Abstract
Collapsin response mediator proteins (CRMPs)/TOAD64/Ulips/DRPs and CRAM have emerged as strong candidates for a role in semaphorin signaling. In this study we identified Fes/Fps (Fes) tyrosine kinase in the CRMP–CRAM complex and investigated whether Fes was involved in semaphorin3A (Sema3A) signaling. In COS-7 cells, the interaction between Fes and plexinA1 (PlexA1) and the tyrosine phosphorylation of PlexA1 by Fes were observed; however, these events were significantly attenuated by co-expression of neuropilin-1 (NP-1). Even with NP-1 co-expression, Sema3A was able to enhance the association of Fes with PlexA1 and Fes-mediated tyrosine phosphorylation of PlexA1, CRAM and CRMP2. Co-expression of Fes with PlexA1 exhibited COS-7 cell contraction activity, indicating that Fes can convert inactive PlexA1 to its active form, whereas combination of Fes/NP-1/PlexA1 or Fes kinase-negative mutants/PlexA1 did not alter cell morphology. Finally, Sema3A-induced growth cone collapse of dorsal root ganglion neurons was suppressed by expression of Fes kinase-negative mutants. Taken together, our findings suggest that Fes links Sema3A signals to CRMP–CRAM, and that NP-1 negatively regulates PlexA1 activation by Fes in resting condition.
Keywords: CRMP/FesFps/neuropilin/plexin/semaphorin
Introduction
Neuronal outgrowth and axonal guidance are important processes that are highly controlled through extracellular signaling. Neuronal outgrowth is directed by attracting and repelling signaling molecules such as the netrins and members of the semaphorin protein family (Kolodkin, 1998). Semaphorins are a large family of secreted or cell-bound signals known to guide axons in developing nervous tissue. A subset of semaphorin interacts with neuropilins, cell surface molecules lacking a signaling-competent cytoplasmic domain (Feiner et al., 1997; He and Tessier-Lavigne, 1997; Kitsukawa et al., 1997; Kolodkin et al., 1997; Fujisawa and Kitsukawa, 1998). Another large family of transmembrane molecules, plexins, bind specifically to semaphorins (Comeau et al., 1998; Winberg et al., 1998). Thus plexins, alone or in association with neuropilins, behave as fully functional semaphorin receptors. Within the neuropilin-1 (NP-1)–plexinA1 (PlexA1) complex, NP-1 is the binding unit for semaphorin3A (Sema3A), while PlexA1 serves as signal transducer. A recent study further revealed that cytoplasmic plexin domains were tyrosine phosphorylated, suggesting that plexins can signal by associating with a protein-tyrosine kinase (PTK) (Tamagnone et al., 1999). However, much less is known about the intracellular events involved in the relay of these multiple signals towards the resulting cellular response.
Collapsin response mediator proteins (CRMPs)/TOAD64/Ulips/DRPs are a family of cytosolic phosphoproteins that are expressed exclusively in the nervous system (Minturn et al., 1995; Hamajima et al., 1996; Wang and Strittmatter, 1996). CRMPs are believed to mediate signals involved in the guidance and outgrowth of neuronal axons, in particular semaphorin-mediated growth cone signaling (Goshima et al., 1995). The CRMP family members reach their highest expression levels in all neurons during their peak periods of axonal growth and are strongly downregulated after that. Importantly, CRMPs bear sequence similarity to a nematode protein, unc-33, the absence of which produces aberrant elongation of axons and uncoordinated Caenorhabditis elegans worm movement (Byk et al., 1996). CRMP2 was first identified by its possible involvement in the Sema3A-induced growth cone collapse in chick dorsal root ganglion (DRG) neurons (Goshima et al., 1995). However, the intermediate candidates, conveying the signals from plexins to CRMP, are as yet unknown. We previously identified a novel CRMP-associated protein, designated CRAM (for CRMP-associated molecule), which belongs to the unc-33 gene family (Inatome et al., 2000). A phylogenetic tree analysis indicates that the CRAM sequence shows the greatest similarity to DHPase and divergence from the four CRMPs, indicating that CRAM is more closely related to DHPase than to the four CRMPs. Thus, CRAM is a new member of the CRMPs. Furthermore, we showed that CRAM is co-immunoprecipitated with an unknown PTK(s). In this study, we identified Fes/Fps (Fes) PTK in CRMP–CRAM complexes purified from rat brain. Therefore, it is critical to investigate the functional relationship between CRMP–CRAM and Fes in semaphorin-mediated signaling.
The Fes proto-oncogene is the normal cellular homolog of the transforming oncogenes found in several avian and feline retroviruses (Hanafusa et al., 1980). The Fes locus encodes a cytoplasmic PTK primarily expressed in hematopoietic cells of the myeloid lineage (Smithgall et al., 1988). Distinct from Src and other non-receptor PTKs, Fes has a long N-terminal unique region, followed by a central SH2 domain and a C-terminal kinase domain with a total mol. wt of 93 kDa. Recent work has shown that Fes exhibits a more widespread expression pattern in both developing neurons and vascular endothelial cells, suggesting broader biological functions for this kinase than originally suspected (Haigh et al., 1996). In addition, transgenic mice overexpressing v-fps showed a neurological disorder that includes a marked trembling, correlated with the expression of v-fps in the brain and a striking bilateral enlargement of the trigeminal nerves (Yee et al., 1989). Thus, expression of Fes in neurons is particularly intriguing; it will be of value to determine whether Fes can mediate signals involved in the guidance and outgrowth of neuronal axons during neuronal development.
In this study, we identified Fes in the CRMP–CRAM complex purified from rat brain and investigated whether Fes was involved in Sema3A and its receptor PlexA1–NP-1 complex-mediated signaling. Here we report that Fes is involved in Sema3A signaling and that NP-1 negatively regulates PlexA1 activation by Fes. The implications of these phenomena in signal transduction are discussed.
Results
Identification of Fes as a CRAM-associated PTK
In a previous study we have shown that immunoprecipitation of CRAM from neonatal rat brain lysates and in vitro kinase reaction resulted in tyrosine phosphorylation of two proteins, of 66 and 95 kDa, as revealed by SDS–PAGE followed by immunoblot analysis with anti-phosphotyrosine antibody (Ab) (Figure 1A). No PTK activity was detected in immunoprecipitates obtained in the presence of 10 µM blocking peptide corresponding to a portion of CRAM (amino acids 468–485). Since CRAM forms a large complex with CRMPs, a similar assay was performed using anti-CRMP2 Ab. As expected, immunoprecipitation of CRMP2 from neonatal rat brain lysates and in vitro kinase reaction resulted in tyrosine phosphorylation of the 66 and 95 kDa proteins (data not shown). Thus, we hypothesized that this 95 kDa protein may be a PTK itself, and may play a role in the regulation of CRMP–CRAM function. To identify the 95 kDa protein, we purified it from rat brain lysates using affinity column chromatography. The purification procedure involved an anti-CRAM Ab affinity column, elution by antigen peptides, in vitro tyrosine phosphorylation and an anti-phosphotyrosine Ab (4G10) affinity column. Purified 95 kDa protein in the final step was analyzed and detected by silver staining or immunoblotting with 4G10 (Figure 1B). Since this protein band disappeared completely in the presence of blocking peptide, it was considered to associate specifically with CRAM. To obtain a sufficient amount of purified 95 kDa protein for amino acid sequence analysis, a large-scale purification was performed from 100 g of rat brains. The purified 95 kDa protein in the final step was concentrated and analyzed by SDS–PAGE and Coomassie Brilliant Blue staining (Figure 1C). The quantity of total purified 95 kDa protein was estimated at 5–10 pmol.
