Abstract
The Fanconi anaemia (FA) nuclear complex (composed of the FA proteins A, C, G and F) is essential for protection against chromosome breakage. It activates the downstream protein FANCD2 by monoubiquitylation; this then forges an association with the BRCA1 protein at sites of DNA damage. Here we show that the recently identified FANCE protein is part of this nuclear complex, binding both FANCC and FANCD2. Indeed, FANCE is required for the nuclear accumulation of FANCC and provides a critical bridge between the FA complex and FANCD2. Disease-associated FANCC mutants do not bind to FANCE, cannot accumulate in the nucleus and are unable to prevent chromosome breakage.
Keywords: chromosome breakage/FANCE/Fanconi anaemia/nuclear complex assembly
Introduction
Genetic predisposition to cancer is commonly precipitated by mutations in genes that result in chromosome breakage. Studies of the inherited chromosomal instability diseases have led to the discovery of some of these genes, and the characterization of their products has profoundly contributed to the understanding of pathways that maintain genetic integrity (Patel et al., 1998; Moynahan et al., 1999; Scully and Livingston, 2000; Kinzler and Vogelstein, 2001). Fanconi anaemia (FA) is an autosomal recessive chromosomal breakage disease that results in developmental abnormalities, growth retardation, bone marrow failure and a marked predisposition to cancer (Auerbach et al., 2001; Joenje and Patel, 2001). The spontaneous chromosome breakage seen in FA cells is greatly potentiated upon exposure to agents that cross-link DNA (Auerbach and Wolman, 1976; Ishida and Buchwald, 1982). However, despite the cloning of the majority of the genes mutated in this disease, their precise function has yet to be elucidated. Recent work has pointed to their participation in a common DNA damage response pathway (Grompe and D’Andrea, 2001).
Currently, eight complementation groups define genes that are mutated in this disease (Joenje et al., 1997), and the recent cloning and characterization of five of them (FANCA, C, G, F and D2) indicates that they all participate in a common pathway that prevents chromosome breakage. The FANCA, C, G and F genes code for proteins that, although conserved in lower vertebrates, do not possess any clearly identifiable motifs or domains (Joenje and Patel, 2001). These four proteins are components of a nuclear complex (Kupfer et al., 1997; Garcia-Higuera et al., 1999; de Winter et al., 2000b) that is required for the monoubiquitylation of a fifth Fanconi protein, FANCD2 (Garcia-Higuera et al., 2001; Timmers et al., 2001). The importance of this modification is vividly illustrated by its induction upon DNA damage, cell cycle progression, and its abrogation in most FA cell lines. A clue as to the significance of this modification has been demonstrated by the localization of the FANCD2 protein, where there is a tight correlation between this modification and its targeting to discrete nuclear foci (Garcia-Higuera et al., 2001). Although much still needs to learned about the nature of these foci, they appear to be sites of DNA damage and the place where the D2 protein associates with the BRCA1 protein, a molecule well known to be implicated in the cellular DNA damage response.
Whilst the above model provides a framework of the FA pathway, many questions remain to be answered. For example, the precise nature of the DNA damage response pathway in FA cells remains unclear. Indeed, the exact composition and mode of assembly of the nuclear complex are unknown. At least one of its components, FANCC, appears to be present predominantly in the cytoplasm (Yamashita et al., 1994; Youssoufian, 1994; Hoatlin et al., 1998; Pang et al., 2001). This finding has stimulated many studies attempting to define a role for this protein in the cytoplasmic compartment (Kruyt et al., 1998; Pang et al., 2000; Cumming et al., 2001). However, FANCC must accumulate in the nucleus in order for the FA complex to form. Indeed, the total exclusion of FANCC from this compartment in cell lines with mutations in FANCB and FANCE suggests that these FA proteins are required for this crucial step (Yamashita et al., 1998). Recent data have suggested that the FA complex could be a monoubiquitin E3 ligase, whose key substrate is FANCD2. However, to date, none of the cloned FA proteins possesses either HECT or RING finger domains, both of which are key signature motifs in proteins that are implicated in terminal ubiquitin transfer (Weissman, 2001). Importantly, it is not known whether the substrate FANCD2 protein is physically linked to the putative nuclear complex, and whether this is an essential prerequisite for its subsequent ubiquitylation.
We show here that the recently cloned FANCE gene product is a nuclear protein and is an integral component of the FA nuclear complex. FANCE functions to target cytoplasmic FANCC to the nucleus as a result of a direct interaction between the two molecules. Furthermore, disease-associated FANCC missense mutants are defective at binding FANCE and are unable to accumulate in the nucleus. In addition, FANCE binds to the FANCD2 protein, thereby providing a necessary link between the potential modifying nuclear complex and its likely substrate. These studies not only intimate how the FA nuclear complex is assembled, but also how it may participate in the activation of FANCD2.
Results
FANCE is a nuclear protein and is part of the FA complex
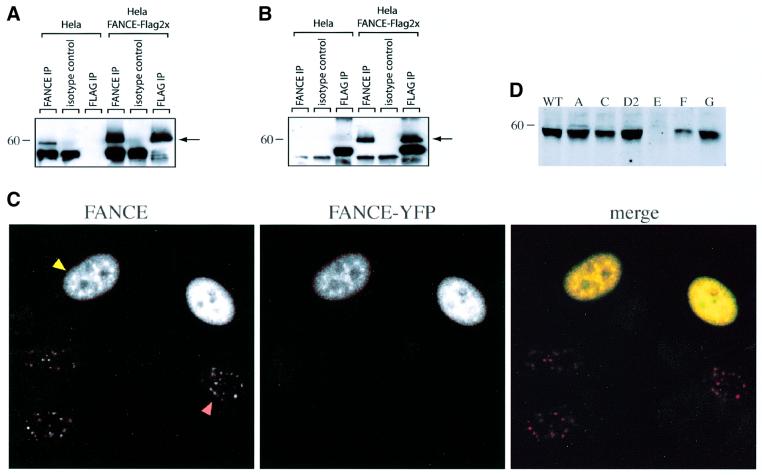
The FANCE gene has recently been cloned and is predicted to code for a 58 kDa nuclear protein. Like the other FA proteins, the putative amino acid sequence of FANCE does not contain any known motifs except for a predicted bipartite nuclear localization signal (de Winter et al., 2000a). Rabbit antiserum was raised to recombinant full-length FANCE protein expressed and purified from baculovirus-infected cells. Affinity-purified antiserum was tested for activity and clearly detected a 58 kDa protein in untransfected HeLa cells; as a positive control, it also recognized Flag epitope-tagged FANCE in a stably transfected HeLa cell line (Figure 1A and B). This antiserum was used to define the localization of FANCE. HeLa cells expressing transiently transfected FANCE tagged at the C-terminus with yellow fluorescent protein (FANCE–YFP) show a clear nuclear YFP signal concentrated in foci overlapping with those identified by the antiserum. Untransfected cells show a signal concentrated in similar nuclear foci, indicating the presence of the endogenous protein (Figure 1C). The nature of these foci remains to be defined but they are present in all HeLa cell nuclei and do not change visibly upon exposure to DNA-damaging agents (data not shown). Immunoblot analysis confirms that the FANCE protein is present in cell lines from the A, C, D, F and G complementation groups, but is absent in the cell line known to be from the E complementation group (Figure 1D).
