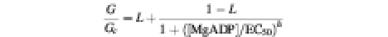Abstract
ATP-sensitive potassium (KATP) channels are composed of an ATP-binding cassette (ABC) protein (SUR1, SUR2A or SUR2B) and an inwardly rectifying K+ channel (Kir6.1 or Kir6.2). Like other ABC proteins, the nucleotide binding domains (NBDs) of SUR contain a highly conserved ‘signature sequence’ (the linker, LSGGQ) whose function is unclear. Mutation of the conserved serine to arginine in the linker of NBD1 (S1R) or NBD2 (S2R) did not alter the ability of ATP or ADP (100 µM) to displace 8-azido-[32P]ATP binding to SUR1, or abolish ATP hydrolysis at NBD2. We co-expressed Kir6.2 with wild-type or mutant SUR in Xenopus oocytes and recorded the resulting currents in inside-out macropatches. The S1R mutation in SUR1, SUR2A or SUR2B reduced KATP current activation by 100 µM MgADP, whereas the S2R mutation in SUR1 or SUR2B (but not SUR2A) abolished MgADP activation completely. The linker mutations also reduced (S1R) or abolished (S2R) MgATP-dependent activation of Kir6.2-R50G co-expressed with SUR1 or SUR2B. These results suggest that the linker serines are not required for nucleotide binding but may be involved in transducing nucleotide binding into channel activation.
Keywords: ABC signature sequence/ABC transporter/KATP channel/nucleotide binding domain/sulfonylurea receptor
Introduction
ATP-binding cassette (ABC) proteins constitute the largest known family of transmembrane proteins (Dean et al., 2001). They serve as ATP-dependent transporters, channels and channel regulators in both prokaryotes and eukaryotes (Higgins, 1992). Many human ABC protein genes play a causative role in disease, including cystic fibrosis (ABCC7), congenital hyperinsulinaemia (ABCC8), retinal degeneration (ABCA4), bile transport disorders (ABCB4), anaemia (ABCB7) and adrenoleukodystrophy (ABCD1), and other ABC proteins are involved in drug resistance (e.g. ABCB1) (Dean et al., 2001).
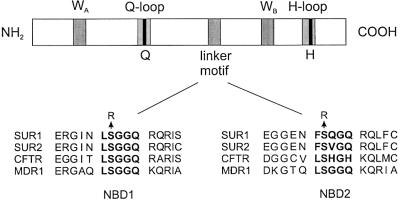
ABC proteins are characterized by the presence of four structural domains: two cytosolic nucleotide binding domains (NBDs) and two integral membrane domains, each containing 6–8 transmembrane α-helices (TMs) (Higgins, 1992). In prokaryotes, these domains are often separate subunits that co-assemble to produce a functional ABC protein, whereas in eukaryotes a single gene usually encodes both NBDs and transmembrane elements. There is considerable homology between the NBDs of ABC proteins: in particular, each NBD contains a highly conserved Walker A (WA) and Walker B (WB) motif, and an intervening linker motif (LSGGQ), unique to ABC proteins, which is known as the ABC signature sequence (Figure 1). The WA and WB motifs are involved in nucleotide binding and hydrolysis, but the function of the linker motif is less clear. As it is conserved throughout all ABC proteins, however, it is thought to play a critical role in their function.
Fig. 1. Schematic showing the structure of the NBD of an archetypal ABC transporter. Below is the sequence alignment of the linker region of the NBDs of human SUR1, SUR2, CFTR and MDR1, showing the extensive sequence conservation.
The crystal structures of a number of bacterial ABC proteins (e.g. MsbA, BtuCD), or their isolated NBDs (eg. HisP, MalK, MJ0796, TAP1), have recently been elucidated, enabling identification of residues that interact directly with ATP (Hung et al., 1998; Diederichs et al., 2000; Chang and Roth, 2001; Gaudet and Wiley, 2001; Yuan et al., 2001; Locher et al., 2002). The NBD monomers have the same basic fold, but the predicted structure of the dimer, which is required for functional activity, differs. Although the WA sequence of MsbA is disordered in the electron density maps, the crystal structure reveals that the linker motif is in direct contact with an intracellular domain that forms an extension of the transmembrane domains (Chang and Roth, 2001). In HisP and MalK, the linker motif lies at some distance from the ATP binding pocket, and is suggested to be located at the membrane interface, facing the membrane subunits (Hung et al., 1998; Diederichs et al., 2000). Comparison of the ATP-bound form of HisP with the MgADP-bound form of MJ0796 has led to the suggestion that the linker motif moves during the ATP hydrolysis step, and is involved in coupling ATPase activity to the transmembrane domains and the transport activity of the protein (Yuan et al., 2001).
In contrast, the linker motif participates in ATP binding to the catalytic domain of Rad50cd, a DNA repair enzyme that shares homology with the NBDs of ABC proteins (Hopfner et al., 2000). A similar configuration has been determined for BtuCD (Locher et al., 2002). The crystal structures of Rad50cd and BtuCD reveal that the overall fold resembles that of HisP, MalK, MJ1267 and MJ0796. However, each ATP-binding site is formed by two subunits, with the ATP molecule lying at the dimer interface, sandwiched between the WA motif of one NBD and the linker motif of the other. The γ-phosphate of ATP binds to the conserved serine and the second glycine in the LSGGQ linker motif. The Rad50cd and BtuCD structures provide an explanation for the functional co-operativity observed between the NBDs of many ABC proteins. However, it is not known which of the published crystal structures most closely resembles the NBDs of eukaryotic ABC proteins. Furthermore, as all the NBDs, with the exception of MsbA and BtuCD, were crystallized in the absence of the transmembrane domains, interactions between the TMs and NBDs remain unclear.
Mutation of the linker residues in bacterial ABC proteins supports the idea that the linker motif is not involved in ATP binding, but rather in ATP hydrolysis or transduction. Thus, mutation of the linker motif in the NBDs of HisP, MalK and HlyB did not impair ATP binding, but abolished both ATPase and transport activity (Shyamala et al., 1991; Koronakis et al., 1995; Schmees et al., 1999). Furthermore, the equivalent mutations in the NBDs of the eukaryotic protein MDR1 did not prevent ATP binding or nucleotide trapping, yet produced a complete loss of drug-stimulated ATPase activity (Bakos et al., 1997; Szakacs et al., 2001) and drug transport (Hoof et al., 1994). Likewise, the CFTR linker mutations G551D and S549R did not change the ATP-binding affinity of purified NBD1 (Qu et al., 1997), whereas G551D reduced the ATPase activity (Li et al., 1996).
Although these results suggest that the linker sequence of ABC proteins does not participate in ATP binding, its role in transduction is less clear. In almost all previous studies, the effects of linker mutations on nucleotide binding were examined in isolated NBDs, without their associated TMs, whereas the functional activity of the ABC protein was examined in the full-length protein (CFTR or MDR1) or transporter complex (histidine permease or maltose transporter). It is possible that the structural and functional properties of the NBDs may differ in the isolated state and the intact complex, complicating analysis of the role of linker. An additional difficulty in the case of the transporters is that the energy of ATP hydrolysis is used to transport the substrate, so it is not easy to distinguish between an impairment of transduction or ATP hydrolysis. Furthermore, most mutational analysis of eukaryotic ABC proteins has been performed on the NBD1 linker, yet recent studies suggest that NBD1 and NBD2 of MDR1, MRP1, CFTR and SUR are functionally distinct (Matsuo et al., 1999; Szabo et al., 1999; Gao et al., 2000; Hou et al., 2000; Matsuo et al., 2000; Nagata et al., 2000; Aleksandrov et al., 2001). Therefore, we cannot exclude the possibility that the roles of the linker motifs in NBD1 and NBD2 are different.
