Abstract
Posttranscriptional gene silencing (PTGS) processes double-stranded (ds) RNAs into 21–25 nucleotide (nt) RNA fragments that direct ribonucleases to target cognate mRNAs. In higher plants, PTGS also generates mobile signals conferring sequence-specific silencing in distant organs. Since PTGS acts as an antiviral system in plants, successful virus infection requires evasion or suppression of gene silencing. Here we report that the 19 kDa protein (p19) of tombusviruses is a potent silencing suppressor that prevents the spread of mobile silencing signal. In vitro, p19 binds PTGS-generated, 21–25 nt dsRNAs and 21-nt synthetic dsRNAs with 2-nt 3′ overhanging end(s), while it barely interacts with single-stranded (ss) RNAs, long dsRNAs or blunt-ended 21-nt dsRNAs. We propose that p19 mediates silencing suppression by sequestering the PTGS-generated 21–25 nt dsRNAs, thus depleting the specificity determinants of PTGS effector complexes. Moreover, the observation that p19-expressing transgenic plants show altered leaf morphology might indicate that the p19-targeted PTGS pathway is also important in the regulation of plant development.
Keywords: dsRNA binding/p19/silencing/suppressor/tombusvirus
Introduction
Accumulation of double-stranded (ds) RNAs in the cytoplasm of eukaryotic cells induces RNA silencing [referred to as posttranscriptional gene silencing (PTGS) in plants or RNA interference (RNAi) in animals], leading to degradation of dsRNA and homologous single-stranded mRNAs (ssRNA). PTGS is thought to be an ancient cellular defense system acting against different molecular parasites including transgenes, viruses and transposons (reviewed in Voinnet, 2001; Zamore, 2001). Identification of genetic components of PTGS showed that related genes are required for gene silencing in filamentous fungi, plants and animals [reviewed in Sharp (2001) and references therein]. Biochemical analysis of Drosophila cell free extracts that retained RNAi activity revealed that dsRNAs are digested to 21–23 nt dsRNAs [ds short interfering RNAs (siRNAs)] by an RNase III-like enzyme complex (DICER) (Bernstein et al., 2001; Nykänen et al., 2001). These siRNAs are proposed to guide another nuclease complex (RISC) to target homologous ssRNAs for degradation (Hammond et al., 2000, 2001; Bernstein et al., 2001; Elbashir et al., 2001a; Nykänen et al., 2001). It was also demonstrated that in Drosophila embryo extract, siRNAs serve as primers to transform the target mRNA into dsRNAs, which are cleaved to new siRNAs (Lipardi et al., 2001).
Although the biochemistry of plant PTGS is poorly understood, many observations suggest that the mechanism of cellular RNA silencing is conserved between plants and animals. For instance, plant PTGS can also be induced by dsRNAs (Tenllado and Diaz-Ruiz, 2001) and it is consistently associated with the accumulation of 21–25 nt sense and antisense RNAs (Hamilton and Baulcombe, 1999). However, in addition to the conserved cell autonomous defense function of gene silencing, in higher plants PTGS has evolved into a whole plant defense system. Sequence-specific mobile signals generated by PTGS can move cell-to-cell and long distances, triggering systemic silencing in distant tissues (Palauqui et al., 1997; Voinnet and Baulcombe, 1997).
In higher plants, transgene expression or viral infection could lead to the induction of PTGS. dsRNAs are proposed to be synthesized from aberrant transgene mRNAs by a plant encoded RNA-dependent RNA polymerase (RdRP) (Dalmay et al., 2000; Mourrain et al., 2000). In virus-infected plants, ds replicative forms of RNA viruses act as PTGS inducers (Voinnet et al., 2000). Virus-induced PTGS is supposed to play an important role in virus control of higher plants (reviewed in Matzke et al., 2001; Voinnet, 2001). This conclusion is supported by the observation that infection of certain viruses including nepoviruses results in a recovery phenotype (Ratcliff et al., 1997). In contrast to the inoculated and first systemic leaves of nepovirus-infected plants, upper leaves are symptom free and have low virus titre. Recovered plants show PTGS-mediated resistance to infection of a second virus sharing homologous sequences with the first pathogen (Ratcliff et al., 1997). Findings that many viruses encode PTGS suppressors further support the proposed antiviral role of gene silencing. Viral PTGS suppressors target different components of PTGS machinery, for instance potyviral HC-Pro-type suppressors restore green fluorescent protein (GFP) expression of posttranscriptionally silenced transgenic plants in both old and new leaves, while Cmv2b-type suppressors rescue green fluorescence only in new emerging leaves (reviewed in Li and Ding, 2001). The 25 kDa protein (p25) of potato virus X (PVX) is the only suppressor described so far that failed to restore posttranscriptionally silenced GFP expression but affects gene silencing. p25 is a systemic silencing suppressor that prevents the spread of transgene- or virus-induced systemic silencing and inhibits the onset of transgene-induced local PTGS. Surprisingly, p25 does not interfere with virus-induced local silencing (Voinnet et al., 2000). Although many virus-encoded PTGS suppressors have been identified, the molecular mechanism of silencing suppression is unknown.
Tombusviruses are positive-stranded RNA viruses with a linear monopartitate genome containing five open reading frames (ORFs) (Russo et al., 1994). The ORF5-encoded 19 kDa protein (p19) is a symptom determinant, suggesting that p19 plays a role in overcoming host defense systems (Dalmay et al., 1993; Scholthof et al., 1995a,b). Consistently, p19 of tomato bushy stunt tombusvirus (TBSV) was identified as a silencing suppressor. Interestingly, p19 restores GFP expression in PTGS-inactivated transgenic plants only in and around the veins of new emerging leaves (Voinnet et al., 1999), even though TBSV accumulates to a high concentration in the whole leaf (Scholthof et al., 1995a). In the absence of p19, cymbidium ringspot tombusvirus (CymRSV) infection results in a PTGS-associated recovery phenotype, further supporting the role of p19 in PTGS suppression (Szittya et al., 2002).
Here we report that p19 inhibits the onset of transgene-induced local and systemic silencing. p19 does not interfere with the virus-induced local silencing, but could prevent the development of virus-induced systemic PTGS. In vitro, p19 specifically binds PTGS-generated 21–25 nt dsRNAs and 21-nt synthetic ds siRNAs with 2-nt 3′ overhanging end(s). We suggest that p19 depletes PTGS-generated 21–25 nt dsRNAs, thus inhibiting the development of transgene-induced silencing and preventing the production of active signal complex. Interestingly, transgenic plants expressing biologically active p19 showed an altered phenotype, suggesting that the p19 targeted PTGS pathway could also play role in developmental regulation.
Results
Ectopic expression of p19 inhibits transgene-induced PTGS
We have analysed the PTGS suppression strategy of tombusviruses in the Nicotiana benthamiana–CymRSV host–virus model system. To test whether CymRSV expresses HC-Pro or Cmv2b-type silencing suppressors, posttranscriptionally silenced GFP transformants of N.benthamiana (Brigneti et al., 1998) were infected with CymRSV. Green fluorescence could not be detected in CymRSV-infected plants (data not shown), suggesting that this tombusvirus did not express HC-Pro or Cmv2b-type PTGS suppressor.
