Abstract
Endonuclease III, encoded by nth in Escherichia coli, removes thymine glycols (Tg), a toxic oxidative DNA lesion. To determine the biological significance of this repair in mammals, we established a mouse model with mutated mNth1, a homolog of nth, by gene targeting. The homozygous mNth1 mutant mice showed no detectable phenotypical abnormality. Embryonic cells with or without wild-type mNth1 showed no difference in sensitivity to menadione or hydrogen peroxide. Tg produced in the mutant mouse liver DNA by X-ray irradiation disappeared with time, though more slowly than in the wild-type mouse. In extracts from mutant mouse liver, we found, instead of mNTH1 activity, at least two novel DNA glycosylase activities against Tg. One activity is significantly higher in the mutant than in wild-type mouse in mitochondria, while the other is another nuclear glycosylase for Tg. These results underscore the importance of base excision repair of Tg both in the nuclei and mitochondria in mammals.
Keywords: base excision repair/DNA glycosylase/endonuclease III/mitochondria/thymine glycol
Introduction
Most living organisms utilize oxygen as their major energy source, but they also suffer from the toxic effects of oxygen on their cells. Oxidative damage to DNA is thought to constitute a major factor in mutagenesis and genome instability (Cerutti, 1985), and evidence has accumulated that the mutations and genome instability thus produced can cause cancer. Furthermore, age-dependent accumulation of oxidative DNA damage in nuclear and mitochondrial DNA suggests that oxidative DNA damage is a causative factor for human aging as well (Ames and Shigenaga, 1992; Wallace, 1995). Living cells have, however, several defense systems against oxidative stress. One of the protection mechanisms is DNA repair, which removes oxidative DNA damage and restores genetic information (Miller, 1998). Many of the DNA repair genes that encode repair enzymes for oxidative damage are conserved from bacteria to humans, as revealed by recent genome projects (Wood et al., 2001), underlying the importance of these systems.
Thymine glycol (Tg) is one of the major lesions caused by oxidative damage and produces a large distortion of the DNA structure (Kao et al., 1993). Consequently, Tg arrests DNA replication (Breimer, 1990) and blocks translesion synthesis (Hatahet and Wallace, 1998), leading to cell death in Escherichia coli, if it not removed by excision repair. Tg is generally repaired by base excision repair (BER) initiated by a DNA glycosylase, and by endonuclease III (endoIII) with an AP lyase activity in E.coli (Demple and Linn, 1980; Katcher and Wallace, 1983; Bailly and Verly, 1987). In addition to Tg, a number of damaged or modified pyrimidine residues or pyrimidine derivatives including 5,6-dihydrothymine, 5-hydroxy-5,6-dihydrothymine, uracil glycol and urea are substrates for endoIII of E.coli (see Wilson et al., 1998). In spite of the importance of repair of Tg, little is known about the repair pathways for Tg in mammals. The accumulation of Tg either in the nuclear and/or in the mitochondrial genome may be a causative factor for aging.
We have previously isolated a mouse homolog of the E.coli nth gene, mNth1, and characterized the enzymatic activity of a recombinant protein, mNTH1, which showed Tg and urea DNA glycosylase as well as AP lyase activity (Sarker et al., 1998). To gain insight into the role of repair for potentially lethal oxidative DNA damage such as Tg in mammals, we decided to generate a mouse model, defective in the mNth1 gene. Here we present the characterization of this mouse mutant. By characterizing cell extracts from mice, we found at least two novel DNA glycosylase activities, which may act as back-up repair systems for nuclear and mitochondrial DNA repair of Tg. Our data suggest the important roles of base excision repair in both organelles of mammals.
Results
Disruption of the mNth1 gene in ES cells and generation of mNth1-knockout mice
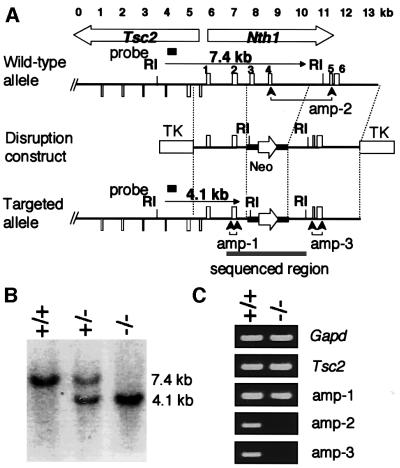
Using sequence data published previously (Sarker et al., 1998; DDBJ/EMBL/GenBank accession No. AB009371), a DNA fragment harboring the mNth1 gene was obtained by PCR of genomic DNA derived from mouse embryonic stem (ES) cells. Figure 1A (upper map) depicts the mouse Nth1 locus, which is located very close to the tuberous sclerosis 2 (Tsc2) gene in a head-to-head orientation. As shown in Figure 1A (middle), we made a disruption construct, in which exons 3 and 4 of the mNth1 gene were replaced by a neomycin-resistant marker. Both exons contained the most evolutionarily conserved helix–hairpin–helix domain with the reactive center for enzymatic activity of endoIII (Thayer et al., 1995). The plan for disruption of mNth1 locus is shown in the bottom map of Figure 1A.
Fig. 1. Targeted disruption of the mNth1 gene. (A) Physical map of the head-to-head-oriented mTsc2-mNth1 locus of the wild type (upper map), mNth1-disruption construct (middle map) and Nth1-disrupted locus (lower map). Exons of mNth1 and a part of Tsc2 genes are indicated by upward and downward boxes, respectively. Exons 2 and 3 were replaced by a Neo marker, which is shown by a bold line with an open arrow indicating the direction of expression in the disruption construct. RI indicates the site for EcoRI. The two TKs show the positions of the thymidine kinase gene for counter-selection. The gray bar in the bottom map shows the sequenced region. Three regions directed for RT–PCR of mNth1 transcripts are indicated by amp-1, amp-2 and amp-3. (B) Genotyping by Southern blot analysis. The EcoRI-digested tail DNA from wild-type (+/+), heterozygous (+/–) and homozygous (–/–) mice was blotted and analyzed by the probe shown in (A). (C) Expression of mTsc2 and mNth1 genes. Typical results of RT–PCR using primer sets for Gapd, Tsc2 and Nth1 at three positions are shown.