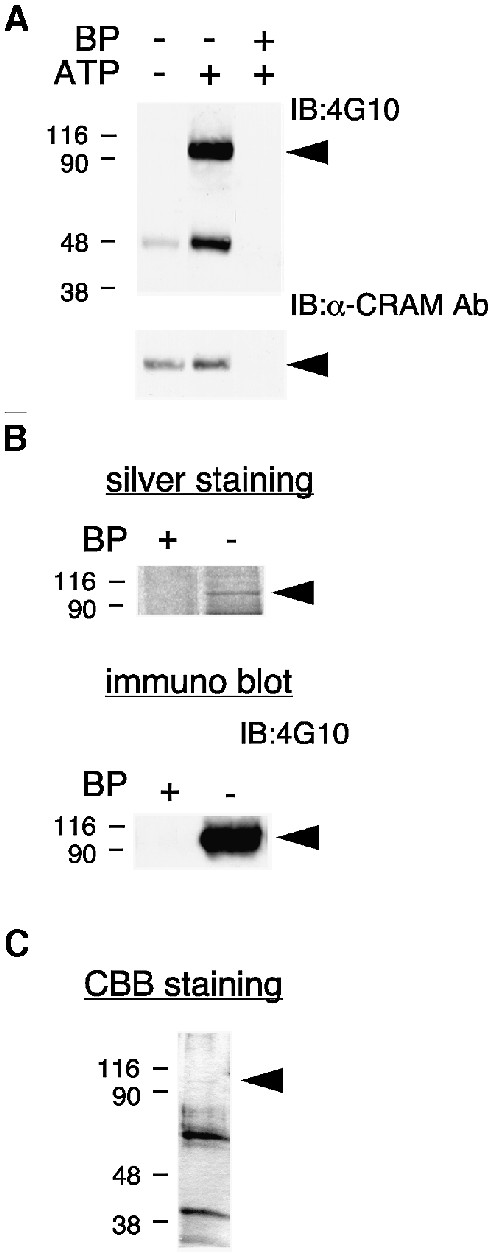
Fig. 1. Isolation of CRAM-associated PTK. (A) Association of PTK activity in the CRMP–CRAM complex. Rat brain lysates were immunoprecipitated with anti-CRAM Ab in the presence or absence of blocking peptide (BP) derived from CRAM protein. Immuno precipitates were incubated with the kinase reaction mixture in the presence or absence of ATP for tyrosine phosphorylation, and then immunoblotted with anti-phosphotyrosine Ab (4G10) or anti-CRAM Ab. (B) Silver staining of purified 95 kDa protein. CRAM-associated proteins were purified from rat brain lysates in the presence or absence of blocking peptide. The 95 kDa protein in the final step was analyzed by silver staining or immunoblotting with 4G10. (C) Large-scale purification of 95 kDa protein. Purified 95 kDa protein in the final step was analyzed by Coomassie Brilliant Blue (CBB) staining. The position of 95 kDa protein is indicated by the arrowheads.
To determine the internal amino acid sequences, an SDS–PAGE gel slice containing the 95 kDa protein was excised, and peptides were generated by digestion with lysyl endopeptidase, separated by reversed-phase high-performance liquid chromatography (HPLC), and subjected to microsequencing as described in Materials and methods. Partial amino acid sequence analysis revealed two peptide fragments. A BLAST homology search of the DDBJ/EMBL/GenBank database of published protein sequences as reference revealed that the two peptide fragments showed 83.3 or 90% identity with peptide sequences from the mouse and human Fes PTK (Table I). Fes is a 93 kDa PTK, but CRAM-associated PTK appears to be larger, at ∼95 kDa. The difference in molecular weight is probably due to phosphorylation-dependent mobility shift. To further confirm that this PTK is identical to Fes, immunological analysis was performed using two epitope-distinct anti-Fes Abs. As shown in Figure 2, in the absence of blocking peptide this CRAM-associated PTK was specifically recognized by both anti-Fes Abs, one directed against the N-terminus (N-19) and the other against the C-terminus (C-19) of Fes. The specificity of the anti-Fes Abs was demonstrated by abolition of the immunoreactive band after blockade with each antigen peptide, respectively (data not shown). In Figure 2, one band is seen above Fes. Since two epitope-distinct anti-Fes Abs recognize this band, this protein may be Fes, as a result of covalent modification such as protein phosphorylation. Together, these results indicated that CRAM-associated PTK was Fes.
Table I. Identification of Fes PTK.
| Sequence | Identification | Identity (%) |
|---|---|---|
| LAEELWYHGAIP | Mouse Fes | 83.3 |
| Human Fes | 83.3 | |
| WVLNHEDVVL | Mouse Fes | 90 |
| Human Fes | 90 |
The purified 95 kDa protein band in SDS–PAGE gels was excised and peptides were generated by digestion with lysyl endopeptidase and separated by reversed-phase HPLC. Two fractions were analyzed using an amino acid sequencer and their peptide sequences were obtained. Each sequence was compared with peptide sequences from the human or mouse Fes amino acid sequence using the DDBJ/EMBL/GenBank database.

Fig. 2. Immunoreactivity with two epitope-distinct anti-Fes Abs. Purified 95 kDa protein described in Figure 1B was immunoblotted either with anti-Fes Ab directed against the N-terminus (N-19) or C-terminus (C-19) of Fes or anti-CRAM Ab in the presence or absence of blocking peptide.
Association and co-localization of Fes and CRAM in P19 cells and DRG neurons
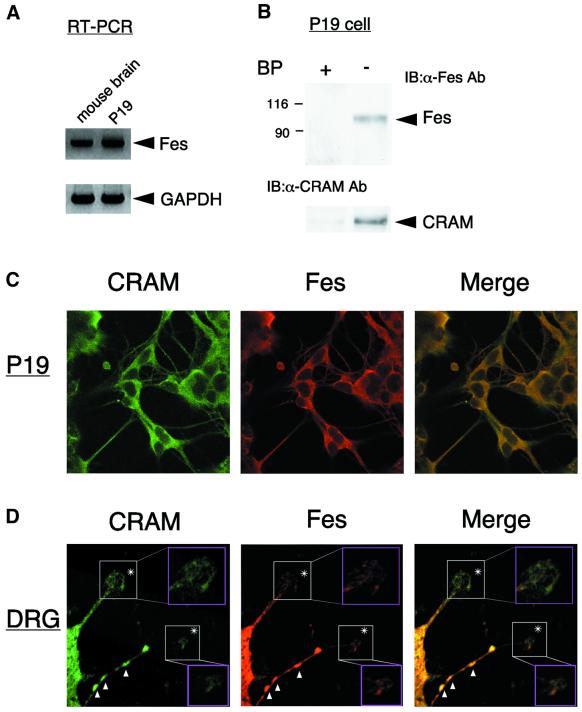
Fes was identified from crude extract of rat brain. Therefore, it was necessary to determine whether the association between Fes and CRAM was also observed in individual cells, such as a cell line or neuronal cells. Fes was initially thought to be expressed in hematopoietic cells of the myeloid lineage, but recent work has shown that Fes exhibits a widespread expression pattern, including in developing neurons. We first examined Fes expression in neuronally differentiated P19 cells as described in Materials and methods. RT–PCR and northern analyses indicated that Fes was expressed in mouse brain and P19 cells (Figure 3A). On the other hand, we have previously reported that CRAM expression was upregulated during neuronal differentiation of P19 cells (Inatome et al., 2000). To detect the association between Fes and CRAM in P19 cells, lysates of P19 cells were immunoprecipitated with anti-CRAM Ab in the presence or absence of blocking peptide. As expected, Fes was detected in CRAM immunoprecipitates in the absence of blocking peptide (Figure 3B). To further confirm this result, we performed double immunofluorescence staining and confocal microscopic analysis of P19 cells and mouse embryonic DRG neurons with anti-CRAM and anti-Fes Abs (N-19). In a previous study, we found that CRAM immunoprecipitates from the membrane fraction, but not the cytosolic fraction, of rat brain homogenate contained a 95 kDa protein (Inatome et al., 2000), suggesting that CRAM associates with Fes at particulate membranes. Consistent with this observation, merged confocal fluorescent images showed complete co-localization of CRAM and Fes in neurites of P19 cells and axon varicosities and growth cones of DRG neurons (Figure 3C and D).
Fig. 3. Co-localization of CRAM and Fes in P19 cells and DRG neurons. (A) Fes expression in mouse brain and P19 cells. RT–PCR and northern blot analysis of Fes mRNA in mouse brain and P19 cells were performed as described in Materials and methods. The positions of Fes and glyceraldehyde phosphate dehydrogenase (GAPDH) as control are indicated. (B) Association of CRAM with Fes in P19 cells. Lysates of P19 cells were immuno precipitated with anti-CRAM Ab in the presence or absence of blocking peptide. Immunoprecipitates were immunoblotted with anti-Fes Ab (N-19). (C and D) Double immunofluorescence staining and confocal microscopic analysis of P19 cells (C) and DRG neurons (D) with anti-CRAM Ab and with anti-Fes Ab (N-19). Merged fluorescent images show co-localization of CRAM and Fes. Asterisks and arrowheads indicate growth cones and axon varicosities, respectively.
In COS-7 cells expressing both Fes and CRAM, double immunofluorescence analysis revealed only a partial co-localization of Fes and CRAM (data not shown). However, we failed to detect a direct interaction between Fes and CRAM using co-immunoprecipitation analysis in COS-7 cells. The failure to detect an interaction between Fes and CRAM in COS-7 cells may be due to the possibility that these interactions are not direct. Thus, CRAM may associate with Fes indirectly at membrane structures in neuronal cells, and an unknown factor(s) or modification, such as phosphorylation, may be involved in the regulation of CRAM and Fes complex formation.
