Abstract
The MLL gene is targeted by chromosomal translocations, which give rise to heterologous MLL fusion proteins and are associated with distinct types of acute lymphoid and myeloid leukaemia. To determine how MLL fusion proteins alter the proliferation and/or differentiation of primary haematopoietic progenitors, we introduced the MLL–AF9 and MLL–ENL fusion proteins into primary chicken bone marrow cells. Both fusion proteins caused the sustained outgrowth of immature haematopoietic cells, which was strictly dependent on stem cell factor (SCF). The renewing cells have a long in vitro lifespan exceeding the Hayflick limit of avian cells. Analysis of clonal cultures identified the renewing cells as immature, multipotent progenitors, expressing erythroid, myeloid, lymphoid and stem cell surface markers. Employing a two-step commitment/differentiation protocol involving the controlled withdrawal of SCF, the MLL–ENL-transformed progenitors could be induced to terminal erythroid or myeloid differentiation. Finally, in cooperation with the weakly leukaemogenic receptor tyrosine kinase v-Sea, the MLL–ENL fusion protein gave rise to multilineage leukaemia in chicks, suggesting that other activated, receptor tyrosine kinases can substitute for ligand-activated c-Kit in vivo.
Keywords: c-Kit/leukaemia/MLL/multipotent progenitor/receptor tyrosine kinases
Introduction
Chromosomal translocations involving the MLL (mixed lineage leukaemia) gene at 11q23 (for review see Ayton and Cleary, 2001) are particularly prevalent in poor prognosis infant leukaemia and are common in secondary, therapy-related acute myeloblastic leukaemia (AML), which arise following treatment of solid tumours with topoisomerase II inhibitors. This indicates that MLL gene fusions initiate leukaemogenesis with a remarkably short latency of ∼3–18 months, suggesting that MLL fusions are powerful transforming agents in vivo, perhaps associated with promotion of genetic instability (Greaves, 1997).
The MLL protein is highly similar to the Drosophila trithorax protein, which is implicated in chromatin remodelling and has been shown to act as a positive maintenance factor controlling expression of members of the Hox gene complexes. Targeted disruption of MLL in mice causes embryonic death, while MLL heterozygous mice show alterations in Hox gene expression and segmental identity (Yu et al., 1995). In human leukaemia, parts of the MLL gene are translocated to a heterogeneous group of >20 genes located on various partner chromosomes, always generating chimaeric fusion proteins. The translocations preserve a common portion of MLL, containing a set of evolutionarily conserved motifs (Caldas et al., 1998) but remove domains of MLL showing strongest similarity to trithorax, including the SET domain. Although specific partner genes are preferentially associated with distinct subtypes of leukaemia, it is unknown whether and how the partner genes contribute to target cell specificity, leukaemic phenotype or variation in the mechanisms of leukaemogenesis (Ayton and Cleary, 2001).
Studies by others have shown that retroviral transduction of MLL–ENL into primitive murine haematopoetic cells (Lavau et al., 1997) gives rise to colonies of proliferating cells that can be grown into leukaemic cell lines. These lines were restricted to the myeloid lineage and gave rise to myelomonocytic leukaemias in SCID or syngeneic mice. Chimaeric mice containing MLL–AF9 introduced by a knock-in strategy developed pre dominantly AML with a latency of between 4 and 9 months (Corral et al., 1996) with rare examples of acute lymphoblastic leukaemia (ALL) emerging after 11 months (Dobson et al., 1999). In humans, however, MLL–ENL is associated primarily with acute lymphoblastic leukaemia.
These studies failed to resolve the lineage and stage of maturity of the target cell for transformation by the MLL fusion proteins. It was also unclear, if and how MLL fusion proteins affected progenitor proliferation, differentiation and/or apoptosis and whether or not they required cooperation with other pathways to cause leukaemia. Here, we address these questions for two MLL fusions, MLL–AF9 and MLL–ENL, in primary avian bone marrow cells, shown previously to represent a useful, heterologous system to study leukaemogenesis by mammalian oncogenes (Beug et al., 1995a; Tran Quang et al., 1997). In contrast to mouse cells, which have a short in vitro lifespan and a strong tendency to form immortalized, aberrant cell lines, primary avian cells resemble human cells in exhibiting a long in vitro lifespan and an exceedingly low rate of immortalization (Beug et al., 1996). We therefore chose the chicken system to test whether MLL fusion proteins would cause sustained, long-term outgrowth of haematopoietic cells and whether or not cooperation with activated c-Kit was required.