Fig. 1. FANCE is a 58 kDa protein and is localized to nuclear foci. (A and B) Detection and analysis of FANCE protein, either immunoprecipitated with FANCE antiserum or with Flag monoclonal, then blotted with the FANCE antiserum (A) or with Flag monoclonal (B). The FANCE antiserum specifically immunoprecipitates and western blots endogenous FANCE, as well as recombinant FANCE with a C-terminal 2×Flag epitope (FANCE-2×Flag) expressed in HeLa cells. The arrows mark the migration of FANCE and FANCE-2×Flag. (C) Subcellular localization of endogenous FANCE as well as transfected FANCE–YFP, as detected by indirect immunofluoresence. FANCE antiserum detects FANCE–YFP transiently expressed in transfected (yellow arrow), as well as native protein in foci of untransfected (red arrow) HeLa cells. The preimmune serum and second layer (Cy5 anti-rabbit γ chain) gave no signal (data not shown). (D) Whole-cell lysate western blot analysis for FANCE expression with FANCE antiserum in nuclear extract from cell lines comprising the A, C, D2, E, F and G complementation groups. FANCE is present in all groups except, as expected, in the FANCE complementation group.
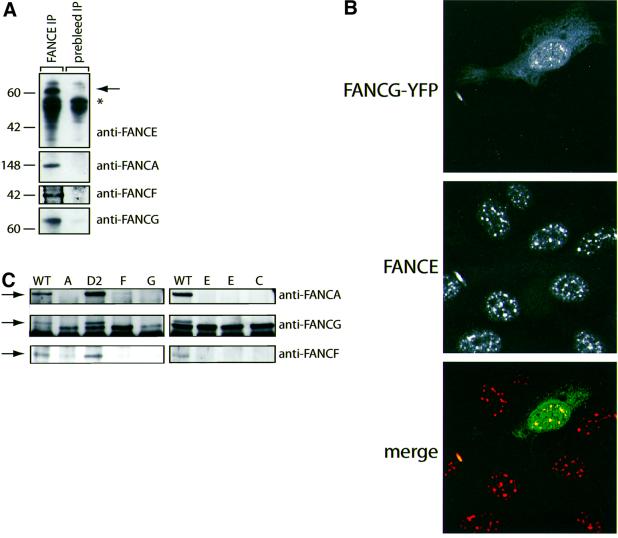
Previous studies have postulated that the FANCE protein is required for the stability of the FA nuclear complex, as this complex is unstable in the E complementation cell line (Yamashita et al., 1998; de Winter et al., 2000b). Consistent with this, FANCA, FANCG and FANCF could all be co-purified with FANCE (Figure 2A). Having shown that the FANCE protein is concentrated in nuclear foci, we then asked whether a well-characterized FA complex protein, FANCG (Garcia-Higuera et al., 1999; Waisfisz et al., 1999a), also co-localizes to these sites. HeLa cells expressing transiently transfected FANCG–YFP gave a nuclear signal that was coincidental with the FANCE antiserum, thereby indicating that FANCE co-localizes in foci with FANCG–YFP (Figure 2B). We next looked to see whether the interactions between FANCE and other FA complex proteins are retained in cell lines from the various FA complementation groups. These interactions are completely abolished when FANCE was purified from cell lines belonging to the A, C, F and G complementation groups (Figure 2C). As expected, in the FANCD2 complementation cell line, all these interactions are preserved (Yamashita et al., 1998; de Winter et al., 2000b). Collectively, these experiments show that FANCE is a nuclear protein that is an essential component of the FA nuclear complex.
Fig. 2. FANCE is part of the FA nuclear complex. (A) Western blot analysis of HeLa nuclear extract immunoprecipitated with anti-FANCE rabbit antiserum and protein A beads. Samples were western blotted with rabbit anti-FANCE, -FANCA, -FANCF and -FANCG antiserum. FANCE co-purifies with FANCA, FANCF and FANCG (the asterisk marks the presence of rabbit γ chain). (B) Co-localization in nuclear foci of transiently transfected FA complex protein FANCG–YFP (green) with endogenous FANCE (red) in HeLa cells. Recombinant FANCG–YFP can be seen to localize to the cytoplasm as well as nuclear foci; in contrast, endogenous FANCE is concentrated in nuclear foci. Upon merging of the layers, the FANCE foci co-localize with the signal from transfected FANCG–YFP. FANCE was detected with FANCE antiserum followed by Cy5 anti-rabbit γ chain. (C) FANCE immunoprecipitations in cell lines from the A, C, D2, E, F and G complementation groups were blotted with anti-FANCA, FANCG and FANCF antisera. The interactions between FA complex proteins and FANCE are disrupted in all except the D2 complementation group.
