer cell lines. Am. J. Pathol. 1985;119:361–366. [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang M., Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041–6048. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
- 12.Arzumanov A., Walsh A.P., Rajwanshi V.K., Kumar R., Wengel J., Gait M.J. Inhibition of HIV-1 Tat-dependent trans-activation by steric block chimeric 2′-O-methyl/LNA oligoribonucleotides. Biochemistry. 2001;40:14645–14654. doi: 10.1021/bi011279e. [DOI] [PubMed] [Google Scholar]
- 13.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 14.Overhoff M., Wünsche W., Sczakiel G. Quantitative detection of siRNA and single-stranded oligonucleotides: relationship between uptake and biological activity of siRNA. Nucleic Acids Res. 2004;32:e170. doi: 10.1093/nar/gnh168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arzumanov A., Stetsenko D.A., Malakhov A.D., Reichelt S., Sorenson M.D., Ravindra Babu B., Wengel J., Gait M.J. A structure-activity study of the inhibition of HIV-1 Tat-dependent trans-activation by mixmer 2′-O-methyl oligoribonucleotides containing locked nucleic acid (LNA), α-LNA or 2′-thio-LNA residues. Oligonucleotides. 2003;13:434–453. doi: 10.1089/154545703322860762. [DOI] [PubMed] [Google Scholar]