The sulfonylurea receptor (SUR) is a member of the ABC protein superfamily that functions as a regulator of the ATP-sensitive potassium (KATP) channel (Ashcroft and Gribble, 1998; Aguilar-Bryan and Bryan, 1999; Seino, 1999). These channels are inhibited by ATP and activated by MgADP, and thereby link cellular metabolism to the membrane potential in tissues such as the pancreatic β-cell, brain, neurones, cardiac muscle and skeletal muscle. KATP channels are heteromultimeric complexes of four SUR and four Kir6.x (Kir6.1 or Kir6.2) subunits. Kir6.x belongs to the inwardly rectifying potassium channel family and forms the channel pore. ATP binding to this subunit closes the channel. SUR functions as a channel regulator, endowing the channel with sensitivity to the stimulatory effects of Mg-nucleotides. There are two isoforms, SUR1 (ABCC8) and SUR2 (ABCC9), with SUR2 being alternatively spliced to give SUR2A and SUR2B, which differ only in their last 42 amino acids.
KATP channels provide a valuable tool for studying the role of the ABC linker motif, because channel activity provides a real-time measurement of NBD function. Furthermore, binding of MgADP to the NBDs of SUR is sufficient to stimulate channel activity, which enables the ATP hydrolysis step to be bypassed. Thus, potassium fluxes through the Kir6.2 pore can be used as a reporter of the transduction step produced by ligand binding to the NBDs of SUR. We have already established a method for measuring nucleotide binding to the NBDs of full-length SUR, which enables binding to each of the two NBDs to be distinguished (Matsuo et al., 1999, 2000). By combining electrophysiological studies with ATP- and ADP-binding measurements, we can determine whether the linker motif of SUR is involved in nucleotide binding or transduction.
The linker motif of NBD1 of SURx is LSGGQ, and thus matches the ABC consensus sequence perfectly, whereas the sequence of the NBD2 linker varies, being LSQGQ in SUR1 and LSVGQ in SUR2 (Aguilar-Bryan et al., 1995; Inagaki et al., 1996; Isomoto et al., 1996) (Figure 1). As in almost all ABC proteins, the serine residue is conserved in both NBDs. In Rad50cd, this invariant serine is involved in ATP binding, and its mutation to arginine abolishes ATP-dependent dimerization (Hopfner et al., 2000). If the structures of HisP, MalK and MJ0796 are correct, this mutation is predicted not to abolish ATP binding, but rather to impair transduction. In this study, therefore, we examined the effects on nucleotide binding, and Mg-nucleotide stimulation of KATP channel activity, of mutating the invariant serine in the linker of NBD1 or NBD2 to arginine.
Results
Functional effects of mutations in the linker domain of SUR
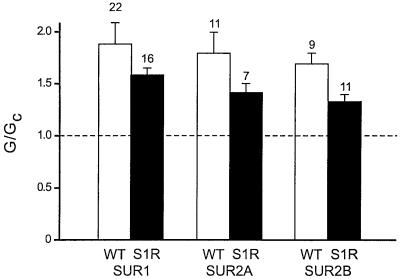
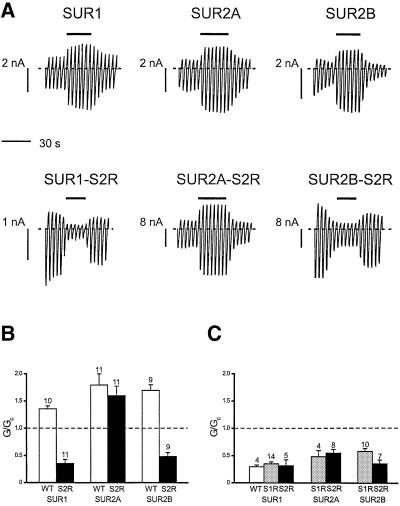
Figure 2 shows that mutation of the invariant serine (S1) in the linker domain of NBD1 of SUR1, SUR2A or SUR2B produced a small but significant decrease in the ability of 100 µM MgADP to stimulate KATP channel activity. The extent of this reduction was similar for all three types of channel. In contrast, the effect of mutating the invariant serine (S2) in the linker domain of NBD2 varied with the type of SUR (Figure 3A). Neither Kir6.2/SUR1-S2R nor Kir6.2/SUR2B-S2R currents were activated by 100 µM MgADP; instead, these channels were inhibited by 65 ± 6.9% (n = 11) and 51 ± 6.8% (n = 9), respectively (Figure 3B). A similar block was observed for wild-type and mutant channels when ADP was tested in the absence of Mg2+ (Figure 3C). As previously reported, the stimulatory effect of MgADP is dependent on Mg2+, and removal of the divalent cation unmasks an inhibitory effect of ADP on Kir6.2 (Gribble et al., 1998). Thus, our results suggest that the stimulatory effect of MgADP on SUR1 and SUR2B is completely abolished by the S2R mutation, whereas the S1R mutation causes only a small decrease in activation. Remarkably, the ability of 100 µM MgADP to stimulate Kir6.2/SUR2A-S2R currents was unimpaired (Figure 3B).
Fig. 2. Mean KATP conductance recorded in the presence of 100 µM MgADP from patches excised from oocytes co-expressing Kir6.2 and either wild-type SUR1, SUR2A or SUR2B (WT, white bars), or SUR1-S1R, SUR2A-S1R or SUR2B-S1R (S1R, black bars). The slope conductance (G) is expressed as a fraction of the mean of that obtained in control solution before and after exposure to MgADP (Gc). The number of patches is given above the bar.
Fig. 3. (A) Macroscopic currents recorded from oocytes co-expressing Kir6.2 and either SUR1, SUR2A, SUR2B, SUR1-S2R, SUR2A-S2R or SUR2B-S2R in response to a series of voltage ramps from –110 to +100 mV. ADP (100 µM) was added to the intracellular solution as indicated by the bars. All solutions contained Mg2+. (B) Mean KATP conductance recorded in the response to 100 µM ADP in the presence of 2 mM Mg2+ from patches excised from oocytes co-expressing Kir6.2 and either wild-type SUR1, SUR2A or SUR2B (WT, white bars), or SUR1-S2R, SUR2A-S2R or SUR2B-S2R (S2R, black bars). The slope conductance (G) is expressed as a fraction of the mean of that obtained in control solution before and after exposure to MgADP (Gc). The number of patches is given above the bar. (C) Mean KATP conductance recorded in the response to 100 µM ADP in the absence of Mg2+ from patches excised from oocytes co-expressing Kir6.2 and either wild-type SUR1 (WT, white bar), or SUR1-S1R, SUR2A-S1R or SUR2B-S1R (S1R, grey bars), or SUR1-S2R, SUR2A-S2R or SUR2B-S2R (S2R, black bars). The slope conductance (G) is expressed as a fraction of the mean of that obtained in control solution before and after exposure to ADP (Gc). The number of patches is given above the bar.
Although MgATP stimulates KATP channel activity via SUR, it also exerts a strong inhibitory effect via the Kir6.2 subunit. To explore the stimulatory effect of MgATP in the absence of this inhibition, we co-expressed wild-type or mutant SUR with a mutant Kir6.2 (R50G), which shows greatly reduced ATP sensitivity (Gribble et al., 1998; Tucker et al., 1998). Because MgADP activation of Kir6.2/SUR2A was largely unaffected by either S1R or S2R mutations, we confined our analysis to Kir6.2/SUR1 and Kir6.2/SUR2B.