Tyrosine phosphorylation of CRAM and CRMPs by Fes
The finding of CRAM and Fes complex formation prompted us to examine whether Fes phosphorylates CRAM and CRMPs. We first examined the tyrosine phosphorylation of CRAM in intact cells. Neuronally differentiated P19 cells were stimulated with or without pervanadate for 10 min, and cell lysates were immunoprecipitated with anti-CRAM Ab and immunoblotted with 4G10. As shown in Figure 4A, tyrosine phosphorylation of CRAM was enhanced by pervanadate treatment. Since the upper band of ∼95 kDa was recognized by anti-Fes Ab (not shown), this band appears to be Fes. This result indicated that CRAM can be phosphorylated on a tyrosine residue(s) in intact P19 cells, and therefore Fes is a potential candidate for phosphorylation of CRAM.

Fig. 4. Tyrosine phosphorylation of CRAM by Fes. (A) Pervanadate treatment enhanced tyrosine phosphorylation of CRAM in P19 cells. P19 cells were stimulated with or without vanadate (100 µM) plus H2O2 (1 mM) for 10 min and cell lysates were immunoprecipitated with anti-CRAM Ab and immunoprecipitates were immunoblotted with 4G10. (B) Tyrosine phosphorylation of CRAM by Fes in COS-7 cells. COS-7 cells were transfected with the indicated expression vectors and total cell lysates were prepared. Each lysate was immunoprecipitated with anti-CRAM Ab or anti-Flag Ab. The immunoprecipitates were immunoblotted with 4G10, anti-CRAM Ab or anti-Flag Ab. The position of CRAM is indicated by the arrowheads. (C) Fes phosphorylates Flag-CRAM in vitro. Purified Flag-CRAM and/or Fes were incubated in the kinase reaction mixture with or without ATP as described in Materials and methods. Samples were immunoblotted with 4G10 or anti-Flag Ab after deprobing.
To examine the tyrosine phosphorylation of CRAM by Fes, we used a COS-7 cell expression system. When COS-7 cells were transfected with CRAM and Fes, a prominent band showing tyrosine phosphorylation of CRAM was observed (Figure 4B). To further substantiate this finding, a similar assay was performed using Flag-tagged CRAM, with the same result as before (Figure 4B). Furthermore, in COS-7 cells expressing CRAM and Fes kinase-negative mutants, the tyrosine phosphorylation of CRAM was not observed (data not shown). These results indicated that CRAM was phosphorylated by Fes in a COS-7 cell expression system. To determine whether Fes can directly phosphorylate CRAM, in vitro reconstitution experiments were performed. Purified Flag-tagged CRAM and Fes were prepared and incubated for 10 min at 30°C in the reaction mixture for tyrosine phosphorylation with or without ATP as described in Materials and methods. In vitro reconstitution experiments revealed that Fes can directly phosphorylate CRAM (Figure 4C).
CRAM forms a large complex composed of CRMPs during neuronal development. Indeed, Fes was also immunoprecipitated with anti-CRMP2 Ab in neonatal mouse whole-brain lysate (not shown). Since CRMPs and CRAM belong to the same gene family, we next examined whether Fes phosphorylates CRMPs. When COS-7 cells were transfected with each of four Flag-tagged CRMPs and Fes, apparent tyrosine phosphorylations of all four CRMPs were observed (Figure 5A). The differences between the extent of tyrosine phosphorylation among four CRMPs were due to the differences in the level of expression of each CRMP. Furthermore, in vitro reconstitution experiments revealed that Fes directly phosphorylated CRMP2 (Figure 5B). These results suggested that CRAM and the four CRMPs may be physiological substrates for Fes during neuronal development.
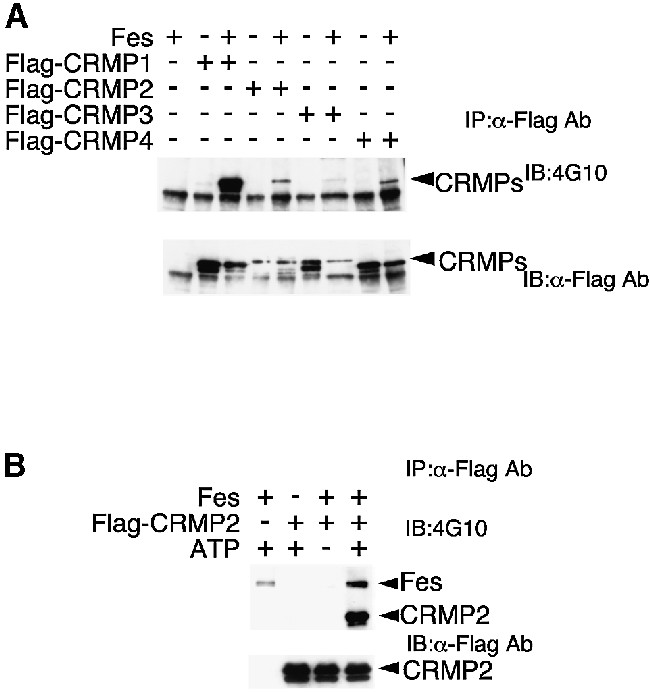
Fig. 5. Tyrosine phosphorylation of CRMPs by Fes. (A) COS-7 cells were transfected with the indicated expression vectors and total cell lysates were prepared. Each lysate was immunoprecipitated with anti-Flag Ab. The immunoprecipitates were immunoblotted with 4G10 or anti-Flag Ab. (B) Fes phosphorylates Flag-CRMP2 in vitro. Purified Flag-CRMP2 and/or Fes were incubated in the kinase reaction mixture with or without ATP as described in Materials and methods. Samples were immunoblotted with 4G10 or anti-Flag Ab after deprobing.
Association of Fes with PlexA1 and negative regulation by NP-1
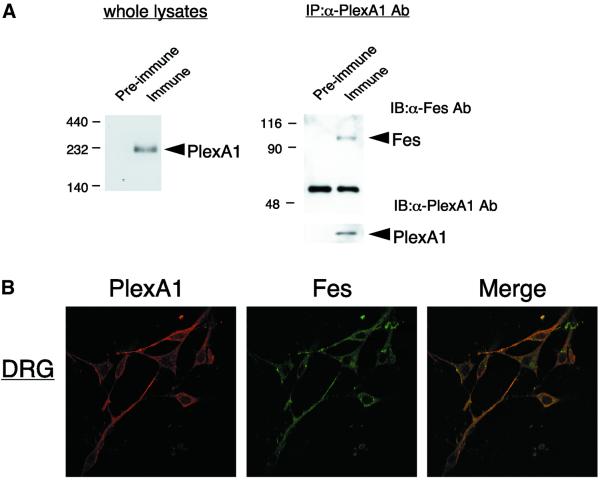
PlexA1 has been identified as a major component of the Sema3A receptor and reported to be tyrosine phosphorylated, suggesting that plexins can signal by associating with a PTK. Therefore, it is critical to examine whether Fes associates with and phosphorylates PlexA1. For this purpose, polyclonal anti-PlexA1 Abs against the cytoplasmic domain of PlexA1 were generated, as described in Materials and methods. Immunoblot analysis of mouse brain lysates revealed that this anti-PlexA1 Ab (immune serum) recognized a 220 kDa band, and pre-immune serum did not react with this band (Figure 6A, left panel), indicating that this anti-PlexA1 Ab could be useful. Lysates of neonatal mouse brain were immunoprecipitated with pre-immune serum as control or immune serum containing anti-PlexA1 Ab. Interestingly, Fes was specifically detected in PlexA1 immunoprecipitates (Figure 6A, right panel). This suggests that Fes associates with PlexA1 in developing neurons.
Fig. 6. Association of Fes with PlexA1. (A) Lysates of neonatal mouse brain were immunoblotted (left) or immunoprecipitated (right) with pre- immune serum or anti-PlexA1 Ab. The immunoprecipitates were immunoblotted with anti-Fes Ab or anti-PlexA1 Ab. The positions of Fes and PlexA1 are indicated by the arrowheads. (B) Double immunofluorescence staining and confocal microscopic analysis of DRG neurons with anti-Fes and anti-PlexA1 Ab (N-19). The merged fluorescent image shows co-localization of Fes and PlexA1.
To further confirm the association of Fes with PlexA1, we performed double immunofluorescence staining and confocal microscopic analysis of DRG neurons with anti-PlexA1 and anti-Fes Abs. The merged fluorescent image showed almost complete co-localization of PlexA1 and Fes in neurites of DRG neurons (Figure 6B).
We next examined the association of Fes with PlexA1 using a COS-7 cell expression system. When COS-7 cells were transfected with PlexA1 and Fes, Fes was detected in PlexA1 immunoprecipitates, suggesting that Fes can directly associate with PlexA1 (Figure 7A, right panel). However, the association of Fes with PlexA1 was blocked by co-expression of NP-1 (Figure 7A, right panel). This result suggests that Fes associates with PlexA1 and that this association is negatively regulated by NP-1 expression.
