Abstract
Convergent studies in human and yeast model systems have shown that some minisatellite loci are relatively stable in somatic cells but not in the germline, and little is known about the mechanism(s) that can destabilize them. Unlike microsatellite sequences, mini satellites are not destabilized by mismatch repair mutations. We report here that the absence of Rad27 and Dna2 functions but not RNase H(35) or Exo1, which play an essential role in the processing of Okazaki fragments during replication, destabilize the human minisatellite CEB1 in mitotically growing Saccharomyces cerevisiae cells, up to 14% per generation in rad27Δ cells. Analysis using minisatellite variant repeat mapping by polymerase chain reaction of the internal structure of 17 variants reveals that the majority of rearrangements in rad27Δ cells are extremely complex contraction events that contain deletions, often accompanied by duplications of motif unit. Altogether, these results suggest that the improperly processed 5′ flap structures that accumulate when replication is impaired can act as a potent stimulator of minisatellite destabilization and can provoke an unexpectedly broad range of mutagenic events. This replication-dependent phenomenom differs from the recombination-induced instability in yeast meiotic cells.
Keywords: instability/minisatellite/mutagenesis/rearrangement/replication
Introduction
All genomes contain repeated DNA sequences that vary in length, complexity and organization. Conventionally, tandem repeat arrays are classified by the length of their constituent repeat unit. Repetitive tracts with repeats of 1–10 bp are termed microsatellites, and tracts with repeats of 10–100 bp are termed minisatellites. Repeated sequences may be found in coding sequences or in elements that regulate the expression of nearby genes. Microsatellite repeats are notoriously prone to size variation and have been implicated in human pathologies (Stevanin et al., 2000). Changes in the lengths of minisatellites are also associated with several disease susceptibilities, such as those associated with the hRAS (Krontiris et al., 1993), insulin (Bennett et al., 1995) and EPM1 (Lalioti et al., 1997) genes. Minisatellite alterations have also been observed in descendants of individuals exposed to low-dosage radiation following the Chernobyl nuclear plant accident (Dubrova et al., 1996).
Despite sharing a repetitive structure, micro- and minisatellites differ in several respects: their base composition, their overall length and the frequency of their occurrence in the genome (Amarger et al., 1998; Vergnaud and Denoeud, 2000). More noteworthy is that they may be distinguished with respect to the mechanism(s) by which they are destabilized. In particular, a feature unique to minisatellites such as MS32, MS205, MS31A, CEB1 and B6-7 is that their spontaneous frequencies of rearrangement in somatic cells are generally two or three orders of magnitude lower than in the germline (Jeffreys et al., 1994, 1999; May et al., 1996; Jeffreys and Neumann, 1997; Tamaki et al., 1999; Buard et al., 2000a). For example, the human minisatellite CEB1, the subject of the present study, exhibits male-specific mutation rates as high as 20% in sperm for some alleles (Buard et al., 1998), but CEB1 alterations are low in lymphocytes (1.8 × 10–4) (Buard et al., 2000a). Moreover, structural analyses of CEB1 and MS32 rearrangements in blood samples have shown that in contrast to the complex rearrangements that occur during gametogenesis, relatively simple intra-allelic events occur in somatic cells (Jeffreys and Neumann, 1997; Buard et al., 2000a), suggesting that minisatellites are destabilized by different processes in somatic cells and in the germline.
Analyses of minisatellite rearrangements in the human germline (Buard and Vergnaud, 1994; Jeffreys et al., 1994; May et al., 1996; Buard et al., 1998; Tamaki et al., 1999) and recent studies of minisatellites in the model system S.cerevisiae (Appelgren et al., 1999; Debrauwère et al., 1999) have strongly suggested that these sequences are targets for a mutagenic process that depends on meiotic recombination. In particular, we demonstrated previously that the preferential meiotic instability of the human CEB1-1.8 and CEB1-0.6 alleles (which consist of 42 and 14 repeat units of 37–43 nucleotides each, respectively) inserted in the yeast genome results from the initiation of meiotic recombination at nearby Spo11-dependent DNA double-strand break (DSB) sites. We concluded that the formation of meiotic rearrangements in human and in yeast meiotic cells is best explained by the DSB-synthesis-dependent strand annealing (SDSA) model, which accounts for the generation of a large variety of motif rearrangements during DSB repair (Buard et al., 1998, 2000b; Debrauwère et al., 1999), a situation that might be stimulated or simply revealed by the repeated structure of the minisatellite and its high degree of internal polymorphism. In contrast, the means by which minisatellite sequences are destabilized in somatic cells is less understood. As in meiotic cells, but at lower frequencies, mitotic rearrangements may result from DSB formation in the vicinity of the minisatellite and faulty repair by the mitotic recombination apparatus. Although the Spo11 nuclease is not mitotically expressed in yeast or in higher eukaryotes, such DSBs may arise from the action of other (presently unidentified) nucleases, or they may arise during the course of replication. Alternatively, studies of bacterial, yeast and mammalian cells (Sia et al., 1997) have clearly demonstrated that changes in the lengths of microsatellites can be stimulated by replication slippage and by inefficient DNA mismatch repair. These findings have raised the possibility that minisatellites are similarly destabilized. However, not in favor of this hypothesis, it has been observed that the mismatch repair mutation msh2 strongly destabilizes repetitive sequences with a short repeat unit (1 or 2 bp), but has a diminished effect on a 5 bp repeat and no effect on a construct containing a triplication of a 20 bp repeat unit (Kokoska et al., 1999). The msh2 and pms1 mutations also have no effect on the stability of the CEB1-0.6 and CEB1-1.8 alleles inserted into the yeast genome (Debrauwère et al., 1999). In contrast, the increased instability of the 20 bp triplication construct in a rad27Δ background suggested that defects in the maturation of Okazaki fragments might destabilize minisatellite loci (Kokoska et al., 1998), although the influence of the rad27Δ mutation on natural minisatellite arrays, which contain longer and less internally conserved repeat units, was not addressed.