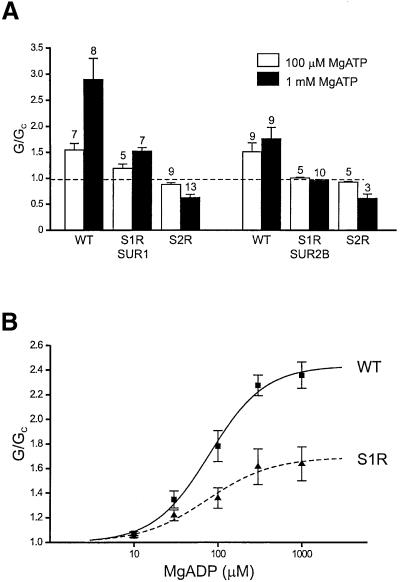
Figure 4A shows that 100 µM MgATP activated Kir6.2-R50G/SUR1 and Kir6.2-R50G/SUR2B to similar extents, but that 1 mM MgATP was significantly more effective on Kir6.2/SUR1. Mutation of the linker serine in NBD1 of either SUR1 or SUR2B substantially decreased this activation. The S2R mutation completely abolished activation of both SUR1- and SUR2B-containing channels, and unmasked an inhibitory effect of MgATP that may be mediated via Kir6.2. In the absence of Mg2+, 1 mM ATP blocked Kir6.2-R50G/SUR1 channels by 43 ± 0.7% (n = 5), which is similar to the extent of block of Kir6.2-R50G/SUR1-S2R and Kir6.2-R50G/SUR2B-S2R channels by 1 mM MgATP. This suggests that the S2R mutation abolishes MgATP activation, whereas the S1R mutation causes a large decrease in activation.
Fig. 4. (A) Mean KATP conductance recorded in response to 100 µM or 1 mM MgATP from patches excised from oocytes co-expressing Kir6.2-R50G and either wild-type or mutant SUR1 or SUR2B as indicated. The slope conductance (G) is expressed as a fraction of the mean of that obtained in control solution before exposure to MgADP (Gc). The number of patches is given above each bar. (B) Mean relationship between KATP channel activation and MgADP concentration for Kir6.2-R50G/SUR1 (black squares; n = 3–5) and Kir6.2-R50G/SUR1-S1R (black triangles; n = 4–8). The slope conductance (G) is expressed as a fraction of the mean of that obtained in control solution before and after exposure to MgADP (Gc). The lines are fitted to the modified Hill equation:
where EC50 is the MgADP concentration that gives half-maximal response and L is the maximal activation. For Kir6.2-R50G/SUR1, L = 2.4, h = 1.3 and EC50 = 79 µM; for Kir6.2-R50G/SUR1-S1R, L = 1.7, h = 1.1 and EC50 = 75 µM.
We also examined the effect of the linker mutations on MgADP activation by co-expressing SUR1-S1R or SUR1-S2R with Kir6.2-R50G. This removes the inhibitory action of MgADP at high nucleotide concentrations and enables construction of the concentration–response relationship for MgADP activation. Figure 4B shows that the potency of MgADP was unaltered by the S1R mutation: EC50 = 75 ± 26 µM (n = 4–8) for Kir6.2-R50G/SUR1-S2R, compared with 79 ± 12 (n = 3–5) for Kir6.2-R50G/SUR1 (not significant). However, the S1R mutation markedly reduced the extent of maximal activation by MgADP, which was 1.7 ± 0.1-fold for Kir6.2- R50G/SUR1-S1R, compared with 2.4 ± 0.1-fold for Kir6.2-R50G/SUR. Thus, the S1R mutation produced a decrease in activation by MgADP of comparable magnitude to that found for MgATP. It was not possible to measure the concentration–response curve for the S2R mutant because even as much as 1 mM MgADP produced no activation of Kir6.2-R50G/SUR1-S2R channels. Thus, neither MgATP nor MgADP was able to activate Kir6.2-R50G/SUR1-S2R channels.
Mutations in other regions of SUR
We also mutated two additional residues that have been shown to interact with the ATP molecule in other ABC transporters: a highly conserved glutamine residue, located between the WA and linker motifs, and a histidine residue that lies downstream of the WB motif (Figure 1). The invariant glutamine makes a direct hydrogen bond to the γ-phosphate of AMPPNP in Rad50cd (Hopfner et al., 2000) and a water-bridged hydrogen bond to the γ-phosphate of ATP in HisP (Hung et al., 1998). In the nucleotide-free structure of Rad50cd (Hopfner et al., 2000) and the MgADP-bound form of MJ0796 (Yuan et al., 2001), this glutamine is positioned further away from the active site. It has therefore been suggested that this region of the protein undergoes a conformational change on ATP hydrolysis. The histidine interacts (via water) with γ-phosphate of ATP in both the HisP and Rad50 structures. In Rad50cd, this water is believed to mount a nucleophilic attack on the γ-phosphate and thus promote ATP hydrolysis.
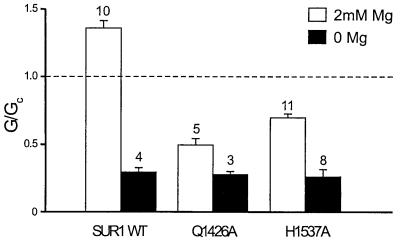
Mutation of Q774A or H888A in NBD1 of SUR1 did not result in functional channels. In contrast, the equivalent mutations in NBD2 (Q1426A and H1537A) did not affect functional expression. Figure 5 shows that in the absence of Mg2+, ADP blocked wild-type and NBD2 mutant channels to similar extents. This is not unexpected because the inhibitory effect of ADP is mediated by interaction with the Kir6.2 subunit (Tucker et al., 1998). However, both the Q1426A and H1537A mutations reduced the ability of 100 µM MgADP to stimulate channel activity, resulting in a 50 ± 5.2% (n = 5) block of Kir6.2/SUR1-Q1426A currents and a 30 ± 2.6% (n = 11) block of Kir6.2/SUR1-H1537A currents (Figure 5). Thus, these residues are likely to be involved in nucleotide binding and/or transduction.
Fig. 5. Mean KATP conductance recorded from patches excised from oocytes co-expressing Kir6.2 and either wild-type SUR1 or SUR containing mutations in the linker of NBD2 (SUR1-Q1426A and SUR1-H1537A). Currents were recorded in the presence of 100 µM ADP with (white bars) or without (black bars) 2 mM Mg2+. The slope conductance (G) is expressed as a fraction of the mean of that obtained in control solution before and after exposure to ADP (Gc). The number of patches is given above the bar.
Binding studies
The ability of mutations in the linker domain of NBD2 of SUR1 and SUR2B to markedly reduce MgADP activation raises the question of whether these mutations impair nucleotide binding, or whether they impair the mechanism involved in transducing binding into channel activation. To answer this question, we examined nucleotide binding to the NBDs of wild-type and mutant SUR1, expressed in COS-7 cells.
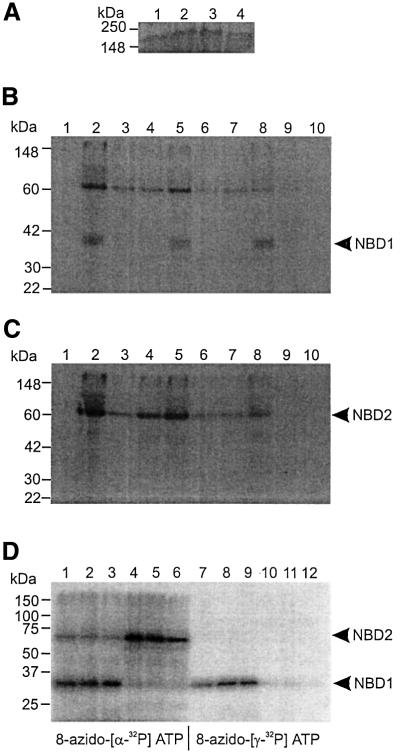
Neither the S1R nor the S2R mutation affected expression levels, as determined by western blotting (Figure 6A). Crude membranes from COS-7 cells expressing wild-type SUR1, SUR1-S1R or SUR1-S2R were incubated with 50 µM 8-azido-[α-32P]ATP, followed by mild trypsin digestion. Labelled tryptic fragments containing NBD1 or NBD2 of wild-type and mutant SUR1 were immunoprecipitated with anti-NBD1 (Figure 6B, lanes 2, 5 and 8) or NBD2 antibody (Figure 6C, lanes 2, 5 and 8), respectively. No change in the pattern of tryptic digestion was observed for the mutant SURs, indicating that the linker mutations do not produce gross conformational changes in the protein.