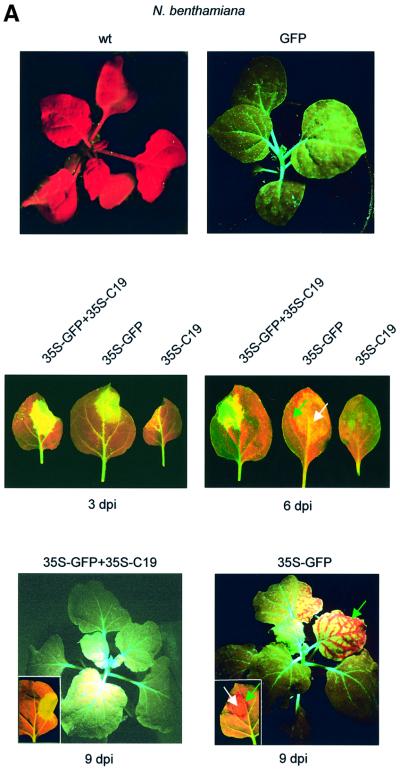
To determine whether CymRSV encodes a systemic silencing suppressor, GFP transformants of N.benthami ana were infiltrated with a culture of an Agrobacterium tumefaciens carrying a 35S–GFP construct, or co-infiltrated with 35S–GFP and another A.tumefaciens expressing each of the CymRSV ORFs. Confirming previous findings (Voinnet and Baulcombe, 1997; Brigneti et al., 1998), infiltration of GFP transformants of N.benthamiana with 35S–GFP alone triggered local and systemic GFP silencing, which manifested in red fluorescence due to the autofluorescence of chlorophyll in the absence of GFP expression. Systemic silencing was induced rapidly, as observed by the formation a red-fluorescent front around the infiltrated area by 5–6 days post-infiltration (d.p.i.) (Figure 1A). By 9–11 d.p.i., 35S–GFP-infiltrated plants also showed systemic silencing in upper leaves (Figure 1A). If CymRSV encodes a systemic silencing suppressor, co-infiltration is expected to prevent the induction of systemic silencing. Co-infiltration of 35S–GFP with ORF1, -2, -3 or -4 did not affect initiation of systemic silencing (data not shown). In contrast, co-infiltration of 35S–GFP with CymRSV ORF5, which encodes p19 (35S–C19), inhibited systemic GFP silencing. A red-fluorescent front did not develop around the infiltrated region and GFP silencing was not detectable in the upper leaves of 35S–GFP+35S–C19 co-infiltrated plants (Figure 1A). This result shows that ectopic expression of CymRSV p19 interferes with either production or spreading of transgene-generated systemic silencing signal.
Fig. 1. Effect of CymRSV p19 on transgene-induced PTGS. (A) Untreated wild-type (wt) and GFP transformants of N.benthamiana (GFP) are shown as control (top panel). GFP transformants of N.benthamiana were infiltrated with 35S–GFP or co-infiltrated with 35S–GFP and 35S-CymRSV p19 (35S–GFP+35S–C19). Infiltrated leaves (middle panels) and plants (bottom panels) are shown. Silencing of GFP is manifested in red fluorescence. Green and white arrows indicate systemic and local PTGS, respectively. Photographs were taken of UV-illuminated plants. (B) GFP mRNA (top panel) and GFP-specific 21–25 nt RNA (bottom panel) levels in leaves of GFP transformants of N.benthamiana infiltrated with 35S–C19 or 35S–GFP, or co-infiltrated with 35S–GFP+35S–C19. RNAs were isolated from the infiltrated areas of treated leaves. The same total RNA in the same amount was used for high and low molecular weight RNA gel blot analyses. Note that 35S–C19 infiltration was used as a control to test background GFP expression in the bacterium-infiltrated cells. Ethidium bromide-stained 25S rRNA is shown as loading contol (middle panel). To estimate the size of small RNAs, labelled 21-nt synthetic RNA as a size marker (M) was run along total RNA isolated at 6 d.p.i. from a 35S–GFP-infiltrated GFP transformant plant, and the blot was then hybridized with GFP probe (bottom panel, left).
To test whether CymRSV p19 could also affect local silencing, GFP expression was monitored in the infiltrated patches of 35S–GFP-infiltrated and 35S–GFP+35S–C19-co-infiltrated leaves of GFP transformant plants. By 1.5–3 d.p.i. there was a modest transient GFP expression, manifested in green fluorescence in 35S–GFP infiltrated tissues. However, starting at 6–7 d.p.i., infiltrated tissues became red fluorescent, indicating the induction of local GFP silencing. In contrast, co-infiltration of 35S–C19 with 35S–GFP dramatically increased GFP expression. By 1.5–3 d.p.i., green fluorescence of the infiltrated tissues was much brighter in 35S–GFP+35S–C19-co-infiltrated leaves than in plants infiltrated with 35S–GFP alone (Figure 1A). Moreover, co-infiltrated leaves remained bright green until infiltrated tissues started to necrotize (10–12 d.p.i.) (data not shown). RNA gel blot analyses of GFP mRNA and 21–25 nt RNA accumulation confirmed that the enhanced and lasting GFP expression of co-infiltrated regions was due to the inhibition of local PTGS. By 1.5 d.p.i., GFP mRNA accumulated to a much higher level in 35S–GFP+35S–C19-co-infiltrated tissues than in 35S–GFP inoculated leaves (Figure 1B, top panel, lane 3 compared with lane 2). GFP transcript levels remained relatively high in co-infiltrated leaves to 9 d.p.i. (Figure 1B, top panel, lanes 3, 6, 9 and 12), while in 35S–GFP-infiltrated leaves GFP mRNA levels decreased to less than background by 6–9 d.p.i. (Figure 1B, top panel, lanes 8 and 11 compared with lanes 7 and 10). GFP-derived 21–25 nt RNAs were detectable by 1.5 d.p.i. in 35S–GFP-infiltrated samples, indicating the early onset of local PTGS (Figure 1B, bottom panel, lane 2). By 3 d.p.i., 21–25 nt RNAs were abundant in 35S–GFP-infiltrated tissues, and they remained so until 9 d.p.i. (Figure 1B, bottom panel, lanes 5, 8 and 11). In contrast, GFP-derived 21–25 nt RNAs were hardly detectable in samples taken from 35S–GFP+35S–C19-co-infiltrated leaves at any time point (Figure 1B, bottom panel, lanes 3, 6, 9 and 12). Interestingly, in RNA gel blot analyses, GFP-derived small RNAs accumulated as two distinct bands (Figure 1B, bottom panel).
Collectively, infiltration studies revealed that p19 targets PTGS step(s), affecting both the transgene-induced local and systemic silencing.
p19 does not influence virus-induced local PTGS but affects systemic silencing
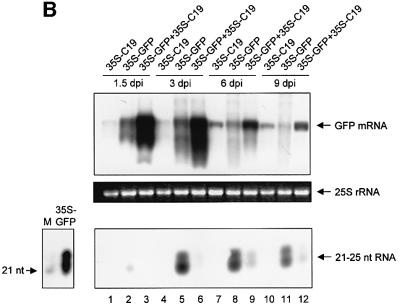
To examine whether p19 also interferes with virus-triggered PTGS, we compared the development of CymRSV-induced silencing in the presence and absence of p19. To test the effect of p19 on virus-induced local PTGS, protoplast transfection assay was carried out with wild-type CymRSV and with a CymRSV mutant (Cym19stop) containing p19-inactivating point mutations (Figure 2A) (Szittya et al., 2002). Consistent with previous reports (Russo et al., 1994), Cym19stop replicated as effectively as CymRSV in N.benthamiana protoplasts (Figure 2B). Twenty-one- to 25-nt RNA accumulation was also comparable in both CymRSV- and Cym19stop-infected protoplasts. Small RNAs were already detectable at 1 d.p.i., and by 3 d.p.i. large amounts of 21–25 nt RNAs accumulated in both CymRSV- and Cym19stop-transfected cells (Figure 2C). Twenty-one- to 25-nt RNAs derived from CymRSV- and Cym19stop-transfected protoplast appeared as single bands, similar to the small RNAs derived from virus-infected leaves (Figures 2C, and 3B and C). Since viral RNAs and silencing-generated 21–25 nt RNAs accumulated to similar levels in CymRSV- and Cym19stop-transfected cells, we concluded that p19 did not affect virus-induced local PTGS.