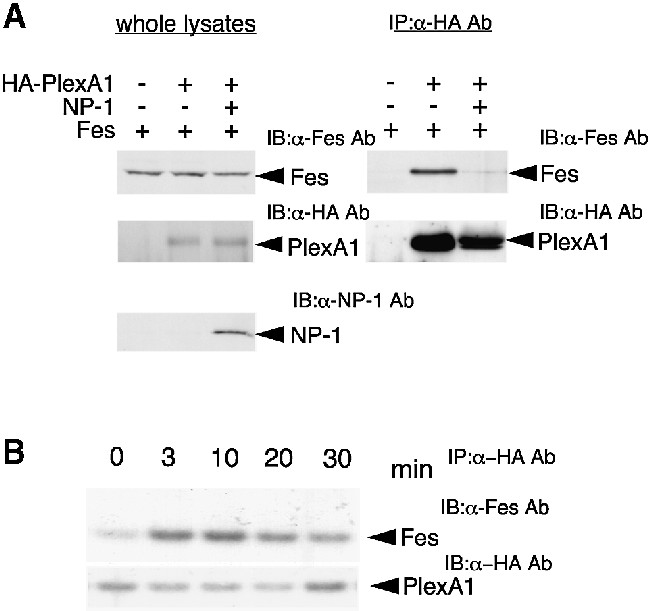
Fig. 7. Association of Fes with PlexA1 and its negative regulation by NP-1 co-expression in COS-7 cells. (A) COS-7 cells were transfected with the indicated expression vectors and total cell lysates were immunoblotted with anti-PlexA1 Ab, anti-NP-1 Ab or anti-Fes Ab, respectively (left). Each lysate was immunoprecipitated with anti-HA Ab and the immunoprecipitates were immunoblotted with anti-Fes Ab or anti-PlexA1 Ab (right). (B) Sema3A enhances association of Fes with PlexA1. COS-7 cells expressing Fes/HA-PlexA1/NP-1 were stimulated with 5 U/ml Sema3A for the indicated times, and cell lysates were immunoprecipitated with anti-HA Ab and then immunoblotted with anti-Fes Ab or anti-HA Ab.
We next examined the effect of Sema3A stimulation on the association of Fes with PlexA1 in COS-7 cells expressing PlexA1/Fes/NP-1. As shown in Figure 7B, Sema3A enhanced the association of Fes with PlexA1. The association reached a maximum level at 10 min and then decreased within 30 min. This result suggested that Sema3A stimulation canceled the negative regulation by NP-1 and permitted the interaction of Fes with PlexA1.
Tyrosine phosphorylation of PlexA1 and inhibition by NP-1 co-expression
We next examined whether Fes phosphorylates PlexA1 in COS-7 cells. Cells were transfected with the indicated expression vectors and total cell lysates were immunoblotted with anti-PlexA1 Ab, anti-NP-1 Ab or anti-Fes Ab (Figure 8A). These results indicated that the expression of each vector was successfully achieved in COS-7 cells. When COS-7 cells were co-transfected with HA-tagged PlexA1 and Fes, a prominent band showing tyrosine phosphorylation of PlexA1 was observed (Figure 8B, lane 3). However, tyrosine phosphorylation of PlexA1 by Fes was significantly reduced by co-expression of NP-1 (Figure 8B, lane 4). This result was correlated with the inhibitory effect of NP-1 on the association of PlexA1 and Fes. Thus, NP-1 appears to inhibit Fes-dependent tyrosine phosphorylation of PlexA1 by blocking the association of PlexA1 and Fes.
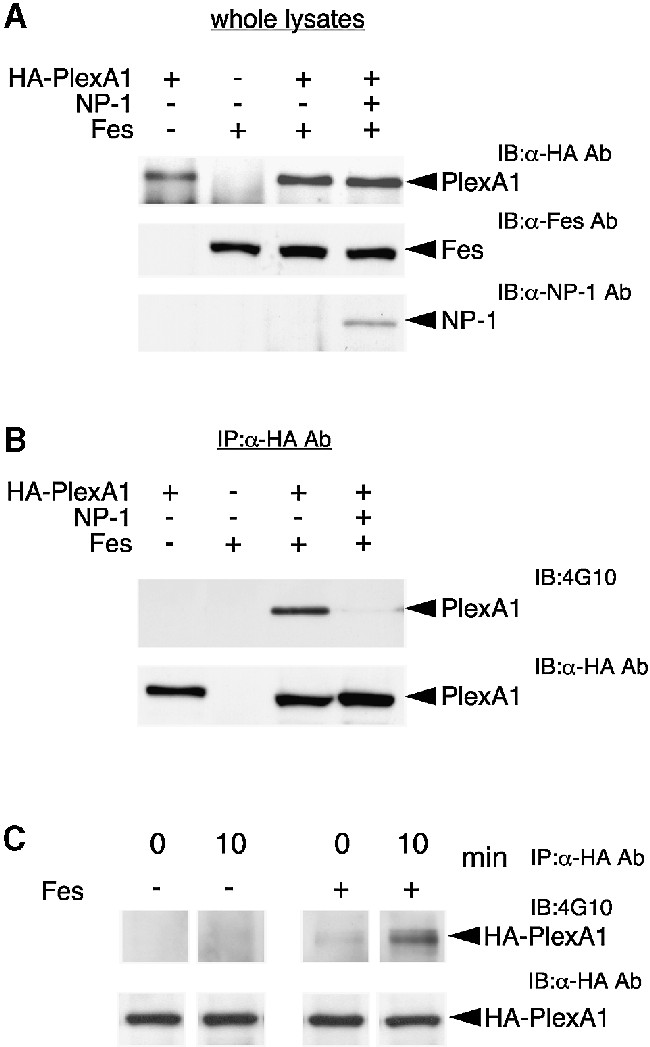
Fig. 8. Tyrosine phosphorylation of PlexA1 by Fes and inhibition by NP-1 co-expression. (A) Expression in COS-7 cells. COS-7 cells were transfected with the indicated expression vectors, and total cell lysates were immunoblotted with anti-PlexA1, anti-NP-1 or anti-Fes Ab. (B) Tyrosine phosphorylation of PlexA1 by Fes. Each lysate was immunoprecipitated with anti-HA Ab. The immunoprecipitates were immunoblotted with anti-phosphotyrosine or anti-HA Ab. (C) Sema3A enhances tyrosine phosphorylation of PlexA1 by Fes. COS-7 cells expressing HA-PlexA1/NP-1 or Fes/HA-PlexA1/NP-1 were stimulated with or without 5 U/ml Sema3A for 10 min, and cell lysates were immunoprecipitated with anti-HA Ab, and then immunoblotted with 4G10 or anti-HA Ab.
Since Sema3A induced the association of Fes with PlexA1, the effect of Sema3A stimulation on tyrosine phosphorylation of PlexA1 by Fes was examined. As shown in Figure 8C, Sema3A induced the tyrosine phosphorylation of PlexA1 in COS-7 cells expressing PlexA1/Fes/NP-1. Taken together, these results suggested that Fes is a candidate PTK that phosphorylates PlexA1 and is closely involved in Sema3A signaling.
Sema3A activates Fes and enhances tyrosine phosphorylation of CRAM and CRMP2
We found that Fes associates with and phosphorylates PlexA1 in response to Sema3A stimulation. We therefore investigated whether Sema3A activates Fes. In vitro kinase assay revealed that Sema3A activated Fes in COS-7 cells expressing Fes/PlexA1/NP-1 (Figure 9A). A time-course study showed that Fes activation induced by Sema3A reached a maximum level at 10 min and then decreased within 20 min (Figure 9A, left panel). To further confirm the Fes activation by Sema3A, tubulin was used as an exogenous substrate to measure Fes-associated kinase activity. As shown in Figure 9A, a similar result was obtained. Consistently, whole-cell tyrosine phosphorylation reached maximum levels at 10 min and then decreased within 20 min (Figure 9A, right panel). In addition, Fes activation was completely suppressed in COS-7 cells expressing Fes, NP-1 and a mutant PlexA1 (amino acids 1–1674), which lacks the cytoplasmic domain (Figure 9B). The equal expression of HA-tagged PlexA1 and HA-tagged mutant PlexA1 was confirmed by immunoblotting with anti-HA Ab (not shown). This result suggested that Sema3A activated Fes in a PlexA1 cytoplasmic domain-dependent manner. Since CRAM and CRMPs were found to be substrates for Fes in vivo and in vitro, the effects of Sema3A stimulation on tyrosine phosphorylation of CRAM and CRMP2 by Fes were examined. As shown in Figure 9C, Sema3A stimulation enhanced Fes-mediated tyrosine phosphorylation of CRAM and CRMP2. These data provide the first evidence for a link between Sema3A signals and CRMP–CRAM. We examined whether PlexA1, CRAM and CRMP2 were tyrosine phosphorylated in response to Sema3A in DRG neurons. In several experiments, the tyrosine phosphoryl ations of PlexA1 and CRMP–CRAM were observed to a small extent, but clear results were not obtained. This may be due to the limited number of primary cultured DRG neurons or the effect of protein tyrosine phosphatase activity associated with PlexA1.