Fig. 6. (A) Western blotting of wild-type and mutant SUR1. Membrane proteins (25 µg) from untransfected COS-7 cells (lane 4) or cells expressing wild-type SUR1 (lane 1), SUR1-S1R (lane 2) or SUR1-S2R (lane 3) were separated by 7% SDS–PAGE. SURs were detected with anti-NBD1 antibody (Matsuo et al., 1999). Experiments were performed in duplicate. (B and C) Photoaffinity labelling of wild-type and mutant SUR1 with 8-azido-[α-32P]ATP. Membrane proteins (25 µg) from untransfected COS-7 cells (lane 1) or cells expressing wild-type SUR1 (lanes 2–4), SUR1-S1R (lanes 5–7) or SUR1-S2R (lane 8–10) were incubated with 50 µM 8-azido-[α-32P]ATP, in the absence of other nucleotide (lanes 1, 2, 5 and 8) or in the presence of 100 µm ADP (lanes 3, 6 and 9) or 100 µm ATP (lanes 4, 7 and 10), for 10 min on ice and then UV irradiated. Photoaffinity-labelled proteins were subject to mild trypsin digestion. The tryptic fragments were immunoprecipitated with anti-NBD1 (Figure 6B) or anti-NBD2 (Figure 6C) antibody and separated by 12% PAGE. A 65 kDa fragment containing NBD2 and a 35 kDa fragment containing NBD1 are indicated. Note that NBD2 was co-immunoprecipitated with NBD1. (D) Photoaffinity labelling of wild-type and mutant SUR1 with 8-azido-[α-32P]ATP or 8-azido-[γ-32P]ATP. Membrane proteins (20 µg) from cells expressing wild-type SUR1 (lanes 1, 4, 7 and 10), SUR1-S1R (lanes 2, 5, 8 and 11) or SUR1-S2R (lane 3, 6, 9 and 12) were incubated with 50 µM 8-azido-[α-32P]ATP (lanes 1–6) or 8-azido-[γ-32P]ATP (lanes 7–12) for 10 min at 37°C and then UV irradiated. Photoaffinity-labelled proteins were subjected to mild trypsin digestion. The tryptic fragments were immunoprecipitated with anti-NBD1 (lanes 1–3 and 7–9) or anti-NBD2 (lanes 4–6 and 10–12) antibody and separated by 12% PAGE. A 65 kDa fragment containing NBD2 and a 35 kDa fragment containing NBD1 are indicated. Note that NBD2 was co-immunoprecipitated with NBD1.
NBD1s of wild-type and mutant SUR1 were photoaffinity labelled to the same extent, indicating that neither the S1R nor S2R mutation affects 8-azido-ATP binding to NBD1. Photoaffinity labelling was also inhibited by cold ATP (Figure 6B, lanes 4, 7 and 10) or ADP (Figure 6B, lanes 3, 6 and 9) to similar extents, indicating that the linker mutations do not affect ATP or ADP binding to NBD1. Mean data are given in Table I. In contrast, NBD2 of SUR1-S2R (Figure 6C, lane 8) was photoaffinity labelled with lower efficiency than either SUR1 or SUR1-S1R (Figure 6C, lanes 2 and 5). Thus, NBD2 of SUR1-S2R (but not SUR1-S1R) binds 8-azido-ATP with lower affinity than that of wild-type SUR1. Photoaffinity labelling of both wild-type and mutant NBD2s was inhibited by ATP (Figure 6C, lanes 4, 7 and 10) or ADP (Figure 6C, lanes 3, 6 and 9), indicating that NBD2s of SUR1-S1R and SUR1-S2R bind both ATP and ADP. At the same concentration as used in the electrophysiological studies (100 µM), MgADP reduced 8-azido-ATP binding to NBD2 of SUR1, SUR1-S1R and SUR1-S2R to similar extents (Table I). Similar data were obtained with 100 µM MgATP. Thus, either the ATP and ADP affinity of NBD2 is unaffected by the S2R mutations (in contrast to azido-ATP), or the binding affinities are reduced but a nucleotide concentration of 100 µM is sufficient to produce maximum binding to both wild-type and mutant SUR. The fact that the EC50 for MgADP activation was unaffected by the S2R mutation (Figure 4B) supports the first hypothesis.
Table I. Percentage of photoaffinity labelling by 8-azido-[α-32P]ATP remaining after displacement with unlabelled ATP or ADP.
| NBD1 (%) |
NBD2 (%) |
|||
|---|---|---|---|---|
| ATP | ADP | ATP | ADP | |
| SUR1 | 12 ± 10 | 21 ± 14 | 52 ± 11 | 45 ± 11 |
| SUR1-S830R | 8 ± 3.7 | 25 ± 9.0 | 43 ± 11 | 33 ± 6.8 |
| SUR1-S1482R | 9 ± 5.1 | 21 ± 11 | 39 ± 14 | 34 ± 8.4 |
Values are expressed as a percentage of 8-azido-[α-32P]ATP bound in the absence of unlabelled ATP (100 µM) or ADP (100 µM). Experiments were performed in the presence of 1 mM Mg2+, as described in Materials and methods.
ATP hydrolysis of SUR
It is suggested that the NBDs of SUR have ATPase activity and that this is required for the stimulatory action of MgATP on the KATP channel (Matsuo et al., 1999, 2000; Bienengraeber et al., 2000; Zingman et al., 2001). Thus, the inability of MgATP to activate KATP channels composed of SUR1-S2R or SUR2B-S2R subunits could result either from defective MgATP hydrolysis or from impaired transduction. To determine whether ATP hydrolysis is affected by the linker mutations, SUR1, SUR1-S1R and SUR1-S2R were photoaffinity labelled with 8-azido-[α-32P]ATP and 8-azido-[γ-32P]ATP. Figure 6D shows that NBD1s of wild-type and mutant SUR1 were photoaffinity labelled with both 8-azido-[α-32P]ATP (lanes 1–3) and 8-azido-[γ-32P]ATP (lanes 7–9). In contrast, NBD2s were photoaffinity labelled with 8-azido-[α-32P]ATP (lanes 4–6) but not with 8-azido-[γ-32P]ATP (lanes 10–12). These results confirm that NBD2 (but not NBD1) hydrolyses ATP (Matsuo et al., 1999, 2000), and further suggest that the S1R and S2R mutations do not abolish this ATPase activity.
Discussion
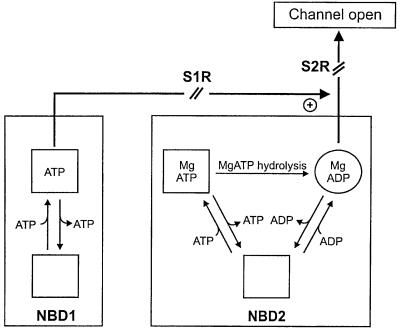
Our results indicate that neither MgATP nor MgADP binding was altered by mutation of the linker serine in either NBD1 or NBD2 of SUR1, nor was MgATP hydrolysis abolished. However, these mutations reduced (S1R) or abolished (S2R) activation of Kir6.2-SUR1 and Kir6.2-SUR2B channels by MgADP and MgATP, indicating that the linker serines play a critical role in transducing nucleotide binding into channel opening (Figure 7). Remarkably, no effect of mutating S2R was observed for Kir6.2-SUR2A channels.
