Abstract
Two-dimensional crystals of the reaction-centre–light-harvesting complex I (RC–LH1) of the purple non- sulfur bacterium Rhodospirillum rubrum have been formed from detergent-solubilized and purified protein complexes. Unstained samples of this intrinsic membrane protein complex have been analysed by electron cryomicroscopy (cryo EM). Projection maps were calculated to 8.5 Å from two different crystal forms, and show a single reaction centre surrounded by 16 LH1 subunits in a ring of ∼115 Å diameter. Within each LH1 subunit, densities for the α- and β-polypeptide chains are clearly resolved. In one crystal form the LH1 forms a circular ring, and in the other form the ring is significantly ellipsoidal. In each case, the reaction centre adopts preferred orientations, suggesting specific interactions between the reaction centre and LH1 subunits rather than a continuum of possible orientations with the antenna ring. This experimentally determined structure shows no evidence of any other protein components in the closed LH1 ring. The demonstration of circular or elliptical forms of LH1 indicates that this complex is likely to be flexible in the bacterial membrane.
Keywords: 2D crystals/electron cryomicroscopy/light-harvesting complex/photosynthesis/Rhodospirillum rubrum
Introduction
The specialized photosynthetic membranes of purple bacteria make fascinating and very approachable systems for exploring general mechanisms of biological energy transduction and the assembly of large multi-component membrane protein complexes. With the tools of genetic manipulation, protein biochemistry, spectroscopic analysis, X-ray crystallography and electron cryomicroscopy, a wealth of data has emerged on the structure, assembly and function of these photosynthetic machines.
In all purple bacteria, a transmembrane potential is generated via the reaction centre (RC), converting light energy into a chemically useful form. Membrane protein complexes (LH1, LH2) bind bacteriochlorophyll (Bchl) and act as light-harvesting antennae. LH1 forms an intimate complex with the RC, at least partially encircling it; LH2 is arranged peripherally around the RC–LH1 ‘core complex’ in an unknown way. There is a rapid and efficient transfer of energy from LH2 to LH1 to the RC (Hess et al., 1995; Hu et al., 1998; Sundström et al., 1999).
One of the major achievements in structural biology has been the description of the X-ray structures of the bacterial RC complex (Deisenhofer et al., 1985; Allen et al., 1987). The RC (Mr ∼ 100 kDa) contains bound Bchls and three membrane-bound protein subunits, L, M and H, with five, five and one transmembrane helices, respectively. The RC receives energy from the LH1 complex, which in turn receives energy from LH2. LH1 and LH2 consist of two types of polypeptide (α and β; ∼50 amino acids each) with a single membrane-spanning α-helix and hydrophilic N- and C-terminal domains. The basic subunit is an αβ heterodimer with two or three molecules of Bchl and one carotenoid bound non-covalently to the protein. The 8.5 Å resolution electron microscopy (EM) projection structure of Rhodospirillum rubrum LH1 was determined by Karrasch et al. (1995), followed by two atomic resolution X-ray structures of different LH2s (McDermott et al., 1995; Koepke et al., 1996). The LH1 map revealed a ring of 16 αβ subunits, whereas the two LH2 structures showed smaller rings of nine and eight subunits, respectively. A 9-fold ring has also been determined by EM for two further species (Savage et al., 1996; Walz et al., 1998), and by atomic force microscopy for the LH2 from Rubrivivax gelatinosus (Scheuring et al., 2001). So far, no high-resolution three-dimensional (3D) structure has been described for any LH1. In the absence of such information, a combination of NMR, mutagenesis and spectroscopy data has been used to model the structure of the LH1 complex from Rhodobacter sphaeroides (Conroy et al., 2000).
Moving on from structures of individual components of the bacterial photosynthetic membrane, it is necessary to describe and understand the structure and assembly of higher order molecular assemblies in order to learn how these structures allow the efficient and rapid transduction of light energy. A number of low-resolution studies have shown that the RC forms an intimate complex with LH1 (Miller, 1982; Stark et al., 1984; Engelhardt et al., 1986; Boonstra et al., 1994; Walz et al., 1998; Jungas et al., 1999). Karrasch et al. (1995) proposed that one RC can be accommodated within the 16-fold LH1 ring of R.rubrum. However, the LH1 crystals in this study were reconstituted from detergent-solubilized αβ subunits, raising the question of whether the intact LH1 antenna surrounding the RC really forms a complete, circular ring of 16 subunits. This becomes especially important in the context of recent publications on ‘square’ RC–LH1 complexes of R.rubrum, and incomplete LH1 rings of R.sphaeroides (Stahlberg et al., 1998; Jungas et al., 1999).
In this paper, we examine the nature of the RC–LH1 interaction in some detail by high-resolution electron crystallography. Part of the fascination in examining the RC–LH1 complex is that the structure of the central component, the RC, is already known to atomic resolution (Deisenhofer et al., 1985; Allen et al., 1987). The RC– LH1 complex provides an excellent model for exploring the degree of specificity and the types of interactions involved in the assembly of higher order membrane protein ‘supercomplexes’. Previous low-resolution studies (Miller, 1982; Stark et al., 1984; Engelhardt et al., 1986; Boonstra et al., 1994; Walz et al., 1998; Jungas et al., 1999) showed an LH1 ring of size similar to that from R.rubrum (Karrasch et al., 1995). However, for all species (including R.rubrum), the precise oligomeric state of the LH1 and the effect of the RC on this state are unknown. In this paper, we report the crystallization of RC–LH1 complexes in very well ordered 2D arrays; we show, for the first time, that association with the RC maintains the 16-fold symmetry of the LH1 antenna and that LH1 forms a closed circular or ellipsoidal complex around the RC. Moreover, we show that there is likely to be a specific interaction between the RC and the LH1 subunits, with the possibility of interaction with any one of the 16 LH1 subunits.