Heterozygous ES cell lines carrying a targeted mNth1 allele were generated by transfection of the construct into E14 ES cells, selection for G418/FIAU resistant clones and Southern blot analysis of resistant ES clones using a probe external to the construct (data not shown). Properly targeted ES cell lines were verified for the absence of detectable chromosomal abnormalities and used for blastocyst injections. Chimeric males were mated to C57Bl/6 females to produce heterozygous offspring, which were further intercrossed to produce homozygous mutant animals. Southern blot analysis of tail DNA from mNth1 +/+, +/– and –/– mice showed the expected 4.1 kb-long EcoRI fragment in the disrupted locus (Figure 1B). We confirmed the correct disruption by sequencing 5.6 kb of genomic DNA covering the target region (the underlined region in Figure 1A, bottom). Figure 1C shows the results of RT–PCR for mGapd encoding glyceraldehyde-3-phosphate dehydrogenase (G3PDH) as control, mTsc2 (a PCR product from a downstream region of the mTsc2 gene, which is not shown in Figure 1A), amp-1 (exon 2 of mNth1), amp-2 (exons 4 and 5 of mNth1) and amp-3 (exons 5 and 6). These data show that there is also no detectable transcript in the region between exons 5 and 6 of the disrupted mNth1 gene, while Tsc2 and exons 1 and 2 are transcribed. Furthermore, we know from sequence analysis in the region shown in Figure 1A that in the rare event that transcription reads through the Neo polyadenylation signal and the mRNA is alternatively spliced (causing exon 2 to be fused to exon 5), the disruption creates a frameshift leading to a protein containing only exons 1 and 2, which can not have any function related to DNA repair (Thayer et al., 1995). Thus, we obtained a mNth1-null mouse.
Nicking activity in crude nuclear extracts
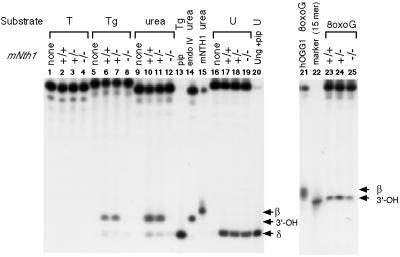
To determine the effects of the disruption on repair activity, crude nuclear extracts were prepared from mNth1 +/+, +/– and –/– mouse lung, and specific nicking activities for Tg and urea were measured (Figure 2). Nicking activity was easily detected in extracts prepared from either wild-type or mNth1 heterozygous mice, in contrast to extracts prepared from mNth1 homozygous mutant mice. Comparing the nicking properties with those of purified recombinant mNTH1 (lane 15) or after piperidine treatment (lane 13), or comparing with the result of urea-containing oligos treated with endoIV (lane 14), the major product of the nicking reaction in wild-type cell extracts contained a 3′-OH end group. Since mNTH1 produces β-elimination products (lane 15) due to its concomitant AP lyase activity, the products with 3′-OH were probably created by processing of the β-elimination products through 3′-phosphodiesterase activity present in the extracts. There was no difference among the extracts prepared from mNth1 +/+, +/– and –/– mice in the nicking activity against uracil or 8-oxo-guanine (8-oxoG) as a substrate. Similar results were obtained in the extracts prepared from liver (data not shown). These data suggest that endoIII-like glycosylase activity is absent or reduced in the extracts of mNth1-defective homozygous mice.
Fig. 2. Nicking activity of mouse extracts against various substrates. Nicking activities in crude nuclear extracts derived from mNth1 (+/+, +/–, –/–) mouse lung tissues were assayed against 19mer (25mer for 8-oxoG) double-stranded DNA harboring T, Tg or urea, or single-strand DNA harboring U. The 5′-end of the substrate strand was labeled with [32P]. Cell extracts of 16 µg were used for T, Tg and 8-oxoG substrate, while 4 µg or 8 µg extracts were used for the urea or U substrates, respectively. As a control, the Tg substrate was treated with piperidine (lane 13) or with mNTH1 protein (lane 15). The urea substrate was treated with E.coli endoIV (lane 14) and the single-strand uracil substrate was treated with Ung and subsequently with piperidine (pip, lane 20). Lane 22 shows the migration of a 15mer labeled at the 5′ end.
Mouse phenotype and sensitivity against oxidative stress
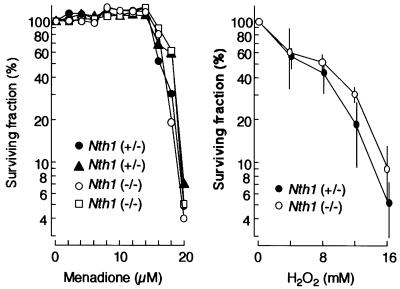
The targeted mNth1 allele segregated with Mendelian ratios, indicating that the inactivation of the gene does not interfere with embryonic development. mNth1 mutant mice looked healthy and showed no overtly abnormal phenotype, even though the oldest animals were now 16 months old. There was no visible change in aging and homozygous mutant mice were fertile. Figure 3 depicts the survival rate of mouse embryonic fibroblasts (MEFs) derived from mNth1 –/– and +/– mice embryos (15 days) after treatment of the cells with menadione or hydrogen peroxide. Both chemicals are known to produce reactive oxygen species, and yeast cells deficient in the yeast homologues of the nth gene are sensitive to these chemicals (Swanson et al., 1999). However, there was no apparent difference between the sensitivities of the cells derived from mNth1 –/– and +/– mice against these chemicals. These data suggest that absence of mNth1 does not cause cellular sensitivity to these agents in our assay.
Fig. 3. Sensitivity of MEFs derived from mNth1 –/– and +/– against oxidative stresses. Surviving fractions after treatment of the embryo cells with menadione (left panel) or hydrogen peroxide (right panel) were determined as described in Materials and methods.
Repair of Tg after X-ray irradiation of mice
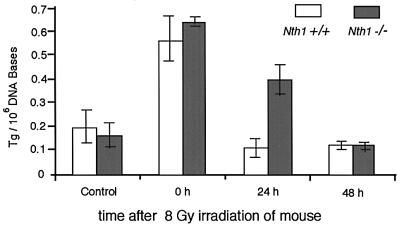
To examine whether Tg lesions are repaired in mNth1 –/– mice, wild-type and mutant animals were irradiated with 8 Gy X-rays. Total liver DNA isolated immediately, or 24 or 48 h after irradiation of intact mice, was analyzed for the Tg content by capillary electrophoresis with a laser-induced fluorescence detection system for antibodies raised against Tg (Le et al., 1998; Xing et al., 2001). Figure 4 depicts the time-dependent disappearance of Tg from mouse liver DNA. While the amount of Tg produced in wild-type mouse liver by X-ray irradiation decreased rapidly and reached the level of non-irradiated mice within one day after irradiation, the amount of Tg in the DNA of the mNth1 –/– mouse decreased more slowly and reached the control level only after 2 days. This suggests that there is still residual genome repair of Tg lesions in mNth1 –/– mice.
