Abstract
Prions are composed of an isoform of a normal sialoglycoprotein called PrPc, whose physiological role has been under investigation, with focus on the screening for ligands. Our group described a membrane 66 kDa PrPc-binding protein with the aid of antibodies against a peptide deduced by complementary hydropathy. Using these antibodies in western blots from two-dimensional protein gels followed by sequencing the specific spot, we have now identified the molecule as stress-inducible protein 1 (STI1). We show that this protein is also found at the cell membrane besides the cytoplasm. Both proteins interact in a specific and high affinity manner with a Kd of 10–7 M. The interaction sites were mapped to amino acids 113–128 from PrPc and 230–245 from STI1. Cell surface binding and pull-down experiments showed that recombinant PrPc binds to cellular STI1, and co-immunoprecipitation assays strongly suggest that both proteins are associated in vivo. Moreover, PrPc interaction with either STI1 or with the peptide we found that represents the binding domain in STI1 induce neuropro tective signals that rescue cells from apoptosis.
Keywords: neuroprotection/prion/PrPc/PrPc ligand/stress-inducible protein 1
Introduction
Prions, the agents of transmissible spongiform encephalopathies (reviewed by Prusiner, 1998), require the expression of a glycosylphosphatidylinositol (GPI)-anchored cell surface sialoglycoproteic homolog (PrPc) to propagate disease (Büeler et al., 1993). Mutations in the gene coding for PrPc are also the cause of hereditary neurological (Prusiner, 1998) and possibly psychiatric (Samaia et al., 1997) disorders. Since clinical manifestations may occur either before or without characteristic PrPc deposits (Collinge et al., 1990; Medori et al., 1992), it has been suggested that loss of PrPc function may concur for the etiology of such diseases (Aguzzi and Weissmann, 1997; Samaia and Brentani, 1998).
Recently, certain biological functions of PrPc have been uncovered. PrPc strongly binds Cu2+ and thus may be involved in both copper metabolism (Brown et al., 1997a) and protection against oxidative stress (Brown et al., 1997b). Since PrPc is localized mainly in synaptosomal fractions, it may serve as a copper buffer in the synaptic cleft or in the re-uptake of copper into the presynaptic terminal (Kretzschmar et al., 2000). Moreover, it is known that PrPc plays a role in the modulation of neuronal survival, both in vivo (Walz et al., 1999) and in vitro (Kuwahara et al., 1999; Chiarini et al., 2002), and is also involved in signal transduction (Mouillet-Richard et al., 2000; Chiarini et al., 2002). We have also shown that PrPc is a specific receptor for the C-terminal domain of the γ-1 chain of extracellular matrix laminin, and is involved in neuronal adhesion and neurite growth (Graner et al., 2000a,b).
To understand further the biological functions of PrPc, binding molecules were sought extensively (reviewed by Martins et al., 2001). In an attempt to characterize a PrPc receptor, we (Martins et al., 1997) designed a peptide that was predicted to bind PrPc on the basis of the complementary hydropathy theory (see for example Bost et al., 1985; Brentani, 1988; Boquet et al., 1995). Theoretically, the peptide should mimic the docking site of PrPc in a ligand. Antibodies raised against this peptide recognize a 66 kDa cell surface antigen, which binds PrPc in vitro. The same antibody prevented toxicity of the human PrPc peptide comprising amino acids 106–126 towards neurons in culture, indicating that the 66 kDa protein may be a receptor for the infectious agent, involved in the pathogenesis of prion diseases. Conversely, the protein might also work in association with PrPc in normal cellular functions.
Here, we report that the PrPc membrane ligand is stress-inducible protein 1 (STI1), a heat shock protein, first described in a macromolecular complex with Hsp70 and Hsp90 chaperone family proteins (Blatch et al., 1997; Lässle et al., 1997). Recombinant PrPc and STI1 showed specific and high affinity binding both in vitro and at the cellular level. The binding site in mouse PrPc confirmed our earlier prediction and spans amino acids 113–128. Furthermore, we identified a domain in the mouse STI1 molecule (amino acids 230–245) with the same hydropathy profile as the predicted PrPc-binding peptide (Martins et al., 1997). This peptide prevented the PrPc–STI1 interaction, indicating that it contains the binding site at the STI1 molecule. We have shown (Chiarini et al., 2002) that PrPc transduces neuroprotective signals when challenged with either the theoretically derived PrPc-binding peptide or with certain antibodies. Here we demonstrate that interactions of PrPc with either STI1 or the STI1 peptide that contains the PrPc-binding site induce neuroprotection.
Results
STI1 is the molecule recognized by antiserum raised against the predicted PrPc-binding peptide (PrR)
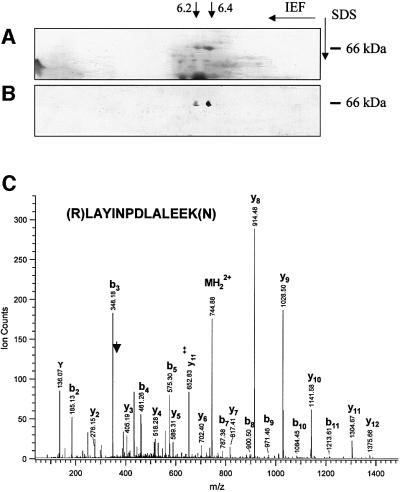
To identify the previously characterized PrPc ligand, we ran two-dimensional gel electrophoresis of a 45–55% ammonium sulfate fraction from whole mouse brain extracts, which partially purifies this ligand (Martins et al., 1997). A 66 kDa doublet with pIs ranging from 6.2 to 6.4 is readily observed after Coomassie Blue staining (Figure 1A), which was also recognized in a western blot (Figure 1B) by antiserum against the predicted PrPc-binding peptide (Martins et al., 1997). Herein, this peptide is referred to as PrR. The same spots were excised from a parallel gel and separately subjected to mass spectrometry. The amino acid sequences deduced for the protein extracted from each of the spots displayed 93–96% identity with the mouse STI1 molecule (Blatch et al., 1997; Lässle et al., 1997). Figure 1C shows the mass spectrometry (MS) spectrum of a doubly charged tryptic peptide from the spot with higher pI. The two spots may represent differential phosphorylation of the molecules, as described (Lässle et al., 1997). Despite the obvious sequence differences, we also tested whether the 66 kDa protein corresponds to the previously reported PrPc-binding laminin receptor (Rieger et al., 1997), which has a similar molecular weight. The relevant spots did not react with a specific monoclonal antibody against the laminin receptor (a kind gift of Dr Sylvie Mènard, Institute of Pathology, Milan University), and its molecular weight was unaffected by treatment with concentrated hydroxylamine (not shown), which cleaves acetyl groups and converts the 67 kDa laminin receptor into the 37 kDa precursor (Buto et al., 1998).
Fig. 1. Two-dimensional gel analysis of the ammonium sulfate 45–55% saturation fraction from total brain extract. (A) Coomassie Blue-stained proteins. (B) Immunoblot of an identical gel reacted with mouse serum against the PrR peptide developed using peroxidase-labeled anti-mouse Ig. Two spots of 66 kDa and pIs of ∼6.2 and 6.4 are recognized specifically, and each was subjected to microsequencing analysis. (C) MS spectrum of a doubly charged tryptic peptide (MH22+ at m/z 744.9) from the spot with the higher pI. A series of fragment ions (b and y ions) were observed due to the breakage of peptide bonds during collision-induced dissociation. The peptide sequence is determined as LAYINPDLALEEK, identifying the protein as mouse stress-inducible protein 1 (accession No. 881485).
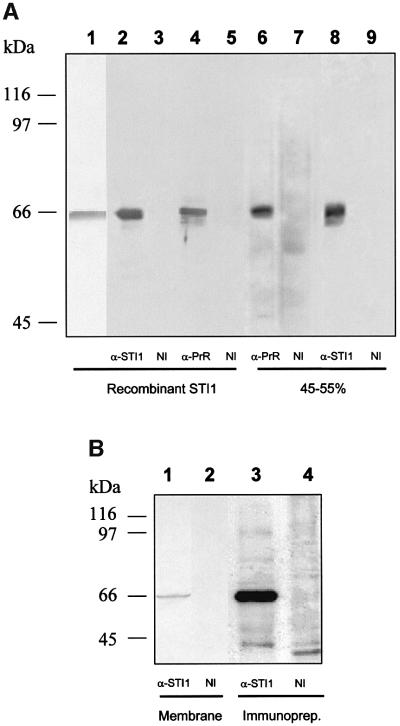
Mouse recombinant STI1 protein was used to generate a rabbit polyclonal antiserum. Figure 2A shows that the recombinant protein has the expected molecular weight (lane 1) and is recognized by the antiserum (lane 2). Furthermore, recombinant STI1 is also recognized by the serum against the PrR peptide (lane 4) which, as shown before (Martins et al., 1997), recognizes a band with the same molecular weight in a 45–55% ammonium sulfate fraction from whole brain extracts (lane 6). Serum raised against STI1 also recognizes a band of similar molecular weight in the 45–55% ammonium sulfate fraction from whole brain extracts (lane 8). Controls with mouse or rabbit non-immune serum did not react in this assay (lanes 3, 5, 7 and 9).