Fig. 2. Effect of p19 on virus-induced local PTGS. (A) Top panel: Cym19stop genomic RNA is shown with ORFs and the molecular masses (kDa) of encoded proteins. The cross shows that 19 kDa protein encoded by ORF5 was inactivated in Cym19stop mutant virus. Bottom panel: western blot of total protein extracted from 35S–C19-infiltrated tissues and from CymRSV-, Cym19stop- and mock-inoculated leaves of N.benthamiana. Samples were isolated at 3 d.p.i. (B) RNA gel blot analyses of viral RNAs extracted from N.benthamiana protoplasts transfected with CymRSV and Cym19stop viruses. ‘G’ indicates genomic RNA, while sg1 and sg2 indicate subgenomic-1 and -2 RNAs, respectively. (C) Accumulation of virus-specific 21–25 nt RNAs in CymRSV- and Cym19stop-transfected protoplasts (right panel). To estimate the size of small RNAs, labelled 21-nt synthetic RNA as a size marker (M) was run along total RNA isolated at 6 d.p.i. from a CymRSV-infected plant, then the blot was hybridized with CymRSV probe (left panel).
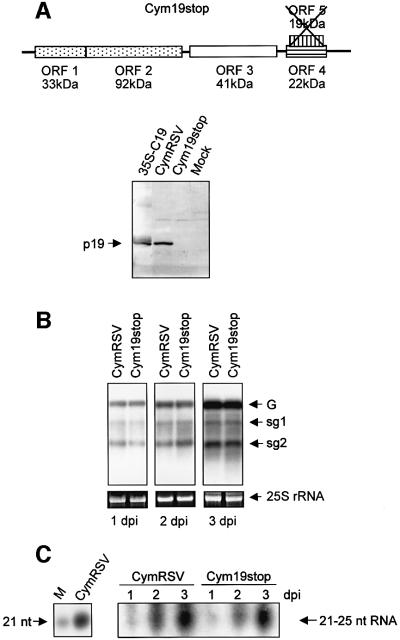
Fig. 3. Effect of p19 on virus-induced systemic PTGS. (A) Recovery phenotype of Cym19stop-infected N.benthamiana. Leaves taken at 30 d.p.i. are shown according to decreasing age, where 1 indicates the first systemic leaf. Note: leaves 5–8 are symptomless, while leaf 1 shows severe symptoms. (B) RNA gel blot analyses of viral RNAs (top panel) and virus-derived, 21–25 nt RNAs (bottom panel) in leaves shown in (A). G indicates genomic RNA, while sg1 and sg2 indicate subgenomic-1 and -2 RNAs, respectively. Numbers below the bottom panel indicate the ratio of 21–25 nt virus-derived RNA/genomic viral RNA calculated by an image analyser. The ratio of 21–25 nt/genomic RNAs from leaf 1 was taken as 100% (1), and others were normalized according to it. (C) Viral RNA (top panel) and virus-derived, 21–25 nt RNA levels (bottom panel) in inoculated (i) and first systemic (s) leaves of two CymRSV- and two Cym19stop-infected plants. Samples were taken at 7 d.p.i. Note: since viral RNA levels are different, unequal total RNA loading was required (especially RNAs derived from Cym19stop-infected first systemic leaves were overloaded) to apply comparable amounts of viral RNAs in each lane (see 25S rRNA controls). For 21–25 nt RNA analyses total RNAs were also applied unequally, while the same amounts of total RNAs were loaded in low molecular weight RNA analysis, which were applied to high molecular weight RNA blots. The ratio of 21–25-nt/genomic RNAs of sample 1 was taken as 100% (1), and others were normalized according to it.
Taking advantage of the fact that p19 is dispensable for virus movement in N.benthamiana, we could also investigate the effect of p19 on virus-induced systemic PTGS. As we have described previously, CymRSV infection led to rapid lethal necrosis (data not shown), whereas Cym19stop inoculation of N.benthamiana plants resulted in a recovery phenotype (Figure 3A). Recovered plants were resistant to a second virus containing homologous sequences to Cym19stop, suggesting that systemic PTGS limited the extent of the virus in the upper symptomless leaves (Szittya et al., 2002). To support the hypothesis that p19 influences systemic silencing further, we have monitored the development of silencing in plants infected with CymRSV and Cym19stop. Virus-induced systemic PTGS models suggest that mobile signals spread ahead of the virus and confer more effective PTGS against the pathogen in the signal-conditioned leaves (Ratcliff et al., 1997). Indeed, younger leaves of Cym19stop-inoculated plants consistently displayed an increased ratio of virus-derived, 21–25 nt RNA/viral genomic RNA. Accordingly, these leaves showed milder symptoms and reduced virus titre (Figure 3A and B). These results indicated that in the absence of p19, systemic PTGS gradually overcame the virus infection suggesting that p19 interfered with the development of virus-induced systemic silencing.
We also compared the development of systemic silencing in CymRSV- and Cym19stop-infected plants. Since CymRSV infection results in rapid necrosis, we could only compare the inoculated and the first systemic leaves of infected plants. Symptom development was similar on both the inoculated and on the first systemic leaves of CymRSV- and Cym19stop-infected plants (data not shown). However, viral RNA levels were lower (Figure 3C, top and middle panels) and 21–25 nt RNA/genomic RNA ratios were slightly higher (Figure 3C, bottom panel) in the inoculated leaves of Cym19stop- than in CymRSV-infected plants. In the first systemic leaves, viral RNA and 21–25 nt RNA levels were dramatically different. Viral RNA levels were much lower (Figure 3C, top and middle panels), whereas 21–25 nt RNAs/genomic RNA ratios were much higher in the first systemic leaves of Cym19stop- than in the CymRSV-infected plants (Figure 3C, bottom panel). These results could suggest that p19 interferes with virus-induced systemic silencing.
These findings that in Cym19stop-infected plants, virus-induced PTGS operates more efficiently in systemic than in inoculated leaves support the model (Ratcliff et al., 1997) that systemic signals generated in the inoculated tissues move to the upper leaves; therefore, these signal-conditioned tissues could silence viral invaders more efficiently than primary infected cells.