Fig. 4. The amount of Tg in the total genome DNA prepared from mouse liver. The amount of Tg/106 DNA bases was measured for wild-type and mNth1 –/– mice after 8 Gy irradiation.
Residual nicking activity
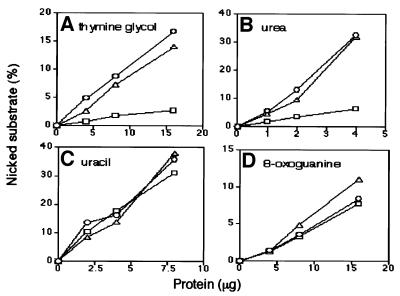
To explain the slow repair of Tg in mNth1 –/– mouse, we looked for residual activity in the mouse cell extracts. To detect any residual activity against Tg and urea in homozygous mutant cell extracts, the dependence of nicking activity on the amount of cell extract was analyzed. As shown in Figure 5, there is a low but detectable nicking activity for Tg and urea in the extracts of mNth1-defective homozygous mouse lung, while the extracts possess wild-type levels of nicking activity for uracil and 8-oxoG. The residual activity in the crude nuclear extracts of homozygous mutant mice is 17% for Tg and 27% for urea compared with wild-type extracts. We speculate that this novel glycosylase activity might substitute for the role of mNTH1 in the mNth1 –/– mouse. Therefore, we further analyzed the residual activity in the mutant mouse liver.
Fig. 5. Dependency of nicking activity on the amount of mouse extracts. The relative amount of nicked substrate was determined by PAGE after treatment of Tg (A), urea (B), uracil (C) and 8-oxoG (D) substrates with various protein concentrations of the extracts. The results for wild-type (circles), heterozygous mutant (triangles) and homozygous mutant mice (squares) are shown.
Nicking activity in the partially purified cell extracts
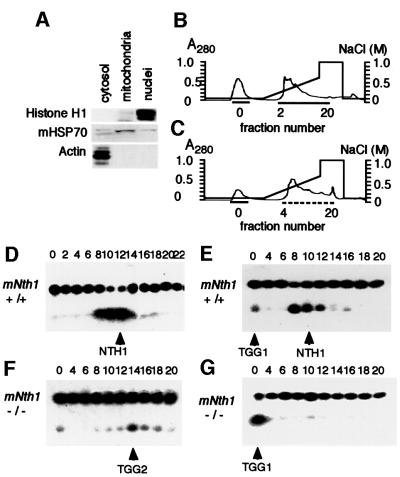
To identify residual activity in the mNth1 –/– mouse, we prepared nuclear and mitochondrial fractions from mNth1 +/+ and –/– mouse liver (Figure 6A) and subjected both extracts to heparin–agarose affinity chromatography. Heparin–agarose-binding (HB) fractions from both organelles of mNth1 –/– mouse liver possess nicking activity for Tg (data not shown). The HB fractions were further fractionated on UNO-S columns for nuclear extracts (Figure 6B) and for mitochondrial extracts (Figure 6C). Wild-type nuclear extracts yielded one clear peak of nuclease activity for Tg (Figure 6D). In the fractionated mitochondrial extracts of wild-type mouse, two peaks of nicking activity for Tg are detected (Figure 6E). The major nicking activity in the nuclear and mitochondrial extracts of wild-type mouse is absent in the extracts of mutant mouse, and, therefore, this is the activity of mNTH1. We designated the novel activity found in the flow-through fraction of mitochondrial extracts as TGG1 (thymine glycol glycosylase 1). In the nuclear extracts of mutant mouse, there are two peaks: one in the flow-through fraction and the other in the fractions around 14 (Figure 6F). The latter major nicking activity was designated as TGG2, while the former minor activity may be the same as that of TGG1, because, so far as analyzed, the biochemical characteristics of this activity coincide very well with those of TGG1. In the mitochondrial fractions of the mutant mouse extracts, only the activity corresponding to TGG1 remains (Figure 6G). These data suggest that TGG1 and TGG2 may be glycosylases for Tg in addition to mNTH1. Although each fraction does not contain an equal amount of protein and, therefore, the nicking activities shown in Figure 6 can not be quantitatively compared with each other, the data suggest that the TGG1 activity may be higher in the mutant mouse than in the wild-type mouse.
Fig. 6. Fractionation of nicking activity against Tg substrate in mNth1 +/+ and –/– mouse liver extracts on UNO-S column. The separation of nuclear and mitochondrial extracts was analyzed by western blotting using antibodies against representative proteins for each organelle (A). Chromatographic separations of proteins by UNO-S column were shown for nuclear (B) and mitochondrial (C) extracts. Nicking activities of UNO-S fractions to 5′-labeled-Tg oligos are shown for nuclear (D) and mitochondrial (E) extracts of wild-type mouse, and for nuclear (F) and mitochondrial (G) extracts of homozygous mutant mouse.
Characterization of TGG activities
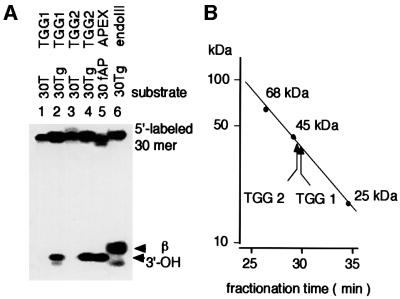
Since Figure 6 does not necessarily indicate that the novel activities are due to Tg-specific glycosylases, the nicking activities of each UNO-S fraction were compared with those of endoIII and human AP endonuclease (Figure 7A). TGG1 and TGG2 prepared from the mitochondrial and nuclear extracts of the mutant mice, respectively, introduced a nick at the site of Tg leaving 3′-OH. These data suggest that both fractions contain AP endonuclease activity besides glycosylase activity to Tg. The molecular weights for TGG1 and TGG2 were determined to be ∼40 kDa (Figure 7B), which is larger than that of mNTH1 (33.6 kDa).