Fig. 2. STI1 is the 66 kDa protein located at the cell membrane recognized by serum against the PrR peptide. (A) Western blot assay of recombinant mSTI1 (lanes 2–5) or ammonium sulfate fractions at 45–55% saturation from brain extracts (lanes 6–9), done with rabbit serum against recombinant mST11 (α-STI1, lanes 2 and 8), serum against PrR peptide (α-PrR, lanes 4 and 6) or non-immune serum (NI, lanes 3, 5, 7 and 9). The recombinant STI1 protein stained with Ponceau is shown in lane 1. (B) Western blot assay from purified membrane fractions from brain extracts (lanes 1 and 2) done with rabbit serum against recombinant mST11 (α-STI1, lane 1) or non-immune serum (NI, lane 2). Cell surface proteins from N2a cells were biotinylated followed by extract preparation and immunoprecipitation with anti-STI1 antibody (α-STI1, lane 3) or non-immune serum (NI, lane 4). The immunoprecipitated material was developed using streptavidin– peroxidase.
STI1 was found either in the cytoplasm (Lässle et al., 1997) or in the Golgi apparatus and small vesicles (Honoré et al., 1992). However, we previously have described that a small fraction of the PrPc ligand was present at the cell surface (Martins et al., 1997). To approach protein localization further, we used membrane preparations from mouse brain and N2a cells, which together with mouse brain extracts were used in the previous study (Martins et al., 1997). Western blots of the mouse brain membrane fraction (Figure 2B, lane 1) showed a specific 66 kDa band recognized by anti-STI1 antibodies. In addition, N2a cell surface proteins were conjugated with biotin and cell lysates were immunoprecipitated with anti-STI1 antibodies. A 66 kDa biotinylated band was labeled with peroxidase-coupled streptavidin in immunoprecipi tated material (lane 3), similar to that recognized in crude brain membrane preparations (lane 1), strongly suggesting that at least part of the STI1 is located at the cell surface.
PrPc binds STI1 with high affinity and independently of copper
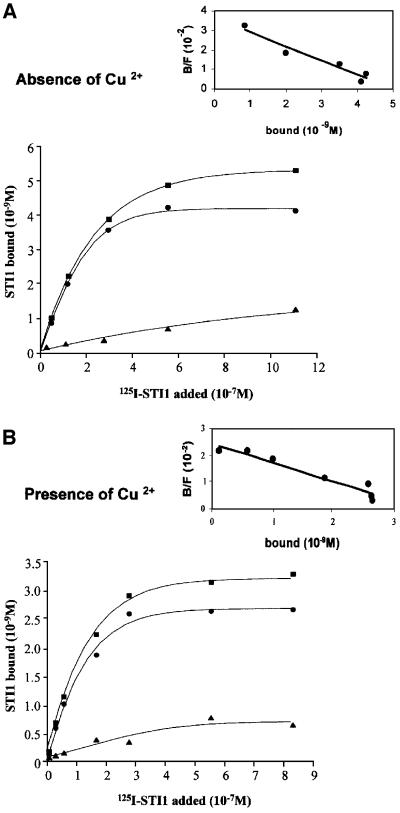
We next measured binding of 125I-labeled His6-STI1 to His6-PrPc. Figure 3 shows curves representative of at least four independent assays carried out with PrPc refolded in either the absence (Figure 3A) or presence of Cu2+ (Figure 3B). The presence of copper ions in the refolded protein was determined by atomic emission spectrometry (data not shown) and confirms the presence of ∼10 Cu2+ atoms bound per His6-PrPc molecule (Brown et al., 1999). PrPc binds STI1 with high affinity, in a saturable manner, and 1.2–1.5 times as much STI1 binds PrPc refolded in the absence than in the presence of Cu2+ ions, although the affinity constants were similar (Kd = 1.4 × 10–7 and 1.2 × 10–7 M in the absence and presence of Cu2+, respectively). Since the amount of His6-PrPc added in both experiments was the same, these data indicate that there is more PrPc able to bind STI1 when the refolding occurs in the absence of Cu2+. One possibility is that due to the extensive process to refold the protein (Wong et al., 2000), a small fraction is degraded.
Fig. 3. STI1 binds PrPc in a saturable and specific manner and independently of Cu2+ incorporation into the PrPc molecule. Representative curves of [125I]His6-STI1 binding to His6-PrPc refolded in the absence (A) or presence (B) of Cu2+. [125I]His6-STI1 was incubated with adsorbed His6-PrPc in the absence (total) or presence of unlabeled STI1 (non-specific). Non-specific binding (triangles) was subtracted from the total binding (squares) to yield His6-PrPc-specific binding to [125I]His6-STI1 (circles). Scatchard plots (inserts) gave Kds of 1.4 × 10–7 and 1.2 × 10–7 M for PrPc refolded in the absence or presence of Cu2+, respectively.
STI1 interacts within amino acids 113–128 of PrPc
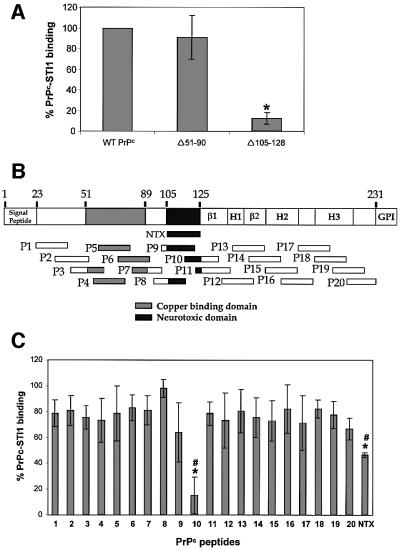
To map the STI1-binding domain in PrPc, we constructed a deletion mutant lacking the region around the neurotoxic domain (Forloni et al., 1993), which contains the previously predicted binding site (Martins et al., 1997). The mouse PrPc mutant Δ105–128 (human 106–129) is unable to bind STI1, while PrPc without the copper-binding domain (PrPc Δ51–90) binds similarly to the wild-type molecule (Figure 4A). We then tested a series of peptides covering the entire PrPc molecule (Figure 4B). Peptide P10, which contains the amino acid sequence spanning residues 113–132 of the mouse sequence, was the most effective competitor of PrPc–STI1 binding, while peptide P9 (amino acids 103–122), which shares residues 113–122 with P10, was also inhibitory, albeit at a concentration 2.5 times higher than that used for P10 (data not shown). Moreover, the human neurotoxic peptide (NTX) (Forloni et al., 1993), which is equivalent to mouse PrPc amino acids 105–125, was able to compete for PrPc–STI1 interaction, but less efficiently than P10. Therefore, amino acids 126–131 present in P10 and absent in both P9 and the NTX peptide appear to be added to the 113–122 domain of PrPc in the interaction with STI1. Since the PrPc deletion mutant Δ105–128 is unable to bind STI1, the data are consistent with the hypothesis that the region comprising amino acids 113–128 of the mouse PrPc, which was the sequence used to draw the PrR peptide (Martins et al., 1997), is a unique binding site for STI1, independent of copper (Figure 4A).
Fig. 4. Mapping the STI1 binding site in PrPc using deletion mutants and synthetic PrPc peptides. (A) Wild-type PrPc and deletion mutants Δ51–90 and Δ105–128 were incubated with [125I]His6-STI1. The binding between wild-type His6-PrPc and [125I]His6-STI1 was set to 100% (control). The results for each PrPc mutant were expressed as percentage binding compared with wild-type. *P <0.01 versus control, single mean Student’s t-test. (B) Twenty mouse PrPc peptides covering the PrPc (23–231) protein sequence were synthesized chemically as a 20mer with 10 overlapping residues. The scheme shows localization of the 20 peptides, the neurotoxic peptide (NTX) and the main PrPc domains: β1 and β2, β-sheet domains; H1, H2 and H3, α-helix domains; GPI, GPI anchor. (C) The synthetic peptides were pre-incubated with [125I]His6-STI1 followed by incubation with adsorbed His6-PrPc. Total binding between His6-PrPc and [125I]His6-STI1 was set to 100%. The results are expressed as the relative percentage of the binding produced by competition with each peptide. *P < 0.01 versus control, single mean Student’s t-test; #P < 0.01 NTX versus P10, Mann– Whitney test.