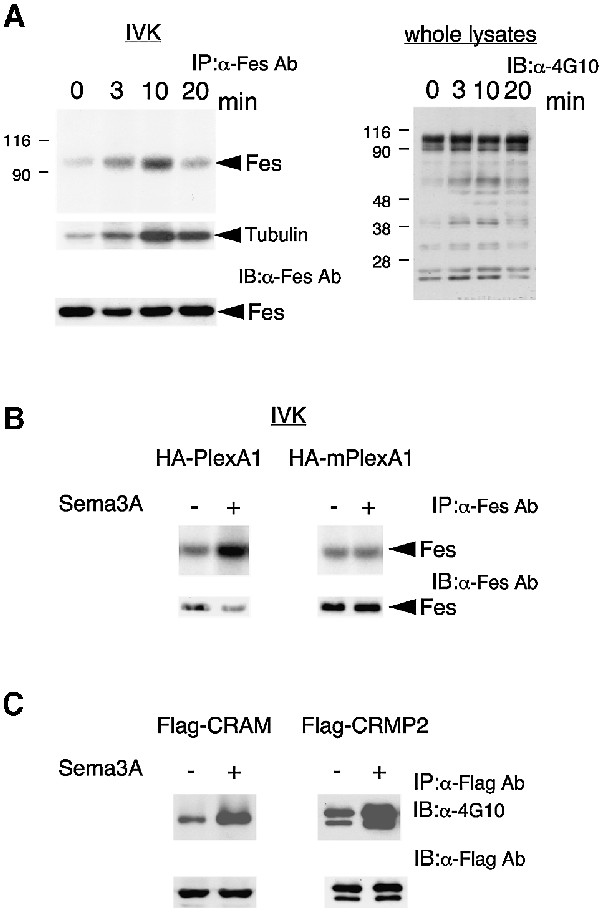
Fig. 9. Sema3A activates Fes and enhances Fes-mediated tyrosine phosphorylation of CRAM and CRMP2. (A) COS-7 cells expressing Fes/PlexA1/NP-1 were stimulated with 5 U/ml Sema3A for the indicated times, and then cell lysates were immunoblotted with 4G10 (right) or immunoprecipitated with anti-Fes Ab (left). The immuno precipitates were processed for immunoprecipitation kinase assay as described in Materials and methods. (B) COS-7 cells expressing Fes/HA-PlexA1/NP-1 or Fes/HA-mPlexA1/NP-1 were stimulated with or without 5 U/ml Sema3A for 10 min. Cell lysates were immunoprecipitated with anti-Fes Ab and processed for immunoprecipitation kinase assay or immunoblotted with anti-Fes Ab. HA-mPlexA1, HA-tagged mutant PlexA1 lacking the cytoplasmic domain. (C) COS-7 cells expressing Fes/PlexA1/NP-1/CRAM or Fes/PlexA1/NP-1/CRMP2 were stimulated with 5 U/ml Sema3A for the indicated times, and then cell lysates were immunoprecipitated with anti-Flag Ab. The immunoprecipitates were immunoblotted with either 4G10 or anti-Flag Ab.
Involvement of Fes in Sema3A signaling
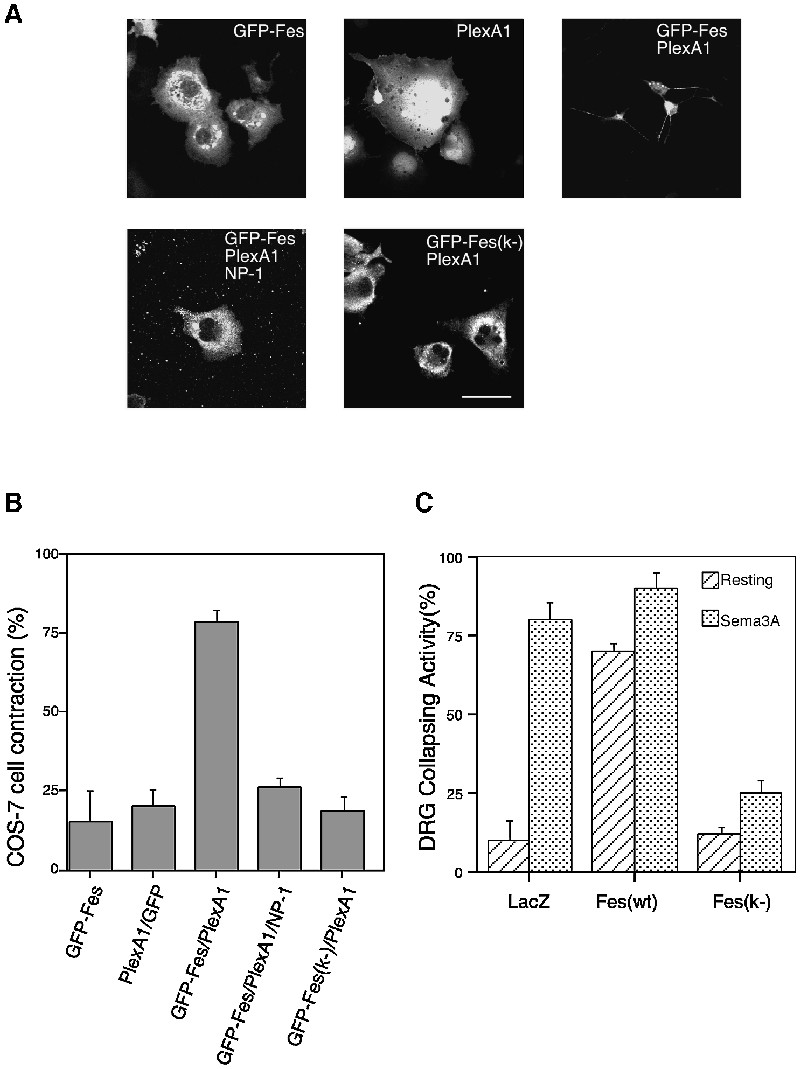
Previous studies demonstrated that COS-7 cells expressing NP-1–PlexA1 exhibited a dramatically reduced cell area after incubation with Sema3A (Takahashi et al., 1999) and that COS-7 cells expressing PlexA1 lacking a sema domain or ectodomain were contracted (Takahashi and Strittmatter, 2001). These results suggest that PlexA1 is inactive in the basal state, but that PlexA1 activated by Sema3A or semaphorin-deleted PlexA1 becomes active and produces cell contraction. These phenomena may reflect actin reorganization of Sema3A signaling, including growth cone collapse. Using this assay system, we examined the effect of Fes on cell contraction mediated by PlexA1 activation. COS-7 cells were transfected with vectors encoding GFP–Fes, GFP/PlexA1, GFP–Fes/PlexA1, GFP–Fes/PlexA1/NP-1 or GFP–Fes(k-)/PlexA1 and visualized by GFP expression 48 h after transfection (Figure 10A). Although PlexA1- or GFP–Fes-expressing COS-7 cells were well spread, PlexA1 and GFP–Fes-co-expressing COS-7 cells were dramatically contracted, whereas combination of Fes/NP-1/PlexA1 or Fes kinase-negative mutants/PlexA1 did not alter the cell morphology. The percentage of transfected cells with contraction is presented in Figure 10B. Therefore, Fes appears to induce PlexA1 activation and this activation might be closely correlated with tyrosine phosphorylation of PlexA1 by Fes. In order to understand the role of Fes in vivo, we next tested whether Fes was necessary for Sema3A-induced growth cone collapse in DRG neurons using virus vector-mediated expression of Fes and Fes kinase-negative mutants. As shown in Figure 10C, expression of Fes promoted growth cone collapse without Sema3A stimulation, whereas Fes dominant-negative mutants significantly suppressed Sema3A-induced growth cone collapse of DRG neurons. Our results are summarized in Figure 11, and a hypothesis presented about the role of Fes in Sema3A signaling.
Fig. 10. Involvement of Fes in Sema3A signaling. (A) Co-expression of Fes and PlexA1 induces COS-7 cell contraction. COS-7 cells were transfected with vectors encoding GFP–Fes, GFP/PlexA1, GFP–Fes/PlexA1, GFP–Fes/PlexA1/NP-1 or GFP–Fes(k-)/PlexA1 and visualized by GFP expression 48 h after transfection. Scale bar, 70 µm. (B) Percentage of COS-7 cell contraction. Cells were transfected with the indicated plasmids as in (A). (C) Effect of Sindbis virus-mediated expression of LacZ (control), Fes(wt) and Fes(k-) on Sema3A-induced growth cone collapse of DRG neurons. The means ± SEM from three experiments are presented.