Results
Purification and spectroscopy
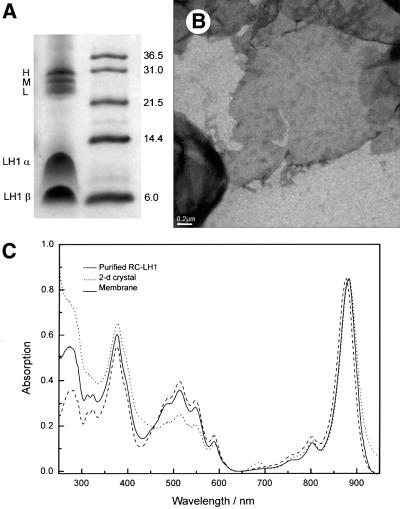
A Schägger gel (Schägger and von Jagow, 1987) of the crystallized complexes (Figure 1A) revealed major bands corresponding to the RC and LH1 polypeptides; no other bands were seen. The absorbance spectrum of the Bchls in crystallized RC–LH1 complexes showed negligible differences from the native membranes, with a maximum at 883 nm in each case (Figure 1C). Prior to crystallization, the solubilized and purified complex had an absorbance maximum at 876 nm; aggregation produces a red shift, which appears to arise from contact between the complexes. A similar phenomenon was noticed for solubilized complexes and 2D crystals of LH2 (Walz et al., 1998). Recently, three-pulse photon echo peak shift (3PEPS) spectroscopy was used to show that LH2 aggregates in the native membrane possess an ∼5 ps decay component, which is absent in the isolated, detergent-solubilized samples. This decay component, which is assigned to inter-complex LH2–LH2 energy transfer, correlates with a red-shifted absorbance maximum for the membranes, when compared with the solubilized complex (Agarwal et al., 2002). The RC:LH1 stoichiometry can be inferred from the absorbance ratios of the near-IR peaks at 800 and 876–883 nm, respectively; inspection of the spectra in Figure 1C shows that this stoichiometry has been maintained during the purification and crystallization procedures.
Fig. 1. (A) Coomassie Blue-stained Schägger gel of 2D crystals of RC–LH1 complex (left column) with subunits labelled. The right column shows the positions of molecular weight standards with molecular weights in kDa. (B) Low-magnification view of a negatively stained 2D crystal. Tetragonal and orthorhombic crystals were indistinguishable at low magnification. (C) Absorbance spectra of chromatophore membranes, purified RC–LH1 complexes and 2D crystals of complexes.
Crystallization of RC–LH1 complexes
Two forms of 2D crystals of RC–LH1 were discovered and often found together in the same crystallization well. 2D crystals of RC–LH1 were found for all crystallization conditions tested. However, the best quality crystals (as judged by optical diffraction) were grown with the lipid dioleoyl phosphatidylcholine (DOPC) solubilized in diheptanoyl phosphatidylcholine (DHPC) with a lipid: protein ratio (LPR) in the range 1.0–1.2 (w/w) and a 10 mM HEPES pH 7.5 dialysis buffer containing 100 mM NaCl. One crystal form, tetragonal, was essentially the same as that described by Walz and Ghosh (1997), but we also found an orthorhombic form. The conditions favoured the growth of vesicular and sheet-like 2D crystals that varied in size from 0.5 to >3 µm (Figure 1B).
EM in negative stain
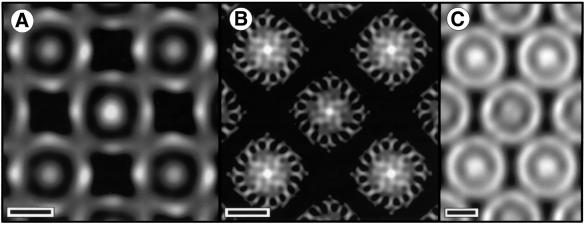
One image of each crystal form was selected for processing to generate a projection map at 20 Å resolution. The tetragonal crystal form with a unit cell of approximate dimensions a = b = 156 Å was comparable to a crystal form obtained by Ghosh and co-workers (Walz and Ghosh, 1997; Stahlberg et al., 1998). The projection map reveals an LH1 ring, with inner and outer diameters of ∼65 and 115 Å, respectively, enclosing a central density attributable to the RC (Figure 2A). We found that individual core complexes could adopt an apparent ‘square’ shape as first described by Stahlberg et al. (1998), depending on the level of image contrast and the nature of stain applied (Figure 2B). This square conformation appears to reflect the way the negative stain distributes over the complexes rather than a novel conformation of the complex (see the next section). A comparable map from one image of the orthorhombic crystal form embedded in stain is shown in Figure 2C. The complex formed crystals with a unit cell of approximate dimensions a = 128 Å, b = 197 Å. This is a crystal form of the RC–LH1 complex that has not been previously reported. The projection map reveals an LH1 ring enclosing the RC, with inner and outer diameters comparable to those of the tetragonal form. In this case, individual core complexes appear to be more ‘circular’.
Fig. 2. EM in negative stain. Scale bar: 50 Å. (A) Fourier-filtered image of a negatively stained tetragonal crystal. Darker regions correspond to regions of higher density (stain). (B) Tetragonal crystal embedded in a glucose–negative stain mixture. (C) Orthorhombic crystal embedded in negative stain.
EM in glucose
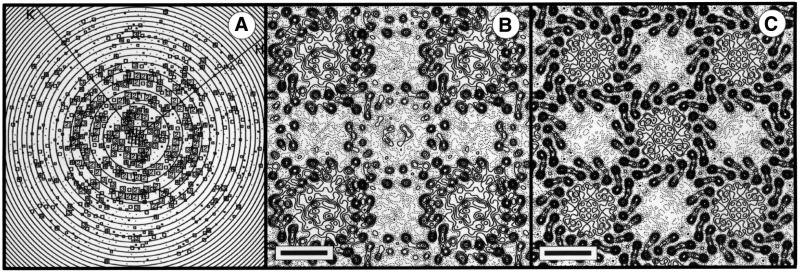
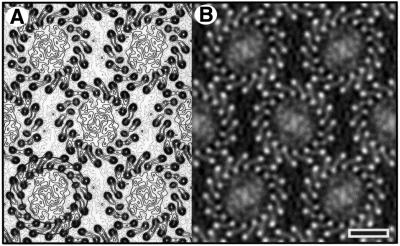
Tetragonal form. A representation of all the Fourier components obtained from one image is shown in Figure 3A. An analysis of symmetry-related phases showed the data to be consistent with p4212 plane group symmetry (Valpuesta et al., 1994). Amplitudes and phases from the five best images were merged and averaged, imposing either p1 or p4212 symmetry with a = b = 165 Å. Origin and phase contrast transfer function (CTF) refinement were performed independently for the two plane groups, with Fourier terms limited to 8.5 Å resolution. Table I shows the average phase residual as a function of resolution for p4212 symmetry, indicating non-random data to 8.5 Å resolution. Amplitudes from individual images were corrected by temperature factors of between 172 and 492 Å2 prior to averaging. The projection map with p1 symmetry (Figure 3B) reveals the α-helical structure of the LH1 antenna, showing both ‘up’ and ‘down’ orientations of the complex. One orientation displays higher contrast than the other. This reflects some differences in penetration of the embedding medium between the side interacting with the carbon support and the side interacting with the air, affecting the overall contrast of the molecular envelope. If lower order terms are excluded from the Fourier synthesis, the contrast appears similar for the two orientations (data not shown).