PrPc interacts with an STI1 domain with a hydropathy profile identical to that of the PrR peptide
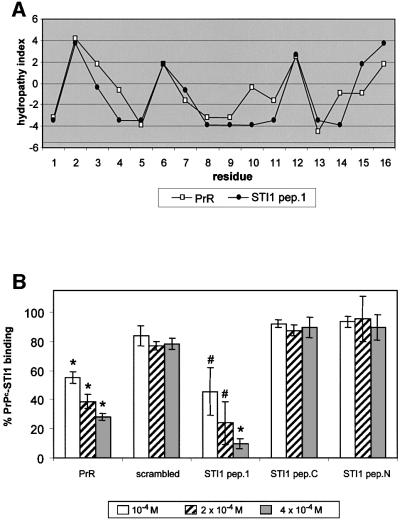
Theoretically, the PrPc docking site within the STI1 molecule should display a similar hydropathic profile to that of the PrR (Martins et al., 1997). We searched for this region using the software HYDROLOG (S.G.Jaquieri, S.M.Zanata and R.R.Brentani, in preparation) that provides a hydropathy index profile of amino acid sequences and searches for a domain with a similar pattern in a given protein. Figure 5A shows the hydropathy profile of an STI1 region with high similarity to PrR. This STI1 peptide (STI1 pep.1), which spans amino acids 230–245, and two other STI1 peptides from either the N- (amino acids 61–76) or the C-terminus (amino acids 422–437), as well as PrR were tested for competition with the PrPc–STI1 interaction. Of the three STI1 peptides, only STI1 pep.1 inhibited the binding of PrPc–STI1 similarly to PrR (Figure 5B). Thus, the region covering amino acids 230–245 in STI1, which has the same hydropathy profile as PrR (Martins et al., 1997), seems to contain the binding site for PrPc.
Fig. 5. Mapping PrPc binding at the STI1 molecule using the complementary hydropathy theory and synthetic peptides. (A) Hydropathy plot of STI1 pep.1 (amino acids 230–245) (filled circles) and the PrR peptide (open squares). (B) Competition of His6-PrPc–[125I]His6-STI1 binding by increasing amounts of the synthetic peptides PrR, scrambled peptide, STI1 pep.1, STI1 N-terminus peptide (STI1 pep.N) or STI1 C-terminus peptide (STI1 pep.C). Total binding between His6-PrPc and [125I]His6-STI1 was set to 100% (control). The results are expressed as the relative percentage of the binding produced by competition with each peptide. #P < 0.04 and *P < 0.01 versus control, single mean Student’s t-test.
Cellular STI1 associates with recombinant PrPc
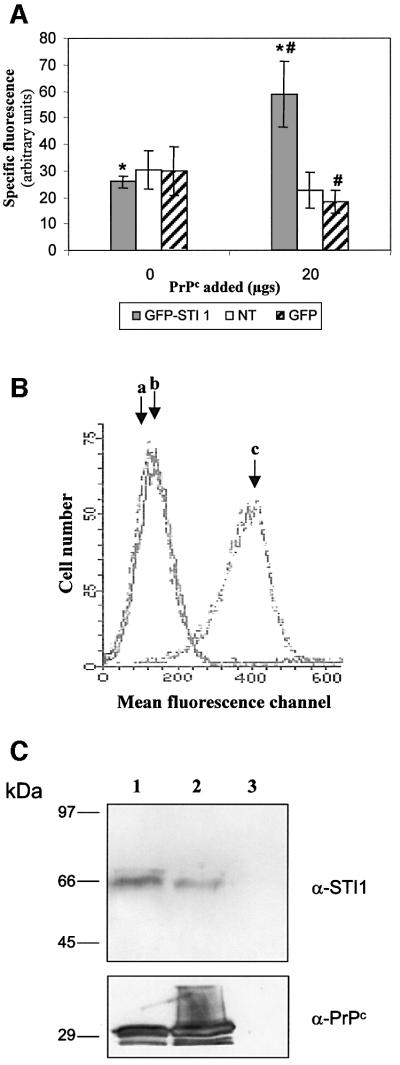
To test whether recombinant PrPc binds STI1 at the cellular level, we used HEK 293T cells overexpressing green fluorescent protein (GFP)–STI1. PrPc binds to the surface of these cells but not to control cells expressing only GFP (Figure 6A). These results indicate that PrPc binding to the cell surface increases after STI1 ectopic expression. We did not detect PrPc binding to non-transfected cells (Figure 6A), probably due to competition between resident and recombinant PrPc for the surface ligands. In fact, recombinant PrPc binds to the surface of primary cultured fibroblasts from PrP0/0 mice (Figure 6B), indicating that the absence of cellular PrPc permits the binding of recombinant PrPc to surface ligands. We next performed pull-down experiments (Rohm et al., 2000; Zanata et al., 2002) using cultured primary fibroblasts from PrP0/0 mice (Büeler et al., 1992) to test whether STI1 was one of these PrPc ligands. Whole cells or cellular extracts were incubated with His6-PrPc followed by affinity chromatography with Ni-NTA–agarose, and resin- bound proteins were assayed by western blot with either anti-STI1 (Figure 6C, top panel) or anti-PrPc antibodies (Figure 6C, bottom panel). Recombinant PrPc binds to cellular STI1 both at the cell surface (lane 1) and in cell-free conditions (lane 2), while STI1 did not associate with the Ni-NTA–agarose resin in the absence of recombinant PrPc (lane 3). The membrane was also re-probed with antibodies against actin and we were unable to detect any reactivity (data not shown). These data indicate that recombinant PrPc specifically binds to cellular STI1.
Fig. 6. Cellular STI1 binds recombinant PrPc. (A) HEK 239T cells transfected with GFP–STI1 or GFP, or non-transfected (NT) were incubated in the absence or presence of 20 mg of His6-PrPc followed by incubation with mouse anti-PrPc or non-immune serum and anti-mouse R-phycoerythrin conjugate. Analyses were carried out using a Becton Dickinson FACScan Cytometer. The specific fluorescence intensity was determined by subtraction of the fluorescence obtained with non-immune serum from that produced with anti PrPc serum. *P <0.01, GFP–STI1 + 20 mg PrPc versus GFP–STI1 without PrPc and #P <0.03, GFP–STI1 + 20 mg PrPc versus GFP + 20 mg PrPc, Mann– Whitney test. (B) Primary fibroblast cultures from PrP0/0 animals were incubated in the absence (b) or presence (c) of His6-PrPc followed by incubation with mouse anti-PrPc (b and c) or non-immune serum (a). (C) Whole cells (lane 1) or cellular extracts (lane 2) from PrP0/0 mice fibroblasts were incubated with His6-PrPc. Whole cells were washed, lysed and the extracts incubated with Ni-NTA–agarose. Extracts from cells without His6-PrPc addition were also incubated with Ni-NTA–agarose (lane 3). The bound material was eluted off the beads and analyzed by western blot using anti-STI1 (α-STI1, upper panel) or anti-PrPc (α-PrPc, lower panel) serum.