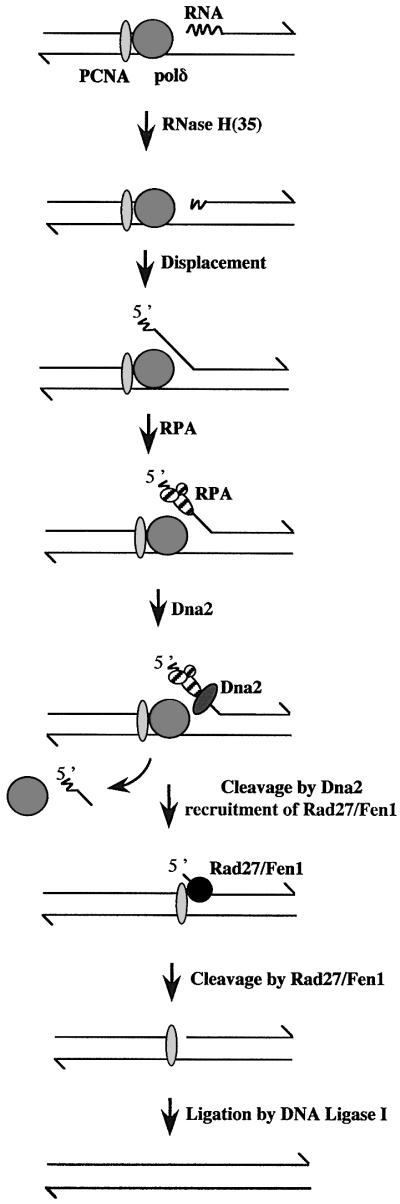
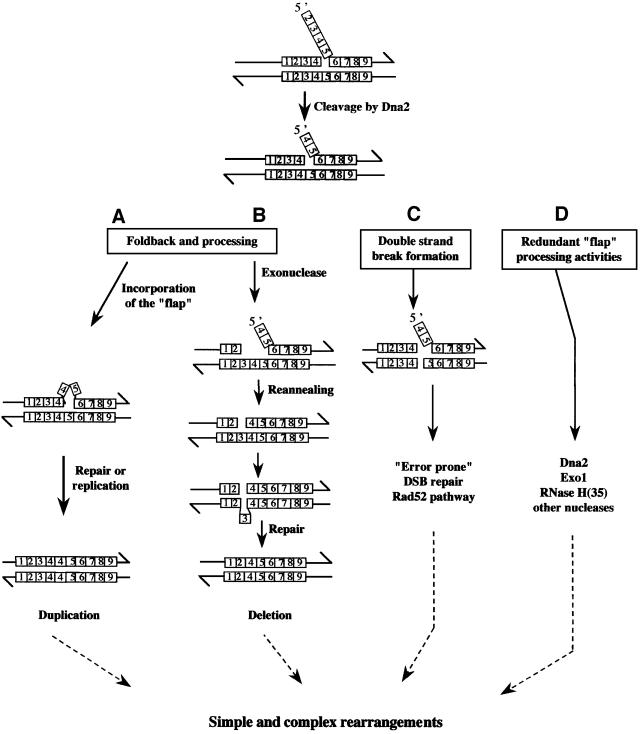
The present work takes advantage of a yeast model system to examine how defects in the processing of Okazaki fragments can affect the stability of the human minisatellite CEB1 in mitotically growing cells. Extensive in vivo and in vitro studies have characterized the process of lagging strand replication, and in particular have provided important clues about the potential mechanism(s) that removes the RNA primer from the 5′ end of each Okazaki fragment (Bambara et al., 1997). Biochemical studies using cell extracts competent for simian virus 40 DNA replication suggested that these primers are removed by the combined action of two nucleases, RNase H1 and Fen1 [the mammalian homologues of the S.cerevisiae proteins RNase H(35) and Rad27, respectively], before Okazaki fragments are joined (Waga and Stillman, 1998). However, genetic analysis in S.cerevisiae has revealed that rnh35 and rad27 null mutations do not confer lethality, suggesting that redundant functions or pathways may contribute to lagging strand synthesis. A current view, based on the interpretation of these genetic data and on in vitro reconstitutions of the processing of replication intermediates (Bae et al., 2001), suggests the model illustrated in Figure 1. Stepwise, the endonuclease RNase H1 [RNase H(35)] is proposed to cleave one nucleotide upstream of the RNA–DNA junction within the Okazaki fragment (Turchi et al., 1994). The 5′ end of this fragment is then displaced as a single strand by the elongation of an upstream Okazaki fragment by the DNA polymerase polδ, thus forming a ‘5′ flap structure’. This key intermediate is then matured by the sequential action of RPA (the single-strand DNA binding heterotrimer) and the Dna2 and Rad27 endonucleases to remove the flap strand and to yield a nick that is sealed by DNA ligase I. Alternatively, at least under certain circumstances (spontaneous failure of this process or in mutant strains), it is also envisaged that: (i) the Dna2 protein can completely process the flap in the absence of Rad27/Fen1 (Bae and Seo, 2000); (ii) the Fen1 protein, which is a 5′–3′ exonuclease in addition to a flap endonuclease (Lieber, 1997), contributes to the removal of the RNA primer with the aid of RNase H1 activity in the absence or reduction of Dna2 activity; and (iii) other nucleases such as the 5′–3′ exonuclease Exo1 (Tishkoff et al., 1997a), which exhibits flap endonuclease activity (Qiu et al., 1999a), are involved. Each of these possibilities thus defines a redundant or alternative pathway, consistent with the observed growth defects of the dna2-1ts and rad27Δ mutants at restrictive temperatures.
Fig. 1. Model for Okazaki fragment maturation, as adapted from Bae et al. (2001) and from Qiu et al. (1999b).
In this context, we have examined the instability of two CEB1 alleles in rad27Δ, dna2-1ts, exo1Δ and rnh35Δ strains, and find that they are unstable in mitotic cells lacking Rad27 or Dna2 activity but not Exo1 or Rnh35 activity. Furthermore, we determined the internal structure of 17 rearranged CEB1 alleles identified in rad27Δ cells and found evidence for a complex pattern of events that produced these rearrangements.
Results
CEB1 is destabilized in rad27Δ cells during vegetative growth
We previously examined the behaviour of the CEB1-0.6 and CEB1-1.8 alleles in wild-type (RAD27) diploids during vegetative growth. For cells grown at 30°C, we found only two changes in size among a total of 1990 colonies analysed from strains either homozygous (CEB1-0.6/CEB1-0.6 or CEB1-1.8/CEB1-1.8) or heterozygous (CEB1-0.6/CEB1-1.8) for CEB1 alleles (Debrauwère et al., 1999), indicating that both alleles are relatively stable in mitotically growing yeast cells, similar to what has been observed for human somatic cells (Buard et al., 2000a). In the present study, we investigated the behaviour of the CEB1 alleles in diploid cells homozygous for a deletion of the RAD27 gene (rad27Δ) or defective for other replication/repair functions, and either homozygous (CEB1-1.8/CEB1-1.8) or heterozygous (CEB1-0.6/CEB1-1.8) for minisatellite constructs. Importantly, due to the temperature-sensitive growth of some of these mutants and consequent effects on the stability of CEB1, all strains used in the present study (Table I) were constructed and systematically propagated at 23°C (a temperature permissive for growth), except during experiments. To establish clonal cultures derived from cells in which rearrangements had occurred and to prevent further rearrangements, cells grown in liquid media at the indicated ‘test’ temperature were always subsequently plated for single colonies at the permissive temperature.
Table I. Genotypes of yeast strains used in this study.