FANCE promotes the nuclear accumulation of FANCC
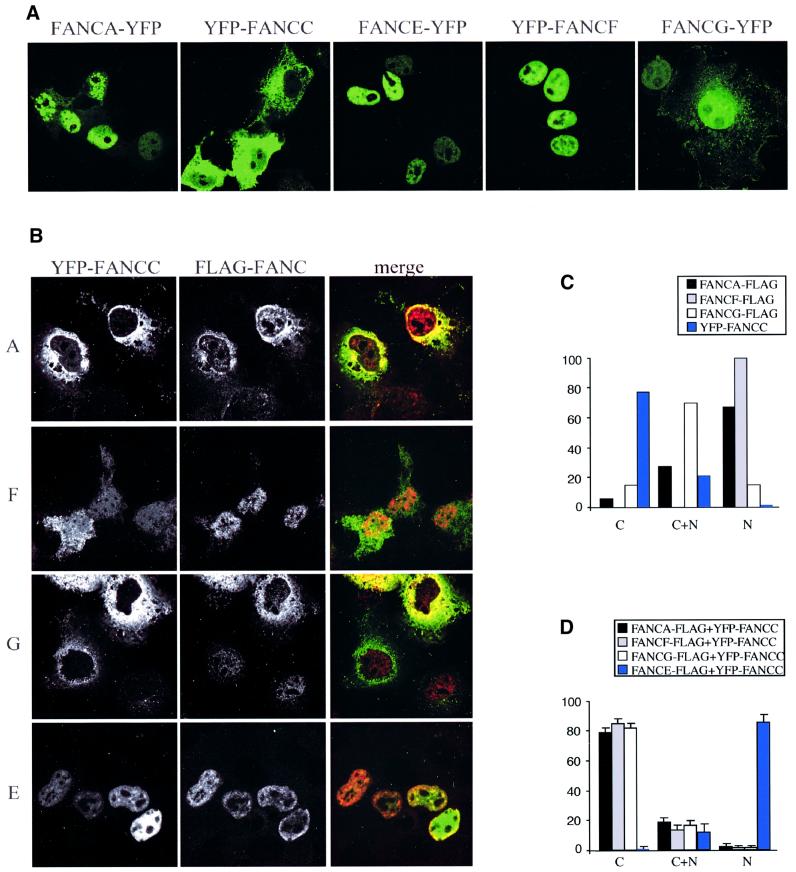
The FA nuclear complex is required for the prevention of chromosome breakage in normal cells. For this complex to form, all the components have to be present in the nucleus, yet FANCC has been shown to be predominantly cytoplasmic (Yamashita et al., 1994; Youssoufian, 1994; Hoatlin et al., 1998). We sought to determine whether YFP-tagged FA complex proteins were capable of entering into the nucleus when transiently expressed in COS cells. This cell line was chosen for the ease with which it can be transfected with two individual expression constructs. All of these proteins, when individually expressed, can accumulate in the nuclear compartment, except for FANCC, which is mainly cytoplasmic (Figure 3A and C). We next investigated whether any of the complex proteins, when co-expressed with FANCC, would enable FANCC to accumulate in the nucleus. Strikingly, the co-expression of only FANCE was sufficient to target FANCC to the nucleus (Figure 3B and D). Indeed, in cells expressing FANCC and FANCE, most of the FANCC was nuclear (Figure 3C and Supplementary data available at The EMBO Journal Online).
Fig. 3. FANCE is essential for the nuclear accumulation of FANCC. (A) Expression of transfected FA complex proteins (FANCA, FANCC, FANCF, FANCG and FANCE) with either N- or C-terminal YFP tag in COS cells. All the FA complex proteins show strong nuclear accumulation except for YFP–FANCC, which is predominantly in the cytoplasmic compartment. (B) Co-expression of YFP–FANCC (green) with C-terminal Flag-tagged FA complex proteins (FANCA, FANCF, FANCG and FANCE, all in red) in COS cells. Only co-expression of FANCE leads to the nuclear accumulation of FANCC. (C) One hundred transfected cells were scored blind for the localization of the Flag tag as well as YFP–FANCC to cytoplasmic (C), cytoplasmic/nuclear (C+N) and nuclear (N) compartments in transfected COS cells. Only FANCC is predominantly in the cytoplasmic compartment. (D) Localization of YFP–FANCC when co-expressed with Flag-tagged FANCA, FANCF, FANCG or FANCE. The scores are expressed as an average from three independent observers. Only FANCE-Flag co-expression leads to FANCC nuclear accumulation.