STI1 associates with cellular PrPc
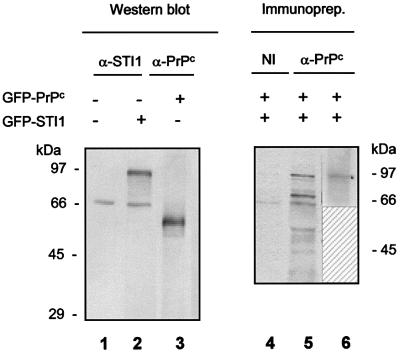
HEK 293T cells were transfected with vectors containing cDNA encoding the fusion proteins GFP–PrPc (Lee et al., 2001a) and GFP–STI1. The cellular STI1 (66 kDa) or the GFP–STI1 protein (96 kDa) were detected by western blots using anti-STI1 antibodies (Figure 7, lanes 1 and 2), while only GFP–PrPc was observed after the reaction with anti-PrPc antibody (Figure 7, lane 3). However, using flow cytometry assays, we observed that HEK 293T cells express PrPc at their surface (Figure 6A). In fact, PrPc is hardly observed in conventional western blots from cell lines (Scott et al., 1988; Cabral et al., 2002). Cells were co-transfected with GFP–PrPc and GFP–STI1 followed by conjugation of surface proteins with biotin and immunoprecipitation using anti-PrPc antibody. The blotting reac tion of the immunoprecipitated material with strepta vidin–peroxidase revealed four major bands (lane 5): one 57–58 kDa band corresponding to GFP–PrPc, and bands of 60, 70 and 96 kDa. The latter band is not observed when GFP–PrPc is transfected alone (data not shown), and reacts with anti-STI1 antibodies (Figure 7, lane 6), indicating that the 96 kDa protein co-immunoprecipitated with PrPc corresponds to GFP–STI1. The blot in lane 6 was also re-probed with a pan antibody to cadherin, which recognizes a 110 kDa isoform, and no reaction was detected (data not shown), indicating specificity for the co-immunoprecipitation reaction. We did not detect the resident STI1, probably because of its low expression at the cell surface. The identity of the 70 kDa band is unknown, and the 60 kDa band is non-specific, since it was also present following immunoprecipitation with non-immune serum (Figure 7, lane 4). These data indicate that PrPc is associated with STI1 at the cellular level, and that at least part of the PrPc–STI1 binding occurs at the cell surface.
Fig. 7. PrPc co-immunoprecipitates with STI1 located at the cell membrane. HEK 293T cells were transfected with GFP–PrPc and/or GFP–STI1 as indicated. Cell extracts were resolved by SDS–PAGE and western blots were done using anti-STI1 (lanes 1 and 2) or anti-PrPc (lane 3) antibodies. Cell surface proteins from transfected cells were biotinylated (lanes 4–6) and immunoprecipitated with anti-PrPc (lanes 5 and 6) or non-immune serum (lane 4). The reactions were developed using streptavidin–peroxidase (lanes 4 and 5) or anti-STI1 antibody followed by anti-rabbit IgG–peroxidase (lane 6). Lane 6 is shown only from 60 kDa upwards, because the antibody used for immunoprecipi tation reacts with the secondary antibody used to develop the western blot.
PrPc–STI1 binding transduces neuroprotective signals
We tested whether STI1 and STI1 pep.1 (the PrPc-binding peptide from the STI1 sequence) would induce neuroprotective responses similar to those induced by the PrR peptide and certain antibodies to PrPc (Chiarini et al., 2002). Similarly to the previous study, we used retinal explants from neonatal rats and mice and tested for protection against cell death induced by the protein synthesis inhibitor anisomycin. In this model, the protein synthesis inhibitor is known to induce death in early post-mitotic cells within the neuroblastic layer (Rehen et al., 1999).
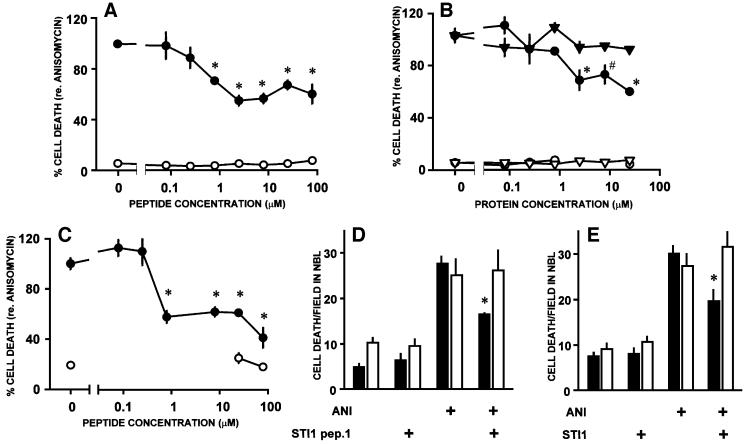
Both STI1 and STI1 pep.1 reduced anisomycin-induced cell death within the neuroblastic layer of retinal explants from neonatal rats. Compared with the PrR (Chiarini et al., 2002), the dose–response curve was shifted to lower concentrations. Thus, whereas the former was effective only at 80 µM or higher (Chiarini et al., 2002), STI1 pep.1 induced significant neuroprotection at 0.8 µM (Figure 8A), while STI1 was effective at 2.5 µM or higher (Figure 8B). No protection was observed with addition of bovine serum albumin (BSA), that has a molecular weight similar to STI1 (Figure 8B). Moreover, similarly to the PrR peptide (Figure 8; Chiarini et al., 2002), a maximum protective effect of 50% was observed with either STI1 pep.1 or STI1 protein upon anisomycin-induced cell death. These may be due to a balance between neuroprotective and pro- degenerative signals generated by PrPc (Chiarini et al., 2002).
Fig. 8. Neuroprotective effects of STI1 and a PrPc-binding STI1 peptide. Cell death was counted within the neuroblastic layer of explants from the retinas of either neonatal rats or mice, maintained in vitro in various conditions. In (A–C), the rate of cell death was normalized with respect to the rate induced by anisomycin alone (100%). (A) Rates of cell death induced by anisomycin (1 µg/ml, filled circles) in retinal explants from neonatal rats, in the presence of various concentrations of the STI1 pep.1. Controls without anisomycin are shown with open circles. (B) Rates of cell death in retinal explants from neonatal rats, incubated with either recombinant STI1 (circles) or BSA (triangles), in either the presence (filled symbols) or absence (open symbols) of anisomycin. (C) Rates of cell death in explants from the retina of neonatal wild-type mice. Symbols as in (A). (D and E) Rates of cell death in retinal explants from neonatal wild-type (filled bars) or PrP0/0 (open bars) mice in various conditions. Note that both the STI1 pep.1 (25 µM, D) and the STI1 protein (8 µM, E) blocked anisomycin-induced cell death in wild-type, but not in PrP0/0 mice. All values are means ± SEM; *P < 0.01 and #P < 0.05 versus anisomycin alone, n ≥3 for each data point. Statistical significance is indicated only for the most relevant among the multiple comparisons tabulated in the Duncan’s test.
To test further whether the neuroprotective response depends on PrPc, we compared the effects of the STI1 pep.1 upon either wild-type or PrP0/0 mice retinal explants. The STI1 pep.1 was effective on wild-type mice retinas at similar concentrations as on rat tissue (Figure 8C). While either the STI1 pep.1 or STI1 protein blocked anisomycin-induced cell death in wild-type retinal tissue, there was no difference between the rates of cell death either in the presence or in the absence of the STI1 pep.1 in explants of the retinas from PrP0/0 mice (Figure 8D and E).
Discussion
We fully characterized a 66 kDa PrPc ligand protein, previously described with the aid of an antibody raised against a PrPc-binding peptide (PrR) designed on the basis of complementary hydropathy (Martins et al., 1997). That antibody was used to isolate the reactive protein, which was identified as ST11 (Lässle et al., 1997), also designated extendin due to its participation in the extention of pseudopodia (Blatch et al., 1995).
Murine STI1 was described as a cytoplasmic protein (Lässle et al., 1997), but its human homolog was also found in the Golgi apparatus and small vesicles in normal cells, and in the nucleolus of SV40-transformed cells (Honoré et al., 1992). We showed that the 66 kDa PrPc ligand was found mainly in the cytoplasm, with a small fraction (∼6%) of the total protein present at the cell membrane (Martins et al., 1997). Indeed, the present work confirmed the presence of STI1 at the cell surface, despite the absence of either a transmembrane domain or a signal peptide for membrane transport (Lässle et al., 1997). In fact, many intracellular proteins such as actin, annexin, nucleolin, cytokeratin 1 and cytokeratin 18, that were expected to be confined to the cytoplasm, are also found at the cell surface where they play specific functions, in particular as receptors for plasma proteins (Semenkovich et al., 1990; Moroianu et al., 1993; Hajjar et al., 1994; Schmaier, 1997; Wells et al., 1997) or for parasites (Magdesian et al., 2001). It has been speculated that these proteins are either projected to the plasma membrane as part of a proteic complex or secreted by a pathway clearly distinct from the classical route through the endoplasmic reticulum and Golgi apparatus (Muesch et al., 1990). We speculate that STI1 is transported to the cell membrane in association with other membrane proteins, which would be consistent with detection in both the Golgi apparatus and small vesicles (Honoré et al., 1992).