Fig. 7. NBD1 of SURx binds ATP or ADP, and has little or no ATPase activity (Bienengraeber et al., 2000; Matsuo et al., 2000). NBD2 of SURx binds MgATP or MgADP depending on their ratio and concentration. Bound MgATP is hydrolysed to MgADP. MgADP activates the KATP channel via NBD2. The S2R mutation of SUR1 and SUR2B abolishes channel activation by Mg-nucleotides completely. In contrast, the S1R mutation in SURx reduces channel activation by MgADP or MgATP, possibly by influencing transduction at NBD2. The S2R mutation of SUR2A does not alter KATP channel activation by MgADP, suggesting that the SUR2A tail prevents (or compensates for) the effect of the S2R mutation on signal transduction.
Role of NBD1 of SUR
Mutation of the linker serine to arginine in NBD1 of SUR1 was without effect on either ATP or ADP binding, suggesting that the linker is not involved in nucleotide binding. The fact that the EC50 for MgADP activation of Kir6.2-R50G/SUR1 was unaltered by the S1R mutation is consistent with this idea. However, the ability of MgADP to enhance channel activity was significantly impaired by the S1R mutation. This effect was more marked when examined using an ATP-insensitive pore mutant that enables the stimulatory action of MgADP on SUR1 to be dissociated from the blocking effect of the nucleotide on Kir6.2. The S1R mutation in SUR1 or SUR2B also suppressed the ability of MgATP to enhance KATP channel activity (to about the same extent as observed for MgADP). These data suggest that the NBD1 linker in SUR1 and SUR2B may play a role in transducing MgADP binding into channel activation.
In SUR2A, MgADP activation appears to be mediated solely by nucleotide binding to NBD2, as mutation of the WA lysine in NBD1 is without effect (Reimann et al., 2000). Although mutation of the WA lysine in NBD1 of SUR1 did abolish MgADP activation of the KATP channel, it also prevented nucleotide binding to both NBD1 and NBD2 (M.Matsuo, unpublished observations). Thus, the effects of the WA lysine mutation cannot be taken as evidence that NBD1 is required for MgADP activation of SUR1. Moreover, the fact that ADP stimulation of the KATP channel requires Mg2+ (Gribble et al., 1997b), yet nucleotide binding to NBD1 is Mg2+ independent (Ueda et al., 1997; Matsuo et al., 2000), demonstrates that NBD1 is not sufficient for channel activation. Taken together, these data suggest that NBD1 of SURx plays a small, but significant, role in KATP channel activation by MgADP, but that it is not as important as NBD2.
Our results confirm that NBD1 of SUR1 has no, or little, ATPase activity (Matsuo et al., 1999, 2000), and reveal that the S1R mutation did not abolish ATP hydrolysis at NBD2 of SUR1. Thus, the S1R mutation is most likely to reduce MgATP activation by impairing the transduction of nucleotide binding into channel activation (Figure 7). Because our method of detecting ATP hydrolysis is not quantitative, we cannot completely exclude the possibility that the S1R mutation in SUR1 reduced (rather than abolished) ATP hydrolysis at NBD2. However, the fact that MgADP activation is also impaired by this mutation favours the idea that transduction is impaired.
Role of NBD2 of SUR
Mutation of the invariant serine in the linker of NBD2 abolished MgADP activation of Kir6.2-SUR1 and Kir6.2-SUR2B channels, but not that of Kir6.2-SUR2A channels. This effect appears to result principally from impaired transduction, because the ability of SUR1 to bind MgADP (at 100 µM) was not reduced. Other mutations (G1484D, G1484R and Q1485H) within the NBD2 linker of SUR1 have also been reported to prevent MgADP activation of the KATP channel (Shyng et al., 1997). As observed for MgADP, binding of MgATP (at 100 µM) was unaltered by the S2R mutation, yet this nucleotide failed to activate Kir6.2-R50G/SUR1-S2R or Kir6.2-R50G/SUR2B-S2R channels. Thus, this effect might arise from either defective MgATP hydrolysis and/or impaired transduction.
Our results demonstrate that NBD2s of wild-type SUR1, SUR1-S1R and SUR1-S2R have ATPase activity, and that this is not abolished by mutation of the linker serine in NBD2. We cannot exclude the possibility that this mutation reduced (rather than abolished) ATP hydrolysis, because (as discussed above) our method of detecting ATP hydrolysis is not quantitative. On balance, however, the evidence favours the idea that the reduced MgATP stimulation we observe derives from a defect in transduction (as is the case for MgADP) (Figure 7).
Structural implications
Our results do not support a role for the linker motif in nucleotide binding, because neither ATP nor ADP binding was prevented by the linker serine mutations. This suggests that the three-dimensional structure of NBD2 of SUR may not resemble that of Rad50cd. On the other hand, Mg-nucleotide activation of SUR1 and SUR2B seems to be impaired by mutation of the linker serine in NBD1, a finding that suggests the NBDs may functionally interact and that NBD1 may influence nucleotide transduction via NBD2 (Figure 7). Such an interaction may indicate that the linker of NBD1 lies close to the ATP molecule bound by the WA and WB motifs of NBD2, and would be consistent with the BtuCD structure. However, an allosteric interaction cannot be ruled out. Mutations in the linker domain of other ABC transporters have also been shown to reduce ATP hydrolysis. Whichever structure is correct, our results demonstrate that the linker is intimately involved in transduction in SUR1 and SUR2B, but is not important for nucleotide binding.
Transduction
How transduction is achieved remains unclear. One possibility is that linker residues interact with the TMs of SUR1, as suggested for other ABC transporters and by the crystal structures of MalK (Diederichs et al., 2000) and MsbA (Chang and Roth, 2001). This could induce a conformational change in the TMs of SUR1 that would be transmitted to the TMs of Kir6.2. Using a trafficking-based assay, it has been shown that TM1 of Kir6.2 interacts with the TMs of SUR1 (Schwappach et al., 2000). This would be consistent with mutagenesis studies of Kir6.2 (Loussouarn et al., 2000), and the crystal structure of the bacterial potassium channel KcsA (Doyle et al., 1998), which suggest that TM2 forms the pore and is surrounded by TM1. One possibility, therefore, is that transduction is via the TMs of SUR1 and TM1 of Kir6.2. An alternative idea is that transduction is mediated via interaction of the linker, directly or indirectly, with the cytosolic domains of Kir6.2. Co-assembly of Kir6.2 and SUR1 required the cytosolic N-terminus of Kir6.2, but not the NBDs of SUR1 (Schwappach et al., 2000). However, it is not necessarily the case that functional interactions between SUR1 and Kir6.2 will parallel the structural interactions needed for co-assembly. Indeed, both the N-terminus (Reimann et al., 1999) and the proximal C-terminus (Giblin et al., 1999; Lorenz and Terzic, 1999) of Kir6.2 are suggested to mediate functional interactions with SUR1.
Remarkably, the S2R mutation did not impair transduction in Kir6.2/SUR2A channels. Because the sequences of SUR2A and SUR2B are identical except for their C-terminal 42 amino acids, this suggests that the ‘tail’ of SUR2 may interact with the linker of NBD2 in the transduction process, and that this interaction differs for SUR2A and SUR2B. One possibility is that the C-terminus and the NBD2 linker co-operate in transducing nucleotide binding into channel stimulation, and that the C-terminus of SUR2A (but not SUR1 or SUR2B) is able to compensate for the effect of the linker mutation, either directly or allosterically. For example, it might influence the local structure of NBD2 such that it is able to tolerate the effect of the serine to arginine mutation at residue 1446. Another idea is that the S2R mutation abolishes the linker function in SUR1 and SUR2B, but not in SUR2A. The possibility that the SUR2A linker (unlike that of SUR1 and SUR2B) is not involved in transduction seems unlikely because this domain is so highly conserved among ABC proteins that it is predicted to have a common function.