| Strain | Relevant genotype |
|---|---|
| ORD2836 | a/α CEB1-0.6 arg4Bg/CEB1-1.8 ARG4 |
| ORD2840 | a/α CEB1-1.8 ARG4/CEB1-1.8 ARG4 |
| ORD6708 | a/α CEB1-0.6 arg4Bg/CEB1-1.8 ARG4 rad27Δ::HIS3/rad27Δ::HIS3 |
| ORD6732 | a/α CEB1-0.6 arg4Bg/CEB1-1.8 ARG4 rnh35Δ::KanMX/rnh35Δ::KanMX |
| ORD6733 | a/α CEB1-0.6 arg4Bg/CEB1-1.8 ARG4 exo1Δ::KanMX/exo1Δ::KanMX |
| ORD6745 | a/α CEB1-1.8 ARG4/CEB1-1.8 ARG4 dna2-1/dna2-1 |
| ORD5336 | a/α adp1::CEB1-1.8 ARG4/adp1::CEB1-1.8 ARG4 rad27Δ::HIS3/rad27Δ::HIS3 |
| ORD6744 | a/α adp1::CEB1-1.8 ARG4/adp1::CEB1-1.8 ARG4 |
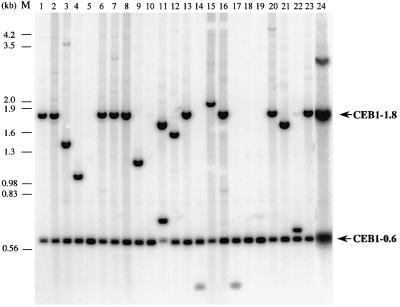
As illustrated in Figure 2, we observed frequent changes in the lengths of the minisatellite constructs in a rad27Δ diploid (ORD6708) that carries both the CEB1-0.6 and CEB1-1.8 alleles. Close examination of the DNA bands derived from cultures grown at 30°C reveals three types of non-parental profile. The first profile (class A; Figure 2, lanes 4, 9 and 15) is characterized by the presence of a parental allele and a novel allele of variable size. This profile, in which the two bands are of equal intensity, indicates that rearrangement of the parental allele occurred during growth at 30°C (the test temperature) and not thereafter at 23°C. We rarely observed the simultaneous loss of both alleles and the appearance of two new bands of equal intensity (lane 11). This low frequency likely reflects the rare occurrence of two independent events. The second profile (class B; Figure 2, lane 24) is characterized by the presence of both parental alleles plus an additional band of novel size but of weaker intensity. This profile, which is likely due to rearrangement during subsequent growth at 23°C, was therefore counted as parental. The third profile, illustrated in Figure 2 (lanes 5 and 10), is characterized by the presence of a single parental band (class C). Altogether, among 102 single colonies derived from ORD6708 cells grown at 30°C, we observed 42 colonies of the class A profile, two of the class B profile and 18 of the class C profile (Table II), indicating that in a majority of these cells (60/102) the CEB1 minisatellites underwent modification in the absence of the RAD27 gene. This proportion contrasts with the frequency of rearrangements observed for an isogenic RAD27 diploid (ORD2836; two events among 576 colonies analysed, both class A) and represents a 170-fold increase in instability. We conclude that the loss of the RAD27 gene function strongly destabilizes the CEB1 minisatellite during vegetative growth at 30°C.
Fig. 2. Detection by Southern blot analysis of CEB1 rearrangements in a rad27Δ diploid ORD6708 grown at 30°C, heterozygous for the CEB1-0.6 and CEB1-1.8 alleles integrated at the ARG4 locus. Each lane contains DNA extracted from a culture grown from an individual colony, digested with AluI and hybridized with a CEB1-0.6 probe. The positions of the parental CEB1 alleles are indicated with arrows. M, the positions of the fragments from the molecular weight marker (λDNA digested by HindIII and EcoRI) are indicated.
Table II. Instability of CEB1 alleles in a rad27Δ/rad27Δ diploid (ORD6708) upon growth at different temperatures.
| Growth conditions |
Minisatellite behaviour |
Frequency of events per cell per generation |
|||||
|---|---|---|---|---|---|---|---|
| Temperature (°C) | Number of generationsa | No change(%) | Rearrangement(%) | Allelic loss (%) | Number of colonies analysed | Rearrangement (%) | Allelic loss (%) |
| 23 | 7.3 | 52 (60.5) | 20 (23.2) | 14 (16.3) | 86 | 3.2 | 2.2 |
| 30 | 7 | 42 (41.2) | 42 (41.2) | 18 (17.6) | 102 | 5.9 | 2.5 |
| 37 | 2 | 39 (72.2) | 15 (27.8) | 0b | 54 | 13.9 | 0 |
The percentage of each class of CEB1 alleles among the total number of CEB1 rearrangements observed at each temperature is indicated in parentheses.
aNumber of generations during 40 h growth at each temperature.
bThe absence of detectable allelic loss at 37°C shown here is likely due to a bias in the sample studied. A larger sample of colonies was analysed phenotypically (see Results).
Destabilization of CEB1 in rad27Δ cells at permissive and restrictive temperatures
As rad27Δ cells are temperature sensitive for growth, we next compared the stability of CEB1 in the same rad27Δ diploid grown at 23°C (a permissive temperature), 30°C (a semi-permissive temperature) or 37°C (a restrictive temperature) (Table II). In all cases, we found that CEB1 is unstable and frequently exhibits alterations in size; the proportion of clones that exhibit the class A profile are 23% at 23°C, 41% at 30°C and 28% at 37°C. These results indicate that CEB1 is rearranged at all temperatures, whether permissive or not for rad27Δ cell growth. However, if the number of rad27Δ cell divisions is taken into account (7.3, 7 and 2 generations at 23, 30 and 37°C, respectively), the adjusted frequency of rearrangement per generation at 37°C (13.9%) is ∼2-fold higher than at 30°C (5.9%) and ∼4-fold higher than at 23°C (3.2%), indicating that the greatest frequency of destabilization of CEB1 per generation occurs at 37°C, a restrictive temperature for the growth of rad27Δ cells.
Preferential rearrangement of the CEB1-1.8 allele in rad27Δ cells
An extended analysis of the alterations observed for the rad27Δ diploid ORD6708 grown at different temperatures is presented in Table III. In all cases, the vast majority of the rearrangements affect only one of the two alleles, almost exclusively the CEB1-1.8 allele. In the total sample of 76 rearrangements identified in this study, only three (4%) involve the shorter CEB1-0.6 allele. This observation contrasts with their behaviour in meiosis, in which the two alleles are similarly unstable (Debrauwère et al., 1999). Table III also indicates the sizes of the rearranged alleles. For all temperatures tested, the majority of the CEB1-1.8 variants are contractions rather than expansions. Overall, an ∼3-fold excess of contractions to expansions was observed (58 versus 18 events, respectively). The range of sizes of rearranged CEB1-1.8 alleles is broad: the shortest CEB1-1.8 variant lost 34–35 motifs from the original 42, and the longest variant gained ∼83 motifs. The smaller set of CEB1-0.6 variants includes one contraction and two expansions. Altogether, we conclude that inactivation of RAD27 preferentially destabilizes the CEB1-1.8 allele independently of the relative rate of rearrangement per generation, and that destabilization produces primarily, shorter variants.
Table III. Expansions and contractions of the CEB1-0.6 and CEB1-1.8 alleles in a rad27Δ/rad27Δ diploid (ORD6708).
| Growth conditions (°C) | Total number of CEB1 rearrangements | Expansions |
Contractions |
||
|---|---|---|---|---|---|
| CEB1-1.8 (%) | CEB1-0.6 (%) | CEB1-1.8 (%) | CEB1-0.6 (%) | ||
| 23 | 20 | 6 (30) | 0 | 13 (65) | 1 (5) |
| 30 | 41a | 5 (12) | 2 (5) | 34 (83) | 0 |
| 37 | 15 | 5 (33) | 0 | 10 (67) | 0 |
The percentage of each class of CEB1 alleles among the total number of CEB1 rearrangements observed at each temperature is indicated in parentheses.
aAn additional cell containing a simultaneous rearrangement of the CEB1-0.6 and CEB1-1.8 alleles was observed (see Figure 2, lane 11), which could not be definitively classified as an expansion or contraction.