Fig. 11. A schematic model of Sema3A signaling mediated by Fes. Sema3A receptors consist of NP-1–PlexA1 complexes. In the resting state, NP-1 blocks the association of Fes with PlexA1 and tyrosine phosphorylation of PlexA1. NP-1 appears to stabilize the inactive conformation of NP-1–PlexA1 complexes. Sema3A binding to NP-1 may cancel this inhibition, and induce a dramatic conformational change in this complex. Presumably, this conformational change of PlexA1 permits Fes to interact with and phosphorylate PlexA1. Furthermore, activated Fes phosphorylates CRMP–CRAM complex. These events may play an important role in actin reorganization and growth cone collapse of Sema3A signaling.
Discussion
Fes links between Sema3A/PlexA1 signals and CRMP–CRAM
The cytoplasmic mediators of semaphorin signaling are poorly understood. Recent studies have revealed that Sema3A receptors consist of NP-1–PlexA1 complexes. Within the NP-1–PlexA1 complex, NP-1 is the binding unit for Sema3A, while PlexA1 serves as signal transducer (Comeau et al., 1998; Winberg et al., 1998; Takahashi et al., 1999; Tamagnone et al., 1999). CRMP was initially identified as a downstream cytoplasmic mediator of Sema3A signaling, but their molecular relationship to semaphorin/plexin has not yet been defined. In the present study, we found that Sema3A activates Fes through PlexA1 and then Fes phosphorylates PlexA1 and CRMP–CRAM. In addition, our data suggest that Fes is required for Sema3A-mediated growth cone collapse of DRG neurons. Thus, our findings provide the first evidence for a link between Sema3A/PlexA1 signals and CRMP–CRAM.
The plexin extracellular domains include three cysteine-rich repeats called MET-related sequence (G-P repeat). These repeats are also found, though in different numbers and with altered spacing, in the MET PTK and MET-related PTKs SEA and RON (Ohta et al., 1995; Maestrini et al., 1996). Because an intrinsic kinase activity of plexins was excluded, their association with receptor or non-receptor PTK has been proposed. Indeed, Tamagnone et al. (1999) reported that cytoplasmic plexin domains were tyrosine phosphorylated in overexpressing cells, although ligand dependency has not yet been shown. Thus, Fes is a candidate component that acts to convey semaphorin/plexin signals. We tested whether other tyrosine kinases (Src, Lyn and Syk) phosphorylate PlexA1 in COS-7 cells. We found that Syk did not phosphorylate PlexA1, but Src and Lyn induced PlexA1 tyrosine phosphorylation in COS-7 cells. However, we could not detect the association of these PTKs with PlexA1. In addition, co-expression of these PTKs and PlexA1 did not induce PlexA1-dependent contraction of COS-7 cells (data not shown). These results suggest that Fes has a distinct role from other PTKs in Sema3A signaling.
Negative action of NP-1 on interaction between Fes and PlexA1
We found that the association of Fes with PlexA1 and tyrosine phosphorylation of PlexA1 by Fes were blocked by the co-expression of NP-1, and that this blockage was canceled by Sema3A stimulation (Figures 7 and 8). This result suggests that NP-1 stabilizes the inactive conformation of NP-1–PlexA1 receptors and Sema3A binding to NP-1 results in a dramatic conformational change in this complex. Presumably, this conformational change of PlexA1 permits Fes to interact with and phosphorylate PlexA1. A recent study has indicated that a mutant PlexA1 lacking the sema domain exhibits constitutive activation and this mutant reduces COS-7 cell area and collapses DRG growth cones, implying that the sema domain of PlexA1 restricts PlexA1 to a basal inactive state by binding to the remainder of the ectodomain (Takahashi and Strittmatter, 2001). However, our results suggest that only the PlexA1 sema domain interaction with the remainder of the ectodomain is not sufficient for blocking Fes interaction with PlexA1. Since the PlexA1 sema domain interacts with both NP-1 and the remainder of the ectodomain, both interactions of the sema domain may strongly stabilize the inactive conformation of NP-1–PlexA1 receptors, which can block Fes interaction with PlexA1. Thus, NP-1 could strengthen the autoinhibition of PlexA1 via its sema domain, and Sema3A–NP-1 interaction may release this inhibition.
We have found that PlexA1 was co-immunoprecipitated with anti-Fes Ab from mouse brain lysate. However, in COS-7 cells, this PlexA1/Fes binding was not observed at resting state in NP-1/PlexA1/Fes-expressing cells. One possibility raised by this result is that all PlexA1 do not always form a complex with NP-1. Indeed, there is no report indicating a close correlation between PlexA1 and NP-1 distribution in brain. In our preliminary study, PlexA1 showed a wider distribution than NP-1 in brain. Therefore, Fes may be associated with PlexA1 that does not bind to NP-1 in the developing brain.
Relationship between Fes and CRMP–CRAM complex
A role for CRMPs and CRAM in the processes of neuronal differentiation is supported by several lines of evidence. In PC12 cells, nerve growth factor (NGF) treatment induces rapid changes in the phosphorylation states of CRMPs, as well as changes in their expression levels (Byk et al., 1996). CRAM expression was upregulated during neuronal differentiation of embryonal carcinoma P19 and PC12 cells (Inatome et al., 2000). Together, CRMPs and CRAM are likely to participate uniquely in neuronal differentiation during development. Since the CRMP/ULIP family members were also identified as phosphoproteins, their function(s) may also be regulated by phosphorylation.
Fes was identified in a CRMP–CRAM complex purified from whole-brain lysates. Indeed, Fes expression was observed not only in myeloid cells, but also in vascular endothelial, epithelial and neuronal cells during early development (Haigh et al., 1996). Consistent with this, our results also showed the expression of Fes in neuronally differentiated P19 cells and DRG neurons (Figure 3). Although Fes function, particularly in neurons, remains largely unknown, transgenic mice overexpressing v-fps showed a neurological disorder that includes a marked trembling, correlated with the expression of v-fps in the brain and a striking bilateral enlargement of the trigeminal nerves (Yee et al., 1989). This result indicates an important role for Fes in neural differentiation and growth. Thus, these results, together with our findings, suggest a close functional relationship among CRMP–CRAM and Fes during neural development.
Most recently, Inagaki et al. (2001) reported that overexpression of CRMP2 in cultured hippocampal neurons led to the formation of supernumerary axons, and that expression of truncated CRMP2 mutants suppressed the formation of primary axon in a dominant manner, suggesting an important role for CRMPs in axon induction and outgrowth. Therefore, it will be interesting to study whether Fes promotes CRMP–CRAM activity to induce axon induction and outgrowth. Further investigations are needed in order to clarify the exact role of Fes in the CRMP–CRAM complex, such as the identification of the site(s) of PlexA1 tyrosine phosphorylation by Fes.
Fes and small GTPases in plexin-mediated signaling
Sema3A is a potent inducer of growth cone collapse by a mechanism that results in the depolymerization of F-actin. However, little is known about the link between the receptors and the structural changes of the cytoskeleton during chemorepulsion. Previous studies have demonstrated that COS-7 cells expressing NP-1–PlexA1 exhibited a dramatically reduced cell area after incubation with Sema3A (Takahashi et al., 1999), and that COS-7 cells expressing PlexA1 lacking the sema domain or ectodomain were contracted (Takahashi and Strittmatter, 2001). This provides further support for the notion that PlexA1 is the transmembrane protein through which Sema3A signals to the neuronal growth cone cytoplasm. Recently, two groups have demonstrated that plexins are able to modulate the activity of small GTPases that are key regulators of the actin cytoskeleton (Rohm et al., 2000; Vikis et al., 2000). These studies also provided evidence that the intracellular domain of plexins binds to specific Rho-like GTPases, allowing for the potential activation of multiple signaling cascades (Rohm et al., 2000). Vikis et al. (2000) have also demonstrated that the cytosolic domain of PlexB1 interacts directly with activated Rac. Thus, Ras GTPase-activating protein-like activity related to plexin and interaction of plexin with small GTPases appear to play critical roles in the regulation of actin cytoskeleton.
In this study, we showed that co-expression of Fes with PlexA1 led to COS-7 cell contraction activity. This result suggests that Fes can convert an inactive PlexA1 to an active form that induces cytoskeletal reorganization. It is possible that this activation might be mediated by tyrosine phosphorylation of PlexA1, but the relationship between PlexA1 activation and cytoskeletal reorganization is unknown. On the other hand, it has been shown that dominant-negative Ras, Rac and Cdc42 inhibited Fes-induced growth of Rat-2 cells, indicating that activation of these small GTP-binding proteins is required for fibroblast transformation by Fes (Li and Smithgall, 1998). Another previous study has also established that BCR is a substrate for Fes (Li and Smithgall, 1996). The human BCR gene encodes a protein with serine/threonine kinase activity and regulatory domains for the small G-proteins Rac and Cdc42. Tyrosine phosphorylation leads to the association of BCR with multiple signaling partners that affect the activity of small GTP-binding proteins. Taken together, these results suggest that Fes may play an important role in plexin-mediated signaling, including actin reorganization and cell contraction, through the regulation of small GTP-binding protein activity.