Fig. 7. Determination of nicking activity and molecular weight of TGG glycosylases. (A) Nicking activities of TGG1 and TGG2 UNO-S fractions to Tg. (B) Estimation of molecular weight for TGG1 and TGG2 activities.
TGG1, a mitochondrial glycosylase for Tg
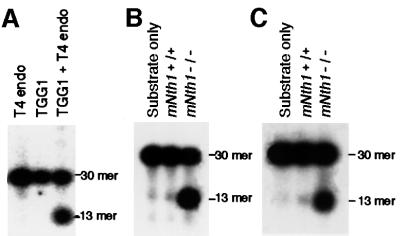
UNO-S fractions containing TGG1 activity were further purified by affinity chromatography on blue-Sepharose columns. TGG1 activity was found in the flow-through fractions (data not shown). The fraction for TGG1 no longer contains AP endonuclease activity and introduced a nick in the Tg substrate only after addition of T4 endonuclease (we used AP lyase activity of this enzyme) to the reaction mixture (Figure 8A). This suggests that TGG1 is a glycosylase for Tg without (or with an extremely weak) AP lyase activity, and the nicking activity may be due to a contaminating activity in the TGG1 fraction from UNO-S column chromatography. Figure 8B and C show comparisons of the glycosylase activity in T4 endonuclease-supplemented flow-through fractions of blue-Sepharose column chromatography between wild-type and homozygous mutant mouse liver for mitochondrial and nuclear extracts, respectively. There were low glycosylase activities in the mitochondrial and nuclear extracts of wild-type mouse liver, while a >20-fold increase in the activity was found in the extracts of mutant mice. The increased Tg-nicking activity in the flow-through fraction of nuclear extracts of mutant mouse may be TGG1, because its biochemical characteristics can not be discriminated from those of TGG1.
Fig. 8. Nicking activity of TGG1 in mitochondrial and TGG1-like activity in nuclear extracts. (A) Nicking activity of TGG1 after application of UNO-S fraction on blue-Sepharose column was obtained only by addition of T4 endonuclease. Comparison of the nicking activity prepared from 3 µg of the T4 endonuclease-supplemented flow-through fractions from blue-Sepharose column for mitochondrial extracts (B) and nuclear extracts (C) between wild-type and homozygous mutant mice.
TGG2 fraction contains novel Tg glycosylase
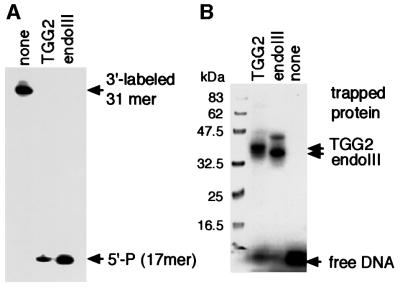
In the case of TGG2 activity from the nuclear extracts, UNO-S fractions with TGG2 activity were applied on a blue-Sepharose column, but the fractions with TGG2 activity still showed the nicking activity with 3′-OH, suggesting that the contaminating AP endonuclease activity could not be separated from TGG2 activity (data not shown). Therefore, we used 3′-labeled substrate containing Tg for a nicking assay. Figure 9A shows that TGG2 fraction prepared by UNO-S chromatography has a nicking activity like bacterial endoIII, an AP lyase activity at the site of Tg. We further confirmed the glycosylase activity to Tg by borohydride trapping assay (Figure 9B). The protein trapped to Tg substrate is slightly slower migrating than endoIII and roughly corresponds to the size determined by gel filtration (Figure 7B). Since trapped mOGG1, another glycosylase with AP lyase activity to 8-oxoG, migrates more slowly in the assay (data not shown), we think that the protein trapped to the Tg substrate is a novel glycosylase for Tg.
Fig. 9. Nicking activity of TGG2. (A) Incision products resulted from β-elimination. PAGE results of 3′-labeled 31mer Tg oligo after incubation with TGG2 fraction or E.coli endoIII are shown. (B) Borohydride trapping analysis of TGG2 fraction and endoIII for Tg oligo.
Discussion
Tg is a potentially lethal DNA lesion produced by ionizing radiation, chemical oxidants and during normal metabolism. The E.coli nth gene encodes the endoIII glycosylase, which initiates BER of Tg as well as other modified pyrimidine residues. In order to understand the biological impact of Tg and other pyrimidine lesions in mammals, we established a mouse mutated in the nth homologue mNth1 by gene targeting. Since the mNth1 gene is very closely located to a tumor suppressor gene, mTsc2, which is responsible for tuberous sclerosis in various organs (Sarker et al., 1998; Kobayashi et al., 2001), only the most conserved region of mNth1 gene was deleted without influencing expression of the Tsc2 gene (Figure 1). In a standard assay for nicking activity in crude nuclear extracts, most of the nicking activity for Tg and urea has disappeared in extracts of the homozygous mutant mouse (Figure 2), indicating that the endoIII homolog mNTH1 represents the major glycosylase against Tg in mouse. This is the first conclusion of this work.
In spite of the absence of the major glycosylase activity against Tg, a homozygous mNth1 mutant mouse was born and neither phenotypical abnormality nor abnormal fertility has been observed in mice over one and a half years of age. MEFs prepared from mNth1-mutated homozygous mouse embryos were not more sensitive to oxidative damage than those prepared from heterozygous mice (Figure 3). In microorganisms, a similar situation has been reported. Escherichia coli nth mutant cells are not more sensitive than wild-type cells (Cunningham and Weiss, 1985). Later, it was found that a functional redundancy of another glycosylase with AP lyase activity, endo nuclease VIII (endoVIII), is present and a double mutant defective in both glycosylases is highly sensitive to hydrogen peroxide (Melamede et al., 1994; Saito et al., 1997). In the yeast Saccharomyces cerevisiae, endoVIII is absent but there are two nth homologues, NTG1 and NTG2, which differ in substrate specificity and subcellular localization (Alseth et al., 1999; You et al., 1999). However, in human and mouse databases, only one nth homologue has so far been found (Aspinwall et al., 1997; Sarker et al., 1998).