It is possible that the linker region may be involved in cross-talk between NBD1 and NBD2. Previous studies have shown that mutation of the WA lysine in NBD1 of SUR1 and SUR2B, but not SUR2A, impairs MgADP activation (Reimann et al., 2000). In this study, we show that mutation of the linker serine in NBD2 abolishes ADP activation of SUR1 and SUR2B, but not SUR2A. These data may suggest that MgATP (or MgADP) binding to NBD1 is required for activation of SUR2B and SUR1, and that this is transduced to NBD2 via the linker of NBD1. Mutation of the NBD2 linker blocks this effect and so impairs nucleotide activation because the linker of NBD2 serves as a sensor of whether nucleotide is bound at NBD1. In SUR2A this cannot be the case because MgADP binding to NBD2 is sufficient to activate the KATP channel.
Relevance to disease
Mutations in SUR1 are a common cause of persistent hypoinsulinaemic hypoglycaemia, a disease characterized by persistent and unregulated insulin secretion even at low plasma glucose concentrations (Sharma et al., 2000). All mutations result in the complete or partial loss of KATP channel activity, either because the channel is not present in the surface membrane, or because it is present but not activated by MgADP. Mutations in both NBD1 and NBD2 of SUR1 have been found, but, to date, no mutations in the NBD2 linker of SUR1 have been described (for a review, see Sharma et al., 2000). However, mutations in the linker of CFTR give rise to cystic fibrosis (Cutting et al., 1990; Kerem et al., 1990; Welsh and Smith, 1993). Thus, linker PHHI mutations of SUR1 may be found in the future. Indeed, many mutations in the same region of SUR1, which abolish MgADP activation of channels pre-blocked by MgATP, have been reported (Nichols et al., 1996; Shyng et al., 1997). Our results raise the possibility that some of these may do so not by altering nucleotide binding/hydrolysis, but by impairing transduction.
Glutamine and histidine mutations
Mutation of the highly conserved glutamine and histidine residues in NBD2 of SUR1 reduced KATP channel activation by MgADP. In the crystal structure of HisP and Rad50cd, those amino acids are suggested to interact with the γ-phosphate of ATP, and the mutation of histidine in MalK (Davidson and Sharma, 1997) and glutamine in MDR1 (Urbatsch et al., 2000) reduces ATPase activity. Mutation of these residues is, therefore, predicted to impair ATP binding and/or hydrolysis. In the crystal structure of MalK, however, the glutamine and histidine do not interact with the ATP molecule, and mutation of the glutamine or histidine residues does not significantly reduce ATPase activity (Walter et al., 1992). We found that MgADP activation was decreased by mutation of either the glutamine or histidine in SUR1, suggesting that MgADP binding (and/or transduction) is affected.
Implications for other ABC proteins
In most bacterial ABC proteins, including the maltose transporter and histidine permease, the two NBDs are identical, and thus their linker regions may be functionally equivalent. Many eukaryotic ABC proteins, however, consist of a single polypeptide containing two transmembrane domains and two NBDs (Figure 1). Recent studies show that NBDs of CFTR, MDR1 and SURs are not equivalent either in terms of sequence, nucleotide binding or function (Matsuo et al., 1999; Szabo et al., 1999; Gao et al., 2000; Hou et al., 2000; Matsuo et al., 2000; Nagata et al., 2000; Aleksandrov et al., 2001). This argues that the linker motifs of the two NBDs may serve different roles. In support of this idea, mutation of serine (this study) or glutamine (Shyng et al., 1997) in the linker motif of NBD2 totally abolished MgADP activation of the KATP channel, whereas the equivalent mutations in NBD1 had a smaller effect. Interestingly, mutations in the linker of either NBD1 or NBD2 impair the function of CFTR and MDR1. This may mean that whereas transduction is mediated primarily via the NBD2 linker in SUR, both linkers are required to transduce nucleotide binding/hydrolysis into changes in CFTR or MDR1 function. It is noteworthy that both NBDs of CFTR and MDR1 exhibit ATPase activity, whereas only NBD2 of SUR has significant ATPase activity (Bienengraeber et al., 2000).
In conclusion, our results suggest that the linker motif in NBD2 of SUR1 and SUR2B (but not SUR2A) is involved in transducing MgADP binding into channel activation. Because mutation of the same residue (S1446) in SUR2A and SUR2B had different effects on channel activation but not on nucleotide binding, the C-terminal 42 residues of SUR2 may modulate this transduction process.
Materials and methods
Molecular biology
Mouse Kir6.2 (DDBJ/EMBL/GenBank accession No. D50581; Sakura et al., 1995), rat SUR2A (DDBJ/EMBL/GenBank accession No. D83598; Inagaki et al., 1996) and rat SUR2B (DDBJ/EMBL/GenBank accession No. D86038; Isomoto et al., 1996) cDNAs were used in this study. Mutagenesis of individual amino acids was performed using the altered sites II system (Promega). We made the following mutations in the NBD1 linker: SUR1-S830R and SUR2-S809R (S1R); SUR1-Q774A, SUR1-H888A. Mutations in the NBD2 linker were: SUR1-S1482R and SUR2-S1446R (S2R); SUR1-Q1426A, SUR1-H1537A. For oocyte expression, cDNAs were cloned in the pBF vector, and capped mRNA prepared using the mMESSAGE mMACHINE large-scale in vitro transcription kit (Ambion, Austin, TX), as described previously (Gribble et al., 1997b). For mammalian cell expression, cDNAs were cloned in the pcDNA3 vector.
Electrophysiology
Female Xenopus laevis were anaesthetized with MS222 (2 g/l, in water). One ovary was removed via a mini-laparotomy, the incision sutured and the animal allowed to recover. Immature stage V–VI oocytes were incubated for 60 min with 1.0 mg/ml collagenase (Sigma; type V) and manually defolliculated. Oocytes were co-injected with ∼0.1 ng of wild-type or mutant Kir6.2 mRNA and ∼2 ng of mRNA encoding either wild-type or mutant SUR1, SUR2A or SUR2B. The final injection volume was 50 nl/oocyte. Isolated oocytes were maintained in Barth’s solution and studied 1–4 days after injection (Gribble et al., 1997a).
Patch pipettes were pulled from borosilicate glass and had resistances of 250–500 kΩ when filled with pipette solution. Macroscopic currents were recorded from giant excised inside-out patches at a holding potential of 0 mV, at 20–24°C (Gribble et al., 1997a). Currents were evoked by repetitive 3 s voltage ramps from –110 to +100 mV and recorded using an EPC7 patch-clamp amplifier (List Electronik, Darmstadt, Germany). They were filtered at 10 kHz, digitized at 0.4 kHz using a Digidata 1200 Interface and analysed using pClamp software (Axon Instruments, Foster City, CA).
The pipette (external) solution contained (mM): 140 KCl, 1.2 MgCl2, 2.6 CaCl2 and 10 HEPES (pH 7.4 with KOH). The intracellular (bath) solution contained (mM): 107 KCl, 2 MgCl2, 1 CaCl2, 10 EGTA and 10 HEPES (pH 7.2 with KOH; final [K+] ∼140 mM). Nucleotides were added as indicated. Rapid exchange of solutions was achieved by positioning the patch in the mouth of one of a series of adjacent inflow pipes placed in the bath. Test solutions were applied in random order unless stated otherwise.
The slope conductance was measured by fitting a straight line to the current–voltage relationship between –20 and –100 mV; the average of five consecutive ramps was calculated in each solution. Data were fit using Microcal Origin software and are presented as mean ± 1 SEM.