Disease-associated FANCC mutants are defective in FANCE binding and nuclear accumulation
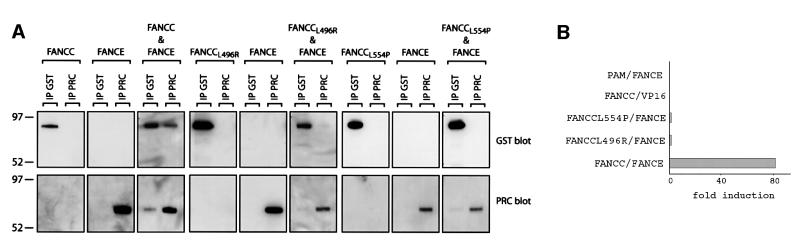
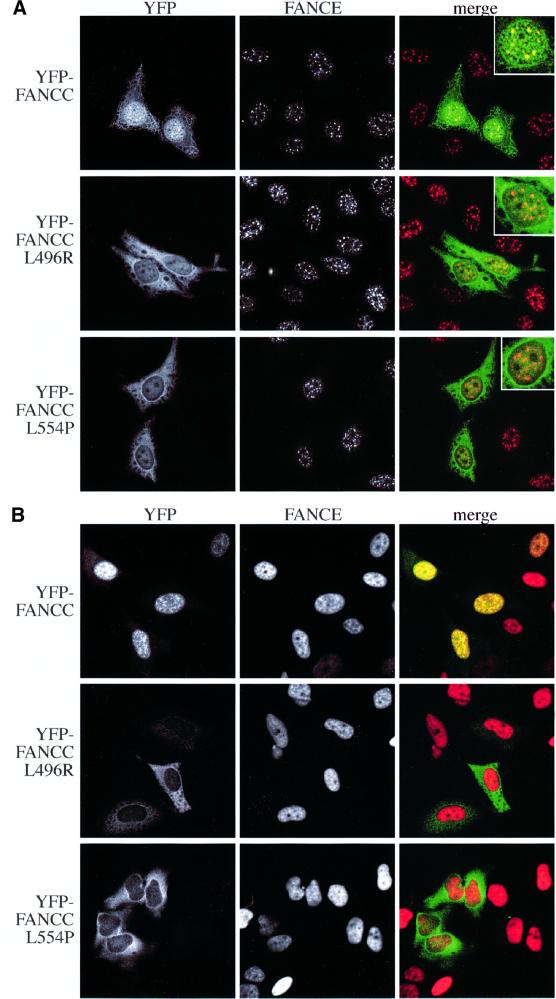
The FANCC protein is known to interact with FANCE in yeast twohybrid interaction studies (Medhurst et al., 2001). We were able to recapitulate these findings by showing that this interaction occurs in mammalian cells by the mammalian two-hybrid (M2H) assay, as well as by co-immunoprecipitation in baculovirus-infected insect cells (Figure 4). There are two documented missense mutant FANCC proteins that are associated with FA: L496R (Waisfisz et al., 1999b) and L554P (Gavish et al., 1993). These mutants are also defective in binding to FANCE when tested in the M2H assay and by co-expression in insect cells (Figure 4). When YFP-tagged wild-type FANCC (YFP–FANCCWT) and FANCC mutants (YFP– FANCCL496R and YFP–FANCCL554P) were individually expressed in HeLa cells, they could be detected in both nuclear and cytoplasmic compartments. This cell line was chosen as it maintains its morphology well, and the availability of a stable FANCE-overexpressing cell line provides an accurate comparison of the effect of FANCE upon FANCC nuclear localization (similar results were obtained in COS and 293 cells; data not shown). Co-staining for endogenous FANCE clearly shows that while the nuclear YFP–FANCCWT co-localizes with FANCE in foci, both the mutants failed to localize to these structures (Figure 5A, see inset). We next repeated the same experiment in HeLa cells stably expressing FANCE-2×Flag. There is a dramatic nuclear accumulation of the YFP–FANCCWT, whereas the mutants are defective in this localization (Figure 5B and Supplementary data). These results show that FANCE promotes FANCC nuclear accumulation because of an interaction between these two molecules. The inability of the mutant FANCC proteins to accumulate in the nucleus provides a molecular explanation of why they are unable to protect against chromosome breakage.
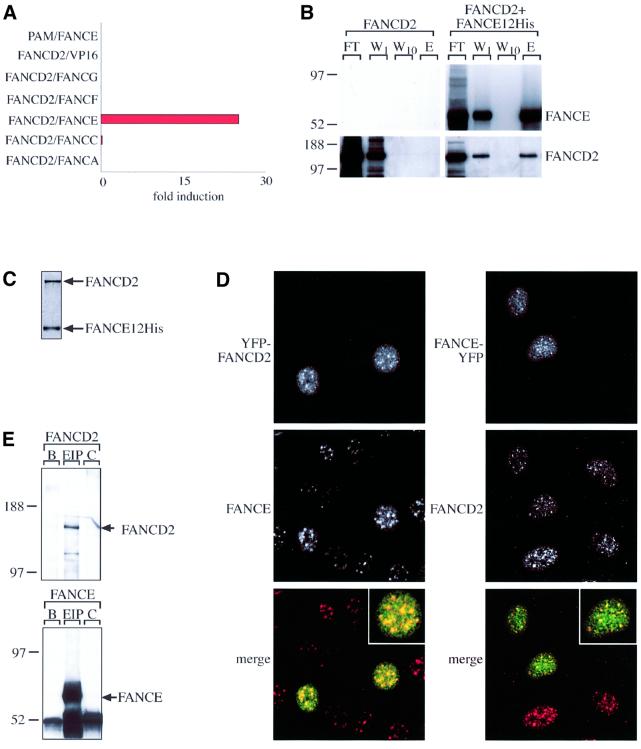
Fig. 4. FANCC mutants are defective in binding FANCE. (A) Co-immunoprecipitation of protein C epitope-tagged FANCE with GST-tagged wild-type (WT) or mutant (L496R and L554P) FANCC in baculovirus-infected insect cells. The gel transfers were western blotted with anti-protein C monoclonal antibody (for the detection of FANCE) or with anti-GST antisera (for the detection of FANCC). The interaction between FANCE and WT FANCC is readily detectable, while that with the mutants is reduced. (B) Mammalian two hybrid interaction (M2H) test showing strong induction of the reporter (luciferase) in cells expressing WT FANCC and FANCE but not in the case of mutant FANCC. Units are expressed as fold induction of luciferase reporter when compared with either gene with empty vector (pAM or VP-16) alone.
Fig. 5. FANCC mutants do not accumulate in the nucleus. (A) Localization of transiently transfected YFP–FANCC (green) and YFP–FANCC mutants in HeLa cells. Endogenous FANCE is present in nuclear foci (red), co-localizing with WT FANCC but not with mutant FANCC (see insets). (B) As (A), but in HeLa cells that stably overexpress FANCE-2×Flag. Wild-type FANCC accumulates in the nucleus, while neither of the mutants do.
FANCE co-localizes with and binds directly to FANCD2
The FANCD2 protein is thought to function downstream of the FA complex, as it is not required for the formation of the complex. Its ‘activation’ and monoubiquitylation require an intact nuclear complex. However, no physical link has yet been demonstrated between FANCD2 and the nuclear complex in order to facilitate this modification (Garcia-Higuera et al., 2001). In searching for such a link, we asked whether any components of the FA complex were able to interact with FANCD2. We found that only FANCE was able to interact with D2 when tested in the M2H assay (Figure 6A). These two proteins also efficiently co-purify with each other when co-expressed in insect cells. Coomassie Blue gel analysis of purified FANCE-His12 shows that FANCE and FANCD2 elute from Ni2+–agarose in the absence of any other proteins, indicating that they interact directly (Figure 6B and C). As both proteins are present in nuclear foci (Garcia-Higuera et al., 2001), we next tested whether they could co-localize; they do. Some of the endogenous D2 foci co-localize with FANCE–YFP, as does YFP–FANCD2 with endogenous FANCE in irradiated HeLa cells (Figure 6D). Next we sought to see whether the interaction could be detected in HeLa nuclear extracts. Immunoprecipitation with FANCE antisera brings down FANCD2; moreover, we were only able to detect the unmodified form of FANCD2 (Figure 6E). Collectively, these results demonstrate that FANCE binds to FANCD2, thereby linking it to the FA nuclear complex.