In summary, our findings indicate the involvement of Fes in semaphorin signaling. The further biological characterization of Fes will contribute to the understanding of the mechanisms underlying axonal guidance during neuronal development.
Materials and methods
Materials
Aprotinin, phenylmethylsulfonyl fluoride (PMSF), protein A–agarose beads, the molecular weight standards, retinoic acid and anti-M2-Flag Ab were purchased from Sigma. Anti-HA Ab was from Convance. Silver stain kit was from Wako. SD rats were from SLC, Inc. (Japan). Anti-Fes polyclonal Ab was from Santa Cruz Biotechnology, Inc. Anti-phosphotyrosine monoclonal Ab 4G10 and Fes (purified enzyme) were from Upstate Biotechnology. Anti-Fes (N-19) and Anti-Fes (C-19) Abs were purchased from Santa Cruz. Anti-CRAM and anti-PlexA1 Abs were produced by immunizing rabbits with synthetic peptides (amino acid residues CRAM 468–485), glutathione S-transferase fusion protein (PlexA1 amino acid residues 1295–1894). Monoclonal anti-neuropilin1 Ab was prepared as described previously (Kitsukawa et al., 1997). NGF (7S) was purchased from Chemicon International, Inc.
Immunoprecipitation and immunoblotting procedures
All procedures were carried out at 4°C. Rat brains or cultured cells were lysed in lysis buffer (20 mM Tris–HCl pH 8.0, 1% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 10 µg/ml aprotinin, 10 µg/ml leupeptin and 1 mM sodium orthovanadate). The lysates (1 mg of protein) from rat brain or cultured cells were clarified by centrifugation at 100 000 g for 10 min and immunoprecipitated with appropriate Ab. Immunoprecipitates were washed three times with lysis buffer, once with 10 mM HEPES–NaOH pH 8.0 containing 0.5 M NaCl and finally with 10 mM HEPES– NaOH pH 8.0. Immunoprecipitates were subjected to either an in vitro kinase assay (see below), or boiled with SDS–PAGE sample buffer for 3 min, separated by SDS–PAGE and transferred to PVDF membranes (Immobilon P; Millipore), followed by detection with the appropriate Ab as described previously (Inatome et al., 2000).
Determination of internal peptide sequence of purified protein
The polyacrylamide gel slice containing ∼95 kDa proteins (∼10 pmol) was excised, and digested with 3 µg of lysyl endopeptidase at 37°C overnight in 100 µl of 20 mM NH4HCO3, 10% (v/v) acetonitrile and 0.1% (v/v) Triton X-100. The peptides were separated with a Perkin Elmer–Applied Biosystems model 173A liquid chromatography system, and then analyzed using a Perkin Elmer–Applied Biosystems model 492 automated amino acid sequencer as described previously (Inatome et al., 2000).
RT–PCR for determination of Fes expression
Total RNAs were isolated from 3-week-old mouse brains and retinoic acid-treated P19 cells, and reverse transcribed using oligo(dT) primers. Primers used for amplification of mouse Fes cDNA were synthesized as follows: primer (sense), 5′-GTCTCACCGATGCCGCTTGCGGAT-3′; primer (antisense), 5′-ACCATTATGGGCTTCTCTTCAGAGCTGTGC-3′. For GAPDH, the following primers were used: primer (sense), 5′-TGAAGGTCGGTGTCAACGGATTTGGC-3′; primer (antisense), 5′-CATGTAGGCCATGAGGTCCACCAC-3′. PCR products were electrophoresed on a 1.0% agarose gel containing ethidium bromide. The sequences of PCR products were confirmed using an ABI-377 sequencer.
Expression constructs
Fes cDNA cloning was performed using 3-week-old mouse brain total RNA as a template, and a 3′ primer (5′-GTCTCACCGATGCCGC TTGCGGAT-3′) and a 5′ primer (5′-ACCATTATGGGCTTCTCTT CAGAGCTGTGC-3′) as specific primers for mouse Fes. Cloning for mouse PlexA1 cDNA was also performed using 3-week-old mouse brain total RNA as a template. Primers used for amplification were synthesized as follows: for mouse PlexA1 (1–2934), a 3′ primer (5′-GCCAATC CAGGTACCTCCAGACAG-3′) and a 5′ primer (5′-ATGCCACTGC CACCTCTGAGCTCT-3′); for mouse PlexA1 (2911–5685), a 3′ primer (5′-GGCTCAGCTGCTCAGGGCCATTGT-3′) and a 5′ primer (5′-CTGTCTGGAGGTACCTGGATTGGC-3′). The amplified products were separated on 0.8% agarose gels, and DNA fragments were recovered using the geneclean kit (Bio101) for subcloning into pCR2.1 TA cloning vector (Invitrogen). To obtain a full-length PlexA1 cDNA, two fragments were successively ligated into pCMV5 expression vectors. Sequencing of all strands of the cloned RT–PCR fragments was performed using an ABI-377 automated DNA sequencer and primers specific for the cloning vector. Fes cDNA, full-length PlexA1 and mutant PlexA1 (amino acids 1–1674) cDNA tagged with HA epitope at the C-terminus, and CRMPs or CRAM cDNAs tagged with Flag epitope at the N-terminus were subcloned into pCMV5 expression vectors. Fes kinase negative was generated by missense mutation K562R as described previously (Craig et al., 1999). Both Fes and Fes kinase-negative cDNAs were subcloned into pEGFP-C1 (Clontech) for green fluorescence. Mouse NP-1 cDNA was subcloned into pCMV5 expression vector. Fes and Fes kinase-negative cDNAs were inserted into the pSinRep5 vector and packaged into virus particles using the Sindbis Expression System (Invitrogen). Routinely, 10 µl of virus solution are more than sufficient to infect all the neurons in 200 µl culture DRG explants.
Cell culture and stimulation
P19 mouse embryonic carcinoma cells were maintained and promoted to differentiation as described previously (Inatome et al., 2000). COS-7 cells were cultured in IMDM (Sigma) containing 10% fetal bovine serum. DRGs were surgically excised from embryonic day 13 mice, trypsinized, treated with 0.01% DNase I and then mechanically dissociated. The cell suspension was plated onto laminin-coated coverslips in MEM medium supplemented with 10% fetal bovine serum and 20 ng/ml NGF. Before addition of Sema3A, the cells were treated with MEM medium (serum free) containing 20 ng/ml NGF for 4 h. Semaphorin stimulation was described previously (Goshima et al., 1995). Semaphorin at 1 U/ml induces the collapse of 50% of growth cones in cultured DRG neurons.
Transfection of plasmid into COS-7 cells
COS-7 cells (1 × 105) were cultured in dishes at ∼50% confluence. Expression vectors (1–2 µg) were mixed with serum-free medium, and transfected using LipofectAMINE plus kit (Gibco-BRL). After 48 h, the medium was changed to serum-free medium for ∼4 h, and the cells were then treated with Sema3A.
In vitro kinase assay
The Fes immunoprecipitates were incubated in a reaction mixture containing 50 mM HEPES–NaOH pH 8.0, 10 µM Na3VO4, 5 mM MgCl2, 5 mM MnCl2 and 1 µM [γ-32P]ATP (200 c.p.m./fmol) for 10 min at 30°C. For an exogenous substrate, 10 µg/ml tubulin was used. The reactions were terminated by boiling for 3 min with SDS sample buffer and subjected to 10% SDS–PAGE followed by autoradiography.
For in vitro reconstitution experiments, immunoprecipitates from COS-7 cells transfected with Flag-tagged CRAM or CRMP2 in expression vector were washed three times with lysis buffer, washed twice with phosphate-buffered saline (PBS) and eluted with Flag epitope peptide. The elution products were incubated with 30 µl of reaction buffer (5 mM MgCl2, 5 mM MnCl2 and 20 mM HEPES pH 7.4) containing 30 µM cold ATP and 10 ng of Fes purified enzyme. After incubation for 10 min at 30°C, the reactions were terminated by the addition of SDS sample buffer and boiled for 3 min, and then subjected to 10% SDS–PAGE followed by immunoblotting using anti-phosphotyrosine Ab (4G10).