We have measured the amount of Tg in genomic DNA from mouse liver after irradiation with 8 Gy of X-rays and followed the kinetics of repair (Figure 4). This result very clearly shows that there is a back-up repair system(s) for Tg in the mNth1-knockout mouse. The back-up repair system(s) functions more slowly than the repair initiated by mNTH1, but is nevertheless able to reduce the number of Tg to the control level. This suggests that the back-up repair is excision repair for the genome overall. The residual nicking activity in the crude nuclear extracts from the mNth1 homozygous mutant mouse (Figure 5) suggests the presence of another glycosylase(s) as a back-up for Tg repair. After two-step chromatography, we found evidence for at least two novel glycosylase activities in extracts from homozygous mutant mice (Figure 6). We have previously shown that mNTH1 has both nuclear and mitochondrial transport signals at the N-terminal region and the encoded protein is transported to either of the organelles (Takao et al., 1998). As expected, we found putative mNTH1 activity both in nuclear and mitochondrial extracts (Figure 6D and E). However, we identified at least two novel nicking activities for Tg in the mNth1 –/– mouse liver.
One novel glycosylase activity (TGG1) was identified in both mutant and wild-type cell extracts. TGG1 was found mainly in mitochondria of wild-type cells, while in mutant extracts, the nicking activity was significantly enhanced, and the activity found in the flow-through fraction of nuclear extract may be TGG1 as well. This suggests that TGG1 activity is enhanced possibly due to accumulation of oxidative DNA damage caused by the absence of mNTH1. It has been reported that repair of Tg was enhanced by pre-irradiation of human cells with X-rays (Le et al., 1998), suggesting that Tg repair may be inducible in human cells. Our data suggest that TGG1 may possess nuclear and mitochondrial transport signals such as mNTH1, and it is inducible. In contrast to mNTH1, TGG1 obtained after blue-Sepharose column chromatography did not show associated AP lyase activity. Although its AP lyase activity might be extremely weak or fragile, we think that this may be the first example of a monofunctional DNA glycosylase for Tg because we never detected any nicking activity in TGG1.
In contrast to the mitochondrial TGG1 fraction, the nuclear TGG2 fraction contains much more AP endonuclease activity, which has not been separated from Tg glycosylase activity yet. We found that the AP endonuclease activity in TGG2 fraction is partly EDTA resistant and anti-APEX antibody resistant, suggesting that AP endonuclease other than EDTA-sensitive APEX (HAP1) of exoIII-type is present in the fraction. Recently, it was reported that AP endonucleases of endoIV-type from E.coli and yeast possess a direct 5′-nicking activity to some pyrimidine base lesions (Ischenco and Saparbaev, 2002; Figure 2, lane 14). Therefore, the TGG2 fraction might contain an endoIV ortholog, but the nicking activity of the TGG2 fraction to 3′-labeled Tg substrate shown in Figure 9 indicates that the TGG2 fraction contains a DNA glycosylase for Tg, and it may possess AP lyase activity as well. Since TGG1 and TGG2 differ significantly in biochemical characteristics, we think that these are two major nicking activities for Tg in addition to mNTH1 in the mitochondria and nucleus of mouse liver cells, respectively. Furthermore, besides TGG1 and TGG2, we recently detected another nicking activity for Tg in the nuclear extracts of the mutant mouse (M.Takao, S.Kanno, K.Kobayashi, Q.-M.Zhang, S.Yonei, G.van der Horst and A.Yasui, manuscript in preparation). These data suggest that there are several back-up glycosylases for Tg in the nucleus of mouse liver cells.
Another possible back-up repair for Tg may be nucleotide excision repair (NER). It has been reported that Tg can be excised from DNA in vitro by NER, though much less efficiently than a UV lesion (Reardon et al., 1997). Since the XPC-HR23B complex of mammalian NER factor can recognize even a small bubble structure with damaged base (Sugasawa et al., 2001), it is possible that part of Tg can be repaired by NER in vivo as well. Furthermore, a replication stop at Tg may be overcome by translesion synthesis in mammalian cells. Data reported here, however, show that BER initiated by mNTH1 plays a major role in the repair of Tg, and there are back-up glycosylases not only in the nucleus but also in the mitochondria. Since mNTH1 accomplishes rapid removal of Tg, it may be important for repair in rapidly growing cells, for example, those in early developmental stages, although we have not yet found any abnormality in the mNth1 –/– homozygous mutant mouse. Furthermore, our data showed that disruption of mNth1 enhanced the back-up glycosylase activity especially for repair in mitochondria. Thus, our results underlined the importance of BER of oxidative pyrimidine base damage both in nuclei and mitochondria of mammalian cells.
Materials and methods
Generation of mNth1 targeting construct
Isogenic genomic mouse DNA was amplified from Ola129-derived E14 ES cell DNA by PCR using LA-taq DNA polymerase (Takara). A 8904 bp fragment of genomic DNA containing the whole mNth1 (mNthl1) gene was isolated by PCR starting at the first cytosine base in the XhoI site in the third intron of the Tsc2 gene. The primer sets were 5′-primer, 5′-CTCGAGGCGAACCTCCAGAATCCTAGGTCAAGGATA and 3′-primer, 5′-GGTTAGCACCAGCTCAGATGCACAGTGTGGGCA GAT. SalI sites were introduced at the 5′ and 3′ ends of the fragments. A neomycin-selectable marker (PGK-neo) was introduced in the same direction as mNth1 expression to replace the fragment between nucleotides 3257 (NheI site) and 5909 (NsiI site) harboring exon 3 and exon 4 of the mNth1 gene. After the three fragments were ligated, the product was digested with SalI and introduced into the XhoI sites of the λ phage vector, λDashII2TK254 (a generous gift from Dr T.Tsuzuki, Kyushu University), harboring two thymidine kinase genes for counter-selection. The recombinant λ DNA was purified and used for transfection into ES cells.
Disruption of mNTH1 in mouse ES cells and generation of mNTH1 knockout mice
As the gene disruption procedure has been reported already (van der Horst et al., 1999), it is only briefly explained here. The targeting vector was transfected into E14 cells [1 × 107 in 400 µl phosphate-buffered saline (PBS)] by electroporation. Electroporated cells were subjected to selection by addition of 200 µg/ml G418 (Geneticin, Gibco) and 0.2 µM fialuridine for counter-selection against randomly integrated DNA. After 9 days, colonies were expanded in 24-well dishes. Duplicate dishes were used for cryopreservation and genotyping. Targeting frequency was ∼5%. After karyotyping and selection for cell lines with 40 chromosomes, targeted ES cells were injected into C57BL/6 blastocysts by standard procedure (Bradley, 1987). Chimeric males were mated with C57BL/6 females and transmission of E-14-derived germ cells was identified by an agouti coat color in the offspring.