Preferential loss of the CEB1-1.8 allele in rad27Δ cells
Among the 102 rad27Δ colonies derived from cells grown at 30°C, we also found 18 class C cases in which the CEB1-1.8 allele was lost (Figure 2). A similar proportion (16%) was observed for cells grown at 23°C, but no losses were observed at 37°C among the samples that were analysed (Table II). We also determined whether these cells retained genetic markers linked in cis to the CEB1 alleles. In the parental diploid, the CEB1-1.8 allele is linked to the ARG4 gene, and the CEB1-0.6 allele is linked to the arg4-bg mutation located at position +1276 of the ARG4 coding region (1498 bp from CEB1 sequences). Since the arg4-bg mutation is recessive, the diploid ARG4/arg4-bg is prototrophic for growth on –Arg plates. Using a simple plating test, we found that all cells that had lost the CEB1-1.8 allele, as determined by Southern blot analysis, were also unable to grow on –Arg plates, indicating that the ARG4 allele had been simultaneously lost or that the arg4-bg allele had become homozygous by gene conversion. We analysed a larger sample of rad27Δ diploids grown at 23, 30 and 37°C; the frequencies of arg– colonies observed were 34/230 at 23°C (15%), 36/231 at 30°C (16%) and 21/337 at 37°C (6%), corresponding to 2–3% per generation, in correlation with the physical loss of the CEB1-1.8 allele observed at 23 and 30°C (Table II). At 37°C, the absence of allelic loss among the 54 colonies analysed at the DNA level is probably due to a bias in the study sample. Altogether, the strong propensity of the CEB1-1.8 allele to be lost or rearranged suggests that both events have the same molecular origin and that each simply represents an alternative outcome of the destabilization of the CEB1-1.8 minisatellite in the absence of Rad27.
MVR–PCR analysis of CEB1-0.6 and CEB1-1.8 rearrangements in rad27Δ cells
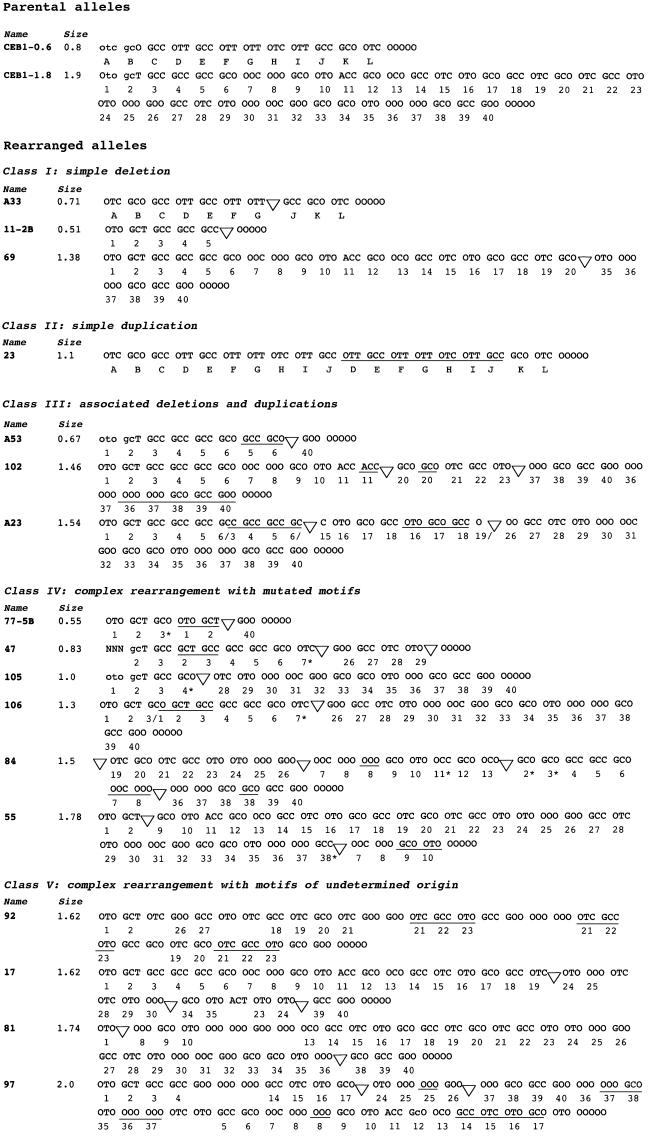
To characterize further the rearrangements observed in rad27Δ diploids, we determined the internal structures of 17 variant alleles (16 contractions and one expansion) by minisatellite variant repeat mapping by polymerase chain reaction (MVR–PCR). This technique takes advantage of the existence of sequence variations among repeat units within the tandem array (Jeffreys et al., 1990). As in a previous study (Buard and Vergnaud, 1994), an analysis of individual CEB1 alleles was performed by using three informative polymorphic sites located within the repeats that, in practice, allow the CEB1 tandem array to be typed precisely. By comparison with the parental CEB1-0.6 and CEB1-1.8 alleles, several classes of mutant allele can be distinguished (Figure 3).
Fig. 3. The structure of 17 CEB1 variant alleles, as characterized by MVR–PCR analysis. All variants are rearranged alleles derived from the rad27Δ diploid ORD6708 grown at 23 or 30°C. The name of the variant is indicated on the left and its size is indicated in kilobases. The variant alleles are grouped in five classes (I–V) according to the type of rearrangement. Each motif is identified by a three letter code representing the three polymorphic sites used for the MVR–PCR, located at positions v4, v17 and v28 within the CEB1 unit motif (Buard and Vergnaud, 1994). For each polymorphic site, two reactions with a specific primer (to detect G or A at the v4 site, C or T at the v17 site, or C or T at the v28 site) were carried out, allowing a pair of complementary ladders to be generated on a gel. Consideration of all six ladders together allows the overall sequence of the allele to be inferred. ‘O’ indicates an absence of amplification with either primer. For both alleles, the last two motifs are represented as ‘OOOOO’ due to the difficulty of typing these motifs by MVR–PCR. ‘N’ indicates ambiguous typing. Lower-case letters indicate that a band of weak intensity was observed after amplification. The sequences of the parental CEB1-0.6 and CEB1-1.8 alleles are expressed as a succession of letters (A–L) and numbers (1–40), respectively, with each letter or number indicating a motif. Duplicated motifs are underlined and deletions are represented by a triangle. Several rearrangements are not fully interpreted, and the relevant repeat motifs were left unassigned. Asterisks indicate motifs for which the cognate parental motif was modified at one of the three polymorphic sites.
The first class, represented by three cases (A33, 11-2B and 69), involves simple deletions of a variable number of contiguous motifs. More specifically, the A33 rearrangement represents a deletion of two motifs from the CEB1-0.6 allele whereas the 11–2B and 69 rearrangements were generated by deletions of 35 and 14 CEB1-1.8 motifs, respectively. The second class is represented by a single case, 23, which exhibits a tandem duplication of seven motifs within the CEB1-0.6 allele. The third class includes three cases (A53, 102 and A23) that are more complex rearrangements, including both deletions and duplications of motifs that overall result in a shorter CEB1-1.8-derived allele. As an example, variant A53 is a CEB1-1.8 derivative with a duplication of a pair of motifs that adjoin a 33-motif deletion. Variant 102 is a more complex CEB1-1.8-derived allele, with three duplications and two deletions, resulting in a total reduction of the equivalent of 12 repeats. The third CEB1-1.8 variant, A23, has two deletions and two duplications, and includes three junction motifs which differ from those of their parental counterparts and probably result from an internal fusion of motifs bordering the junction. Intriguingly, in all three of these variants, the duplicated motifs are adjacent to deletions.
The fourth class, represented by six cases (77-5B, 47, 105, 106, 84 and 55), consists of more complex rearrangements, including duplications and/or deletions of motifs, but each variant contains one or several repeats (labelled with an asterisk in Figure 3) that are not present or recognizable in the parental alleles and that cannot be explained as a fusion of motifs. For example, variant 105 can be interpreted as a deletion of CEB1-1.8 motifs 5 to 27, associated with a modification of border motif 4, which changed from GCC to GCO. This C to O change at the third position of the repeat unit signifies either a substitution (or a deletion) of this C nucleotide. Or, more likely, it results from a nearby change in sequence that prevented the specific primer from amplifying this repeat, so that this site was therefore scored as a ‘O’ (Tamaki et al., 1992). Among these six variants, we identified eight motifs with such internal changes. Importantly, we noticed that all but one of these changes occur at the junction of a deletion or duplication, strongly suggesting that these anomalies are not due to experimental ambiguity but rather arose from mutagenic events associated with the deletion or duplication of nearby repeat tracts.