Fig. 6. FANCE binds to and co-localizes with the downstream FA protein FANCD2. (A) M2H test indicating that FANCE, but not any of the other FA complex proteins, interacts with FANCD2 (activation domain-fused FA complex protein tested against DNA-binding domain-fused FANCD2). (B) FANCD2 co-elutes with His12-tagged FANCE on a nickel-NTA matrix when captured from lysates of baculovirus-infected insect cells expressing both proteins. Insect cells expressing only FANCD2 were used as a control. FT, flow-through; W1, first wash; W10, tenth wash; E, elution with imidazole. Gels were transferred and blotted with anti-FANCE antiserum or anti-FANCD2 antisera. (C) Coomassie Blue stain of FANCD2/FANCE-His12 complexes purified by Ni2+–agarose chromatography from baculovirus-infected insect cells expressing both proteins. Both proteins co-purify efficiently. (D) Co-localization of transiently expressed YFP–FANCD2 (green) with endogenous FANCE (red) or endogenous FANCD2 (red) with transiently expressed FANCE–YFP (green), in irradiated (6 h post-3 Gy) HeLa cells. In the left-hand panel, transfected YFP–FANCD2 (top) as well as endogenous FANCE (middle) localize to nuclear foci. Merging of the images shows co-localization for some of these foci. The right-hand panel shows the same experiment, but this time with the tagging combination reversed, i.e. FANCE is now YFP tagged and transfected, and it is endogeneous FANCD2 that is being assayed. Top, transfected FANCE–YFP. Middle, endogenous FANCD2 (detected by rabbit anti-FANCD2 antisera followed by anti-rabbit Cy5 antibody). Bottom, merged image. (E) Endogenous FANCE co-purifies with endogenous FANCD2 in HeLa nuclear extract. HeLa nuclear extract (4 mg) was incubated with protein A–Sepharose beads without antibody (B), with rabbit anti-FANCE antisera (EIP) and with control preimmune antisera (C). The samples were western blotted with anti-FANCD2 antisera (top) and anti-FANCE antisera (bottom). The arrows indicate the migration of FANCD2 and FANCE.
Discussion
The recently identified FANCE gene product is an integral part of the FA nuclear complex, not only enabling its assembly, but also linking this complex to FANCD2. The striking effect of the FANCE protein in promoting FANCC nuclear accumulation demonstrates the inter dependence of the proteins in the complex. Many previous studies have shown that the FANCC protein is present predominantly in the cytoplasm (Yamashita et al., 1994; Youssoufian, 1994; Hoatlin et al., 1998), leading to the suggestion that it has a distinct cytosolic function in this compartment. FANCC can bind to cytoplasmic proteins like GSTP1, an enzyme implicated in the detoxification of reactive oxygen radicals (ROS) (Cumming et al., 2001). While our studies do not exclude such a function, they show that FANCE is a rate-limiting factor for FANCC nuclear trafficking. It will be interesting to determine whether the FANCC-interacting cytoplasmic enzymes can be co-transported, with this protein perhaps enabling the detoxification of nuclear ROS.
How might FANCE promote the nuclear accumulation of FANCC? We can envisage two models that could explain this. The FANCE protein has a predicted nuclear localization signal sequence (de Winter et al., 2000a), which is absent in FANCC. Binding of the two proteins to each other might enable FANCE to carry FANCC into the nucleus (Figure 7A). Alternatively, it is possible that the two proteins enter the nucleus independently and their interaction in the nucleus leads to retention of FANCC (Figure 7B). Our experiments do not distinguish between these potential mechanisms and further studies are under way to test which of these models is correct. The inability of the mutant FANCC proteins to accumulate in the nucleus clearly correlates with reduced FANCE binding, thereby explaining why these mutations result in FA. Understanding the mechanism of the nuclear transport of FANCC will not only improve our understanding of how this large multi-protein nuclear complex is assembled, but may also provide insights for therapeutic intervention in FA patients.
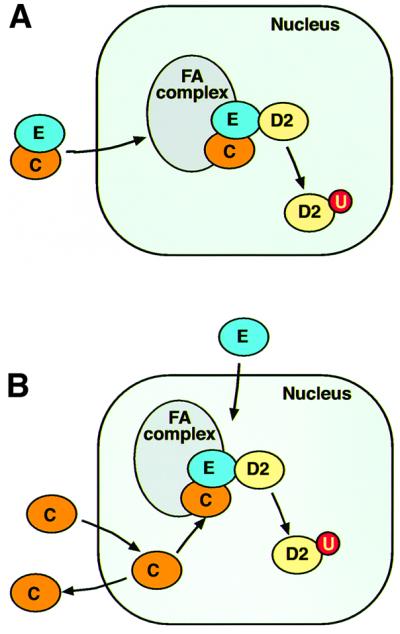
Fig. 7. A model for the assembly of the FA nuclear complex. (A) The FANCE protein carries FANCC into the nucleus and assembles the FA complex. (B) FANCE and FANCC enter the nucleus independently and FANCC is retained in this compartment upon complex assembly.
The FA nuclear complex is required for the conjugation of a single ubiquitin molecule to FANCD2. This ‘activation’ step also correlates with the localization of FANCD2 to nuclear foci, where other DNA damage response proteins reside (Garcia-Higuera et al., 2001). How might this complex facilitate FANCD2 monoubiquitylation? The complex itself might function as a ubiquitin ligase, but, at present, none of the known FA proteins contains motifs that are implicated in terminal ubiquitin transfer. Alternatively, the complex may be a co-factor in FANCD2 modification. In either case, the complex must somehow be able to sense DNA damage and relay this signal to FANCD2. Our finding that the FANCE protein can bind to FANCD2 shows that at least one component of this complex is physically linked to this substrate.