Fig. 3. (A) Representation of amplitudes of Fourier components calculated for one image of a glucose-embedded tetragonal crystal. Numbers and box sizes correspond to the spot IQ value, with spots of the highest signal-to-noise ratio having an IQ of 1 (Henderson et al., 1986). Spots are shown to a resolution of 1/6 Å–1. (B) 8.5Å resolution projection map calculated from five averaged images of glucose-embedded tetragonal crystals, assuming p1 symmetry. Contouring is at 0.5 r.m.s. density with density above mean (protein) represented by solid contours. Scale bar: 50 Å. (C) As (B), with p4212 symmetry applied.
Table I. Mean phase residuals in resolution shells for merged images in p4212 and p22121.
| Resolution range (Å) | No. of spots |
Mean phase residual (°) (random = 45°) |
SEM (°) |
|||
|---|---|---|---|---|---|---|
| Tetragonal | Orthorhombic | Tetragonal | Orthorhombic | Tetragonal | Orthorhombic | |
| ∞–15 | 51 | 110 | 21.0 | 29.1 | 3.3 | 2.4 |
| 15–10.5 | 53 | 109 | 16.7 | 26.4 | 2.6 | 2.2 |
| 10.5–8.5 | 52 | 94 | 38.5 | 35.0 | 3.3 | 2.6 |
| ∞–8.5 | 156 | 333 | 25.4 | 29.4 | 1.9 | 1.3 |
Each complex can be seen to be composed of a central mass (the RC) enclosed by a 16-subunit LH1 ring. The LH1 ring has a projection structure similar to that reported by Karrasch et al. (1995) for LH1 crystals embedded in ice. Figure 3C shows the equivalent map with p4212 symmetry averaging applied and the central RC density clearly contrasted against the background. The 16-fold symmetry of the LH1 complex is clearly evident. Whilst the central RC density has an imposed 4-fold symmetry, the RC density at a high radial distance from the 4-fold origin gives some indication of even higher symmetry. Previous analysis has shown that there could only be room for one RC within the complex (Karrasch et al., 1995), and so any symmetry in the RC density must reflect the superposition of single RCs in different orientations over all the complexes imaged (Stahlberg et al., 1998; Walz et al., 1998).
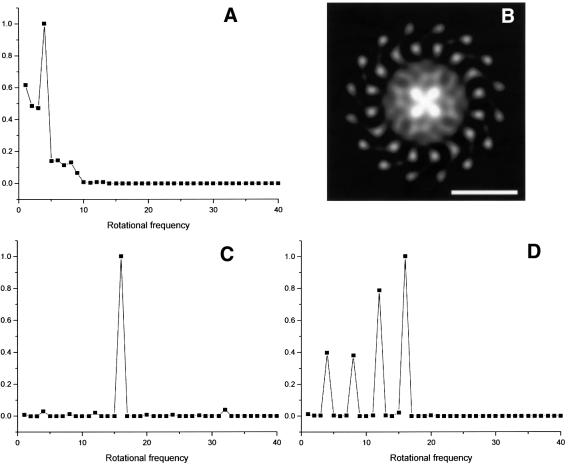
Analysis of rotational symmetry in tetragonal form. Although the observed image phases were consistent with p4212 symmetry, we considered the possibility that this only reflected the symmetry arising from the highly contrasted 16-fold LH1 rings related by crystallographic screw axes in the crystal plane. As a test of the crystallographic symmetry of the RCs, we initially merged and refined the data from individual images assuming no crystallographic symmetry. Figure 4A shows the rotational power spectrum for the central density of one of the two crystallographically independent complexes of the p1 projection (Figure 3B) out to a radius of 28 Å (thereby excluding any LH1 density) (Crowther and Amos, 1971). This indicates a significant 4-fold rotational symmetry. Figure 4B shows a 4-fold rotationally filtered view of the complex in which four lobes of density are visible, projecting from the centre. We computed the density correlation coefficients between the two crystallographically independent RC densities of the p1 projection (after 4-fold rotational averaging of the density) within the program suite IMAGIC (van Heel et al., 1996). Com parison of densities out to a radius of 28 Å gave the highest cross-correlation coefficient (55%) when the mirror image of one RC was matched with the unreflected image of the other RC, without any rotation, and a translation of less than one pixel. This is consistent with the RC densities being related by an in-plane 2-fold screw axis independently of the LH1 densities. In other words, the RC density obeys the crystallographic p4212 symmetry.
Fig. 4. (A) Rotational power spectrum (Crowther and Amos, 1971) of one of the central RC densities extracted from Figure 3B. Density from the centre of the complex to a radius of 28 Å was analysed. (B) Four-fold rotationally filtered density of one complex extracted from Figure 3B. Whiter regions correspond to higher density. Scale bar: 50 Å. (C) Rotational power spectrum of an entire RC–LH1 complex extracted from Figure 3B. (D) Rotational power spectrum of an outer ring of density of one RC extracted from Figure 3B (radius of 22–39 Å), not including any LH1 density.
The rotational power spectrum of the p4212 averaged RC–LH1 complex is dominated by the 16-fold symmetry of the outer LH1 ring (Figure 4C). Interestingly, a rotational power spectrum of a ring of density between 22 and 39 Å radius (Figure 4D) (i.e. not including any LH1 density) shows 8-, 12- and 16-fold harmonics, with the 16-fold being relatively strong. This indicates a possible 16-fold symmetry for the RC density in addition to the crystallographic 4-fold symmetry.