It is also known that STI1 is phosphorylated by casein kinase II (CK-II), with unknown consequences (Longshaw et al., 2000). However, CK-II is one of the few protein kinases present at the outer leaflet of the plasma membrane (Walter et al., 1996), and PrPc can both be phosphorylated by and increase the activity of CK-II (Meggio et al., 2000; Negro et al., 2000). The role of phosphorylation in PrPc– STI1 binding and signal transduction will be addressed in future studies.
Besides the cell membrane, PrPc is also found in the Golgi apparatus and recycling endosomes (Lee et al., 2001a,b). However, PrPc is subject to ubiquitylation and degradation by the proteasome (Yedidia et al., 2001) and it may also enter the cytoplasmic compartment through normal quality control pathways (Ma and Lindquist, 2001). Wild-type PrPc cannot be detected in the cytoplasm unless a proteasome inhibitor is used and therefore should not contact the cytoplasmic form of STI1. Conversely, a mutant PrPc (D117N), which is associated with a spongiform encephalopathy, accumulates in the cytoplasm and co-localizes with Hsp70 (Ma and Lindquist, 2001), which is found in a complex with STI1 and Hsp90 (Lässle et al., 1997). Due to both the fact that exposure to a cytoplasmic environment in vivo favors formation of a PrPsc-like conformation (Ma and Lindquist, 1999), and to the chaperoning activity of the STI1-associated Hsp70 and Hsp90 (Lässle et al., 1997), cytoplasmic STI1 may participate in the process of PrPc conversion to PrPsc. Moreover, due to variation among species, in particular to the PrPc-binding domain (STI1 amino acids 230–245) in mouse and human molecules (Honoré et al., 1992; Lässle et al., 1997), we speculate that STI1 may also correspond to Prusiner’s proposed protein X (Telling et al., 1995), which would be consistent with the idea that the species barrier to prion infection is related to the variability of protein X among species.
Several molecules associate with PrPc in vitro, such as heparin, chaperones Hsp60 and BiP, glial fibrillary acidic protein (GFAP), Nrf-2 (a NF-E2-related factor), apolipoprotein 1, Bcl-2 and the 37/67 kDa laminin receptor (reviewed by Martins et al., 2001). Dystroglycan (Keshet et al., 2000) and neural cell adhesion molecules (N-CAMs) also bind PrPc (Schmitt-Ulms et al., 2001). However, there is little evidence of physiological relevance for these interactions. Nonetheless, we have found that PrPc binds laminin (Graner et al., 2000a), an extracellular matrix protein with an important role in cell development and differentiation (Beck et al., 1990). Indeed, the PrPc–laminin complex affects neuronal cell adhesion, neurite formation and maintenance (Graner et al., 2000a,b).
The 37/67 kDa laminin receptor (Rieger et al., 1997; Gauczynski et al., 2001; Hundt et al., 2001) may play a role in the internalization of 20–50% of the membrane-bound PrPc in association with heparan sulfate proteoglycan (Hundt et al., 2001). Interestingly, a 37/67 kDa laminin receptor-binding site at the PrPc molecule maps to amino acids 161–179 (Gauczynski et al., 2001) distinct from the STI1-binding domain (amino acids 113–128, Figure 4). If indeed STI1 plays a role in PrPc internalization, as previously suggested (Martins et al., 1997), it is possible that the association of PrPc with both molecules may have an additive effect. It is also possible that binding of PrPc to the 37/67 kDa laminin receptor may be enhanced by copper ions, since the associated heparan sulfate proteoglycan binds to a PrPc copper-binding domain (Brown et al., 1997a), which is important for internalization mediated by this metal (Pauly and Harris, 1998; Lee et al., 2001a). Conversely, PrPc–STI1 binding is not affected by the presence of copper either associated with PrPc (Figures 3 and 4) or in the binding reaction (data not shown). In addition, the internalization of PrPc may be involved in switching off signals trigged by the PrPc–STI1 interaction.
We showed (Chiarini et al., 2002) that PrPc transduces neuroprotective signals, elicited by either the PrR peptide or by certain antibodies, thereby rescuing retinal neurons from apoptosis throughout a cAMP/PKA pathway. This was confirmed here by the efficient neuroprotection provided by either the STI1 pep.1 that mimics PrR, or the whole STI1 molecule. These data show that PrPc–STI1 interactions are likely to have a functional impact upon sensitivity to cell death within the nervous tissue. In addition, association of PrPc with STI1 does not exclude its interaction with laminin (unpublished data), indicating that PrPc can be part of a macromolecular complex formed between the cell surface and extracellular proteins, and composed at least of laminin, PrPc and STI1 (Martins et al., 2002), which transduces both cytoprotective and differentiation signals.
Materials and methods
Two-dimensional polyacrylamide gel electrophoresis
Two-dimensional gels were run as previously described (Görg et al., 1995). Samples of 1.5 mg were applied directly to Immobilin DryStrip gel, pH range 3–10 (Amersham Pharmacia), and the second dimension was carried out using an 8–18% linear acrylamide gradient gel (ExcelGel SDS, Amersham Pharmacia). Proteins from two identical gels were either stained with Coomassie Blue or transferred to nitrocellulose membranes that were immunoblotted with serum against PrR peptide (Martins et al., 1997). After matching the Coomassie-stained two-dimensional gel spot map with the immunoblotting membrane as a guide, the corresponding PrPc ligand spots were carefully excised and subjected to mass spectrometric analysis.
Mass spectrometric analysis
The in-gel digestion procedure is similar to that described in Huang et al. (1999). Molecular masses of tryptic peptides were determined by analyzing 1 µl of unseparated digest using MALDI-TOF MS (Voyager DESTR, Perspective Biosystems, Framingham, MA) (Huang et al., 1999). Peptide masses were submitted to database searching using the MS-Fit program (http://propector.ucsf.edu) (Clauser et al., 1999). Peptide sequencing using tandem MS was performed on a prototype QqoaTOF mass spectrometer (Sciex, Toronto, Canada) equipped with a nanoelectrospray ion source (Protana A/S, Odense, Denmark). The fragment ion masses were submitted to the MS-Tag program (http://propector. ucsf.edu) for unambiguous protein identification.
Immunoblotting analyses
Immunoblotting assays were done in mouse brain and cell line extracts and in membrane fractions as previously described, using polyclonal antibodies: anti-PrR peptide raised in mice (1:1000) (Martins et al. 1997), anti-recombinant mSTI1 raised in rabbits (purified IgG, 0.1 µg/ml) (Bethyl Co) and anti-recombinant PrPc raised in PrP0/0 mice (Lee et al., 2001a) (1:1000). Mouse non-immune serum or rabbit non-immune purified IgG were used as negative controls.
Expression and purification of PrPc
The expression vector containing the cDNA fragment encoding amino acids 23–231 of the mouse PrPc protein cloned in the BamHI–EcoRI restriction sites of pRSET (Invitrogen™) was kindly provided by Ralph Zahn (Institut für Molekularbiologie und Biophysik, Eidgenössische Technische Hochschule, Switzerland). Expression, purification and elution of His6-PrPc were performed as previously described (Zahn et al., 1997).
Construction expression and purification of mSTI1
Two oligonucleotides were used as primers: 5′-CCGCTCGAGGAGCAGGTGAATGAGCTAAAGGA-3′ (with an XhoI restriction site) and 5′-CGGGGTACCTCACCGAATTGCGATGAGACCC-3′ (with a KpnI restriction site) for PCR to amplify base pairs +56 to +1687 of the mouse STI1 cDNA (DDBJ/EMBL/GenBank accession No. U27830). The fragment was amplified using Tth (Thermus thermophilus, Amersham) and cloned using XhoI and KpnI restriction sites into pTrc-A His (Invitrogen™) vector. Sequencing analysis was performed (ABI-Pharmacia) to check the integrity of the amplified region. Protein expression was induced by 1.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h in Escherichia coli DH-5α cells (Stratagene) containing the expression vector His6-STI1. Cells were resuspended in lysis buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl and 10 mM imidazole) and lysed in a French press. Protein was purified using Ni-NTA–agarose (Qiagen) in accord with the manufacturer’s instructions.
Construction of murine PrPc deletion mutants
PrPc mutants were constructed using the recombinant PCR technique (Ausubel et al., 1993). To construct PrPc mutants, we amplified cDNA fragments employing pRSET-A PrPc (23–231) (Zahn et al., 1997) with internal primers (Δ51–90R, 5′-TACCCCCTCCTGGGTAACGGTTGC CTCC-3′; Δ51–90F, 5′-AACCGTTACCCAGGAGGGGGTACCCATA ATC-3′; Δ105–128R, 5′ GCTCATGGCGCTCCCCAGTGGTTTGCTG GGCTTGTTCC 3′; Δ105–128F, 5′-GGAACAAGCCCAGCAAACCA CTGGGGAGCGCCATGACG-3′) and external primers R, 5′-AGAG AATTCTCAGCTGGATCTTCTCCCGTC-3′; and F, 5′-GAGGGATCC AAAAAGCGGCCAAAG-3′. The PCR fragments were cloned into BamHI and EcoRI restriction sites in the same vector (pRSET-A; Invitrogen). Sequencing analysis were performed to check for the deletion. The expression and purification of these proteins were done as previously described (Zahn et al., 1997).