The localization of FANCE to nuclear foci suggests a potential site where FANCD2 modification might occur because some of these sites co-localize with endogenous FANCD2. However, unlike FANCD2 foci, these are constitutive and not visibly induced by cell cycle progression or DNA damage. Much needs to be done to define further the nature of FANCE foci, particularly in terms of underlying DNA structures as well as what other proteins may be present in them. It is nonetheless conceivable that the complex and FANCD2 may be present in the same foci upon DNA damage, where they might co-operate in a possible holoenzyme complex involved in DNA metabolism. The interaction between FANCE and FANCD2 was readily detectable in insect cells, such that they efficiently co-purify with each other, but in HeLa cell nuclear extract, this interaction, although detectable, was much less vigorous. We do not know the reason for this, but it could imply either that the interaction is weak and disrupted by the extraction, or that it is regulated such that there are few FANCE–FANCD2 complexes present at a given time. Finally, although the interaction can occur in insect cells, we do not know whether, in mammalian cell nuclei, it requires an intact FA nuclear complex or indeed another unidentified FA protein.
Materials and methods
Construction of cDNA plasmids
C-terminus YFP- or epitope-tagged (His12, PRC-His12 and 2×Flag) FANCE, A, G, F and D2 cDNA. The 3′ end was adapted to remove the stop codon and replaced with an in-frame XbaI site TCTAGA; these were cloned into a BlueScript vector where the epitope tag cDNA (2×Flag, PRC-His12, YFP) had been cloned into the XbaI–NotI site. After sequencing, these were then cloned into the Gateway destination vector pENTR3C (Invitrogen) and the vector Peak8 (Edge Biosystems) for the expression of FANCE–YFP, FANCE-2×Flag, FANCA–YFP, FANCA-2×Flag, FANCG–YFP, FANCG-2×Flag, FANCF–YFP and FANCF-2×Flag. All FA cDNA expression constructs also have a 5′-adapted SalI site after a Kozak sequence, ACCATG, enabling the generation of in-frame fusions at the 5′ end.
GST-tagged FANCC and FANCC mutants. To generate GST-tagged wild-type FANCC and FANCC mutants, cDNAs were generated by PCR to contain an adapted SalI 5′ end, cloned into pENTR3C, and transferred by Gateway cloning to the baculovirus expression vector pDEST20 (Invitrogen).
N-terminal YFP fusions of FA proteins. To generate N-terminal YFP fusions of FANCC, FANCC mutants, FANCD2 and FANCF, SalI-adapted 5′ cDNAs were cloned directly into the vector Peak8 (HindIII–NotI) as a three-way ligation consisting of a HindIII–SalI EYFP fragment (generated by PCR where the 3′ end had an in-frame SalI site replacing the stop codon) and the relevant cDNA as a SalI–NotI fragment.
Mammalian two-hybrid construct. To generate mammalian two-hybrid constructs, the SalI–NotI-adapted FANCE, FANCC, FANCC mutants, FANCG, FANCA, FANCF and FANCD2 were cloned into the vectors pVP16-activation domain and pM-gal4 DNA-binding domain (both from Clontech).
Recombinant protein and antibody production
The FANCE and FANCD2 rabbit antisera were produced by immunization of rabbits with full-length recombinant FANCE and FANCD2, each with a C-terminal His12 tag. Briefly, cDNAs coding for adapted FANCE and FANCD2 were cloned into the Gateway destination vector pDEST8 (Invitrogen); insect cell infection and protein expression were carried out following the manufacturer’s instructions. Recombinant protein was purified on nickel-NTA–agarose and eluted with imidazole. Each rabbit received four immunizations with native purified protein, spiked with SDS–PAGE gel slices of denatured antigen. Fourth bleed antiserum was affinity purified on a column, where the antigen was previously immobilized using an Amino-Link kit (Pierce), as per the manufacturer’s instructions. The specificity of each antiserum was validated by testing for recognition of recombinant protein, as well as endogenous protein in cell lysates from the complementation groups.
Cell lines
The following cell lines were used in this study: HSC-72 (FA-A), HSC-536 (FA-C), VU-120 (FA-D2), EUFA-410 and EUFA-622 (FA-E), EUFA-698 (FA-F) and EUFA-143 (FA-G). NV-012 lymphoblasts were from a healthy control individual.
Immunoprecipitations and western blotting
Cell lines used for immunoprecipitations were washed in phosphate-buffered saline (PBS) and harvested into NETN buffer (20 mM Tris pH 8, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1 mM AEBSF), lysed by shearing three times through a 19G needle, and clarified at 15 700 g for 20 min at 4°C. Total protein concentrations for the soluble fraction were measured by the Bradford method. All immunoprecipitations were performed at 4°C, or on ice. One to two milligrams of total soluble protein in 1 ml of NETN buffer (except where mentioned otherwise) were mixed gently with 1–2 µg of antibody for 1 h at 4°C. Immune complexes were captured with 5 mg of pre-equilibrated protein A–Sepharose CL-4B (Amersham-Pharmacia) for a further hour, collected at 800 g for 5 min, and washed four times in 1 ml of NETN buffer at 800 g for 5 min each time. Beads were treated with SDS–PAGE dye, boiled for 10 min, and samples analysed by PAGE followed by immunoblotting on nitrocellulose as described below. For the FANCE–FANCD2 interaction in HeLa cells, the nuclear extract was made according to the method described by Dignam et al. (1983). For each immunoprecipitation, HeLa nuclear lysates containing 4 mg of total protein were treated with 1% NP-40, 10 µg/ml DNase I (Sigma), 1 mM AEBSF (Melford Laboratories) and mammalian protease inhibitors (Sigma), incubated for 30 min at room temperature, then centrifuged at 16 200 g for 30 min at 4°C. Clarified lysates were immunoprecipitated as described, except in nuclear resuspension buffer (20 mM HEPES pH 7.9, 10 µM ZnSO4, 2 mM MgCl2, 2 mM dithiothreitol, 100 mM KCl, 10% glycerol, 1 mM AEBSF), and washed in the same buffer containing 1% NP-40.