Orthorhombic form. An analysis of symmetry-related phases showed the data to be consistent with p22121 plane group symmetry (Valpuesta et al., 1994). Amplitudes and phases from the eight best images were merged and averaged, imposing either p1 or p22121 symmetry. Origin and CTF refinement was performed independently for the two plane groups, with Fourier terms limited to 8.5 Å resolution. Table I shows the average phase residual as a function of resolution for p22121 symmetry, indicating non-random data to 8.5 Å resolution. Again we see two oppositely oriented RC–LH1 complexes, with a 16-subunit LH1 ring (Figure 5). The central RC density, which has imposed 2-fold symmetry, does not show any marked higher symmetry in the way that the RC in the tetragonal crystal form does, suggesting a preference for only two orientations relative to the crystal lattice, with the consequent 2-fold averaging of the central density.
Fig. 5. (A) 8.5 Å resolution projection map calculated from eight averaged images of glucose-embedded orthorhombic crystals, assuming p22121 symmetry. Exact circles have been drawn through the innermost α- and β-polypeptide densities. Contouring is at 0.5 r.m.s. density with density above mean (protein) represented by solid contours. Scale bar: 50 Å. (B) A grey level representation of the density in (A).
In this crystal form, the negative staining pattern does not give the impression of a square RC–LH1 complex (Figure 2C), but, remarkably, the surrounding LH1 is also not circular. Figure 5A shows how the density peak positions for the putative α-helices of the subunits deviate from an exact circular assembly. These peak positions describe an ellipse whose major and minor axes differ by ∼11%.
Discussion
Subunit structure and symmetry
The 8.5 Å resolution projection structures of the R.rubrum RC–LH1 complex (Figures 3C and 5) are sufficiently detailed to clearly show 16 LH1 αβ subunits arranged in a closed ring around the central RC density. The rotational power spectrum of the tetragonal form (Figure 4C) shows an unambiguous 16-fold symmetry; no significant difference can be observed between the densities representing the crystallographically independent LH1 subunits, leading to the conclusion that the RC is surrounded by 16 identical protein subunits. There is no evidence in these projection maps for any other minor protein component in the outer LH1 ring. In both maps, the area enclosed within the LH1 ring is occupied by significant positive density (overall mean unit cell density is zero); this density corresponds to the RC, as proposed by earlier workers for RC–LH1 complexes from a number of species (Miller, 1982; Stark et al., 1984; Engelhardt et al., 1986; Boonstra et al., 1994; Karrasch et al., 1995; Walz and Ghosh, 1997; Stahlberg et al., 1998; Walz et al., 1998). Although the LH1 ring is only able to accommodate one RC, the RC density apparently has a significant 4-fold rotational symmetry in the tetragonal form and 2-fold symmetry in the orthorhombic form. By treating RC–LH1 complexes in negatively stained tetragonal 2D crystals as single particles, Stahlberg et al. (1998) showed that individual RCs adopted one of four possible orientations with respect to the crystal axes; the 4-fold symmetry we see in our analysis simply arises from the superposition of all RCs in these four possible orientations. However, the rotational power spectrum of the RC density at higher radius, in addition to showing a 4-fold component, shows an even stronger 16-fold harmonic (Figure 4D), suggesting that there must also be a 16-fold symmetry to the RC density, not resolved at lower radius. The propensity of the RC to adopt up to 16 discrete orientations suggests a specific interaction binding the RC within the antenna ring. It is likely that the crystal contacts perturb the 16-fold symmetry of the LH1 so as to favour four major occupancy RC orientations, with a further 12 minor occupancy orientations to give 16 in total.
An analysis for the orthorhombic form indicated only two major orientations for the RC and very low occupancy indeed of other minor orientations (data not shown). What the structure of this orthorhombic form clearly demonstrates is the ability of the LH1 ring to adopt circular and elliptical conformations. Inspection of the RC complex in projection shows that it possesses long and short axes, so it might be expected that the LH1 ring could distort to pack round the RC, when subjected to external packing constraints in the orthorhombic crystal lattice.
Comparison with the LH1 ring in the absence of RC
The 8.5 Å projection data for LH1 from R.rubrum (Karrasch et al., 1995), together with the present data on RC–LH1, provide a valuable opportunity to assess the effect of binding one membrane protein to another, in this case the association of a RC with a light-harvesting complex. It should be noted that there was a difference in the way that the two complexes, LH1 and RC–LH1, were prepared. In the study of Karrasch et al., the 2D crystals had been grown from dissociated subunits prepared from a carotenoidless mutant of R.rubrum; in the present study, the RC–LH1 complex contained carotenoids, and was not dissociated at any step of the purification or crystallization, so it is likely that the 16-fold arrangement of subunits represents the in vivo structure. Despite this difference between LH1 and RC–LH1 complexes in terms of preparation, the peaks of density of the α- and β-chains have, within experimental uncertainty, the same coordinates in both the LH1 map of Karrasch et al. (1995) and our tetragonal map (Figure 3C), suggesting that there has been no significant conformational change within the individual subunits upon incorporation of the RC. However, the elliptical arrangement of LH1 subunits in the orthorhombic crystal form (Figure 5) shows that a conformational change in the entire LH1 assembly is possible. This points to a flexibility in the whole LH1 ring assembly, which may lead to fluctuations in the degree of ellipticity, as has been inferred from fluorescence studies of LH2 complexes immobilized on a mica surface (Bopp et al., 1999).
Comparison with other RC–LH1 complexes
Our p4212 crystals display the same characteristics as those of a carotenoidless mutant analysed at lower resolution by Walz and Ghosh (1997) and Stahlberg et al. (1998), with similar unit cell dimensions and four high-occupancy orientations for the RC with respect to the crystal axes. Moreover, maps from crystals embedded in negative stain (Figure 2A) or negative stain–glucose mixtures (Figure 2B) can give a rather ‘square’ appearance to the RC–LH1 complexes, depending on the degree of staining. However, negative stain tends to contrast mainly the external surfaces of 2D crystals, and this square-like appearance most likely arises from the accumulation of stain in ‘pools’ arranged in the square crystal lattice. In our images of unstained crystals where the internal protein structure is revealed, the LH1 appears to show little deviation from a true circle in the tetragonal crystal form (Figure 3C). There is no evidence of any square-like distortion of the LH1 induced by either an additional protein component or by crystal packing, as had been suggested by earlier work (Stahlberg et al., 1998). Moreover, we have discovered a new orthorhombic crystal form, which when embedded in negative stain always appears to contain ‘circular’ complexes (Figure 2C). Subsequent higher resolution analysis does indicate a significant deviation from circular symmetry, but into a 2-fold symmetric elliptical form, not as a square (Figure 5).