Binding studies
Membranes from COS-7 cells expressing wild-type or mutant SUR1 were prepared as described (Ueda et al., 1997). Crude membranes, containing similar amounts of wild-type SUR1, SUR1-S830R and SUR1-S1482R, as determined by western blotting, were incubated with 50 µM 8-azido-[α-32P]ATP or 50 µM 8-azido-[γ-32P]ATP (ICN Biomedicals) in the presence or absence of 100 µM ATP or 100 µM ADP in 3 µl of TEM buffer (40 mM Tris–HCl pH 7.5, 0.1 mM EGTA and 1 mM MgSO4) containing 2 mM ouabain. Proteins were UV irradiated in a Stratalinker UV cross-linker (at 254 nm, 1 J/cm2) on ice. Ice-cold TEM buffer was then added to the sample and the supernatant was removed after centrifugation (15 000 g for 5 min at 4°C). Pellets were resuspended in 40 mM Tris–HCl pH 7.5 buffer containing 0.1 mM EGTA, 250 mM sucrose and 10 µg/ml trypsin to a concentration of 10 µg of membrane proteins/µl, and incubated for 5 min at 37°C. One hundred microlitres of RIPA buffer (20 mM Tris–HCl pH 7.5, 1% Triton X-100, 0.1% SDS and 1% sodium deoxycholate) containing protease inhibitor cocktail (Calbiochem) were added to the samples to terminate proteolysis, and membrane proteins were solubilized for 30 min. After centrifugation for 15 min at 15 000 g, tryptic fragments were immunoprecipitated from the supernatant using an antibody raised against NBD1 or NBD2 of hamster SUR1, as described previously (Matsuo et al., 1999). Samples were electrophoresed on 12% SDS–polyacrylamide gel and autoradiographed.
Acknowledgments
Acknowledgements
This work was supported by grants from the Royal Society, the Wellcome Trust, Novo Nordisk A/S, Toyobo Biotechnology Foundation, Japan Society for the Promotion of Science and Japan Ministry of Education, Science, Sports and Culture. M.D. is a Robert Turner Visiting Scholar. F.M.A. is the Royal Society GlaxoSmithKline Research Professor.
References
- Aguilar-Bryan L., Nichols,C.G., Wechsler,S.W., Clement,J.P.,IV, Boyd,A.E.,III, Gonzalez,G., Herrera-Sosa,H., Nguy,K., Bryan,J. and Nelson,D.A. (1995) Cloning of the β-cell high-affinity sulfonylurea receptor: a regulator of insulin secretion. Science, 268, 423–426. [DOI] [PubMed] [Google Scholar]
- Aguilar-Bryan L. and Bryan,J. (1999) Molecular biology of adenosine triphosphate-sensitve potassium channels. Endocr. Rev., 20, 101–135. [DOI] [PubMed] [Google Scholar]
- Aleksandrov L., Mengos,A., Chang,X., Aleksandrov,A. and Riordan,J.R. (2001) Differential interactions of nucleotides at the two nucleotide binding domains of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem., 276, 12918–12923. [DOI] [PubMed] [Google Scholar]
- Ashcroft F.M. and Gribble,F.M. (1998) Correlating structure and function in ATP-sensitive K+ channels. Trends Neurosci., 21, 288–294. [DOI] [PubMed] [Google Scholar]
- Bakos E., Klein,I., Welker,E., Szabo,K., Muller,M., Sarkadi,B. and Varadi,A. (1997) Characterization of the human multidrug resistance protein containing mutations in the ATP-binding cassette signature region. Biochem. J., 323, 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienengraeber M., Alekseev,A.E., Abraham,M.R., Carrasco,A.J., Moreau,C., Vivaudou,M., Dzeja,P.P. and Terzic,A. (2000) ATPase activity of the sulfonylurea receptor: a catalytic function for the KATP channel complex. FASEB J., 14, 1943–1952. [DOI] [PubMed] [Google Scholar]
- Chang G. and Roth,C.B. (2001) Structure of MsbA from E.coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science, 293, 1793–1800. [DOI] [PubMed] [Google Scholar]
- Cutting G.R., Kasch,L.M., Rosenstein,B.J., Zielenski,J., Tsui,L.C., Antonarakis,S.E. and Kazazian,H.H.,Jr (1990) A cluster of cystic fibrosis mutations in the first nucleotide-binding fold of the cystic fibrosis conductance regulator protein. Nature, 346, 366–369. [DOI] [PubMed] [Google Scholar]
- Davidson A. and Sharma,S. (1997) Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J. Bacteriol., 179, 5458–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Rzhetsky,A. and Allikmets,R. (2001) The human ATP-binding cassette (ABC) transporter superfamily. Genome Res., 11, 1156–1166. [DOI] [PubMed] [Google Scholar]
- Diederichs K., Diez,J., Greller,G., Muller,C., Breed,J., Schnell,C., Vonrhein,C., Boos,W. and Welte,W. (2000) Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J., 19, 5951–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral,J., Pfuetzner,R.A., Kuo,A., Gulbis,J.M., Cohen,S.L., Chait,B.T. and MacKinnon,R. (1998) The structure of the potassium channel: molecular basis of K+ conductance and selectivity. Science, 280, 69–77. [DOI] [PubMed] [Google Scholar]
- Gao M., Cui,H.R., Loe,D.W., Grant,C.E., Almquist,K.C., Cole,S.P.C. and Deeley,R.G. (2000) Comparison of the functional characteristics of the nucleotide binding domains of multidrug resistance protein 1. J. Biol. Chem., 275, 13098–13108. [DOI] [PubMed] [Google Scholar]
- Gaudet R. and Wiley,D.C. (2001) Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J., 20, 4964–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giblin J.P., Leaney,J.L. and Tinker,A. (1999) The molecular assembly of ATP-sensitive potassium channels. J. Biol. Chem., 274, 22652–22659. [DOI] [PubMed] [Google Scholar]
- Gribble F.M., Ashfield,R., Ämmäla,C. and Ashcroft,F. (1997a) Properties of cloned ATP-sensitive K+ currents expressed in Xenopus oocytes. J. Physiol., 498, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble F.M., Tucker,S.J. and Ashcroft,F.M. (1997b) The essential role of the Walker A motifs of SUR1 in K-ATP channel activation by Mg-ADP and diazoxide. EMBO J., 16, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble F.M., Tucker,S.J., Haug,T. and Ashcroft,F.M. (1998) MgATP activates the β cell KATP channel by interaction with its SUR1 subunit. Proc. Natl Acad. Sci. USA, 95, 7185–7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C.F. (1992) ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol., 8, 67–113. [DOI] [PubMed] [Google Scholar]
- Hoof T., Demmer,A., Hadam,M.R., Riordan,J.R. and Tümmler,B. (1994) Cystic fibrosis-type mutational analysis in the ATP-binding cassette transporter signature of human P-glycoprotein MDR1. J. Biol. Chem., 269, 20575–20583. [PubMed] [Google Scholar]
- Hopfner K.P., Karcher,A., Shin,D.S., Craig,L., Arthur,L.M., Carney,J.P. and Tainer,J.A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell, 101, 789–800. [DOI] [PubMed] [Google Scholar]
- Hou Y.-X., Cui,L., Riordan,J.R. and Chang,X.-B. (2000) Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. J. Biol. Chem., 275, 20280–20287. [DOI] [PubMed] [Google Scholar]
- Hung L.W., Wang,I.X., Nikaido,K., Liu,P.Q., Ames,G.F. and Kim,S.H. (1998) Crystal structure of the ATP-binding subunit of an ABC transporter. Nature, 396, 703–707. [DOI] [PubMed] [Google Scholar]