Nitrocellulose transfers from SDS–PAGE were blocked with 5% fat-free milk powder (Marvel) dissolved in PBS (MPBS). Blocking was for 1 h at room temperature, or overnight at 4°C. All antibody blotting was for 1 h at room temperature using antibodies at the concentrations described below, in 5% MPBS, unless specified otherwise. Filters were washed with PBS + 0.05% Tween-20 for 10 min per wash, three times between antibody incubations, and four times following incubations. Anti-protein C–horseradish peroxidase (HRP) blots were blocked and blotted with antibody in 5% fat-free milk in TBS + 1 mM CaCl2, and washes were with TBS + 0.05% Tween-20 + 1 mM CaCl2.
For western blotting, affinity purified anti-FANCE and anti-FANCD2 antibodies were used at 1:1000 dilution. IgG purified anti-FANCA, anti-FANCF and anti-FANCG were used at 1:100 in 5% bovine serum albumin in PBS. Mouse monoclonal anti-flag M2 (Sigma) was used at 1:2000 dilution. Mouse monoclonal anti-GST (B14)–HRP (Santa Cruz Biotech) was used at 1:1000 dilution. Mouse monoclonal anti-protein C–HRP (Boehringer Mannheim) was used at 1:1000 in 5% MTBS + 10 mM CaCl2. Secondary detection antibodies were goat anti-rabbit IgG (H+L)–HRP (human and mouse adsorbed) (Southern Biotechnology), or HRP-conjugated rabbit anti-mouse immunoglobulins (Dako), both used at 1:2000 dilution, where necessary. Enhanced chemiluminescence was used to visualize blotted bands.
Immunofluoresence and microscopy
HeLa and COS-7 cells were cultured in Dulbecco’s modified Eagle’s medium and 10% fetal calf serum on poly-l-lysine-coated coverslips. Transfections were carried out using Superfect (Qiagen) according to the manufacturer’s instructions. After 24 h, the coverslips were fixed with methanol at –20°C, and stained with antibodies as described below. FANCE and FANCD2 affinity-purified primary antibodies were used at 1 µg/ml. Appropriate Alexa 488- (Molecular Probes) and Cy5- (Jackson) coupled secondary antibodies were used at 1:500 dilution. All imaging was performed using a Bio-Rad 200 scanning confocal microscope. Images were imported into Adobe Photoshop 6.0 and adjusted to use the full range of pixel intensities.
Protein interaction methods
The mammalian two-hybrid system. CHO cells (0.1 × 106) were plated onto 6-well plates. After 24 h, they were transfected with the following: 5 µg of each of the following plasmids; interacting pairs consisting of fusions of either Gal4 DNA-binding domain (pM; Clontech) or activation domain (pVP16; Clontech), together with a reporter plasmid, where luciferase is driven by a Gal4 promoter, G5E1bLUC (a gift from Dr Richard Baer, Columbia University, New York). In addition, a transfection control reporter plasmid was also added (pRL-CMV; Promega); this drives the constitutive expression of Renilla luciferase. Cells were harvested 48 h post-transfection and analysed according to the manufacturer’s instructions (Promega dual luciferase reporter kit); induction of reporters was carried out with an Orion microplate luminometer (Berthold Detection Systems). Each experimental data set is expressed as an average of triplicate samples. First, a ratio of the induction of the transfection control reporter with that obtained from the Gal4-inducible promoter is calculated. This is quantitated as fold induction over ratios obtained from controls consisting of vector alone, with either test fusion with Gal4 or VP-16.
FANCE–FANCC interaction. The FANCE–FANCC interaction in insect cells was probed as follows: 50 ml of insect cells were infected with baculovirus to express full-length protein C-His12-tagged FANCE, glutathione S-transferase (GST)-tagged FANCC, GST-tagged FANCC-L496R, or GST-tagged FANCC-L554P individually, or to co-express them in pairs as illustrated. Lysates were made in NCTN buffer (NETN without EDTA, supplemented with calcium: 20 mM Tris pH 8, 150 mM NaCl, 10 mM CaCl2, 0.1% NP-40, 1 mM AEBSF) containing 10 µg/ml DNase I, sonicated and clarified at 16 200 g. Supernatants were filtered through 0.22 µm filters, and protein levels were determined by the Bradford method. In each case, lysates containing 0.5 mg of total protein were pre-cleared on protein G–Sepharose for 1 h at 4°C before immunoprecipitation with the relevant antibodies (1–2 µg of mouse anti-GST for FANCC or anti-PRC for FANCE). The samples were washed, eluted, resolved by SDS–PAGE and blotted as described above.
FANCE–FANCD2 interaction. The FANCE–FANCD2 interaction in insect cells was probed as follows: 50 ml of baculovirus-infected insect cells were used to express untagged full-length FANCD2, or to co-express both untagged full-length FANCD2 and FANCE–protein C-His12 together. Lysates were made in a lysis buffer (20 mM Tris pH 8, 500 mM NaCl, 10 mM imidazole, 10% glycerol, 1 mM AEBSF, 0.1% β-mercaptoethanol) containing 10 µg/ml DNase I, sonicated and clarified at 16 200 g. In each case, clarified lysate containing 4 mg of total protein was incubated with 100 µl bed volume of nickel-NTA Superflow resin (Qiagen) overnight at 4°C. Beads were subsequently washed 10 times in the same lysis buffer, but containing 20 mM imidazole. FANCE protein was eluted in lysis buffer supplemented with 200 mM imidazole by incubation for 10 min at 4°C. Samples from washes, eluates and beads were treated with SDS–PAGE dye for gel analyses and western blotting as described above.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Julian Sale for many stimulating discussions, and Michael Neuberger, Roger Williams, Olga Persic, Kevin Hiom and Alex Betz for critically reviewing the manucript as well as for experimental advice. The FANCD2 cDNA was a kind gift of Dr A.D’Andrea. M.J. is supported by an MRC training fellowship, F.G. by a Royal Society Dorothy Hodgkin Fellowship, and K.J.P. is a recipient of an MRC Senior Clinical Fellowship.