The fifth class groups the remaining complex alleles (92, 17, 81 and 97), which contain one to four contiguous CEB1 repeats that cannot be traced to a parental array. We qualified these blocks of repeats and the overall rearranged alleles as being of ‘unknown origin’.
In conclusion, most of these CEB1 variants (14/17) contain one or several duplications, which is also a specific mutational signature of the absence of Rad27 function (Tishkoff et al., 1997b). Finally, we found no evidence of chimerism between the parental CEB1-0.6 or CEB1-1.8 alleles among the variants, suggesting that all rearrangements resulted from intra-allelic events.
Position effect and CEB1 destabilization
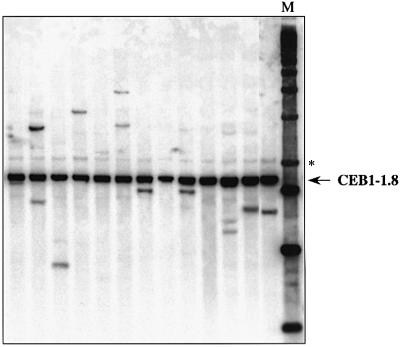
We previously reported that the CEB1-1.8 allele is unstable in meiosis when it is inserted at the ARG4 locus (chromosome VIII) but not at the ADP1 locus (chromosome III) (Debrauwère et al., 1999). To determine whether this position effect also holds for vegetatively growing cells, we compared the mitotic behaviour of the CEB1-1.8 allele inserted at both loci. For a RAD27 diploid strain homozygous for the CEB1-1.8 allele at the ADP1 locus (ORD6744) grown at 23°C, we observed no changes in allele length among 576 independent colonies, as assessed by pool analysis. This result indicates that in the wild-type background, the CEB1-1.8 allele is as stable in mitotically growing cells at the ADP1 locus as at the ARG4 locus. In contrast, for an isogenic rad27Δ diploid (ORD5336) grown at 23°C, we observed 21 rearrangements among 288 colonies (7.3%) (Figure 4), indicating that in the absence of Rad27 activity the CEB1-1.8 allele is also destabilized at the ADP1 locus (χ2 = 43, P< 0.001). However, the frequency of rearrangement is significantly lower at the ADP1 locus than at the ARG4 locus: 3.6% per CEB1-1.8 allele for ORD5336 (7.3% divided by two) compared with 26.7% per CEB1-1.8 allele for ORD6708 (19/71 colonies; χ2 = 58, P < 0.001). The ratio of contractions to expansions is similar at both locations: 13 contractions and eight expansions were observed at ADP1, and 13 contractions and six expansions were observed at ARG4.
Fig. 4. Destabilization of the CEB1-1.8 allele inserted at the ADP1 locus in the rad27Δ diploid ORD5336, grown at 23°C. Each lane contains DNA prepared from a pool of 10 individual cultures, digested by AluI and hybridized with a CEB1-0.6 probe. The CEB1-1.8 parental allele is indicated with an arrow. An additional band marked with an asterisk is presumably due to partial digestion by AluI. M, molecular weight marker (1 kb DNA ladder; Gibco-BRL).
CEB1 is destabilized in the dna2-1ts mutant, but not in the rnh35Δ and exo1Δ mutants
To determine how other replication/repair mutations affect minisatellite stability, we examined the CEB1-1.8 allele in diploids homozygous for the dna2-1ts mutation or for a deletion of the EXO1 or RNH35 genes.
The reported restrictive temperature for growth of dna2-1ts mutants is 33°C (Budd and Campbell, 1995). In our strain background, homozygous dna2-1ts diploids (ORD6745) die at 33°C, since they cannot resume growth upon subsequent plating at 23°C. We analysed CEB1 at the restrictive temperatures of 30, 31 and 32°C, at which 29, 17 and 9% of the cells are viable, respectively. Under these conditions, changes in length of the CEB1-1.8 allele were observed in four out of 384 colonies grown at 30°C, in six out of 333 colonies at 31°C and in nine out of 576 colonies at 32°C (Figure 5). Since these three values are not statistically different (χ2 = 0.77, P < 1.0), the average frequency of CEB1-1.8 rearrangement per cell is ∼1.5%. In comparison, the frequency of rearrangement of the CEB1-1.8 allele in a wild-type strain (ORD2840) is significantly lower (χ2 = 11, P < 0.001), as no rearranged alleles were observed among 742 colonies analysed. Concerning the other mutants, we identified two rearranged alleles among 552 rnh35Δ colonies (ORD6732, 30°C) and none among 576 exo1Δ colonies (ORD6733, 30°C). These frequencies are not significantly different from those of wild-type strains. Altogether, these results indicate that the dna2-1ts mutation destabilizes the CEB1 minisatellite in vegetatively growing cells, but less so than does the rad27 deletion.
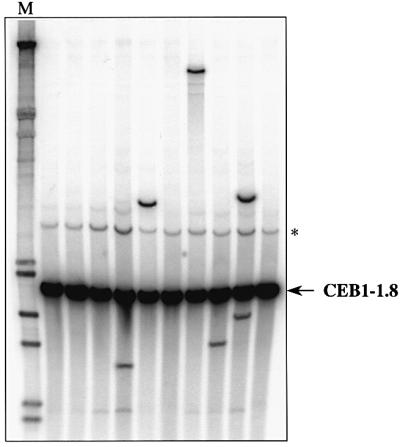
Fig. 5. Destabilization of the CEB1-1.8 allele inserted at the ARG4 locus in the dna2-1ts diploid ORD6745, grown at 31°C. Each lane contains DNA prepared from a pool of 24 individual cultures and treated as described in the legend to Figure 4. An additional band marked with an asterisk is presumably due to partial digestion by AluI. M, molecular weight marker (λDNA digested by HindIII and EcoRI; Appligene).
Discussion
In this study, we have determined the fates of the CEB1-0.6 and CEB1-1.8 human minisatellite alleles in several mutant strains deficient in replication and/or repair. Our main findings concerning the rearrangements we observed are: (i) the rad27Δ and dna2-1ts mutations both destabilize these complex tandem arrays in mitotically growing cells, whereas the rnh35Δ and exo1Δ deletions have no effect; (ii) CEB1 destabilization is less pronounced in dna2-1ts cells at a restrictive temperature than in rad27Δ cells; (iii) in contrast to what is observed for meiosis, CEB1 is unstable in rad27Δ mitotic cells at both the ARG4 and ADP1 loci, although it is more unstable at ARG4 (26.7% compared with 7.3% of cells are rearranged at 23°C, respectively); (iv) most rad27Δ rearrangements preferentially involve the CEB1-1.8 allele; and (v) the CEB1-1.8 allele is also frequently lost in rad27Δ cells (17%). Each of these observations provides insights into the potential mechanism by which minisatellite sequences are destabilized in somatic cells. Below, we first discuss the results regarding CEB1 instability, and then we place our findings in the context of other studies of repetitive sequence instability in yeast and human cells.