DNA and RNA analysis
Genotyping of ES cells or tail DNA was performed by Southern blot analysis using tail DNA digested with EcoRI and probed with a 597-bp DNA fragment prepared by PCR and located outside the targeting construct as shown in Figure 1. To analyze the expression of the mNth1, mGa3dh and mTsc2 genes, the following primers were used for RT–PCR (where Fw and Rev are forward and reverse primers, respectively): amp-1 (exon 2): Fw: GCCACAGGCCCGTGAGACATCCACGGAGAA, Rev: GCACTTGCATCATAGCAGTGCTCG; amp-2 (exons 4–6); Fw: GGGACATCCCTGCTTCCGTGGCTG, Rev: TGAGGCAAGCCTG ACATCGAGGAT; amp-3 (exon 5 and 6); Fw: GTGGACACA CATGTGCACAGAATAG, Rev: TTAGAGATCCTGGGCAGCAG GGC; Tsc2: Fw: GGGCACCATTAAGCAGGGCCAGTTC, Rev: CTGGCGGAGACGCTTAATGTGGCGG; Ga3dh: Fw: CCTTGGCTC CTGCATGTGCCCAAT, Rev: CATGTAGGCCATGAGGTCCACCAC. PCR was monitored by a real-time PCR-measuring system (Light Cycler, Roche).
Sensitivity analysis of mouse embryonic cells against oxidative stress
MEFs of litter-mates obtained by crossing a mNth1 –/– mouse and a mNth1 +/– mouse were analyzed. The embryo (15 days) was dissected and cultured for 4 days. Cells were further sub-cultured in 96-well plates with ∼1000 cells per well for 24 h. Menadione was added to the culture medium at the indicated concentration and cells were cultivated for 24 h, while hydrogen peroxide was added to the medium for 15 min followed by washing and re-culturing the cells with fresh medium for 24 h. Surviving cells were counted by the Cell Counting Kit-8 (Dojindo Molecular Technologies, Japan), which measures the amount of formazan produced from a tetrazolium derivative, 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt, by cellular dehydrogenase activity.
X-ray irradiation of mice and measurement of Tg in mouse liver genomic DNA
Mice were irradiated through an aluminum plate (1 mm depth) with 1.6 Gy/min by an X-ray irradiation apparatus (Hitachi MBR1520R). Total genomic DNA was prepared from the livers of the mice using GenElute (Sigma-Aldrich). To determine the amount of Tg, we used a method described previously (Le et al., 1998; Xing et al., 2001) with minor modifications. Each DNA sample (2 µl of 100 µg/ml solution) was added to 13.3 µl of Tris–glycine buffer (25 mM Tris and 192 mM glycine pH 8.3). The DNA was denatured by heating the sample at 95°C for 5 min followed by cooling on ice. Two milliliters of anti-Tg antibody (kindly provided by Dr S.A.Leadon, University of North Carolina School of Medicine) and 2 µl of 5 mg/ml human IgG (Sigma-Aldrich) were then added to the DNA sample and the mixture was incubated on ice for 10 min. Fluorescent-labeled AlexaFluor 546 F(ab′)2 fragment of goat anti-mouse IgG (H+L) (0.7 µl, 40 µg/ml, Molecular Probes, Eugene, OR) was added, and the mixture was incubated at room temperature for a further 3 h. Control mixtures containing all the reagents except the DNA were prepared similarly. Antibody–DNA complexes were separated from the antibodies and analyzed by capillary electrophoresis with laser-induced fluorescence (CE–LIF) detection, using a laboratory-built CE–LIF instrument with a fused silica capillary. The samples were injected into the capillary and separated by electrophoresis using the Tris–glycine buffer as running buffer. A computer was used to control the instrument and to collect data. Data obtained from three independent measurements were shown with standard deviations.
Preparation of crude nuclear cell extracts
Crude nuclear cell extracts were prepared from lungs of +/+, +/– and –/– mNth1 mice by homogenization in buffer A [10 mM HEPES–KOH pH 7.7, 0.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol (DTT)]. The pellet obtained by centrifugation (2000 g, 10 min) of the extracts was suspended in buffer B (20 mM HEPES–KOH pH 7.7, 0.5 mM MgCl2, 420 mM NaCl, 1 mM DTT, 0.2 mM PMSF, 0.2 mM EDTA, 25% glycerol) and centrifuged again (14 000 g, 10 min). The supernatant (nuclear extract) was dialyzed against buffer C (40 mM HEPES–KOH pH 7.7, 50 mM KCl, 2 mM DTT). After 3 h of dialysis, extracts were frozen in liquid nitrogen and kept at –80°C.
In vitro assay of nicking activity in crude cell extracts
5′-32P-labeled oligos harboring various modified bases (except for uracil, see Table I) were annealed with their complementary strands. Double- or single-stranded substrate (0.015 pmol) was mixed with cell extracts (16 µg for T, Tg and 8-oxo-G, 4 µg for urea and 8 µg for uracil reactions) in buffer D [40 mM HEPES–KOH, 100 mM KCl, 0.5 mM EDTA, 0.5 mM DTT, 0.2 mg/ml bovine serum albumin (BSA)] and incubated for 1 h at 37°C. After phenol extraction of the reaction mixture, DNA was precipitated and analyzed by PAGE. For uracil, precipitated DNA was treated with 10% piperidine (90°C for 30 min) prior to PAGE analysis to incise abasic sites. For control experiments with purified proteins, recombinant mNTH1 (10 ng), hOGG1 (10 ng), endoIV (2 ng) or Ung (1 U) were used in a reaction with 0.1 pmol of the substrate DNA. Buffer E (20 mM HEPES–KOH pH 8.0, 0.25 mM EDTA, 0.25 mM DTT, 50 mM KCl, 0.1 mg/ml BSA) was used for mNTHL1. Buffer F (50 mM Tris–HCl pH 7.5), buffer G (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 50 mM NaCl) and buffer H (20 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DTT) were used for hOGG1, for endoIV and for Ung, respectively. mNTH, hOGG1 and endoV were prepared as described previously (Sarker et al., 1998; Masaoka et al., 1999; Asagoshi et al., 2000). Escherichia coli uracil glycosylase (Ung) and T4 polynucleotide kinase were purchased from Amersham Pharmacia and New England BioLabs, respectively.