We have attempted, in an approximate way, to model this 2-fold symmetric RC density using atomic coordinates derived from the R.sphaeroides structure. We have assumed that contrast will arise mainly from protein embedded within the transmembrane region, but rather than attempt to model the density of the surround ing medium, we have calculated an 8.5 Å projection map from coordinates of atoms in a vacuum. A projection perpendicular to the putative membrane plane was calculated. Rotational and translational alignment of this density, followed by 2-fold averaging, gave a correlation coefficient between model and experimental density of 0.7 for the best fit. In this orientation, the four RC light subunit helices LA, LB, LC and LE are superimposed on four relatively dense regions separated by ∼10 Å in the RC map (marked in red in Figure 6A). The long axis of the RC model coincides approximately with the major axis of the elliptical LH1 ring. The pseudo 2-fold axis of the RC running through the Fe and between the ‘special pair’ of Bchls lies within 2–3 Å of the crystallographic 2-fold axis. With the uncertainties that arise from calculating model density at low resolution in such a complex environment of lipid, protein, pigment and glucose, the uncertainty in orientation must be at least ±3°. However, within this degree of uncertainty, the planes of the Bchl porphyrin rings make an angle of ∼43° with the major axis of the LH1 ring.
Fig. 6. (A) Density for one RC–LH1 complex extracted from Figure 5B. Four prominent density peaks separated by ∼10 Å are marked in red. The 2-fold symmetry-equivalent peaks are not marked. The straight line represents the approximate major axis of the elliptical LH1 ring. (B) Representation of the transmembrane helices of the R.sphaeroides RC aligned to give a best fit with the experimental density. Running from high radius to the centre of the complex, red helices form an arc in the sequence corresponding to LA, LB, LC, LE, LD; likewise, blue helices form an arc in the sequence MA, MB, MC, ME, MD; the yellow helix corresponds to HA; green, ‘special pair’ Bchls; orange, QB. This view is from the cytoplasmic face. The model image was generated with Molscript (Kraulis, 1991) and Raster3D (Merrit and Bacon, 1997). Scale bar: 50 Å.
There are few data on the detailed nature of the interactions of the RC with the LH1, although cross-linking experiments suggest that L, M and H subunits all interact in some way with LH1 subunits (Peters et al., 1983). In our model, within the membrane, the only RC transmembrane helices that could interact with the putative LH1α helices would be LA, MA and possibly LB and MB; all other helices are much further than 10 Å from their nearest LH1 neighbours. A near-straight line can be drawn linking the centres of gravity of two diametrically opposed LH1α subunits and the LA and MA helices. This would be consistent with a helix–helix packing arrangement of LA and MA against nearest-neighbour LH1α subunits.
This study represents the highest resolution view, to date, of any RC–LH1 complex from a purple bacterium. The previous study by Karrasch et al. (1995), which indicated that R.rubrum forms a closed LH1 ring, raised the question of how quinone transfer takes place between the apparently fully encircled RC and the spatially distant cytochrome bc1 complex during cyclic electron flow. The projection maps of the RC–LH1 complex (Figures 3C and 5) would appear to confirm that the RC is indeed completely enclosed by 16 identical LH1 subunits, so that the mechanism of quinone transfer remains unclear. This is in the context of work on the PufX polypeptide from R.sphaeroides and Rhodobacter capsulatus, which has shown that it facilitates the transport of reducing equivalents from the RC to the bc1 complex (Farchaus et al., 1990, 1992; Lilburn et al., 1992). It has been shown that PufX is unnecessary in strains that cannot assemble a normal LH1 complex (McGlynn et al., 1994, 1996). It has been suggested that PufX might affect the structure of the LH1 so as to allow the free diffusion of quinone, perhaps even substituting for one or more LH1 subunits (Bopp et al., 1999). Moreover, Jungas et al. (1999) have proposed that the functional photosynthetic unit in R.sphaeroides is made up of a dimeric RC–LH1–PufX assembly with the two RCs only partially encircled by LH1 subunits, this open assembly presumably allowing the free movement of quinone from RC to bc1 complex. However, the resolution was not sufficient to reveal individual LH1 subunits. To date, no such assembly has been seen in R.rubrum and no equivalent of the pufX gene has been found. It has been proposed that the functional RC–LH1 complex might incorporate an additional small (4 kDa) polypeptide component, Ω (Ghosh et al., 1994), and that this might form a quinone channel (Walz and Ghosh, 1997; Stahlberg et al., 1998). However, no amino acid sequence is available for this polypeptide and, within current experimental uncertainty, all 16 subunits seen in the projection of the LH1 ring (Figures 3C and 5A) appear identical. An EM analysis of the structure in three dimensions and/or a specific labelling experiment might help to clarify this issue. In the absence of any evidence yet for a ‘quinone channel’ or fixed opening in the LH1 ring, the alternative possibilities of a close approach of quinone molecules either side of the LH1 or ‘breathing motions’ in LH1 could still be entertained, at least for quinone transfer in R.rubrum (Walz and Ghosh, 1997). Certainly, the LH1 is not a rigid assembly, as shown by the circular and elliptical forms found in this study. Fluctuations in ellipticity have also been observed in the case of LH2, and it has been suggested that there might even be a partial dissociation of the antenna assembly (Bopp et al., 1999). Could such a dissociation take place in LH1, thereby allowing free diffusion of the quinone?
Conclusion
This study represents the highest resolution view of any purple bacterial RC–LH1 complex to date, with the RC apparently enclosed by 16 identical LH1 subunits in an arrangement that can adopt a circular or elliptical form, showing that the LH1 assembly is likely to be flexible in the bacterial membrane. However, we have found no evidence that the LH1 can adopt a square form and no evidence for any other protein component in the antenna. The RC appears to make specific interactions with the LH1 rather than adopting a continuum of possible orientations within the antenna ring. The quality of the 2D crystals obtained may be sufficient to allow us to pursue a structure determination of the LH1 in three dimensions to a relatively high resolution, although the RC density will represent the superposition of a number of different orientations.