p19 binds ds siRNAs with 2-nt 3′overhanging end(s)
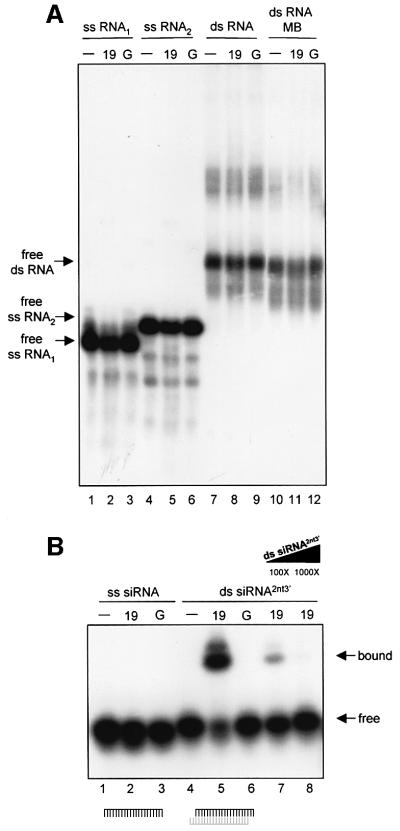
It is widely believed that RNAs provide sequence specificity for the signal complex (Voinnet et al., 2000; Mallory et al., 2001). If p19 binds the RNA component of the signal complex, as was speculated for Cmv2b (Guo and Ding, 2002), it would explain the systemic silencing suppression effect of p19. Therefore, gel mobility shift experiments were carried out to test the RNA binding capacity of p19. CymRSV p19 as a glutathione S-transferase (GST) fusion protein (GST–19) was expressed in Escherichia coli [Havelda et al., 1998; Supplementary data (A) available at The EMBO Journal Online], and then used for RNA binding assays. Since precursor molecules of 21–25 nt RNAs were suggested to serve as specificity determinants for the signal complex (Voinnet et al., 2000; Mallory et al., 2001), we tested the affinity of p19 to longer RNAs. We have found that p19 did not bind long ssRNAs (Figure 4A, lanes 2 and 5) and failed to form complexes with long dsRNAs with either extended ss overhanging 5′ ends or with blunt ends (Figure 4A, lanes 8 and 11).
Fig. 4. Gel mobility shift assay of p19 binding to RNAs. Purified GST–19 fusion protein (19) was tested for RNA binding. Labelled RNA (–), and labelled RNA incubated with GST (G) were used as negative controls. (A) p19 does not bind long RNAs. In vitro-transcribed ssRNAs [ss RNA1 (116 nt) and ss RNA2 (132 nt)] or annealed complementary in vitro transcripts (ds RNA) were used as labelled probes for p19 binding experiments. Blunt-ended long dsRNAs (ds RNA MB) were generated by mung bean nuclease treatment. Note that long dsRNA shows a non-specific upper band, presumably due to specific structures of the molecule. (B) p19 binds ds siRNAs. Synthetic 21-nt RNAs were radioactively labelled and used as ss (ss siRNA) probes for protein binding experiments. Complementary 21-nt RNAs, which form ds siRNAs with 2-nt 3′ overhanging ends, were labelled, annealed and used as ds (ds siRNA2nt3′) probes for protein binding assays. The molar excess of unlabelled competitor is shown under the triangle. (C) p19 requires 2-nt 3′ overhang for efficient binding. Labelled RNA duplexes with a blunt end, and a 2-nt 3′ overhang (ds siRNA2nt3′+blunt) with blunt ends (ds siRNAblunt) and with 4-nt 3′ overhangs (ds siRNA4nt3′), were used as probes for binding assays. (D) p19 binds PTGS-generated dsRNAs. Total RNA derived from CymRSV-infected N.benthamiana was separated in denaturing or in native PAGE, then short RNAs were isolated. Denaturing PAGE-isolated RNAs were denatured and used as ss (ss RNAPTGS-d) probes, or were renatured and used as ds (ds RNAPTGS-d) probes. Native PAGE-isolated RNAs were used as ds (ds RNAPTGS-n) probes or were denatured and used as ss (ss RNAPTGS-n) probes for binding experiments. Note that free ss RNAPTGS-d and ss RNAPTGS-n levels decreased slightly with 19, presumably due to partial renaturation during the binding reaction. (E) Comparative binding experiments were carried out to define p19 binding affinity to PTGS-generated dsRNAs. ds siRNA2nt3′, ds RNAPTGS-n and ds siRNAblunt (50 pg each) were labelled and used for binding experiments with increasing amount of 19.
We have also analysed whether p19 binds 21–25 nt RNAs. Synthetic RNAs (21 nt long) were used as ss siRNAs, while 21-nt RNA duplexes with 2-nt 3′ overhanging ends were used as ds siRNA probes for RNA binding tests. Gel mobility shift assays showed that GST–19 formed complexes efficiently with ds siRNAs, but failed to bind ss siRNA (Figure 4B, lanes 5 and 2). p19 dsRNA binding is sequence independent, since ds siRNA was not homologous with the virus. To test whether p19 RNA binding is influenced by the 3′ ends of dsRNAs, we also examined the affinity of p19 to 21-nt RNA duplexes with blunt-ends, or with 4-nt 3′ overhangs. These RNA duplexes conferred significantly weaker mRNA degradation in Drosophila extracts than ds siRNAs with 2-nt 3′ overhanging ends (Elbashir et al., 2001b). Interestingly, p19 bound RNA duplexes with blunt-ends or with 4-nt 3′ overhangs fairly weakly compared with ds siRNAs with 2-nt 3′ overhangs (Figure 4C, compare lane 1 to lanes 6 and 9) [Supplementary data (B)]. These data suggest that p19 is a specific dsRNA binding protein, which requires 2-nt 3′ overhangs for efficient binding.
To assess whether the presence of a single 2-nt 3′ overhang is sufficient for p19 ds siRNA binding, we tested the RNA binding affinity of p19 to 21-nt RNA duplex with a blunt end and a 2-nt 3′ overhanging end. As Figure 4C (lane 3) shows, p19 shifted this RNA duplex efficiently, indicating that p19 interacts with a single 2-nt 3′ overhanging end of short dsRNAs.
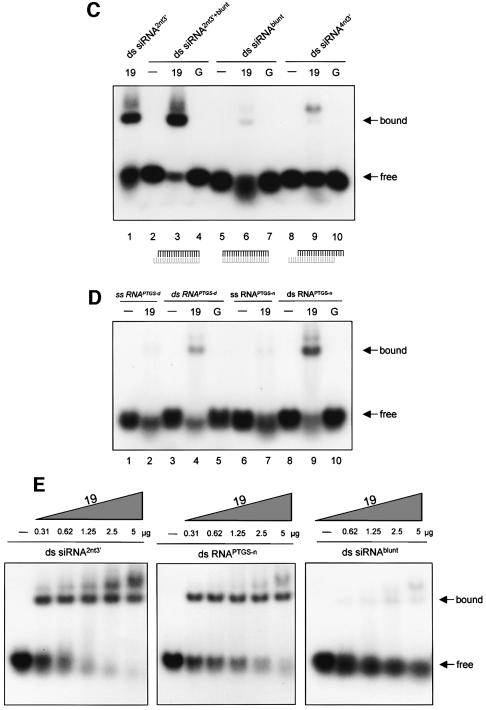
p19 binds PTGS-generated, 21–25 nt dsRNAs
Accumulation of sense and antisense 21–25 nt RNAs is a characteristic feature of plant PTGS. To test whether p19 also binds these PTGS-generated short RNAs, 21–25 nt RNAs were purified from CymRSV-infected leaves. RNAs (21–25 nt) were isolated from denaturing polyacrylamide gel, then heat-denatured and used as ssRNAs, or were renatured and used as dsRNAs in RNA binding assays. Consistent with the result of synthetic RNA binding experiments, the mobility of ssRNAs was barely affected, whereas PTGS-generated 21–25 nt dsRNAs were (partially) shifted by the GST–19 (Figure 4D, compare lane 4 with lane 2). These results showed that p19 failed to form complexes with silencing-produced 21–25 nt ssRNAs and suggested that p19 bound PTGS-generated 21–25 nt dsRNAs. During renaturation, however, artificial RNA duplexes, especially thermodynamically favoured blunt-ended dsRNAs, could be formed (Nykänen et al., 2001). Moreover, in CymRSV-infected leaves, ∼80% of 21–25 nt RNAs were derived from plus-strand viral RNAs, therefore complete shift could not be expected (Szittya et al., 2002). To enrich PTGS-generated natural RNA duplexes in the isolated RNA fraction, we separated total RNA extracted from CymRSV-infected leaves by native polyacrylamide gel electrophoresis (PAGE), and the short RNA fraction was isolated. To assess the size of extracted molecules, isolated RNAs were labelled and run along a size marker in denaturing gel. We confirmed that native PAGE-isolated RNAs were molecules of 21–25 nt in length (data not shown). Hybridization to plus- and minus-strand viral transcripts revealed that 21–25 nt RNAs isolated from native PAGE were almost symmetric, and plus- and minus-strand-derived RNAs were approximately equally represented [Supplementary data (C)]. These findings showed that separation in native PAGE enriched a symmetrical fraction of PTGS-generated, 21–25 nt RNAs.