Table I. Oligonucleotides used in the experiments.
| Name | Sequence |
|---|---|
| 19T | 5′-ACAGACGCCATCAACCAGG-3′ |
| 19Tg | 5′-ACAGACGCCATgCAACCAGG-3′ |
| 19Urea | 5′-ACAGACGCCAUreaCAACCAGG-3′ |
| 19U | 5′-ACAGACGCCAUCAACCAGG-3′ |
| 19comp | 5′-CCTGGTTGATCGCGTCTGT-3′ |
| 25oxoG | 5′-CATCGATAGCATCCToxoGCCTTCTCTC-3′ |
| 25comp | 5′-GAGAGAAGGCAGGATGCTATCGATG-3′ |
| 15mer | 5′-CATCGATAGCATCCT-3′ |
| 30T | 5′-CTCGTCAGCATCTTCATCATACAGTCAGTG-3′ |
| 30Tg | 5′-CTCGTCAGCATCTTgCATCATACAGTCAGTG-3′ |
| 30fAP | 5′-CTCGTCAGCATCTfAPCATCATACAGTCAGTG-3′ |
| 30comp | 5′-CACTGACTGTATGATGAAGATGCTGACGAG-3′ |
Preparation of oligos containing Tg, urea and 8-oxoG has been reported previously (Asagoshi et al., 2000). 30mer Tg oligo and 30mer f-AP oligo containing an AP site analog [3-hydroxy-2-(hydroxylmethyl)-tetrahydrofyran] were generous gift from Dr S.Iwai (Tokyo University) and prepared as reported previously (Kanno et al., 1999).
Partial purification of Tg glycosylase activity from liver extracts and in vitro assay
Nuclear and mitochondrial fractions were prepared from mouse liver by the methods reported previously (Pedersen et al., 1978; Croteau et al., 1997) and homogenized in extraction buffer J (50 mM Tris–HCl pH 7.5, 0.1% NP-40, 2 mM DTT, protease inhibitor cocktail purchased from Boehringer) with 0.3 M NaCl. Nuclear, cytosol and mitochondrial fractions were identified by antibodies against representative proteins of each fraction, histone H1 (Santa Cruz), actin (Santa Cruz) and mitochondrial heat shock protein 70 (Affinity BioReagents), respectively. After centrifugation (100 000 g, 30 min), supernatants were recovered and dialyzed against buffer K (50 mM Tris–HCl pH 7.5, 2 mM DTT, 1 mM EDTA, 10% glycerol) with 0.1 M NaCl overnight. The dialyzed extracts were centrifuged (100 000 g, 30 min) and applied to a heparin–agarose column (Sigma-Aldrich) equilibrated with buffer J with 0.1 M NaCl. After washing the column with buffer K with 0.1 M NaCl, a HB fraction was eluted by applying buffer K with 0.8 M NaCl and dialyzed against buffer K with 0.1 M NaCl. The dialyzed sample was applied to a UNO-S column in an HPLC system (Bio–Rad) and eluted with a linear gradient of 0.1–0.6 M NaCl. Fractions with nicking activity were pooled and dialyzed against buffer K with 0.1 M NaCl and applied further onto a blue-Sepharose column (HiTrap Blue HP, Amersham Pharmacia Biotech). Fractions were eluted by a step gradient of buffer K containing 0.05, 0.5, 1.0 and 2.0 M NaCl. A Tg substrate (0.015 pmol of double-stranded 30 Tg in Table I) was used for the nicking assay in a 10 µl reaction mixture containing EDTA buffer (20 mM HEPES–KOH pH 7.5, 1 mM EDTA, 1 mM DTT, 50 mM NaCl). Protein (3 µg) from the blue-Sepharose column fraction was mixed with substrate DNA for 60 min at 37°C for one reaction. Escherichia coli endoIII was prepared from E.coli cells by the method reported previously (Katcher and Wallace, 1983).
Borohydride trapping assay
Labeled Tg oligo was mixed with column fraction in 50 mM sodium phosphate buffer (pH 7.5) containing 10 mM EDTA and 100 mM NaBH4. After incubation at 37°C for 5 min, the reaction was stopped by boiling with SDS sample buffer and analyzed by SDS–PAGE.
Determination of molecular weight
UNO-S column fractions containing DNA glycosylase activity for Tg were applied to a Superose 12 HR 10/30 column (Amersham Pharmacia Biotech) in buffer K with 0.15 M NaCl and eluted at 0.5 ml/min. The molecular weight of the nicking activity was calibrated with 68 kDa BSA, 45 kDa hen egg albumin and 25 kDa chymotrypsinogen A.
Acknowledgments
Acknowledgements
We thank Dr Shirley McCready for editing this paper and Drs T.Tuzuki and S.Iwai for providing us with phage vector for gene targeting construct and with DNA substrate containing Tg, respectively. This work was supported by Grant-in-Aid for Scientific Research on Priority Areas no. 12143201 and no. 13480162 (to A.Y.), the Netherlands Organization for Scientific Research (NWO; TF004), the Dutch Cancer Foundation (EUR 98-1774) and the NIH (AG17242-02).
References
- Alseth I., Eide,L., Pirovano,M., Rognes,T., Seeberg,E. and Bjoras,M. (1999) The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol., 19, 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames B.N. and Shigenaga,M.K. (1992) Oxidants are a major contributor to aging. Ann. N. Y. Acad. Sci., 663, 85–96. [DOI] [PubMed] [Google Scholar]
- Asagoshi K., Odawara,H., Nakano,H., Miyano,T., Terato,H., Ohyama,Y., Seki,S. and Ide,H. (2000) Comparison of substrate specificities of Escherichia coli endonuclease III and its mouse homologue (mNTH1) using defined oligonucleotide substrates. Biochemistry, 39, 11389–11398. [DOI] [PubMed] [Google Scholar]
- Aspinwall R. et al. (1997) Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl Acad. Sci. USA, 94, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V. and Verly,W.G. (1987) Escherichia coli endonuclease III is not an endonuclease but a β-elimination catalyst. Biochem. J., 242, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A. (1987) Production and analysis of chimaeric mice. In Robertson,E.J. (ed.) Teratocarcinomas and Embryonic Stem Cells, A Practical Approach. IRL press, Oxford, UK, pp. 113–151.