PrPc–mSTI1 binding assay
A 4 µg aliquot of His6-PrPc or PrPc deletion mutants Δ51–90 or Δ105–128 was immobilized in polystyrene wells (Dynex Technologies) and non-specific sites blocked with 1% BSA for 2 h at room temperature. Increasing concentrations of [125I]His6-TI1 (labeled as described in Martins et al., 1997) with a specific activity of 7 × 105 c.p.m./µg were added to the wells and incubated for 16 h at 4°C. After extensive washing, incorporated radioactivity was measured and originated the total binding curve. In parallel, 4 µg of His6-PrPc was incubated with [125I]His6-STI1 plus a 25-fold excess of unlabeled His6-STI1, which generated non-specific binding. The specific binding curve was obtained by subtraction of non-specific from total values. Equilibrium dissociation constants (Kds) were obtained from Scatchard plots (Scatchard, 1949).
Competition assay using PrPc synthetic peptides
Synthetic mouse PrPc peptides obtained from amino acid sequence 23–231 (Neosystem, France or INFAR, Brazil) at the concentration of 3 × 10–5 M: P1 (23–42), P2 (33–52), P3 (43–62), P4 (53–72) P5 (63–82), P6 (73–92), P7 (83–102), P8 (93–112), P9 (103–122), P10 (113–132), P11 (123–142), P12 (133–152), P13 (143–162), P14 (153–172), P15 (163–182), P16 (173–192), P17 (183–203), P18 (194–213), P19 (204– 223), P20 (214–231) and neurotoxic peptide (NTX; KTNMKHMAGAAAAGAVVGGLG) were pre-incubated with 10–8 M [125I]His6-mSTI1 for 3 h at room temperature. Then, the reagents were added to the wells containing 4 µg of adsorbed His6-PrPc and incubated for 16 h at 4°C. After extensive washing, incorporated radioactivity was determined using a gamma counter.
Competition assay using mSTI1 synthetic peptides
Synthetic peptides: mSTI1 pep.1 (amino acids 230–245, ELGNDAYKKKDFDKAL), PrR (HVATKAPHHGPCRSSA), scrambled PrR peptide (KSRGHVHCHAPAPATS) and two other mSTI1 peptides pNH2 (amino acids 61–76 GCKTVDLKPDWGKGYS) and PCOOH (amino acids 422–437 QLEPTFIKGYTRKAAA) were synthesized chemically (Neosystem, France or INFAR, Brazil). Increasing amounts of synthetic peptides (from 10–4 to 4 × 10–4 M) were pre-incubated with 4 µg of His6-PrPc immobilized in polystyrene wells for 3 h at room temperature. Next, 10–8 M [125I]His6-STI1 was added and incubated for 16 h at 4°C. After extensive washing, radioactivity was determined by using a gamma counter.
Construction of GFP–PrPc and GFP–STI1 vectors
PrPc protein was cloned in vector pEGFP-C1 (Clontech) as previously described (Lee et al., 2001a) and the entire mouse STI1 open reading frame, obtained as described above, was cloned in KpnI–SalI restriction sites on the pEGFP-C1 vector.
Detection of PrPc binding to the cell surface by flow cytometry assay
A total of 106 HEK 293T cells (non-transfected and transfected with GFP–PrPc or GFP) or primary fibroblast cultures from PrP0/0 animals (MEFs) (Büeler et al., 1992) were pre-incubated in the absence or presence of 20 or 9 µg, respectively, of His6-PrPc for 1 h at 4°C, then cells were washed and incubated with anti-PrPc serum (Lee et al., 2001a) or non-immune serum (1:200) for 1 h at 4°C. After three washes, cells were incubated with anti-mouse IgG conjugated to R-phycoerythrin (HEK 293T) or fluorescein isothiocyante (FITC) (MEFs) (1:80) for 1 h at 4°C. Analyses were carried out using a Becton Dickinson FACScan Cytometer, and data acquisition from 10 000 cells was performed with the Consort 32 system, Lysis II software (Becton Dickinson).
His tag pull-down
A total of 107 cells from PrP0/0 MEFs (Büeler et al., 1992) were incubated with 90 µg of His6-PrP for 1 h at 4°C, washed and lysed with ice-cold phosphate-buffered saline (PBS), 1% NP-40 plus complete protease inhibitor cocktail (Roche). Alternatively, cell extracts were first prepared as described above and then incubated with 90 µg of His6-PrPc for 1 h at 4°C. Both preparations were incubated with 30 µl of packed Ni-NTA–agarose beads for 1 h at room temperature. Beads were then washed with 1.5 ml of 10 mM Tris–HCl, 100 mM NaH2PO4, 25 mM imidazole, 1% NP-40. Bound material was eluted with Laemmli buffer at 100°C and analyzed by western blotting using anti-STI1 or anti-PrPc antibodies, followed by anti-rabbit or anti-mouse IgG peroxidase. Reactions were developed using the ECL kit (Amersham, Co).
Cell transfection, surface labeling and immunoprecipitation
HEK 293T cells were transfected by calcium phosphate co-precipitation as previously described (Püschel et al., 1995). After 48–72 h of culture, transfected cells were biotinylated using EZ-Link-Sulfo-NHS-biotin according to the manufacturer’s instructions (Pierce), lysed in 1% NP-40 in PBS plus complete protease inhibitor cocktail (Roche) and centrifuged for 30 min at 10 000 g. Supernatants were pre-cleared with mouse non-immune serum or rabbit irrelevant IgG mixed with protein A/G–Sepharose (Sigma) and immunoprecipitated as previously described (de Souza and Brentani, 1992) using mouse anti-PrPc or rabbit anti-STI1 antibodies. Sepharose beads were washed, and bound proteins were eluted with Laemmli buffer at 100°C and analyzed by western blot using using anti-STI1 or anti-PrPc antibodies as described above. Immunoprecipitation of non-transfected biotin-labeled N2a cells proceeded in a similar way.
Neuroprotection experiments
Explants from the retinas of rats, and wild-type and PrP0/0 mice (Büeler et al., 1992) were cultured as previously described (Chiarini et al., 2002). Following treatment, the tissue was fixed by immersion in 4% paraformaldehyde in phosphate buffer pH 7.2 for at least 40 min, followed by 20% sucrose in the same buffer. Frozen sections were stained with neutral red.
Anisomycin at 1 µg/ml was added to tissue culture alone or together with either STI1 pep. 1 (ELGNDAYKKKDFDKAL) (Neosystem, Immunograde) or recombinant STI1 protein at the beginning of a 24 h incubation period. Cell death induced by anisomycin in the neuroblastic layer of the retina was detected as condensed, pyknotic profiles as described (Chiarini et al., 2002). In each experiment, at least three microscopic fields delimited by an eyepiece graticule of 0.0148 mm2 were counted in each of three explants per group. For more details, see Chiarini et al. (2002).
Statistical analyses
Each experiment was done in triplicate and mean values represent at least three independent experiments. The statistical significance of peptide inhibition assays and mutant PrPc proteins was tested by single mean Student’s t-test, and cell surface PrPc binding by Mann–Whitney test. Quantification of cell death was tested statistically by analysis of variance followed by planned comparisons using Duncan’s multiple range test.
Acknowledgments
Acknowledgements
We thank Dr Sylvie Mènard from the Institute of Pathology, Faculty of Medicine, Milan University, Italy for the antibody against the 37/67 kDa laminin receptor, Dr Ralph Zahn from the Institut für Molekularbiologie und Biophysik, ETH, Switzerland for the PrPc expression vector, and Dr Andreas W.Püschel from the Institut für Allgemeine Zoologie und Genetik, Westfälische Wilhelms-Universität, Germany for helpful suggestions. This work was supported by grants from FAPESP (99/07124-8), FAPERJ (to R.L.) and CNPq (to R.L.). Fellowships from FAPESP to S.M.Z., M.H.L., A.F.M., G.N.M.H., A.R.O.F., A.L.B.C. and K.S.L. and from FAPERJ to L.B.C. are gratefully acknowledged.