- Inagaki N., Gonoi,T., Clement,J.P.,IV, Wang,C.-Z., Aguilar-Bryan,L., Bryan,J. and Seino,S. (1996) A family of sulfonylurea receptors determines the pharmacological properties of ATP-sensitive K+ channels. Neuron, 16, 1011–1017. [DOI] [PubMed] [Google Scholar]
- Isomoto S., Kondo,C., Yamada,M., Matsumoto,S., Higashiguchi,O., Horio,Y., Matsuzawa,Y. and Kurachi,Y. (1996) A novel sulfonylurea receptor forms with BIR (Kir6.2) a smooth muscle type ATP-sensitive K+ channel. J. Biol. Chem., 271, 24321–24324. [DOI] [PubMed] [Google Scholar]
- Kerem B. et al. (1990) Identification of mutations in regions corresponding to the two putative nucleotide (ATP)-binding folds of the cystic fibrosis gene. Proc. Natl Acad. Sci. USA, 87, 8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis E., Hughes,C., Milisav,I. and Koronakis,V. (1995) Protein exporter function and in vitro ATPase activity are correlated in ABC-domain mutants of HlyB. Mol. Microbiol., 16, 87–96. [DOI] [PubMed] [Google Scholar]
- Li C., Ramjeesingh,M., Wang,W., Garami,E., Hewryk,M., Lee,D., Rommens,J.M., Galley,K. and Bear,C.E. (1996) ATPase activity of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem., 271, 28463–28468. [DOI] [PubMed] [Google Scholar]
- Locher K.P., Lee,A.T. and Rees,D.C. (2002) The E.coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science, 296, 1091–1098. [DOI] [PubMed] [Google Scholar]
- Lorenz E. and Terzic,A. (1999) Physical association between recombinant cardiac ATP-sensitive K+ channel subunits Kir6.2 and SUR2A. J. Mol. Cell. Cardiol., 31, 425–434. [DOI] [PubMed] [Google Scholar]
- Loussouarn G., Makhina,E.N., Rose,T. and Nichols,C.G. (2000) Structure and dynamics of the pore of inwardly rectifying KATP channels. J. Biol. Chem., 275, 1137–1144. [DOI] [PubMed] [Google Scholar]
- Matsuo M., Kioka,N., Amachi,T. and Ueda,K. (1999) ATP binding properties of the nucleotide binding folds of SUR1. J. Biol. Chem., 274, 37479–37482. [DOI] [PubMed] [Google Scholar]
- Matsuo M., Tanabe,K., Kioka,N., Amachi,T. and Ueda,K. (2000) Different binding properties and affinities for ATP and ADP among sulfonylurea receptor subtypes, SUR1, SUR2A and SUR2B. J. Biol. Chem., 275, 28757–28763. [DOI] [PubMed] [Google Scholar]
- Nagata K., Nishitani,M., Matsuo,M., Kioka,N., Amachi,T. and Ueda,K. (2000) Nonequivalent nucleotide trapping in the two nucleotide binding folds of the human multidrug resistance protein MRP1. J. Biol. Chem., 275, 17626–17630. [DOI] [PubMed] [Google Scholar]
- Nichols C.G., Shyng,S.-L., Nestorowicz,A., Glaser,B., Clement,J.P.,IV, Gonzalez,G., Aguilar-Bryan,L., Permutt,M.A. and Bryan,J. (1996) Adenosine diphosphate as an intracellular regulator of insulin secretion. Science, 272, 1785–1787. [DOI] [PubMed] [Google Scholar]
- Qu B.-H., Strickland,E.H. and Thomas,P.J. (1997) Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J. Biol. Chem., 272, 15739–15744. [DOI] [PubMed] [Google Scholar]
- Reimann F., Tucker,S.J., Proks,P. and Ashcroft,F.M. (1999) Involvement of the N-terminus of Kir6.2 in coupling to the sulphonylurea receptor. J. Physiol., 518, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann F., Gribble,F.M. and Ashcroft,F.M. (2000) Differential response of KATP channels containing SUR2A or SUR2B subunits to nucleotides and pinacidil. Mol. Pharmacol., 58, 1318–1325. [DOI] [PubMed] [Google Scholar]
- Sakura H., Ämmäla,C., Smith,P.A., Gribble,F.M. and Ashcroft,F.M. (1995) Cloning and functional expression of the cDNA encoding a novel ATP-sensitive potassium channel subunit expressed in pancreatic β-cells, brain, heart and skeletal muscle. FEBS Lett., 377, 338–344. [DOI] [PubMed] [Google Scholar]
- Schmees G., Stein,A., Hunke,S., Landmesser,H. and Schneider,E. (1999) Functional consequences of mutations in the conserved ‘signature sequence’ of the ATP-binding cassette protein MalK. Eur. J. Biochem., 266, 420–430. [DOI] [PubMed] [Google Scholar]
- Schwappach B., Zerangue,N., Jan,Y.N. and Jan,L.Y. (2000) Molecular basis for KATP assembly: transmembrane interactions mediate association of a K+ channel with an ABC transporter. Neuron, 26, 155–167. [DOI] [PubMed] [Google Scholar]
- Seino S. (1999) ATP-sensitive potassium channels: A model of heteromultimeric potassium channel/receptor assemblies. Annu. Rev. Physiol., 61, 337–362. [DOI] [PubMed] [Google Scholar]
- Sharma N., Crane,A., Gonzalez,G., Bryan,J. and Aguilar-Bryan,L. (2000) Familial hyperinsulinism and pancreatic β-cell ATP-sensitive potassium channels. Kidney Int., 57, 803–808. [DOI] [PubMed] [Google Scholar]
- Shyamala V., Baichwal,V., Beall,E. and Ames,G.F.-L. (1991) Structure–function analysis of the histidine permease and comparison with cystic fibrosis mutations. J. Biol. Chem., 266, 18714–18719. [PubMed] [Google Scholar]
- Shyng S.-L., Ferrigni,T. and Nichols,C.G. (1997) Regulation of KATP channel activity by diazoxide and MgADP. J. Gen. Physiol., 110, 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo K., Szakacs,G., Hegedus,T. and Sarkadi,B. (1999) Nucleotide occlusion in the human cystic fibrosis transmembrane conductance regulator. J. Biol. Chem., 274, 12209–12212. [DOI] [PubMed] [Google Scholar]
- Szakacs G., Ozvegy,C., Bakos,E., Sarkadi,B. and Varadi,A. (2001) Role of glycine-534 and glycine-1179 of human multidrug resistance protein (MDR1) in drug-mediated control of ATP hydrolysis. Biochem. J., 356, 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S.J., Gribble,F.M., Proks,P., Trapp,S., Ryder,T.J., Haug,T., Reimann,F. and Ashcroft,F.M. (1998) Molecular determinants of KATP channel inhibition by ATP. EMBO J., 17, 3290–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Inagaki,N. and Seino,S. (1997) MgADP antagonism to Mg2+-independent ATP binding of the sulfonylurea receptor SUR1. J. Biol. Chem., 272, 22983–22986. [DOI] [PubMed] [Google Scholar]
- Urbatsch I.L., Gimi,K., Wilke-Mounts,S. and Senior,A.E. (2000) Investigation of the role of glutamine-471 and glutamine-1114 in the two catalytic sites of P-glycoprotein. Biochemistry, 39, 11921–11927. [DOI] [PubMed] [Google Scholar]
- Walter C., Wilken,S. and Schneider,E. (1992) Characterization of site-directed mutations in conserved domains of MalK, a bacterial member of the ATP-binding cassette (ABC) family. FEBS Lett., 303, 41–44. [DOI] [PubMed] [Google Scholar]
- Welsh M.J. and Smith,A.E. (1993) Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell, 73, 1251–1254. [DOI] [PubMed] [Google Scholar]
- Yuan Y.-R., Blecker,S., Martsinkevich,O., Millen,L., Thomas,P.J. and Hunt,J.F. (2001) The crystal structure of the MJ0796 ATP-binding cassette: implications for the structural consequences of ATP hydrolysis in the active site of an ABC-transporter. J. Biol. Chem., 276, 32313–32321. [DOI] [PubMed] [Google Scholar]
- Zingman L.V., Alekseev,A.E., Bienengraeber,M., Hodgson,D., Karger,A.B., Dzeja,P.P. and Terzic,A. (2001) Signaling in channel/enzyme multimers: ATPase transitions in SUR module gate ATP-sensitive K+ conductance. Neuron, 31, 233–245. [DOI] [PubMed] [Google Scholar]