Immunofluorescence microscopy
COS-7 cells transiently co-expressing cDNAs or P19 or DRG culture cells were fixed with 4% paraformaldehyde in PBS for 10 min, washed twice with PBS, permeabilized with 0.2% Triton X-100 in PBS for 10 min, washed three times with PBS and blocked with 3% bovine serum albumin in PBS, all at room temperature. For double staining, the cells were incubated with appropriate Abs for 2 h at room temperature, washed three times with 0.2% Triton X-100 in PBS, and then with appropriate secondary Ab (FITC-labeled goat anti-rabbit IgG, Cy3-labeled goat anti-rabbit IgG, Cy3-labeled goat anti-mouse IgG, FITC-labeled donkey anti-goat IgG) for 1 h. The samples were washed as before, mounted using SlowFade-Light (Molecular Probes), and analyzed using an OPTIPHOT-2 (NIKON) and MRC-1024 confocal imaging system (Bio-Rad).
Acknowledgments
Acknowledgements
We are very grateful to Dr H.Fujisawa for providing mouse NP-1 cDNA and anti-NP-1 monoclonal Ab, and Dr M.Nonaka for providing mouse and human DRPs/CRMPs cDNA. We also thank Dr S.Jahangeer for critically reading the manuscript. This work was supported by Grant-in-Aids for Scientific Research (B), Scientific Research on Priority Areas (A) from the Ministry of Education, Science, Sports and Culture, Japan.
References
- Byk T., Dobransky,T., Cifuentes-Diaz,C. and Sobel,A. (1996) Identification and molecular characterization of Unc-33-like phosphoprotein (Ulip), a putative mammalian homolog of the axonal guidance-associated unc-33 gene product. J. Neurosci., 16, 688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau M.R. et al. (1998) A poxvirus-encoded semaphorin induces cytokine production from monocytes and binds to a novel cellular semaphorin receptor, VESPR. Immunity, 8, 473–482. [DOI] [PubMed] [Google Scholar]
- Craig A.W., Zirngibl,R. and Greer,P. (1999) Disruption of coiled-coil domains in Fer protein-tyrosine kinase abolishes trimerization but not kinase activation. J. Biol. Chem., 274, 19934–19942. [DOI] [PubMed] [Google Scholar]
- Feiner L., Koppel,A.M., Kobayashi,H. and Raper,J.A. (1997) Secreted chick semaphorins bind recombinant neuropilin with similar affinities but bind different subsets of neurons in situ. Neuron, 19, 539–545. [DOI] [PubMed] [Google Scholar]
- Fujisawa H. and Kitsukawa,T. (1998) Receptors for collapsin/semaphorins. Curr. Opin. Neurobiol., 8, 587–592. [DOI] [PubMed] [Google Scholar]
- Goshima Y., Nakamura,F., Strittmatter,P. and Strittmatter,S.M. (1995) Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature, 376, 509–514. [DOI] [PubMed] [Google Scholar]
- Haigh J., McVeigh,J. and Greer,P. (1996) The fps/fes tyrosine kinase is expressed in myeloid, vascular endothelial, epithelial and neuronal cells and is localized in the trans-Golgi network. Cell Growth Differ., 7, 931–944. [PubMed] [Google Scholar]
- Hamajima N., Matsuda,K., Sakata,S., Tamaki,N., Sasaki,M. and Nonaka,M. (1996) A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene, 180, 157–163. [DOI] [PubMed] [Google Scholar]
- Hanafusa T., Wang,L.H., Anderson,S.M., Karess,R.E., Hayward,W.S. and Hanafusa,H. (1980) Characterization of the transforming gene of Fujinami sarcoma virus. Proc. Natl Acad. Sci. USA, 77, 3009–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z. and Tessier-Lavigne,M. (1997) Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell, 90, 739–751. [DOI] [PubMed] [Google Scholar]
- Inagaki N., Chihara,K., Arimura,N., Menager,C., Kawano,Y., Matsuo,N., Nishimura,T., Amano,M. and Kaibuchi,K. (2001) CRMP-2 induces axons in cultured hippocampal neurons. Nat. Neurosci., 4, 781–782. [DOI] [PubMed] [Google Scholar]
- Inatome R., Tsujimura,T., Hitomi,T., Mitsui,N., Hermann,P., Kuroda,S., Yamamura,H. and Yanagi,S. (2000) Identification of CRAM, a novel unc-33 gene family protein that associates with CRMP3 and protein-tyrosine kinase(s) in the developing rat brain. J. Biol. Chem., 275, 27291–27302. [DOI] [PubMed] [Google Scholar]
- Kitsukawa T., Shimizu,M., Sanbo,M., Hirata,T., Taniguchi,M., Bekku,Y., Yagi,T. and Fujisawa,H. (1997) Neuropilin–semaphorin III/D-mediated chemorepulsive signals play a crucial role in peripheral nerve projection in mice. Neuron, 19, 995–1005. [DOI] [PubMed] [Google Scholar]
- Kolodkin A.L. (1998) Semaphorin-mediated neuronal growth cone guidance. Prog. Brain Res., 117, 115–132. [DOI] [PubMed] [Google Scholar]
- Kolodkin A.L., Levengood,D.V., Rowe,E.G., Tai,Y.T., Giger,R.J. and Ginty,D.D. (1997) Neuropilin is a semaphorin III receptor. Cell, 90, 753–762. [DOI] [PubMed] [Google Scholar]
- Li J. and Smithgall,T.E. (1996) Co-expression with BCR induces activation of the FES tyrosine kinase and phosphorylation of specific N-terminal BCR tyrosine residues. J. Biol. Chem., 271, 32930–32936. [DOI] [PubMed] [Google Scholar]
- Li J. and Smithgall,T.E. (1998) Fibroblast transformation by Fps/Fes tyrosine kinases requires Ras, Rac and Cdc42 and induces extracellular signal-regulated and c-Jun N-terminal kinase activation. J. Biol. Chem., 273, 13828–13834. [DOI] [PubMed] [Google Scholar]
- Maestrini E. et al. (1996) A family of transmembrane proteins with homology to the MET-hepatocyte growth factor receptor. Proc. Natl Acad. Sci. USA, 93, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minturn J.E., Fryer,H.J., Geschwind,D.H. and Hockfield,S. (1995) TOAD-64, a gene expressed early in neuronal differentiation in the rat, is related to unc-33, a C. elegans gene involved in axon outgrowth. J. Neurosci., 15, 6757–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Mizutani,A., Kawakami,A., Murakami,Y., Kasuya,Y., Takagi,S., Tanaka,H. and Fujisawa,H. (1995) Plexin: a novel neuronal cell surface molecule that mediates cell adhesion via a homophilic binding mechanism in the presence of calcium ions. Neuron, 14, 1189–1199. [DOI] [PubMed] [Google Scholar]
- Rohm B., Ottemeyer,A., Lohrum,M. and Puschel,A.W. (2000) Plexin/neuropilin complexes mediate repulsion by the axonal guidance signal semaphorin 3A. Mech. Dev., 93, 95–104. [DOI] [PubMed] [Google Scholar]
- Smithgall T.E., Yu,G. and Glazer,R.I. (1988) Identification of the differentiation-associated p93 tyrosine protein kinase of HL-60 leukemia cells as the product of the human c-fes locus and its expression in myelomonocytic cells. J. Biol. Chem., 263, 15050–15055. [PubMed] [Google Scholar]
- Takahashi T. and Strittmatter,S.M. (2001) Plexina1 autoinhibition by the plexin sema domain. Neuron, 29, 429–439. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Fournier,A., Nakamura,F., Wang,L.H., Murakami,Y., Kalb,R.G., Fujisawa,H. and Strittmatter,S.M. (1999) Plexin– neuropilin-1 complexes form functional semaphorin-3A receptors. Cell, 99, 59–69. [DOI] [PubMed] [Google Scholar]
- Tamagnone L. et al. (1999) Plexins are a large family of receptors for transmembrane, secreted and GPI-anchored semaphorins in vertebrates. Cell, 99, 71–80. [DOI] [PubMed] [Google Scholar]
- Vikis H.G., Li,W., He,Z. and Guan,K.L. (2000) The semaphorin receptor plexin-B1 specifically interacts with active Rac in a ligand-dependent manner. Proc. Natl Acad. Sci. USA, 97, 12457–12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.H. and Strittmatter,S.M. (1996) A family of rat CRMP genes is differentially expressed in the nervous system. J. Neurosci., 16, 6197–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg M.L., Noordermeer,J.N., Tamagnone,L., Comoglio,P.M., Spriggs,M.K., Tessier-Lavigne,M. and Goodman,C.S. (1998) Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell, 95, 903–916. [DOI] [PubMed] [Google Scholar]
- Yee S.P., Mock,D., Maltby,V., Silver,M., Rossant,J., Bernstein,A. and Pawson,T. (1989) Cardiac and neurological abnormalities in v-fps transgenic mice. Proc. Natl Acad. Sci. USA, 86, 5873–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]