Materials and methods
Growth of R.rubrum and purification of core complexes
Cultures of R.rubrum strain S1 were grown under anaerobic heterotrophic conditions at 30°C in M22 medium. Cells were harvested in the late exponential phase by centrifugation at 3300 g at 4°C. Harvested cells were washed with buffer A (10 mM Tris–HCl, 1 mM EDTA pH 7.9) and stored at –70°C. Cells were suspended in buffer A with a few grains of DNase and MgCl2, and disrupted by French pressing. Cells were spun on a two-layer density gradient (15%, 40% sucrose in buffer A) for 4 h at 100 000 g. Intracytoplasmic membrane was recovered from just above the 40% sucrose layer. The intracytoplasmic membrane fraction was centrifuged at 150 000 g for 1 h at 4°C, and resuspended and solubilized in 40 mM DHPC + buffer A at 4°C. Insoluble material was removed by centrifugation (150 000 g for 1 h at 4°C). A DEAE–Sepharose ion-exchange column was equilibrated with buffer A + 3 mM DHPC. Solubilized core complexes were loaded onto the column. The column was washed stepwise with buffer A, 3 mM DHPC, 0–120 mM NaCl. A 100 ml gradient of 120–300 mM NaCl in 3 mM DHPC containing buffer A was used to elute the complex. The ratio of A875 to A280 was used as an estimate of the relative purity, and the A875 to A800 ratio was used as a relative estimate of RC content. Fractions with an A875 to A280 ratio of >2.0 were pooled and concentrated in a Centricon-50 centrifugal filter. The concentrated sample was further purified on a Superdex-200 size-exclusion column. In this case, the column was equilibrated and run with 3 mM DHPC, 50 mM NaCl in buffer A. Fractions with A875 to A800 ratios of 2.0–2.2 were used for 2D crystallization trials. Protein concentrations were determined by the BCA assay method (Pierce and Warriner Ltd, Chester, UK). Gel electrophoresis was performed according to Schägger and von Jagow (1987).
Crystallization
Purified RC–LH1 was mixed with lipids at LPR weight ratios ranging from 0.4 to 1.4. Lipids tested were DOPC solubilized in DHPC and mixed Escherichia coli lipids solubilized in decyl maltoside. Samples were diluted with the dialysis buffer to be used in the crystallization trial to give a final protein concentration of 0.5 mg/ml. Crystallization was attempted by the slow removal of detergent using a home-built continuous-flow dialysis machine (Engel et al., 1992; Jap et al., 1992). Temperature was controlled through a programmable device (ITC 502; Oxford Instruments, UK) powering a resistive pad mounted under the heating block, with ambient temperature being maintained by recirculated water passing through a manifold mounted under the heating block. During the first 2 h of dialysis, the temperature was increased from 20 to 35°C, where it was held for 6 h. The temperature was then decreased gradually back to 20°C over a 2 h period, where it was then held for a further 6 h. This temperature profile was repeated four times in total, giving a total dialysis time of 64 h.
EM
Reconstituted samples were examined in 0.75% (w/v) uranyl formate on a Philips CM100 electron microscope. Electron micrographs were recorded at a nominal magnification of 50 000× and underfocus of 4000–7000 Å. For high-resolution microscopy, crystals were embedded in 1% (w/v) glucose instead of negative stain or mixtures of 1% glucose and 1% uranyl formate in a ratio of 9:1. In the majority of cases, data were recorded from freshly prepared grids. However, we found that rapid freezing in liquid ethane (Bellare et al., 1988) and storing under liquid nitrogen could also preserve grids. High-resolution micrographs were recorded on a Philips CM200 FEG electron microscope equipped with a field emission gun. Grids were mounted on an Oxford cold stage and cooled with liquid nitrogen to a temperature between –175 and –180°C after insertion into the microscope. For EM of previously frozen grids, the grids were mounted on a pre-cooled stage prior to insertion in the microscope. Low-dose images were recorded at 200 kV from nominally untilted crystals. The nominal magnification was 50 000×, exposure time was 0.7–1.0 s with a beam of 1–1.5 µm diameter at the specimen in ‘flood beam’ mode. For ‘spot scan’ imaging (Downing and Glaeser, 1986; Bullough and Henderson, 1987), a beam diameter of 0.3 µm and exposure time of 0.4 s were used in a raster scan, using the Philips software. Flood beam and spot scan exposures amounted to an electron dose of 5–10 electrons/Å2 at the specimen. All images were recorded on Kodak SO-163 film and developed in concentrated Kodak D19 developer for 12 min. Films were fixed with Ilford fixer for 5 min and washed.
Image processing
Low resolution. Selected areas were digitized as 1000 × 1000 pixel arrays in steps of 21 µm on a Zeiss SCAI densitometer. Image processing and merging of image amplitudes and phases followed procedures described previously (Henderson et al., 1986; Havelka et al., 1993; Crowther et al., 1996). Structure factors were not corrected for the effects of the phase and amplitude contrast transfer functions.
High resolution. Selected areas were digitized as 4000 × 4000 pixel arrays in steps of 7 µm and processed as above. The program ALLSPACE was used to determine the crystal symmetry (Valpuesta et al., 1994). We discovered two crystal forms: a tetragonal p4212 form and an orthorhombic p22121 form. In the early stages of processing, we assumed only p1 symmetry, although there was no attempt to distinguish the two possible hands predicted for projections of crystals with this symmetry, and in the case of the tetragonal crystal we assumed that a and b were equivalent, since the uncertainties in inter-image phase residuals were greater than any difference found for each possible hand or axis assignment. The phase origins of five tetragonal form images or eight orthorhombic form images were refined against each other at successively higher resolutions out to 8.5 Å by minimizing the average phase residual (ORIGTILTK). Defocus and astigmatism were refined for each image by comparison with averaged phases from all the other images using the program CTFSEARCH (Crowther et al., 1996). We estimated the Fourier amplitudes from the amplitudes measured in image transforms. However, these amplitudes were significantly attenuated by the fall-off arising from the CTF, astigmatism and image or specimen drift. The program SCALIMAMP3D was used to correct for the effects of the CTF and secondly to apply a resolution-dependent scale factor separately to the amplitudes from each image by comparison of the amplitudes with a reference data set (Havelka et al., 1995). Amplitudes and phases from the images were averaged as described previously (Henderson et al., 1986). Average structure factors, weighted by their figure of merit (Bullough and Tulloch, 1990), were used to calculate p1 projection maps.