In earlier work, the v-ski oncogene was found to cooperate with activated c-Kit in transformation of avian multipotent progenitor cells capable of growing for >100 generations in culture (Larsen et al., 1993; Beug et al., 1995a). Since multipotent progenitors were the likely target cell for transformation by MLL fusion proteins, we employed similar conditions favouring the proliferation of multipotent avian progenitors during retroviral transfer of MLL–AF9 or MLL–ENL, thus generating a relatively easy and rapid transformation assay for individual MLL fusion proteins. The in vitro-transformed cells have a significantly enhanced lifespan of >50 generations and require the c-Kit ligand, SCF, for survival and/or sustained proliferation. More importantly, isolated clonal cultures display clear features of multipotent progenitors displaying erythroid, myeloid and lymphoid markers. In response to a two-step commitment/differentiation induction protocol, these cells differentiate into mature cell types of the myeloid or erythroid lineage. Finally, MLL–ENL cooperated with the weakly oncogenic receptor tyrosine kinase v-Sea to cause multilineage leukaemia in chicks.
Results
Infection of chicken bone marrow cells with MLL–AF9 and MLL–ENL
Avian bone marrow cells were infected in vitro with retroviral constructs expressing Myc-tagged MLL–AF9 and MLL–ENL fusion proteins and expanded in media containing SCF plus various cytokines, known to support proliferation of avian multipotent cells (multipotent cell medium, see Materials and methods and Beug et al., 1995a). In this medium, MLL–AF9- and MLL–ENL-infected populations grew exponentially (with doubling times of 36–48 h) for >3 months (>50 generations) without undergoing senescence. In contrast, uninfected or empty vector-infected control cultures disintegrated after transient outgrowth (7–14 days) of predominantly erythroid progenitors (Figure 1A; Hayman et al., 1993). Cytospin analysis showed that the proliferating cultures consisted of immature blast-like cells plus partially mature and mature macrophages, neutrophils, eosinophils and erythroid cells (data not shown).
Fig. 1. Growth of MLL–AF9- and MLL–ENL-infected cultures. (A) MLL–AF9- (solid circles) and MLL–ENL (open squares)-infected mass cultures, as well as control cultures infected with empty vector (+ and ×), were cultivated in multipotent cell medium and cumulative cell numbers determined daily by counting in an electronic counter (see Materials and methods). (B) Four clones verified for single MLL–ENL integration sites: D8 (triangles), D11 (diamonds), B8 (circles) and IIC10 (squares) were cultured in multipotent cell medium containing (+SCF) or lacking (–SCF) SCF. Cumulative cell numbers were determined as above.
To investigate whether MLL fusion proteins transformed a multipotent progenitor or committed progenitors from diverse lineages, bone marrow cells infected with MLL–AF9- and MLL–ENL-expressing viruses or empty control vectors were plated in methylcellulose in the presence of appropriate drug selection. MLL–AF9- and MLL–ENL-infected cells yielded diffuse, macroscopic colonies, which reached a size of 1.5–3 mm in diameter within 21 days. After isolation and expansion in multipotent cell medium, the clones showed sustained, exponential proliferation with no obvious signs of senescence (Figure 1B). Uninfected or empty vector control cells only gave rise to small colonies, which ceased to increase in size after 3–4 days and then disintegrated.
Since MLL–ENL-transformed cells were easier to generate, we focused on colonies induced by this construct for further analysis. After suitable expansion, nine individually isolated MLL–ENL methocel colonies were analysed for the presence of the appropriate, intact MLL–ENL cDNA by Southern blot analysis (Figure 2B). Two cultures contained a truncated MLL–ENL cDNA of equal size (∼3 kb), in addition to the full-length cDNA. The remaining seven clones with an intact MLL–ENL cDNA were then analysed for numbers of retroviral integration sites. All contained a predominant, apparently single copy of the provirus (Figure 2C). To exclude that different culture conditions might offer a growth advantage for contaminating subclones not revealed in the multipotent cell populations, analysis of proviral integration sites was repeated after differentiation induction. Four clones (B8, IIC10, D8 and D11) clearly retained a single proviral integration site after maturing into erythrocytes or macrophage-like cells (see below; Figure 2D). All further analyses for growth factor requirement, cell surface markers and differentiation potential were therefore carried out with these verified clones.