Defects in Okazaki fragment processing destabilize the CEB1 minisatellite
Extensive in vivo and in vitro studies have characterized the process of lagging strand replication and have provided important clues about the mechanisms that remove the initiator RNA primer from the 5′ end of each Okazaki fragment (Bambara et al., 1997; Bae et al., 2001). A key intermediate is a 5′ flap structure, which is matured by the sequential action of the Dna2 and Rad27 endonucleases and is removed before ligation (Figure 1). The destabilizing effects of the dna2-1ts and rad27Δ mutations on CEB1 may be due to the absence of or reduction in the activity of either protein, allowing improperly processed 5′ flaps to accumulate, which then initiate CEB1 rearrangements. Several hypotheses can be proposed to account for the lower rate of instability of CEB1 in dna2-1ts cells than in rad27Δ cells. First, as the dna2-1ts point mutation confers temperature sensitivity, it is possible that at subrestrictive temperatures (30–32°C), a large proportion of cells retain sufficient Dna2 activity to process 5′ flap structures. Secondly, at subrestrictive growth temperatures, a majority of dna2-1ts cells is inviable, raising the possibility that those survivor cells that can complete replication are not representative of the entire cell population. Thirdly, the total or partial absence of Dna2 activity can be partially compensated for by other activities that are not associated with rearrangements, such as those of the Rad27 and RNase H(35) proteins, consistent with the observation that dna2-1ts mutants undergo normal levels of spontaneous mutagenesis and that they have only a slightly elevated level (2.5-fold) of dinucleotide tract instability compared with wild-type cells (Budd and Campbell, 2000). Finally, the aberrant DNA structures that form in dna2-1ts and rad27Δ cells might be different, and in the absence of Rad27 activity they might be processed by a more error-prone pathway than in the absence of Dna2 activity. The stability of CEB1 in the rnh35Δ and exo1Δ cells suggests that unprocessed Okazaki fragments do not accumulate in these cells.
Preferential instability of the CEB1-1.8 allele
Two related features of CEB1 rearrangements in rad27Δ cells provide insight into the factors that play a role in destabilization. First, previous studies that examined the rate of rearrangement of repeats of various length in rad27Δ strains concluded that sequences that have small repeat units (mono- and dinucleotide repeats) are more likely to be rearranged and larger repeats are less likely (Kokoska et al., 1998). The longest tandem array studied was a triplication of a 20-nucleotide sequence that exhibited a low rate of alteration in the wild-type background (7.4 × 10–5) but which was destabilized 11-fold in the absence of Rad27 activity. By comparison, the present study revealed that both CEB1 alleles are rearranged more frequently in the wild-type background (∼4 × 10–3) and that in the absence of Rad27, instability increases 170-fold. Furthermore, ∼95% of the rearrangements involve the CEB1-1.8 allele and very few involve the shorter CEB1-0.6 allele, although both alleles contain repeat units of similar size (37–43 nucleotides) and are inserted at identical positions on chromosome VIII. This bias may simply reflect the overall size of the two alleles, which contain 14 (CEB1-0.6) and 42 (CEB1-1.8) repeats, and the frequency of rearrangement may therefore correlate with the probability that an Okazaki fragment or a flap structure forms within the minisatellite array. Alternatively, this bias may be due to features that differentiate the two alleles, such as base composition, or the potential to form secondary structure or internal polymorphisms. These factors may explain the disparate results that concern the relationship between minisatellite array size and human germline instability: for the MS32 and MS205 minisatellites, mutation rates appear to be independent of array length (Jeffreys et al., 1994; May et al., 1996), whereas for CEB1 (Buard et al., 1998) and B6-7 (Tamaki et al., 1999), instability increases with the size of the tandem array. Since it is easy to isolate CEB1 alleles of altered size in rad27Δ cells, the relationship between minisatellite size and frequency of rearrangement could be addressed.
The second noteworthy feature of CEB1 destabilization in rad27Δ cells is the frequent loss of the CEB1-1.8 allele without the appearance of an allele of novel size. We have not yet determined whether this apparent loss results from conversion of the CEB1-0.6 allele, a large deletion of the ARG4 region, and/or a complete loss of chromosome VIII, but the high frequency of loss and rearrangement of the CEB1-1.8 allele raises the hypothesis that both phenomena are mechanistically related. One possibility is that the loss events may result from aborted rearrangements. In this case, the proportion of rad27Δ cells that initiate rearrangement would then equal the proportion of cells that exhibit a change in the parental CEB1 alleles upon growth at 30°C, namely as high as 60%.
The mutational signature of CEB1 rearrangements in rad27Δ cells
In the present study, we observed that most CEB1-1.8 rearrangements in rad27Δ cells are contractions (57/73), with losses of 2–35 repeats from the 42 parental repeats. The CEB1-1.8 expansions are also diverse in size, with increases in length of 2–83 repeats. We also characterized the internal structure of 17 variant alleles. A minority (4/17) exhibit a simple deletion or duplication of contiguous repeats (classes I and II), but most (13/17) are complex and include several deletions and duplications (classes III, IV and V). Interestingly, the frequent occurrence of duplicated motifs is a signature of the spontaneous mutation profile of rad27Δ cells (Tishkoff et al., 1997b). In addition, we observed several rearrangements (classes IV and V) that contain stretches of repeats of unknown origin and novel motifs, raising the possibility that the rearrangement process provoked by the absence of Rad27 activity is associated with additional mutagenic events. Altogether, the remarkable complexity of the CEB1 rearrangements in rad27Δ strains suggests that there is either a multistep mechanism or several simultaneous mechanisms that can profoundly alter the pattern of repeat units, thereby creating major rearrangements.
Mechanisms of formation of the rearrangements
Assuming that the primary effect of the absence of the Dna2 and Rad27 activities is an accumulation of 5′ flap structures, we suggest that rearrangements arise by several possible mechanisms during their processing. One mechanism, illustrated in Figure 6A, proposes that an uncleaved or incompletely cleaved flap strand containing repeat motifs reanneals with the template in various registers, thus creating a duplication of several contiguous motifs. In a second process, 3′–5′ exonuclease activity exposes a single-stranded region upstream of the 5′ flap structure to which it can reanneal, thereby generating simple deletions (Figure 6B). Such single-stranded DNA has been observed in telomeric repeat regions of rad27Δ cells grown at 37°C (Parenteau and Wellinger, 1999). In cells lacking Rad27, we more frequently observed complex events not easily explained by these two simple models. As summarized in Figure 6C, the flap may be the target of other processing activities and, in particular, a substrate for other nucleases whose activity may entrain the formation of secondary lesions such as DSBs (located in or near minisatellite sequences). In this scenario, rearrangements would result from faulty recombinational repair of DSBs. This possibility is supported by three observations: first, the increased rates of recombination of rad27Δ mutants (Tishkoff et al., 1997b); secondly, the synthetic lethality of the rad27Δ mutation in combination with mutations in genes of the Rad52 recombinational repair pathway (Tishkoff et al., 1997b; Symington, 1998; Debrauwère et al., 2001); and thirdly, the meiotic instability of CEB1, which depends on Spo11-induced DSBs and which is best explained by a SDSA model (Debrauwère et al., 1999). The last model, proposed in Figure 6D, is the maturation of the unprocessed flap structure in rad27Δ cells by one or several other nucleases, such as Dna2, Exo1 or RNase H(35), which possess flap processing activities but may imperfectly process these DNA structures. In all cases, the rearrangements appear to be composed of motifs derived from a single parental allele, and therefore these intra-allelic events probably resulted from either intramolecular or intersister interactions.