References
- Aguzzi A. and Weissmann,C. (1997) Prion research: the next frontiers. Nature, 389, 795–798. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,K., Struhl,K., Kingston,R.E., Moore,D.D., Seidman,J.G. and Smith,J.A. (eds) (1993) Current Protocols in Molecular Biology. Wiley Interscience, New York.
- Beck K., Hunter,I. and Engel,J. (1990) Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J., 4, 148–160. [DOI] [PubMed] [Google Scholar]
- Blatch G.L., Lassle,M., Takatori,T., Grandhi,T., Kundra,V. and Zetter,B.R. (1995) Molecular characterization of extendin: a protein localized in extending pseudopodia. Proc. Am. Assoc. Cancer Res., 36, 68. [Google Scholar]
- Blatch G.L., Lassle,M., Zetter,B.R. and Kundra,V. (1997) Isolation of a mouse cDNA encoding mSTI1, a stress-inducible protein containing the TPR motif. Gene, 194, 277–282. [DOI] [PubMed] [Google Scholar]
- Boquet D., Dery,O., Frobert,Y., Grassi,J. and Couraud,J.Y. (1995) Is hydropathic complementarity involved in antigen–antibody binding? Mol. Immunol., 32, 303–308. [DOI] [PubMed] [Google Scholar]
- Bost K.L., Smith,E.M. and Blalock,J.E. (1985) Similarity between the corticotropin (ACTH) receptor and a peptide encoded by an RNA that is complementary to ACTH mRNA. Proc. Natl Acad. Sci. USA, 82, 1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentani R.R. (1988) Biological implications of complementary hydropathy of amino acids. J. Theor. Biol., 135, 495–499. [DOI] [PubMed] [Google Scholar]
- Brown D.R. et al. (1997a) The cellular prion protein binds copper in vivo. Nature, 390, 684–687. [DOI] [PubMed] [Google Scholar]
- Brown D.R., Schulz-Schaeffer,W.J., Schmidt,B. and Kretzschmar,H.A. (1997b) Prion protein-deficient cells show altered response to oxidative stress due to decreased SOD-1 activity. Exp. Neurol., 146, 104–111. [DOI] [PubMed] [Google Scholar]
- Brown D.R., Wong,B.S., Hafiz,F., Clive,C., Haswell,S.J. and Jones,I.M. (1999) Normal prion protein has an activity like that of superoxide dismutase. Biochem. J., 344, 1–5. [PMC free article] [PubMed] [Google Scholar]
- Büeler H., Fischer,M., Lang,Y., Bluethmann,H., Lipp,H.P., DeArmond,S.J., Prusiner,S.B., Aguet,M. and Weissmann,C. (1992) Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature, 356, 577–582. [DOI] [PubMed] [Google Scholar]
- Büeler H., Aguzzi,A., Sailer,A., Greiner,R.A., Autenried,P., Aguet,M. and Weissmann,C. (1993) Mice devoid of PrP are resistant to scrapie. Cell, 73, 1339–1347. [DOI] [PubMed] [Google Scholar]
- Buto S. et al. (1998) Formation of the 67-kDa laminin receptor by acylation of the precursor. J. Cell. Biochem., 69, 244–251. [DOI] [PubMed] [Google Scholar]
- Cabral A.L., Lee,K.S. and Martins,V.R. (2002) Regulation of PrPc gene expression depends on chromatin conformation. J. Biol. Chem., 277, 5675–5682. [DOI] [PubMed] [Google Scholar]
- Chiarini L.B., Freitas,A.R.O., Zanata,S.M., Brentani,R.R., Martins,V.R. and Linden,R. (2002) Cellular prion protein transduces neuroprotective signals. EMBO J., 21, 3317–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauser K.R., Baker,P. and Burlingame,A.L. (1999) Role of accurate mass measurement (±10 p.p.m.) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem., 71, 2871–2882. [DOI] [PubMed] [Google Scholar]
- Collinge J. et al. (1990) Prion dementia without characteristic pathology. Lancet, 336, 7–9. [DOI] [PubMed] [Google Scholar]
- de Souza S.J. and Brentani,R. (1992) Collagen binding site in collagenase can be determined using the concept of sense–antisense peptide interactions. J. Biol. Chem., 267, 13763–13767. [PubMed] [Google Scholar]
- Forloni G., Angeretti,N., Chiesa,R., Monzani,E., Salmona,M., Bugiani,O. and Tagliavini,F. (1993) Neurotoxicity of a prion protein fragment. Nature, 362, 543–546. [DOI] [PubMed] [Google Scholar]
- Gauczynski S. et al. (2001) The 37 kDa/67 kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J., 20, 5863–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görg A., Boguth,G., Obermaier,C., Posch,A. and Weiss,W. (1995) Two-dimensional polyacrylamide gel electrophoresis with immobilized pH gradients in the first dimension (IPG-Dalt): the state of art and the controversy of vertical versus horizontal systems. Electrophoresis, 16, 1079–1086. [DOI] [PubMed] [Google Scholar]
- Graner E. et al. (2000a) Cellular prion protein binds laminin and mediates neuritogenesis. Brain Res. Mol. Brain Res., 76, 85–92. [DOI] [PubMed] [Google Scholar]
- Graner E., Mercadante,A.F., Zanata,S.M., Martins,V.R., Jay,D.G. and Brentani,R.R. (2000b) Laminin-induced PC-12 cell differentiation is inhibited following laser inactivation of cellular prion protein. FEBS Lett., 482, 257–260. [DOI] [PubMed] [Google Scholar]
- Hajjar K.A., Jacovina,A.T. and Chacko,J. (1994) An endothelial cell receptor for plasminogen/tissue plasminogen activator. I. Identity with annexin II. J. Biol. Chem., 269, 21191–21197. [PubMed] [Google Scholar]
- Honoré B., Leffers,H., Madsen,P., Rasmussen,H.H., Vandekerckhove,J. and Celis,J.E. (1992) Molecular cloning and expression of a transformation-sensitive human protein containing the TPR motif and sharing identity to the stress-inducible yeast protein STI1. J. Biol. Chem., 267, 8485–8491. [PubMed] [Google Scholar]
- Huang L., Shen,M., Chernushevich,I., Burlingame,A.L.,Wang,C.C. and Robertson,C.D. (1999) Identification and isolation of three proteasome subunits and their encoding genes from Trypanosoma brucei. Mol. Biochem. Parasitol., 102, 211–223. [DOI] [PubMed] [Google Scholar]
- Hundt C. et al. (2001) Identification of interaction domains of the prion protein with its 37 kDa/67 kDa laminin receptor. EMBO J., 20, 5876–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet G.I, Bar-Peled,O., Yaffe,D., Nudel,U. and Gabizon,R. (2000) The cellular prion protein colocalizes with the dystroglycan complex in the brain. J. Neurochem., 75, 1889–1897. [DOI] [PubMed] [Google Scholar]
- Kretzschmar H.A., Tings,T., Madlung,A., Giese,A. and Herms,J. (2000) Function of PrP(C) as a copper-binding protein at the synapse. Arch. Virol., Suppl., 16, 239–249. [DOI] [PubMed] [Google Scholar]
- Kuwahara C. et al. (1999) Prions prevent neuronal cell-line death. Nature, 400, 225–226. [DOI] [PubMed] [Google Scholar]
- Lässle M., Blatch,G.L., Kundra,V., Takatori,T. and Zetter,B.R. (1997) Stress-inducible, murine protein mSTI1. Characterization of binding domains for heat shock proteins and in vitro phosphorylation by different kinases. J. Biol. Chem., 272, 1876–1884. [DOI] [PubMed] [Google Scholar]
- Lee K.S., Magalhães,A.C., Zanata,S.M., Brentani,R.R., Martins,V.R. and Prado,M.A.M. (2001a) Internalization of mammalian fluorescent cellular prion protein and N-terminal deletion mutants in living cells. J. Neurochem., 79, 79–87. [DOI] [PubMed] [Google Scholar]
- Lee K.S., Martins,V.R., Zanata,S.M., Brentani,R.R. and Prado,M.A.M. (2001b) Internalization of a mammalian fluorescent prion protein involves classical endocytic organelles. J. Neurochem., 78 Suppl., 19. [DOI] [PubMed] [Google Scholar]