As Figure 4D shows, short RNAs isolated from native PAGE were efficiently bound by the GST–19, while mobility of denatured short RNAs was barely affected (compare lane 9 with lane 7). Comparative binding studies revealed that p19 bound native PAGE-isolated RNAs much more efficiently than 21-nt RNA duplexes with blunt ends, but less efficiently than ds siRNAs with 2-nt 3′ overhangs (Figure 4E). Since p19 interacts weakly with ssRNAs or blunt-ended RNA duplexes, but binds ds siRNAs with 2-nt 3′ overhang(s) and native PAGE-isolated short RNAs efficiently, we concluded that symmetrical RNA fraction isolated from native PAGE mainly contained 21–25 nt dsRNAs with 3′ overhanging ends. Thus, silencing produced a short RNA fraction, which could be bound by p19 and will be referred to as PTGS-generated, 21–25 nt dsRNAs. Since RNase III-like nucleases generate RNA duplexes with 3′ overhanging ends, these findings support the previous suggestion that 21–25 nt dsRNAs are also processed by a DICER-related enzyme in plants (Bernstein et al., 2001).
Transgenic expression of p19 induces an altered plant phenotype
To explore whether a PTGS pathway that is targeted by p19 affects plant development, we tried to establish CymRSV p19 transformant N.benthamiana lines. Although several transgenic plants expressed CymRSV p19 transgene at the RNA level, western blot analyses failed to detect p19. However, we were able to obtain transgenic N.benthamiana expressing p19 that was derived from a related tombusvirus, artichoke mottled crinkle virus (AMCV). To test whether AMCV p19 acts in a similar way to CymRSV p19, silencing suppression assays were also carried out with AMCV p19. We have found that the effect of AMCV and CymRSV p19 was identical on PTGS. AMCV, like CymRSV, failed to restore the GFP expression of PTGS-silenced transgenic plants (data not shown). Moreover, ectopic expression of AMCV p19 also inhibited the development of transgene-induced local and systemic silencing [Supplementary data (D)] (data not shown). Therefore, we conclude that the p19 proteins of CymRSV and AMCV are similar silencing suppressors, presumably operating identically (see Discussion).
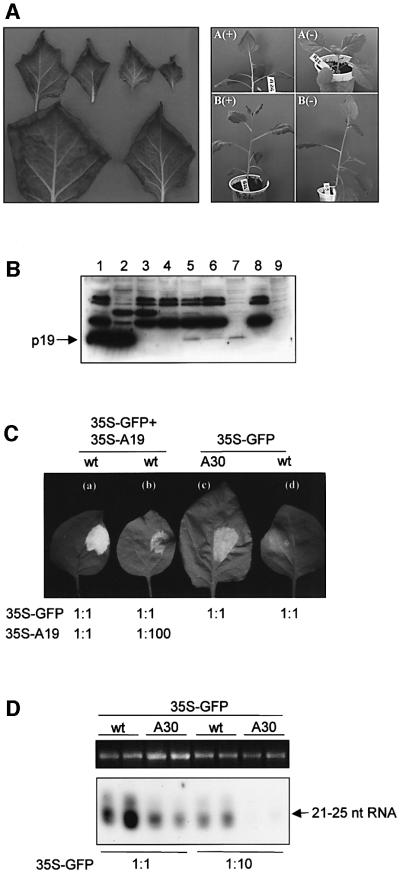
Six regenerated (R0) plants that expressed AMCV p19 mRNA were grown to set seeds. All R0 plants showed a similar abnormal phenotype characterized by curling of the leaf margin (curled down) (Figure 5A). Two transgenic lines (A and B), which showed 3:1 Mendelian segregation for the altered to wild-type phenotype in the R1 generation, were further characterized. Southern blot analyses [Supplementary data (E)] showed that the transgene was integrated as a single copy into different locations. We tested 30 R1 plants of each line and found a perfect correlation between the curled phenotype and the expression of p19 mRNA [Supplementary data (F)]. Western blot analysis confirmed the accumulation of p19 in transgenic plants. Low level of p19 accumulation was revealed in total as well as in the soluble protein fraction of transgenic A30, but not in A22 segregant plant (Figure 5B, lanes 5 and 7 compared with lanes 4 and 9).
Fig. 5. Characterization of the AMCV p19 transgenic plants. (A) Phenotype of transgenic AMCV p19 plants. Left panel: isolated curled leaves are shown according to decreasing age. Right panel: R1 plants of line A and B expressing (+) or not expressing (–) the AMCV p19 gene. (B) Western blot of total (lanes 1–5), soluble S30 (lanes 7 and 9) and raw protein P1 fractions (lanes 6 and 8) extracted from N.benthamiana infected with AMCV (lane 1), PVX/AMCV p19 (lane 2) or PVX (lane 3), or from the A22 non-transgenic (lanes 4, 8 and 9) and A30 transgenic (lanes 5–7) R1 plants of the AMCV p19 A line. (C) p19 transgenic plants show PTGS suppression activity. To estimate silencing suppressor capacity of transgenic p19, wild-type plants were co-infiltrated with 35S–GFP+35S–A19 at regular concentration (1:1) (a) or a 100-fold dilution of 35S–A19 solution (1:100) (b), while 35S–GFP concentration was added in regular concentration (1:1). AMCV p19 (A30) transgenic (c) and wild-type (d) plants were infiltrated with 35S–GFP at a density used in previous experiments (showed as 1:1). Photos were taken at 3 d.p.i. (D) Accumulation of GFP-specific 21–25 nt RNAs in GFP-infiltrated tissues of wild-type and AMCV p19 transgenic N.benthamiana. RNAs were isolated from tissues infiltrated with 35S–GFP at a density used in previous experiments (1:1) or diluted (1:10) at 3 d.p.i. As a loading control, equivalent amounts of total RNA that were used for low molecular weight RNA gel blot analyses were separated in agarose gel and ethidium bromide-stained.
Since 35S–GFP infiltration triggers local GFP silencing even in the absence of a stably integrated transgene (Johansen and Carrington, 2001), we could test whether transgenically expressed AMCV p19 retained the PTGS suppressor activity. As shown in Figure 5C, green fluorescence was stronger in 35S–GFP-infiltrated p19 transformants than in wild-type plants. Consistently, significantly less 21–25 nt RNA accumulated in p19 transformants than in wild-type plants (Figure 5D), confirming that transgenically expressed p19 retained silencing suppression activity.