References
- Auerbach A.D. and Wolman,S.R. (1976) Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature, 261, 494–496. [DOI] [PubMed] [Google Scholar]
- Auerbach A.D., Buchwald,M. and Joenje,H. (2001) The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York, NY.
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Cumming R.C., Lightfoot,J., Beard,K., Youssoufian,H., O’Brien,P.J. and Buchwald,M. (2001) Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nat. Med., 7, 814–820. [DOI] [PubMed] [Google Scholar]
- de Winter J.P. et al. (2000a) Isolation of a cDNA representing the Fanconi anemia complementation group E gene. Am. J. Hum. Genet., 67, 1306–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter J.P. et al. (2000b) The Fanconi anemia protein FANCF forms a nuclear complex with FANCA, FANCC and FANCG. Hum. Mol. Genet., 9, 2665–2674. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I., Kuang,Y., Naf,D., Wasik,J. and D’Andrea,A.D. (1999) Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol. Cell. Biol., 19, 4866–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I., Taniguchi,T., Ganesan,S., Meyn,M.S., Timmers,C., Hejna,J., Grompe,M. and D’Andrea,A.D. (2001) Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell, 7, 249–262. [DOI] [PubMed] [Google Scholar]
- Gavish H., dos Santos,C.C. and Buchwald,M. (1993) A Leu554-to-Pro substitution completely abolishes the functional complementing activity of the Fanconi anemia (FACC) protein. Hum. Mol. Genet., 2, 123–126. [DOI] [PubMed] [Google Scholar]
- Grompe M. and D’Andrea,A. (2001) Fanconi anemia and DNA repair. Hum. Mol. Genet., 10, 2253–2259. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D.P. (1999) Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 448-449.
- Hoatlin M.E., Christianson,T.A., Keeble,W.W., Hammond,A.T., Zhi,Y., Heinrich,M.C., Tower,P.A. and Bagby,G.C.,Jr (1998) The Fanconi anemia group C gene product is located in both the nucleus and cytoplasm of human cells. Blood, 91, 1418–1425. [PubMed] [Google Scholar]
- Ishida R. and Buchwald,M. (1982) Susceptibility of Fanconi’s anemia lymphoblasts to DNA-cross-linking and alkylating agents. Cancer Res., 42, 4000–4006. [PubMed] [Google Scholar]
- Joenje H. and Patel,K.J. (2001) The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet., 2, 446–457. [DOI] [PubMed] [Google Scholar]
- Joenje H. et al. (1997) Evidence for at least eight Fanconi anemia genes. Am. J. Hum. Genet., 61, 940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler K.W. and Vogelstein,B. (2001) Introduction. In Scriver,C.R.E.A. (ed.), The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York, NY, pp. 675–676.
- Kruyt F.A., Hoshino,T., Liu,J.M., Joseph,P., Jaiswal,A.K. and Youssoufian,H. (1998) Abnormal microsomal detoxification implicated in Fanconi anemia group C by interaction of the FAC protein with NADPH cytochrome P450 reductase. Blood, 92, 3050–3056. [PubMed] [Google Scholar]
- Kupfer G.M., Naf,D., Suliman,A., Pulsipher,M. and D’Andrea,A.D. (1997) The Fanconi anaemia proteins, FAA and FAC, interact to form a nuclear complex. Nat. Genet., 17, 487–490. [DOI] [PubMed] [Google Scholar]
- Medhurst A.L., Huber,P.A., Waisfisz,Q., de Winter,J.P. and Mathew,C.G. (2001) Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum. Mol. Genet., 10, 423–429. [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu,J.W., Koller,B.H. and Jasin,M. (1999) Brca1 controls homology-directed DNA repair. Mol. Cell, 4, 511–518. [DOI] [PubMed] [Google Scholar]
- Pang Q., Fagerlie,S., Christianson,T.A., Keeble,W., Faulkner,G., Diaz,J., Rathbun,R.K. and Bagby,G.C. (2000) The Fanconi anemia protein FANCC binds to and facilitates the activation of STAT1 by γ interferon and hematopoietic growth factors. Mol. Cell Biol., 20, 4724–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q., Keeble,W., Christianson,T.A., Faulkner,G.R. and Bagby,G.C. (2001) FANCC interacts with Hsp70 to protect hematopoietic cells from IFN-γ/TNF-α-mediated cytotoxicity. EMBO J., 20, 4478–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.J. et al. (1998) Involvement of Brca2 in DNA repair. Mol. Cell, 1, 347–357. [DOI] [PubMed] [Google Scholar]
- Scully R. and Livingston,D.M. (2000) In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature, 408, 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers C. et al. (2001) Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell, 7, 241–248. [DOI] [PubMed] [Google Scholar]
- Waisfisz Q. et al. (1999a) A physical complex of the Fanconi anemia proteins FANCG/XRCC9 and FANCA. Proc. Natl Acad. Sci. USA, 96, 10320–10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waisfisz Q. et al. (1999b) Spontaneous functional correction of homozygous Fanconi anaemia alleles reveals novel mechanistic basis for reverse mosaicism. Nat. Genet., 22, 379–383. [DOI] [PubMed] [Google Scholar]
- Weissman A.M. (2001) Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol., 2, 169–178. [DOI] [PubMed] [Google Scholar]
- Yamashita T., Barber,D.L., Zhu,Y., Wu,N. and D’Andrea,A.D. (1994) The Fanconi anemia polypeptide FACC is localized to the cytoplasm. Proc. Natl Acad. Sci. USA, 91, 6712–6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Kupfer,G.M., Naf,D., Suliman,A., Joenje,H., Asano,S. and D’Andrea,A.D. (1998) The Fanconi anemia pathway requires FAA phosphorylation and FAA/FAC nuclear accumulation. Proc. Natl Acad. Sci. USA, 95, 13085–13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H. (1994) Localization of Fanconi anemia C protein to the cytoplasm of mammalian cells. Proc. Natl Acad. Sci. USA, 91, 7975–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]