- Longshaw V.M., Dirr,H.W., Blatch,G.L. and Lassle,M. (2000) The in vitro phosphorylation of the co-chaperone mSTI1 by cell cycle kinases substantiates a predicted casein kinase II-p34cdc2-NLS (CcN) motif. Biol. Chem., 381, 1133–1138. [DOI] [PubMed] [Google Scholar]
- Ma J. and Lindquist,S. (1999) De novo generation of a PrPSc-like conformation in living cells. Nature Cell Biol., 1, 358–361. [DOI] [PubMed] [Google Scholar]
- Ma J. and Lindquist,S. (2001) Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc. Natl Acad. Sci. USA, 98, 14955–14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdesian M.H., Giordano,R., Ulrich,H., Juliano,M.A., Juliano,L., Schumacher,R.I., Colli,W. and Alves,M.J. (2001) Infection by Trypanosoma cruzi. Identification of a parasite ligand and its host cell receptor. J. Biol. Chem., 276, 19382–19389. [DOI] [PubMed] [Google Scholar]
- Martins V.R., Graner,E., Garcia-Abreu,J., Souza,S.J., Mercadante,A.F., Veiga,S.S., Zanata,S.M., Moura Neto,V. and Brentani,R.R. (1997) Complementary hydropathy identifies a cellular prion protein receptor. Nature Med., 3, 1376–1382. [DOI] [PubMed] [Google Scholar]
- Martins V.R., Mercadante,A.F., Cabral,A.L., Freitas,A,R.O. and Castro,R.M. (2001) Insights into the physiological function of cellular prion protein. Braz. J. Med. Biol. Res., 34, 585–595. [DOI] [PubMed] [Google Scholar]
- Martins V.R., Linden,R., Prado,M.A.M., Walz,R., Sakamoto,A.C., Izquierdo,I. and Brentani,R.R. (2002) Cellular prion protein: on the road for functions. FEBS Lett., 512, 25–28. [DOI] [PubMed] [Google Scholar]
- Medori R., Montagna,P., Tritschler,H.J., LeBlanc,A., Cortelli,P., Tinuper,P., Lugaresi,E. and Gambetti,P. (1992) Fatal familial insomnia: a second kindred with mutation of prion protein gene at codon 178. Neurology, 42, 669–670. [DOI] [PubMed] [Google Scholar]
- Meggio F., Negro,A., Sarno,S., Ruzzene,M., Bertoli,A., Sorgato,M.C. and Pinna,L.A. (2000) Bovine prion protein as a modulator of protein kinase CK2. Biochem. J., 352, 191–196. [PMC free article] [PubMed] [Google Scholar]
- Moroianu J.W., Fett,J.W., Riordan,J.F. and Vallee,B.L. (1993) Actin is a surface component of calf pulmonary artery endothelial cells in culture. Proc. Natl Acad. Sci. USA, 90, 3815–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet-Richard S., Ermonval,M., Chebassier,C., Laplanche,J.L., Lehmann,S., Launay,J.M. and Kellermann,O. (2000) Signal transduction through prion protein. Science, 289, 1925–1928. [DOI] [PubMed] [Google Scholar]
- Muesch A., Hartmann,E., Rohde,K., Rubartelli,A., Sitia,R. and Rapoport,T.A. (1990) A novel pathway for secretory proteins? Trends Biochem. Sci., 15, 86–88. [DOI] [PubMed] [Google Scholar]
- Negro A., Meggio,F., Bertoli,A., Battistutta,R., Sorgato,M.C. and Pinna,L.A. (2000) Susceptibility of the prion protein to enzymic phosphorylation. Biochem. Biophys. Res. Commun., 271, 337–341. [DOI] [PubMed] [Google Scholar]
- Pauly P.C. and Harris,D.A. (1998) Copper stimulates endocytosis of the prion protein. J. Biol. Chem., 273, 33107–33110. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B. (1998) Prions. Proc. Natl Acad. Sci. USA, 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Püschel A.W., Adams,R.H. and Betz,H. (1995) Murine semaphorin D/collapsin is a member of a diverse gene family and creates domains inhibitory for axonal extension. Neuron, 14, 941–948. [DOI] [PubMed] [Google Scholar]
- Rehen S.K., Neves,D.D., Fragel-Madeira,L., Britto,L.R. and Linden,R. (1999) Selective sensitivity of early postmitotic retinal cells to apoptosis induced by inhibition of protein synthesis. Eur. J. Neurosci., 11, 4349–4356. [DOI] [PubMed] [Google Scholar]
- Rieger R., Edenhofer,F., Lasmezas,C.I. and Weiss,S. (1997) The human 37-kDa laminin receptor precursor interacts with the prion protein in eukariotic cells. Nature Med., 3, 1383–1388. [DOI] [PubMed] [Google Scholar]
- Rohm B., Ottemeyer,A., Lohrum,M. and Püschel,A.W. (2000) Plexin/neuropilin complexes mediate repulsion by the axonal guidance signal semaphorin 3A. Mech. Dev., 93, 95–104. [DOI] [PubMed] [Google Scholar]
- Samaia H.B. and Brentani,R.R. (1998) Can loss-of-function prion-related diseases exist? Mol. Psychiatry, 3, 196–197. [DOI] [PubMed] [Google Scholar]
- Samaia H.B., Mari,J.J., Vallada,H.P., Moura,R.P., Simpson,A.T.G. and Brentani,R.R. (1997) A prion-linked psychiatric disorder. Nature, 390, 241. [DOI] [PubMed] [Google Scholar]
- Scatchard G. (1949) The attraction of proteins for small molecules and ions. Ann. NY Acad. Sci., 51, 660–672. [Google Scholar]
- Schmaier A.H. (1997) Contact activation: a revision. Thromb. Haemostasis, 78, 101–107. [PubMed] [Google Scholar]
- Schmitt-Ulms G. et al. (2001) Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J. Mol. Biol., 314, 1209–1225. [DOI] [PubMed] [Google Scholar]
- Scott M.R., Butler,D.A., Bredesen,D.E., Walchli,M., Hsiao,K.K. and Prusiner,S.B. (1988) Prion protein gene expression in cultured cells. Protein Eng., 2, 69–76. [DOI] [PubMed] [Google Scholar]
- Semenkovich C.F., Ostlund,R.E., Olson,M.O.J. and Yang,J.W. (1990) A protein partially expressed on the surface of HepG2 cells that binds lipoproteins specifically is nucleolin. Biochemistry, 29, 9708–9713. [DOI] [PubMed] [Google Scholar]
- Telling G.C., Scott,M., Mastrianni,J., Gabizon,R., Torchia,M., Cohen,F.E., DeArmond,S.J. and Prusiner,S.B. (1995) Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell, 83, 79–90. [DOI] [PubMed] [Google Scholar]
- Walter J., Schnolzer,M., Pyerin,W., Kinzel,V. and Kubler,D. (1996) Induced release of cell surface protein kinase yields CK1- and CK2-like enzymes in tandem. J. Biol. Chem., 271, 111–119. [DOI] [PubMed] [Google Scholar]
- Walz R., Amaral,O.B., Rockenbach,I.C., Roesler,R., Izquierdo,I., Cavalheiro,E.A., Martins,V.M. and Brentani,R.R. (1999) Increased sensitivity to seizures in mice lacking cellular prion protein. Epilepsia, 40, 1679–1682. [DOI] [PubMed] [Google Scholar]
- Wells M.J., Hatton,M.W., Hewlett,B., Podor,T.J., Sheffield,W.P. and Blajchman,M.A. (1997) Cytokeratin 18 is expressed on the hepatocyte plasma membrane surface and interacts with thrombin–antithrombin complexes. J. Biol. Chem., 272, 28574–28581. [DOI] [PubMed] [Google Scholar]
- Wong B.S., Venien-Bryan,C., Williamson,R.A., Burton,D.R., Gambetti,P., Sy,M.S., Brown,D.R. and Jones,I.M. (2000) Copper refolding of prion protein. Biochem. Biophys. Res. Commun., 276, 1217–1224. [DOI] [PubMed] [Google Scholar]
- Yedidia Y., Horonchik,L., Tzaban,S., Yanai,A. and Taraboulos,A. (2001) Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J., 20, 5383–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn R., von Schroetter,C. and Wuthrich,K. (1997) Human prion proteins expressed in Escherichia coli and purified by high-affinity column refolding. FEBS Lett., 417, 400–404. [DOI] [PubMed] [Google Scholar]
- Zanata S.M., Hovatta,I., Rohm,B. and Puschel,A.W. (2002) Antagonistic effects of Rnd1 and RhoD GTPases regulate receptor activity in semaphorin 3A-induced cytoskeletal collapse. J. Neurosci., 22, 471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]