Discussion
In this study we have analysed the PTGS counterdefensive strategy of tombusviruses. Here we show that p19 is an efficient silencing suppressor. We also demonstrate that p19 specifically binds PTGS-generated 21–25 nt dsRNAs, showing a biochemical activity that can be linked to silencing suppression.
p19-mediated PTGS suppression
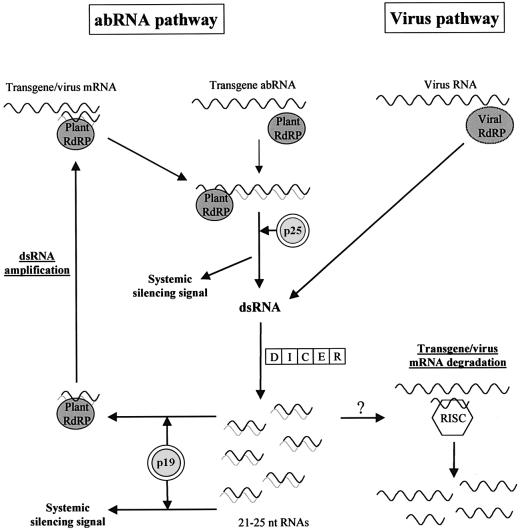
Current plant PTGS models suggest that a DICER-like enzyme digests long dsRNAs to 21–25 nt RNAs, after which these short RNAs provide specificity to the effector complexes of gene silencing (Matzke et al., 2001; Vance and Vaucheret, 2001; Voinnet, 2001; Waterhouse et al., 2001). Transgene- and virus-induced PTGS differ mainly in the generation of dsRNAs. Double-stranded replicative RNA forms of plant RNA viruses are synthesized by viral-encoded RdRP, while transgene-derived dsRNAs are generated from aberrant transgenic transcripts via a PTGS pathway, which involves a plant-encoded RdRP and other endogenous proteins [referred to in this discussion as the aberrant RNA (abRNA) pathway] (Dalmay et al., 2000, 2001; Mourrain et al., 2000). Systemic silencing signals of both transgene- and virus-induced PTGS might be produced in the abRNA pathway, suggesting that the abRNA pathway also recruits viral RNAs (Figure 6) (Voinnet et al., 2000).
Fig. 6. A model for p19-mediated PTGS suppression. The dsRNA amplification cycle was integrated into current plant PTGS models. The replication cycle of plant RNA viruses results in dsRNAs. Aberrant transgenic mRNAs might be converted into dsRNAs by host-encoded RdRP without 21–25 nt dsRNAs. Effective transformation of transgene mRNAs or viral RNAs into dsRNA by plant RdRP might require PTGS-generated, 21–25 nt dsRNAs as primers. p19 sequesters 21–25 nt dsRNAs, thus inactivating the dsRNA amplification and intensification of the abRNA pathway. Consequently, p19 interferes with transgene-induced local silencing. Generation of systemic signal is prevented by the inactivation of the abRNA pathway, if precursors of 21–25 nt RNAs are involved in mobile complex. Alternatively, p19 prevents the incorporation of 21–25 nt dsRNAs into the systemic complex. Further studies are required to clarify whether p19 can influence RISC activity or not (symbolized by ‘?’).
p19 inhibits transgene-induced local and systemic silencing. Moreover, p19 could interfere with virus-induced systemic PTGS while it does not affect virus-triggered local silencing. PVX-encoded p25 also interferes with transgene-induced local PTGS and prevents the development of both transgene- and virus-induced systemic silencing (Voinnet et al., 2000). These findings suggest that p19 and p25 target the same PTGS pathway. It was speculated that p25 interacts with a host-encoded protein involved in the abRNA pathway, thus suppressing transgene-induced local silencing and preventing the generation of silencing signals (Figure 6) (Voinnet et al., 2000). Although it is possible that p19 inhibits the abRNA pathway by interacting with a host-encoded protein, we suggest a model in which p19 suppresses silencing by binding to PTGS-generated small dsRNAs.
In animal systems, 21-nt RNA duplexes with 2-nt 3′ overhanging ends incorporate into the effector complexes of silencing machinery and provide sequence specificity for them (Lipardi et al., 2001; Nykänen et al., 2001). Here we showed that the silencing-specific 21–25-nt RNAs isolated from native PAGE were symmetrical and could be bound efficiently by p19, indicating that virus-induced PTGS generated 21–25 nt RNA duplexes with 3′ overhangs. If, in plant cells, these 21–25 nt RNA duplexes incorporate into the effector complexes of PTGS as reported for Drosophila, p19 would compete for these short dsRNAs with the different silencing effector complexes, including the RdRP, the signal, and the RISC complex.
In Drosophila embryo extract, silencing-competent ds siRNAs serve as primers, presumably to an RdRP complex, to transform target mRNAs into dsRNAs (Lipardi et al., 2001). If plant RdRP also requires 21–25 nt dsRNAs as guides or primers to convert transgenic and viral mRNAs (or abRNAs) into dsRNA efficiently, p19 and the RdRP complex would compete for the PTGS-generated, 21–25 nt dsRNAs. Since p19 accumulates in the virus-infected or 35S–C19-infiltrated cells to very high level (Havelda et al., 1998; Figure 2A), it might deplete the PTGS-generated, 21–25 nt dsRNA fraction. p19 could therefore inactivate the plant RdRP-mediated dsRNA generation, thus suppressing transgene-induced local silencing. Consistent with the dsRNA depletion model, a high level of p19 was required for complete suppression of transgene-induced local silencing in 35S–GFP infiltration experiments (Figure 5C and D).
p19 suppresses transgene-induced systemic silencing and interferes with the development of virus-induced systemic PTGS. If a virus- and transgene-derived mobile signal is produced in the abRNA pathway, inactivation of host-encoded RdRP would also result in the inhibition of transgene- and virus-induced systemic silencing. Alternative PTGS models suggest that 21–25 nt RNAs confer sequence specificity for the signal (Hamilton and Baulcombe, 1999; Waterhouse et al., 2001). Our model for p19-mediated systemic silencing suppression is also consistent with these suggestions. If 21–25 nt dsRNAs are required for the formation of active mobile complex, p19-mediated 21–25 nt dsRNA depletion would result in the inactivation of systemic silencing as well as the abRNA pathway (Figure 6).
Virus-induced local silencing occurred similarly in CymRSV- or in Cym19stop-infected protoplasts. These findings suggest that p19 does not interfere with the RISC complex. Alternatively, cellular PTGS could not reduce significantly the accumulation of tombusvirus RNA accumulation (Figure 6).
p19 also binds RNA duplexes with a blunt end and a 2-nt 3′ overhanging end. In animal systems, DICER acts as an exonuclease, digesting stepwise from the ends of long dsRNAs (Elbashir et al., 2001a), and therefore might produce long dsRNAs with 2- nt 3′ overhangs. Although it is possible that p19 competes with DICER-related protein for the 2-nt 3′ overhanging ends of long dsRNAs, the high level of 21–25 nt RNAs in CymRSV-infected cells suggests that p19 fails to suppress DICER-like activity.
p19 might also act as silencing suppressor in other hosts. Since p19 siRNA binding does not require host factors in vitro and these short RNAs are specificity determinants of RNAi effector complexes, p19 could be used to inhibit RNAi in heterologous systems, including those of Drosophila, worms or mammals.