Further refinement of image phases was carried out subsequently, assuming p4212 or p22121 symmetry. Projection maps were calculated using the corrected and averaged image amplitudes and phases, weighted by their figures of merit as described previously (Bullough and Tulloch, 1990), with phases set to 0 or 180° for the centrosymmetric reflections.
Analysis of non-crystallographic symmetry. For both p1 maps, crystallographically independent RC–LH1 complexes were masked within a circle or ring of inner and outer diameters sufficient to include either the entire RC–LH1 density or the RC density alone. The masked density was subjected to a rotational power analysis (Crowther and Amos, 1971). Similar analyses were performed on crystallographically averaged complexes, including the additional analysis of a ring including only the higher radius RC density.
Modelling of the RC density. A projection map of the transmembrane domain of the R.sphaeroides RC was calculated to 8.5 Å resolution from a set of atomic coordinates (Chang et al., 1991) with an overall temperature factor of 200 Å2 applied. The projection was calculated in a direction approximately perpendicular to the putative membrane plane. The 2-fold averaged projection density was aligned with the central RC density of the experimental p22121 map using programs in the IMAGIC software suite (van Heel et al., 1996).
Acknowledgments
Acknowledgements
P.A.B. and C.N.H. gratefully acknowledge financial support from the BBSRC. We would like to thank Lynne Kennedy-Johnson for excellent technical assistance.
References
- Agarwal R., Rizvi,A.H., Prall,B.S., Olsen,J.D., Hunter,C.N. and Fleming,G.R. (2002) The nature of disorder and inter-complex energy transfer in LH2 at room temperature: a three pulse photon echo peak shift study. J. Phys. Chem B, 18, 624–632. [Google Scholar]
- Allen J.P., Feher,G., Yeates,T.O., Komiya,H. and Rees,D.C. (1987) Structure of the reaction center from Rhodobacter sphaeroides R-26: the cofactors. Proc. Natl Acad. Sci. USA, 84, 5730–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare J.R., Davis,H.T., Scriven,L.E. and Talmon,Y. (1988) Controlled environment vitrification system: an improved sample preparation technique. J. Electron Microsc. Tech., 10, 87–111. [DOI] [PubMed] [Google Scholar]
- Boonstra A.F., Germeroth,L. and Boekema,E. (1994) Structure of the light-harvesting antenna from Rhodospirillum molischianum studied by electron microscopy. Biochim. Biophys. Acta, 1184, 227–234. [Google Scholar]
- Bopp M.A., Sytnik,A., Howard,T.D., Cogdell,R.J. and Hochstrasser,R.M. (1999) The dynamics of structural deformations of immobilized single light-harvesting complexes. Proc. Natl Acad. Sci. USA, 96, 11271–11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough P.A. and Henderson,R. (1987) Use of spot-scan procedure for recording low-dose micrographs of beam-sensitive specimens. Ultramicroscopy, 21, 223–229. [Google Scholar]
- Bullough P.A. and Tulloch,P.A. (1990) High-resolution spot-scan electron microscopy of microcrystals of an α-helical coiled-coil protein. J. Mol. Biol., 215, 161–173. [DOI] [PubMed] [Google Scholar]
- Chang C.H., el-Kabbani,O., Tiede,D., Norris,J. and Schiffer,M. (1991) Structure of the membrane-bound protein photosynthetic reaction centre from Rhodobacter sphaeroides.Biochemistry, 30, 5352–5360. [DOI] [PubMed] [Google Scholar]
- Conroy M.J., Westerhuis,W.H., Parkes-Loach,P.S., Loach,P.A., Hunter,C.N. and Williamson,M.P. (2000) LH1β reveals two helical domains separated by a more flexible region: structural consequences for the LH1 complex. J. Mol. Biol., 298, 83–94. [DOI] [PubMed] [Google Scholar]
- Crowther R.A. and Amos,L.A. (1971) Harmonic analysis of electron microscope images with rotational symmetry. J. Mol. Biol., 60, 123–130. [DOI] [PubMed] [Google Scholar]
- Crowther R.A., Henderson,R. and Smith,J.M. (1996) MRC image processing programs. J. Struct. Biol., 116, 9–16. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J., Epp,O., Miki,K., Huber,R. and Michel,H. (1985) X-ray structure analysis at 3 Å resolution of a membrane protein complex: folding of the protein subunits in the photosynthetic reaction centre from Rhodopseudomonas viridis. Nature, 318, 618–624. [DOI] [PubMed] [Google Scholar]
- Downing K.H. and Glaeser,R.M. (1986) Improvement in high resolution image quality of radiation-sensitive specimens achieved with reduced spot size of the electron beam. Ultramicroscopy, 20, 269–278. [DOI] [PubMed] [Google Scholar]
- Engel A., Hoenger,A., Hefti,A., Henn,C., Ford,R.C., Kistler,J. and Zulauf,M. (1992) Assembly of 2-D membrane protein crystals: dynamics, crystal order, and fidelity of structure analysis by electron microscopy. J. Struct. Biol., 109, 219–234. [DOI] [PubMed] [Google Scholar]
- Engelhardt H., Engel,A. and Baumeister,W. (1986) Stoichiometric model of the photosynthetic unit of Ectothiorhodospira halochloris. Proc. Natl Acad. Sci. USA, 83, 8972–8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farchaus J.W., Gruenberg,H. and Oesterhelt,D. (1990) Complementation of a reaction center-deficient Rhodobacter sphaeroides pufLMX deletion strain in trans with pufBALM does not restore the photosynthesis-positive phenotype. J. Bacteriol., 172, 977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farchaus J.W., Barz,W.P., Grunberg,H. and Oesterhelt,D. (1992) Studies on the expression of the pufX polypeptide and its requirement for photoheterotrophic growth in Rhodobacter sphaeroides.EMBO J., 11, 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Ghosh-Eicher,S., Di Berardino,M. and Bachofen,R. (1994) Protein phosphorylation in Rhodospirillum rubrum—characterization of a water-soluble B873 protein-kinase and a new component of the B873 complex, Ω, which can be phosphorylated. Biochim. Biophys. Acta, 1184, 28–36. [Google Scholar]