Fig. 6. Proposed mechanisms for the generation of simple or complex rearrangements due to the accumulation of unprocessed 5′ flap structures in rad27Δ strains. The minisatellite repeat units are indicated by small boxes on each DNA strand and are numbered for clarity. The pathways shown in (A) and (B) illustrate mechanisms of reannealing of the 5′ flap with the template in various registers, leading to duplications or deletions of motifs, respectively [adapted from Kokoska et al. (1998), see text]. In the (C) pathway, processing of the flap structure leads to the formation of DNA double-strand breaks that are not faithfully repaired by recombinational repair, as proposed by Tishkoff et al. (1997b). In (D), the 5′ flap structure is processed by one or several alternative nucleases, such as Dna2, Exo1 or RNase H(35).
The diverse features of minisatellite instability in human and model systems suggest a multiplicity of destabilization and rearrangement mechanisms
An important question about minisatellites is whether they are destabilized by different mechanisms under different circumstances—in different organisms, cell types, chromosomal locations or genetic backgrounds. Our studies in yeast demonstrate that instability arises from two different sources. The first source, operating in meiosis, is the initiation of homologous recombination in the vicinity of the minisatellite (Debrauwère et al., 1999). The second source, as reported here, is an impairment in the maturation of Okazaki fragments during mitotic replication. These two stimulatory mechanisms have distinct consequences for the rearrangement process that deserve further comment. First, they differentially affect the CEB1-0.6 and CEB1-1.8 alleles. During meiosis, both alleles are similarly destabilized because Spo11-dependent DSBs occur with equal frequency on both homologous chromosomes (Debrauwère et al., 1999). In contrast, in the rad27Δ background, the CEB1-1.8 allele is preferentially destabilized (∼24-fold more so than the CEB1-0.6 allele), suggesting that some intrinsic feature of the minisatellite, for example its total length as discussed above, is at work. Secondly, in yeast meiosis, the variable distribution of meiotic DSBs throughout the genome may explain the variable frequency of destabilization of the same minisatellite inserted at different loci. In contrast, the homogeneous and widespread distribution of small Okazaki fragments during replication may destabilize minisatellites regardless of chromosomal location, although more subtle position effects may modify the extent of rearrangement, as shown here. Thirdly, different initiation mechanisms produce different types of rearrangements. In yeast meiosis, the majority of the CEB1 rearrangements are simple deletions or duplications, whereas complex structures with associated duplications and deletions are more rare (Debrauwère et al., 1999). By contrast, the absence of Rad27 in mitotic cells gives rise primarily to complex events and, more enigmatically, events that produce a significant number of single motif changes and structural alterations that are uninterpretable. MVR–PCR analysis of the rare CEB1 events identified in human blood cells showed that they consisted of simple intra-allelic duplications and deletions (Buard et al., 2000a). A comparison with the complexity of the rearrangements observed in yeast rad27Δ cells suggests that the spontaneous somatic instability of CEB1 in human cells is not due to a rad27Δ-like phenomenon. Currently, there is insufficient information about the structure of the rare spontaneous CEB1 events that occur in wild-type yeast strains during vegetative growth to determine whether they resemble those observed in somatic human cells or in yeast rad27Δ mutants. However, the complex CEB1 structures that appear in the absence of Rad27 in mitotically growing yeast cells are strikingly similar to those generated by CEB1 and the human B6-7 minisatellites in the germline (Buard et al., 1998, 2000a; Tamaki et al., 1999). This resemblance suggests the possibility that in addition to recombinationally related events during meiotic prophase, spontaneous replication defects that occur during gameto genesis may also contribute to the burst of minisatellite rearrangements recovered in gametes.
In conclusion, the large variety of mutational profiles that have been described in human and yeast cells indicates that minisatellite repeat sequences can be destabilized by numerous mechanisms, which have begun to be uncovered.
Materials and methods
Media and yeast strains
Standard media were used for vegetative growth and sporulation of S.cerevisiae cells. The origin and the relevant genotype of yeast strains (S288C background) are indicated in Table I. The rad27Δ mutation was introduced by crosses with the isogenic strains FW2612 (MATα, rad27Δ::HIS3) or FW2617 (MATa, rad27Δ::HIS3) (Tishkoff et al., 1997b). To minimize the risk of rearrangement of the CEB1 alleles during construction, all strains were constructed and grown exclusively at 23°C. The rnh35Δ and the exo1Δ mutations were introduced into CEB1-bearing strains by crosses with the corresponding rnh35Δ::KanMX and exo1Δ::KanMX haploids (Winzeler et al., 1999), and the dna2-1ts mutation by crosses with the 3X1549A strain (Budd and Campbell, 1995).
Identification of CEB1 rearrangements
To examine the behaviour of CEB1 alleles during vegetative growth, we inoculated YPD to 2 × 105cells/ml (5 ml) and incubated the cultures at 23, 30 or 37°C constantly over a period of 40 h. Single cells were plated on YPD and incubated at 23°C to limit additional rearrangements during subsequent growth. Variations in the length of the CEB1 alleles were investigated by Southern blot analysis. Genomic DNA was prepared from cultures inoculated with individual colonies and grown overnight in YPD liquid media at 23°C. For pool analysis, used only when the frequency of CEB1 rearrangements at the tested temperature was low, individual colonies were inoculated into 96-well megaplaques and incubated at 23°C for 2 days. DNA was extracted from cultures pooled from 10–24 wells. DNA was digested with AluI and the resulting fragments were separated by electrophoresis in 0.8% agarose gels and transferred under vacuum (Appligene) onto Hybond N+ membranes (Amersham). The membranes were hybridized with a radiolabelled CEB1-0.6 PCR fragment, which also allowed the CEB1-1.8 allele to be visualized. Under these experimental conditions, a gain or loss of two or more CEB1 repeat units can be detected.
Structural analysis of CEB1 alleles by MVR–PCR
Analysis of the internal structure of parental and new CEB1 alleles was performed by MVR–PCR. MVR–PCR typing was performed by isolation of individual alleles; for this purpose, genomic DNA from diploids containing a rearranged allele and a parental allele was amplified with primers flanking the minisatellite (P15: 5′-CGACCTGAGGTAATT ATAACCCGGGCCCTAT-3′; P20: 5′-TCTCAGAGTTCTGTGCTTCG CTGACCTTTT-3′), at a final concentration of 1 µM each, in a total volume of 15 µl using the buffer described in Jeffreys et al. (1990). Reaction conditions were: 15 s at 95°C, 15 s at 68°C and 3 min at 72°C for 26 cycles, and the reaction was terminated by 10 min at 72°C in a Perkin-Elmer GeneAmp PCR system 9700. To increase the quantity of amplified DNA, 12 identical PCR reactions were performed in parallel. These PCR products were pooled, precipitated and electrophoresed in 0.8% agarose gels in order to separate the individual alleles. Variant alleles were purified with the QIAquick Gel Extraction Kit (Qiagen), diluted in water to a concentration of 0.1–0.5 pg/µl, and 1 µl was submitted to MVR–PCR typing as described previously (Buard and Vergnaud, 1994).
Acknowledgments
Acknowledgements
We thank members of our laboratory and G.Vergnaud for technical advice, materials and helpful discussions, S.Loeillet, R.Kolodner, C.Soustelle, D.Tishkoff and J.Campbell for providing strains and plasmids, and K.Smith for English correction and for helpful discussion. This study was supported by the Association de la Recherche contre le Cancer (ARC), the Délégation Générale de l’Armement (DGA/DSP/STTC) and the Institut Curie, Paris. J.L. was supported by a post-doctoral fellowship from the Ligue Nationale contre le Cancer. H.D. was supported by a graduate student fellowship from the Ministère de l’Education Nationale, de la Recherche et de la Technologie (MENRT) and the ARC, and J.B. by the Fondation pour la Recherche Médicale.