The PTGS counterdefensive strategy of tombusviruses is conserved
p19-mediated systemic silencing suppression appears to be a conserved PTGS counterdefensive strategy for tombusviruses. Supporting this conclusion, p19 encoded by CymRSV and AMCV acts identically in PTGS suppression assays. Furthermore, TBSV p19 was also shown to prevent the onset of transgene-induced local silencing (Voinnet, 2001). Finally, the PTGS-generated recovery phenotype of Cym19stop-infected plants strongly resembles the phenotype of N.benthamiana infected with other p19 inactivated tombusviruses (Russo et al., 1994; Scholthof et al., 1995b). Conservation of the p19-mediated systemic suppression strategy suggests that p19-related proteins target a conserved element of PTGS machinery. PTGS-generated 21–25 nt dsRNAs, the putative targets of p19, are consistently central elements of the silencing machinery.
PTGS as an efficient whole-plant antiviral system
The wide occurrence of PTGS suppressors in plant viruses supports the argument that PTGS is an important antiviral system in higher plants. It was speculated that effective cellular or systemic PTGS could confine viral pathogens to very few cells contributing to non-host resistance (Voinnet et al., 2000; Dalmay et al., 2001). In contrast, CymRSV, even in the absence of its silencing suppressor, could accumulate to a high level in transfected cells and spread effectively in N.benthamiana, suggesting that PTGS is unable to localize virus infection in the inoculated tissues. However, PTGS as a whole-plant defense system efficiently limited the extent of Cym19stop virus infection, resulting in a recovery phenotype.
The p19-targeted PTGS pathway could have a role in plant development
An increasing number of observations support the theory that PTGS is involved in the regulation of plant development. For instance, transgenic plants expressing HC-Pro or the endogenous silencing suppressor protein (rgs-CaM), which interacts with HC-Pro, develop a differentiated tumour at the junction of the stem and the root (Anandalakshmi et al., 2000). Moreover, silencing mutant ago-1 (Fagard et al., 2000) also show developmental abnormality. Here we demonstrated that low-level expression of p19 silencing suppressor altered leaf morphology in transgenic plants. In addition to leaf curling, some severely affected plants also showed a delayed timing in the appearance of developed secondary stems (Figure 5A, right panel). Although, it is possible that developmental abnormalities in transgenic plants are not related to silencing suppressor activity of p19, these findings could indicate that the p19-targeted PTGS pathway plays a role in plant development. Constitutive expression of p19 might interfere with the abRNA pathway of gene silencing. Presumably, elimination of aberrant transcripts by the abRNA pathway is important for normal plant growth or certain genes are directly regulated by abRNA pathway-mediated inactivation.
Alternatively, the effect of p19 on other short RNA-mediated cellular process caused the mutant phenotype of transgenic plants. In animals, DICER also generates short ssRNAs, which regulate developmental timing by controlling the translation of other genes [reviewed in Ambros (2001) and references therein]. If this short ssRNA-mediated developmental control also occurs in plants, and if these molecules are derived from dsRNA precursors generated by a DICER-related enzyme, p19 could deplete the precursor dsRNAs. Consequently, ssRNA-mediated translational control would be compromised in p19 transgenics, leading to an altered phenotype.
Materials and methods
Plant materials and A.tumefaciens infiltration
Transgenic N.benthamiana carrying the GFP ORF was described previously (Brigneti et al., 1998). For preparing AMCV p19 transgenic plants, N.benthamiana leaf disks were transformed according to Tavazza et al. (1988) with A.tumefaciens LBA 4404 harbouring the plasmid pBI-NA. The A.tumefaciens infiltration method was carried out according to Voinnet et al. (2000). For co-infiltration, equal volumes of indicated A.tumefaciens cultures (OD600 = 1) were mixed before infiltration.
PTGS suppression assay and GFP imaging
The PTGS suppression assay was carried out as described by Voinnet et al. (1999). Visual detection of GFP fluorescence was performed using a 100 W, hand-held, long-wave ultraviolet (UV) lamp (Black Ray model B 100AP; UV products, Upland, CA).
Plasmid constructs
The infectious cDNA clones of CymRSV (Dalmay et al., 1993), Cym19stop mutant (Szittya et al., 2002) and AMCV (Tavazza et al., 1994) were as described previously.
The NcoI–AvaI fragment corresponding to nucleotides 3902–4545 of AMCV genomic sequence was blunt-ended with Klenow polymerase and cloned into the SmaI site of pSP65 (Promega, Madison, WI), obtaining pSP-AN. The SacI–BamHI fragment of pSP-AN was cloned into the SacI–BamHI sites of the plant expression binary vector pBI121.1, replacing the β-glucuronidase sequence with the AMCV p19 sequence and obtaining pBI-NA.
Plasmid PVX/AMCV p19 was obtained by cloning the T4 DNA polymerase-repaired SacI–BamHI fragment of pSP-AN in the T4 DNA polymerase-repaired SalI site of pP2c2s (Chapman et al., 1992).
35S–GFP plasmid was described previously (Brigneti et al., 1998). Suppression activity of different CymRSV ORFs and AMCV ORF5 were tested in BIN61S. BIN61S is a BIN19 derivative obtained by inserting the HindIII–EcoRI fragment of pff19 (Timmermans et al., 1990) carrying the CaMV 35S promoter-polyA terminator cassette. CymRSV and AMCV p19 inserts were isolated from previously described PVX19 (Szittya et al., 2002) and PVX/AMCV p19 constructs with NheI–SalI, and cloned into XbaI–SalI-digested BIN61S vector. CymRSV ORF1, -2, -3 and -4 were PCR amplified using primers corresponding to the appropriate first and last 22 nt. PCR fragments were cloned into SmaI-digested BIN61S. Cloning of GST–19 was as described previously (Havelda et al., 1998).
To obtain pRNA1 and pRNA2, the EcoRV–XbaI fragment corresponding to nucleotides 4062–4143 of CymRSV genomic sequence was cloned into pBluescript KS and SK vectors, respectively.
In vitro RNA transcription and plant inoculation
In vitro transcription of CymRSV, Cym19stop and AMCV RNAs, and inoculation of RNA transcripts onto N.benthamiana were performed as described previously (Dalmay et al., 1993; Tavazza et al., 1994).
Protoplast preparation and inoculation
Protoplasts were isolated from N.benthamiana and transfected with in vitro-synthesized transcripts of genomic RNAs as described previously (Dalmay et al., 1993).
Protein extraction and analysis
Total proteins, S30 and P1 fractions were obtained essentially as described by Berna et al. (1985), omitting the filtration step. Proteins were electrophoresed through 12% SDS–PAGE and probed with anti-p19 (CymRSV) rabbit polyclonal antibodies. Secondary antibody was a donkey anti-rabbit biotinylated antibody (Amersham-Pharmacia, Piscataway, NJ) used at 1:5000 dilution. Detection was performed using streptavidin–horseradish peroxidase conjugate (1:2000) and ECL-Plus reagents (Amersham-Pharmacia).
RNA gel blot analysis
The same total RNA extract was used for high and low molecular weight RNA gel blot analysis. RNA extraction and RNA gel blot analysis was carried out as described previously (Szittya et al., 2002). PCR fragments labelled with random priming method were used for northern analyses of high molecular weight RNAs. Radioactively labelled in vitro transcripts corresponding to the positive strand of virus RNA and the antisense strand of GFP were used as probes for northern analyses of low molecular weight RNAs. Labelling was carried out according to Szittya et al. (2002).