- Breimer L.H. (1990) Molecular mechanisms of oxygen radical carcinogenesis and mutagenesis: the role of DNA damage. Mol. Carcinog., 3, 188–197. [DOI] [PubMed] [Google Scholar]
- Cerutti P.A. (1985) Pro-oxidant states and tumor promotion. Science, 227, 375–381. [DOI] [PubMed] [Google Scholar]
- Croteau D.L., ap Rhys,C.M.J., Hudson,E.K., Dianov,G.L., Hansford, R.G. and Bohr,V.A. (1997) An oxidative damage-specific endonuclease from rat liver mitochondria. J. Biol. Chem., 272, 27338–27344. [DOI] [PubMed] [Google Scholar]
- Cunningham R.P. and Weiss,B. (1985) Endonuclease III (nth) mutants of E. coli. Proc. Natl Acad. Sci. USA, 82, 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B. and Linn,S. (1980) DNA N-glycosylase and UV repair. Nature, 287, 203–208. [DOI] [PubMed] [Google Scholar]
- Hatahet Z. and Wallace,S.S. (1998) Translesion DNA synthesis. In Nickoloff,J.A. and Hoekstra,M.F. (eds) DNA Damage and Repair. Vol. 1. Humana Press, Totowa, NJ, pp. 229–262.
- Ischenco A.A. and Saparbaev,M.K. (2002) Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature, 415, 183–187. [DOI] [PubMed] [Google Scholar]
- Kanno S., Iwai,S., Takao,M. and Yasui,A. (1999) Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage. Nucleic Acids Res., 27, 3096–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao J.Y., Goljer,I., Phan,T.A. and Bolton,P.H. (1993) Characterization of the effects of a thymine glycol residue on the structure, dynamics, and stability of duplex DNA by NMR. J. Biol. Chem., 268, 17787–17793. [PubMed] [Google Scholar]
- Katcher H.L. and Wallace,S.S. (1983) Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry, 22, 4071–4081. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Minowa,O., Sugitani,Y., Takai,S., Mitani,H., Kobayashi,E., Noda,T. and Hino,O. (2001) A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc. Natl Acad. Sci. USA, 98, 8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le X.C., Xing,J.Z., Lee,J., Leadon,S.A. and Weinfeld,M. (1998) Inducible repair of thymine glycol detected by an ultrasensitive assay for DNA damage. Science, 280, 1066–1069. [DOI] [PubMed] [Google Scholar]
- Masaoka A., Terato,H., Kobayashi,M., Honjo,A., Ohyama,Y. and Ide,H. (1999) Enzymatic repair of 5-formyluracil. Excision of 5-formyluracil site-specifically incorporated into oligonucleotide substrates by AlkA protein (Escherichia coli 3-methyladenine DNA glycosylase II). J. Biol. Chem., 274, 25136–25143. [DOI] [PubMed] [Google Scholar]
- Melamede R.J., Hatahet,Z., Kow,Y., Ide,H. and Wallace,S.S. (1994) Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry, 33, 1255–1264. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1998) The ‘GO’ system in Escherichia coli. In Nickoloff,J.A. and Hoekstra,M.F. (eds) DNA Damage and Repair. Vol. 1. Humana Press, Totowa, NJ, pp. 97–105.
- Pedersen P.L., Greenawalt,J.W., Reynafarje,B., Hullihen,J., Decker, G.L., Soper,J.W. and Bustamente,E. (1978) Preparation and characterization of mitochondria and sub-mitochondrial particles of rat liver and liver-derived tissues. Methods Cell Biol., 20, 411–481. [DOI] [PubMed] [Google Scholar]
- Reardon J.T., Bessho,T., Kung,H.C., Bolton,P.H. and Sancar,A. (1997) In vitro repair of oxidative DNA damage by human nucleotide excision system: possible explanation for neurodegeneration in Xeroderma pigmentosum patients. Proc. Natl Acad. Sci. USA, 94, 9463–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Uraki,F., Nakajima,S., Asaeda,A., Ono,K., Kubo,K. and Yamamoto,K. (1997) Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J. Bacteriol., 179, 3783–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker A.H. et al. (1998) Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J. Mol. Biol., 282, 761–774. [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Okamoto,T., Shimizu,C., Iwai,S. and Hanaoka,F. (2001) A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev., 15, 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R.L., Morey,N.J., Doetsch,P.W. and Jinks-Robertson,S.J. (1999) Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 2929–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M., Aburatani,H., Kobayashi,K. and Yasui,A. (1998) Mitochondrial targeting of human DNA glycosylases for repair of oxidative damage. Nucleic Acids Res., 26, 2917–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst G.T.J. et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature, 398, 627–630. [DOI] [PubMed] [Google Scholar]
- Wallace D.C. (1995) Mitochondrial DNA mutations in human disease and aging. In Esser,K. and Martin,G.M. (eds) Molecular Aspects of Aging. Wiley, Chichester, England, pp. 163–178.
- Wilson D.M. III, Engelward,B.P. and Samson,L. (1998) Prokaryotic base excision repair. In Nickoloff,J.A. and Hoekstra,M.F. (eds) DNA Damage and Repair. Vol. 1. Humana Press, Totowa, NJ, 29–64.
- Wood R.D., Mitchel,M., Sgouros,J. and Lindahl,T. (2001) Science, 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- Xing J.Z., Carnelly,T., Lee,J., Watson,W.P., Booth,E., Weinfeld,M. and Le,X.C. (2001) Assay for DNA damage using immunochemical recognition and capillary electrophoresis. In Mitchelson,K.R. and Cheng,J. (eds) Methods in Molecular Biology. Vol. 162. Capillary Electrophoresis of Nucleic Acids. Vol. 1. Introduction to the Capillary Electrophoresis of Nucleic Acids. Humana Press, Totowa, NJ, pp. 419–428. [DOI] [PubMed]
- You H.J. et al. (1999) Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry, 38, 11298–11306. [DOI] [PubMed] [Google Scholar]