- Havelka W.A., Henderson,R., Heymann,J.A. and Oesterhelt,D. (1993) Projection structure of halorhodopsin from Halobacterium halobium at 6 Å resolution obtained by electron cryo-microscopy. J. Mol. Biol., 234, 837–846. [DOI] [PubMed] [Google Scholar]
- Havelka W.A., Henderson,R. and Oesterhelt,D. (1995) Three-dimensional structure of halorhodopsin at 7 Å resolution. J. Mol. Biol., 247, 726–738. [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin,J.M., Downing,K.H., Lepault,J. and Zemlin,F. (1986) Structure of purple membrane from Halobacterium halobium— recording, measurement and evaluation of electron-micrographs at 3.5 Å resolution. Ultramicroscopy, 19, 147–178. [Google Scholar]
- Hess S., Chachisvilis,M., Timpmann,K., Jones,M.R., Fowler,G.J., Hunter,C.N. and Sundström,V. (1995) Temporally and spectrally resolved subpicosecond energy transfer within the peripheral antenna complex (LH2) and from LH2 to the core antenna complex in photosynthetic purple bacteria. Proc. Natl Acad. Sci. USA, 92, 12333–12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Damjanovic,A., Ritz,T. and Sculten,K. (1998) Architecture and mechanism of the light-harvesting apparatus of purple bacteria. Proc. Natl Acad. Sci. USA, 95, 5935–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jap B.K., Zulauf,M., Scheybani,T., Hefti,A., Baumeister,W., Aebi,U. and Engel,A. (1992) 2D crystallization: from art to science. Ultramicroscopy, 46, 45–84. [DOI] [PubMed] [Google Scholar]
- Jungas C., Ranck,J.L., Rigaud,J.L., Joliot,P. and Vermeglio,A. (1999) Supramolecular organization of the photosynthetic apparatus of Rhodobacter sphaeroides. EMBO J., 18, 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrasch S., Bullough,P.A. and Ghosh,R. (1995) The 8.5 Å projection map of the light-harvesting complex I from Rhodospirillum rubrum reveals a ring composed of 16 subunits. EMBO J., 14, 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepke J., Hu,X., Muenke,C., Schulten,K. and Michel,H. (1996) The crystal structure of the light-harvesting complex II (B800–850) from Rhodospirillum molischianum.Structure, 4, 581–597. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) Molscript—a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lilburn T.G., Haith,C.E., Prince,R.C. and Beatty,J.T. (1992) Pleiotropic effects of pufX gene deletion on the structure and function of the photosynthetic apparatus of Rhodobacter capsulatus.Biochim. Biophys. Acta, 1100, 160–170. [DOI] [PubMed] [Google Scholar]
- McDermott G., Prince,S.M., Freer,A.A., Hawthornthwaite-Lawless,A.M., Papiz,M.Z., Cogdell,R.J. and Isaacs,N.W. (1995) Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria Nature, 374, 517–525. [Google Scholar]
- McGlynn P., Hunter,C.N. and Jones,M.R. (1994) The Rhodobacter sphaeroides PufX protein is not required for photosynthetic competence in the absence of a light harvesting system. FEBS Lett., 349, 349–353. [DOI] [PubMed] [Google Scholar]
- McGlynn P., Westerhuis,W.H., Jones,M.R. and Hunter,C.N. (1996) Consequences for the organization of reaction center-light harvesting antenna 1 (LH1) core complexes of Rhodobacter sphaeroides arising from deletion of amino acid residues from the C terminus of the LH1α polypeptide. J. Biol. Chem., 271, 3285–3292. [DOI] [PubMed] [Google Scholar]
- Merrit E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 504–524. [DOI] [PubMed] [Google Scholar]
- Miller K.R. (1982) Three-dimensional structure of a photosynthetic membrane. Nature, 300, 53–55. [Google Scholar]
- Peters J., Takemoto,J. and Drews,G. (1983) Spatial relationships between the photochemical-reaction center and the light-harvesting complexes in the membrane of Rhodopseudomonas capsulata.Biochemistry, 22, 5660–5667. [Google Scholar]
- Savage H., Cyrklaff,M., Montoya,G., Kuhlbrandt,W. and Sinning,I. (1996) Two-dimensional structure of light harvesting complex II (LHII) from the purple bacterium Rhodovulum sulfidophilum and comparison with LHII from Rhodopseudomonas acidophila. Structure, 4, 243–252. [DOI] [PubMed] [Google Scholar]
- Schägger H. and von Jagow,G. (1987) Tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem., 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Scheuring S., Reiss-Husson,F., Engel,A., Rigaud,J.L. and Ranck,J.L. (2001) High-resolution AFM topographs of Rubrivivax gelatinosus light-harvesting complex LH2. EMBO J., 20, 3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlberg H., Dubochet,J., Vogel,H. and Ghosh,R. (1998) Are the light-harvesting I complexes from Rhodospirillum rubrum arranged around the reaction centre in a square geometry? J. Mol. Biol., 282, 819–831. [DOI] [PubMed] [Google Scholar]
- Stark W., Kühlbrandt,W., Wildhaber,H., Wehrli,E. and Mühlethaler,K. (1984) The structure of the photoreceptor unit of Rhodopseudomonas viridis. EMBO J., 3, 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström V., Pullertis,T. and van Grondelle,R. (1999) Photosynthetic light-harvesting: reconciling dynamics and structure of purple bacterial LH2 reveals function of photosynthetic unit. J. Phys. Chem., 103, 2327–2346. [Google Scholar]
- Valpuesta J.M., Carrascosa,J.L. and Henderson,R. (1994) Analysis of electron microscope images and electron diffraction patterns of thin crystals of φ29 connectors in ice. J. Mol. Biol., 240, 281–287. [DOI] [PubMed] [Google Scholar]
- van Heel M., Harauz,G. and Orlova,E.V. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol., 116, 17–24. [DOI] [PubMed] [Google Scholar]
- Walz T. and Ghosh,R. (1997) Two-dimensional crystallization of the light-harvesting I-reaction centre photounit from Rhodospirillum rubrum. J. Mol. Biol., 265, 107–111. [DOI] [PubMed] [Google Scholar]
- Walz T., Jamieson,S.J., Bowers,C.M., Bullough,P.A. and Hunter,C.N. (1998) Projection structures of three photosynthetic complexes from Rhodobacter sphaeroides: LH2 at 6 Å, LH1 and RC–LH1 at 25 Å. J. Mol. Biol., 282, 833–845. [DOI] [PubMed] [Google Scholar]