Preparation of RNA probes for gel mobility shift assay
PTGS-produced, 21–25 nt RNAs were isolated from denaturing PAGE and labelled as described previously (Szittya et al., 2002). To isolate PTGS-generated 21–25 nt dsRNAs, total RNA extract was separated in 8% native PAGE, then ethidium bromide-stained 21–25 nt RNA fraction was cut and eluted in KT buffer [0.5 M NH4OAc, 10 mM Mg(OAc)2, 1 mM EDTA, 0.1% SDS]. After precipitation, 21–25 nt RNAs were dephosphorylated, pelleted and labelled in a 10-µl reaction volume in the presence of [γ-32P]ATP and RNasin, with 8 U T4 polynucleotide kinase. Synthetic siRNAs (listed in Supplementary table I) were labelled with T4 PNK as described above.
RNA1 and RNA2 probes were generated by in vitro RNA transcription in the presence of [α-32P]UTP from linearized pRNA1 and pRNA2, respectively. For gel mobility shift assay, in vitro-transcribed RNA probes were separated and isolated from denaturing polyacrylamide gel as described previously (Szittya et al., 2002).
Mobility shift assay
To obtain RNA duplexes, corresponding short or long ssRNAs were annealed in 100 mM potassium acetate, 30 mM HEPES–KOH pH 7.4 and 2 mM magnesium acetate buffer (Elbashir et al., 2001a). Binding reactions were performed in 83 mM Tris–HCl pH 7.5, 0.8 mM MgCl2, 66 mM KCl, 100 mM NaCl and 10 mM DTT. Each sample contained 5U RNasin. GST and GST–19 proteins were expressed and purified according to manufacturer protocols (Amersham–Pharmacia–Biotech). Purified proteins (∼1 µg) were added to samples last. Labelled RNA (1–3 ng) was used for binding assays, unless otherwise indicated in the Figure legends. Binding was performed at room temperature for 18–20 min, stopped by adding dyes, and loaded onto a 5% native PAGE. Gels were dried and autoradiographed.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to David Baulcombe for kindly providing GFP plants and A.tumefaciens carrying the 35S–GFP construct. We thank Phillip D.Zamore and Thomas Tuschl for kindly providing siRNAs. We thank Brandan Bell, Lóránt Lakatos and Zoltán Havelda for helpful comments, and László Szabó for quantitation of RNA by 2.0, Soft-Imaging Software GmbH. This research was supported by grants from the Hungarian Scientific Research Fund (OTKA) (31929) and the Ministry of Education (FKFP0442/1999).
References
- Ambros V. (2001) Dicing up RNAs. Science, 293, 811–813. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi R., Marathe,R., Ge,X., Herr,J.M.,Jr, Mau,C., Mallory,A., Pruss,G., Bowman,L. and Vance,V.B. (2000) A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science, 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Berna A., Godefroy-Colburn,T. and Stussi-Garaud,C. (1985) Preparation of an antiserum against an in vitro translation product of alfalfa mosaic virus RNA 3. J. Gen. Virol., 66, 1669–1678. [Google Scholar]
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Brigneti G., Voinnet,O., Li,W.X., Ji,L.H., Ding,S.W. and Baulcombe,D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J., 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chapman S., Kavanagh,T. and Baulcombe,D.C. (1992) Potato virus X as a vector for gene expression in plants. Plant J., 2, 549–557. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Rubino,L., Burgyán,J., Kollár,Á. and Russo,M. (1993) Functional analysis of cymbidium ringspot virus genome. Virology, 194, 697–704. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Hamilton,A., Rudd,S., Angell,S. and Baulcombe,D.C. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Horsefield,R., Braunstein,T.H. and Baulcombe,D.C. (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J., 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001a) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001b) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M., Boutet,S., Morel,J.B., Bellini,C. and Vaucheret,H. (2000) AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene in plants, quelling in fungi, and RNA interference in animals. Proc. Natl Acad. Sci. USA, 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.S. and Ding,S.W. (2002) A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J., 21, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe,D.C. (1999) A novel species of small antisense RNA in post-transcriptional gene silencing. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- Havelda Z., Szittya,Gy. and Burgyán,J. (1998) Characterization of the molecular mechanism of defective interfering RNA mediated symptom attenuation in tombusvirus infected plants. J. Virol., 72, 6251–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.K. and Carrington,J.C. (2001) Silencing on the spot. Induction and suppression of RNA silencing in an Agrobacterium-mediated transient expression system. Plant Physiol., 126, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.X. and Ding,S.W. (2001) Viral suppressors of RNA silencing. Curr. Opin. Biotechnol., 12, 150–154. [DOI] [PubMed] [Google Scholar]
- Lipardi C., Wei,Q. and Paterson,B.M. (2001) RNAi as random degradative PCR. siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell, 107, 297–307. [DOI] [PubMed] [Google Scholar]
- Mallory A.C. et al. (2001) HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell, 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A., Matzke,A.J., Pruss,G.J. and Vance,V.B. (2001) RNA-based silencing strategies in plants. Curr. Opin. Genet. Dev., 2, 221–227. [DOI] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Nykänen A., Haley,B. and Zamore,P.D. (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell, 107, 309–321. [DOI] [PubMed] [Google Scholar]
- Palauqui J.C., Elmayan,T., Pollien,J.M. and Vaucheret,H. (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J., 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Harrison,B.D. and Baulcombe,D.C. (1997) A similarity between viral defense and gene silencing in plants. Science, 276, 1558–1560. [DOI] [PubMed] [Google Scholar]
- Russo M., Burgyán,J. and Martelli,P.G. (1994) Molecular biology of Tombusviridae. Adv. Virus Res., 44, 381–428. [DOI] [PubMed] [Google Scholar]
- Scholthof H.B., Scholthof,K.-B.G. and Jackson,A.O. (1995a) Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell, 7, 1157–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof H.B., Scholthof,K.-B.G., Kikkert,M. and Jackson,A.O. (1995b) Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology, 213, 425–438. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (2001) RNA interference—2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- Szittya G., Molnár,A., Silhavy,D., Hornyik,C. and Burgyán,J. (2002) Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell, 14, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazza R., Ordas,R.J., Tavazza,M., Ancora,G. and Benvenuto,E. (1988) Genetic transformation of Nicotiana clevelandii using a Ti plasmid derived vector. J. Plant Physiol., 133, 640–644. [Google Scholar]
- Tavazza M., Lucioli,A., Calogero,A., Pay,A. and Tavazza,R. (1994) Nucleotide sequence, genomic organization and synthesis of infectious transcripts from a full-length clone of artichoke mottled crinkle virus. J. Gen. Virol., 75, 1515–1524. [DOI] [PubMed] [Google Scholar]
- Tenllado F. and Diaz-Ruiz,J.R. (2001) Double-stranded RNA-mediated interference with plant virus interaction. J. Virol., 75, 12288–12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans M.C., Maliga,P., Vieira,J. and Messing,J. (1990) The pFF plasmids: cassettes utilising CaMV sequences for expression of foreign genes in plants. J. Biotechnol., 14, 333–344. [DOI] [PubMed] [Google Scholar]
- Vance V.B. and Vaucheret,H. (2001) RNA silencing in plants—defense and counterdefense. Science, 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet., 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet O. and Baulcombe,D.C. (1997) Systemic signalling in gene silencing. Nature, 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto,Y. and Baulcombe,D.C. (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses. Proc. Natl Acad. Sci. USA, 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Lederer,C. and Baulcombe,D.C. (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell, 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang,M.B. and Finnegan,E.J. (2001) Role of short RNAs in gene silencing. Trends Plant Sci., 6, 297–301. [DOI] [PubMed] [Google Scholar]
- Zamore P.D. (2001) RNA interference: listening to the sound of silence. Nat. Struct. Biol., 8, 746–750. [DOI] [PubMed] [Google Scholar]