References
- Amarger V. et al. (1998) Analysis of distribution in the human, pig and rat genomes points toward a general subtelomeric origin of minisatellite structures. Genomics, 52, 62–71. [DOI] [PubMed] [Google Scholar]
- Appelgren H., Cederberg,H. and Rannug,U. (1999) Meiotic interallelic conversion at the human minisatellite MS32 in yeast triggers recombination in several chromatids. Gene, 239, 29–38. [DOI] [PubMed] [Google Scholar]
- Bae S.H. and Seo,Y.S. (2000) Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem., 275, 38022–38031. [DOI] [PubMed] [Google Scholar]
- Bae S.H., Bae,K.H., Kim,J.A. and Seo,Y.S. (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature, 412, 456–461. [DOI] [PubMed] [Google Scholar]
- Bambara R.A., Murante,R.S. and Henricksen,L.A. (1997) Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem., 272, 4647–4650. [DOI] [PubMed] [Google Scholar]
- Bennett S.T. et al. (1995) Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nature Genet., 9, 284–292. [DOI] [PubMed] [Google Scholar]
- Buard J. and Vergnaud,G. (1994) Complex recombination events at the hypermutable minisatellite CEB1 (D2S90). EMBO J., 13, 3203–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J., Bourdet,A., Yardley,J., Dubrova,Y. and Jeffreys,A.J. (1998) Influences of array size and homogeneity on minisatellite mutation. EMBO J., 17, 3495–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J., Collick,A., Brown,J. and Jeffreys,A.J. (2000a) Somatic versus germline mutation processes at minisatellite CEB1 (D2S90) in humans and transgenic mice. Genomics, 65, 95–103. [DOI] [PubMed] [Google Scholar]
- Buard J., Shone,A.C. and Jeffreys,A.J. (2000b) Meiotic recombination and flanking marker exchange at the highly unstable human minisatellite CEB1 (D2S90). Am. J. Hum. Genet., 67, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E. and Campbell,J.L. (1995) A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl Acad. Sci. USA, 92, 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E. and Campbell,J.L. (2000) The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res. DNA Repair, 459, 173–186. [DOI] [PubMed] [Google Scholar]
- Debrauwère H., Buard,J., Tessier,J., Aubert,D., Vergnaud,G. and Nicolas,A. (1999) Meiotic instability of human minisatellite CEB1 in yeast requires DNA double-strand breaks. Nature Genet., 23, 367–371. [DOI] [PubMed] [Google Scholar]
- Debrauwère H., Loeillet,S., Lin,W., Lopes,J. and Nicolas,A. (2001) Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl Acad. Sci. USA, 98, 8263–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrova Y.E., Nesterov,V.N., Krouchinsky,N.G., Ostapenko,V.A., Neumann,R., Neil,D.L. and Jeffreys,A.J. (1996) Human minisatellite mutation rate after the Chernobyl accident. Nature, 380, 683–686. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J. and Neumann,R. (1997) Somatic mutation processes at a human minisatellite. Hum. Mol .Genet., 6, 129–136. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Neuman,R. and Wilson,W. (1990) Repeat unit sequence variation in minisatellite: a novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell, 60, 473–485. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Tamaki,K., MacLeod,A., Monckton,D.G., Neil,D.L. and Armour,J.A.L. (1994) Complex gene conversion events in germline mutation at human minisatellites. Nature Genet., 6, 136–145. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. et al. (1999) Human minisatellites, repeat DNA instability and meiotic recombination. Electrophoresis, 20, 1665–1675. [DOI] [PubMed] [Google Scholar]
- Kokoska R.J., Stefanovic,L., Tran,H.T., Resnick,M.A., Gordenin,D.A. and Petes,T.D. (1998) Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol. Cell. Biol., 18, 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska R.J., Stefanovic,L., Buermeyer,A.B., Liskay,R.M. and Petes,T.D. (1999) A mutation of the yeast gene encoding PCNA destabilizes both microsatellite and minisatellite DNA sequences. Genetics, 151, 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krontiris T.G., Devlin,B., Karp,D.D., Robert,N.J. and Risch,N. (1993) An association between the risk of cancer and mutations in the HRAS1 minisatellite locus. N. Engl. J. Med., 329, 517–523. [DOI] [PubMed] [Google Scholar]
- Lalioti M.D., Scott,H.S., Buresi,C., Rossier,C., Bottani,A., Morris,M.A., Malafosse,A. and Antonarakis,S.E. (1997) Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature, 386, 847–851. [DOI] [PubMed] [Google Scholar]
- Lieber M.R. (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. BioEssays, 19, 233–240. [DOI] [PubMed] [Google Scholar]
- May C.A., Jeffreys,A.J. and Armour,J.A. (1996) Mutation rate heterogeneity and the generation of allele diversity at the human minisatellite MS205 (D16S309). Hum. Mol. Genet., 5, 1823–1833. [DOI] [PubMed] [Google Scholar]
- Parenteau J. and Wellinger,R.J. (1999) Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol., 19, 4143–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Qian,Y., Chen,V., Guan,M.X. and Shen,B. (1999a) Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal and mutation avoidance. J. Biol. Chem., 274, 17893–17900. [DOI] [PubMed] [Google Scholar]
- Qiu J.Z., Qian,Y., Frank,P., Wintersberger,U. and Shen,B.H. (1999b) Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol., 19, 8361–8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia E.A., Jinks-Robertson,S. and Petes,T.D. (1997) Genetic control of microsatellite stability. Mutat. Res., 383, 61–70. [DOI] [PubMed] [Google Scholar]
- Stevanin G., Durr,A. and Brice,A. (2000) Clinical and molecular advances in autosomal dominant cerebellar ataxias: from genotype to phenotype and physiopathology. Eur. J. Hum. Genet., 8, 4–18. [DOI] [PubMed] [Google Scholar]
- Symington L.S. (1998) Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res., 26, 5589–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki K., Monckton,D.G., MacLeod,A., Neil,D.L., Allen,M. and Jeffreys,A.J. (1992) Minisatellite variant repeat (MVR) mapping: analysis of ‘null’ repeat units at D1S8. Hum. Mol. Genet., 1, 401–406. [DOI] [PubMed] [Google Scholar]
- Tamaki K., May,C.A., Dubrova,Y.E. and Jeffreys,A.J. (1999) Extremely complex repeat shuffling during germline mutation at human minisatellite B6.7. Hum. Mol. Genet., 8, 879–888. [DOI] [PubMed] [Google Scholar]
- Tishkoff D.X., Boerger,A.L., Bertrand,P., Filosi,N., Gaida,G.M., Kane,M.F. and Kolodner,R.D. (1997a) Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with Msh2. Proc. Natl Acad. Sci. USA, 94, 7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff D.X., Filosi,N., Gaida,G.M. and Kolodner,R.D. (1997b) A novel mutation avoidance mechanism dependent on Saccharomyces cerevisiae RAD27 is distinct from DNA mismatch repair. Cell, 88, 253–263. [DOI] [PubMed] [Google Scholar]
- Turchi J.J., Huang,L., Murante,R.S., Kim,Y. and Bambara,R.A. (1994) Enzymatic completion of mammalian lagging-strand DNA replication. Proc. Natl Acad. Sci. USA, 91, 9803–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnaud G. and Denoeud,F. (2000) Minisatellites: mutability and genome architecture. Genome Res., 10, 899–907. [DOI] [PubMed] [Google Scholar]
- Waga S. and Stillman,B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem., 67, 721–751. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A. et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]