Fig. 2. Molecular analysis of MLL–ENL-infected cultures. (A) Schematic representation of the CRNCM-MLL–ENL retroviral vector used in this study. (B) The presence of full-length MLL–ENL cDNA in nine multipotential methocel clones was detected by Southern blot analysis, digesting genomic DNA with EcoRI and hybridizing with a human MLL–ENL probe (1.25 kb BamHI–BstXI fragment, see A). (C and D) Monoclonal origin of MLL–ENL-infected clonal cultures. To identify clones containing single retroviral integration sites, genomic DNA from nine clones grown in multipotential medium (C) and from four clones after terminal differentiation (D) into myeloid (left panel) or erythroid cells (right panel; see Figure 4 and Materials and methods) was subjected to Southern blot analysis after digestion with BamHI and hybridization with the same human MLL–ENL probe as in (B). Genomic DNA from the chicken T-cell line MSB1 was used as control for cross-reactive endogenous chicken MLL.
In both mass cultures and clones, the presence of MLL–AF9 and MLL–ENL mRNA was verified by RT–PCR of the fusion junction region (data not shown). However, using available anti-MLL or anti-Myc-tag antibodies, we could not detect MLL–ENL fusion proteins in our cells due to extremely low protein expression levels, as reported by others (Lavau et al., 1997).
MLL–ENL-transformed bone marrow cells require SCF for proliferation
To determine whether MLL–ENL required cooperation with ligand-activated c-Kit for apoptosis protection and/or sustained renewal, the verified MLL–ENL-infected clones were cultured in media containing or lacking SCF. All cultures proliferated exponentially in the presence of SCF, but underwent either apoptosis or partial differentiation after SCF withdrawal (Figure 1B). Since the numbers of immature and spontaneously differentiating cells varied in the different cultures, there was some variation in the rate of apoptosis and persistence of maturing cells, the latter being less sensitive to SCF withdrawal (data not shown). In conclusion, MLL–ENL transforms immature progenitors that require SCF for both survival and renewal, i.e. sustained proliferation without apparent differentiation.
MLL–ENL clones express markers of multipotent progenitors and lineage-restricted erythroid, myeloid and B-lymphoid cells
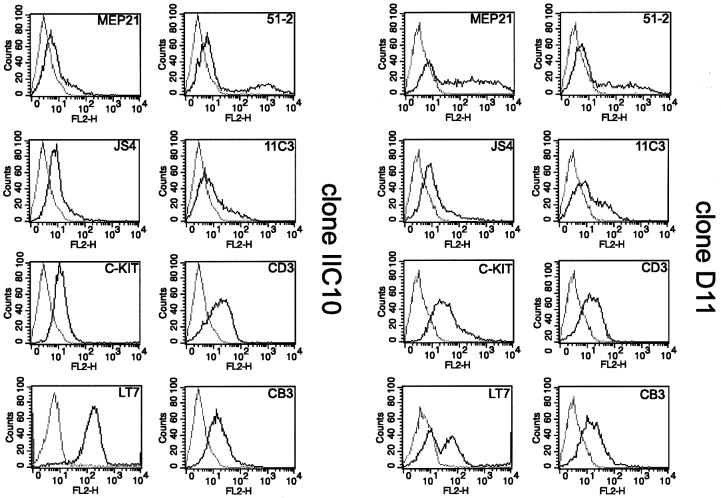
Next, we tried to verify the multipotent nature of the MLL–ENL-transformed clones by FACS analysis for cell surface antigens characteristic of multipotent and lineage-restricted cells. In particular, we wanted to determine whether or not MLL-transformed clones would include progenitors with lymphoid lineage markers. Results for clones IIC10 and D11 are shown in Figure 3.
Fig. 3. MLL–ENL-infected cultures express multilineage cell surface markers. Cell surface marker profiles for MLL–ENL clones IIC10 and D11 as determined by FACS analysis. For a detailed description of antigens detected by the panel of antibodies used, see Results.
In agreement with their dependence on SCF (Figure 1B) these clones contained cells expressing avian c-Kit (Vainio et al., 1996). Cells expressing the myelomonocytic marker 51-2 and the erythroid marker JS4 (Beug et al., 1995a) were also present in both clones. A subset of cells expressed the multipotent cell markers MEP26 (data not shown) and MEP21 (Figure 3). MEP21 represents thrombomucin, distantly related to CD34 and typical of E26 avian leukaemia virus-transformed, blastoderm-derived multipotent progenitors (Graf et al., 1992; McNagny et al., 1997). The cells also reacted with the 4.5.A.5 antibody, which stains immature lymphohaematopoietic progenitors (Hayman et al., 1982; data not shown), and 11C3 (Ody et al., 1999), which recognizes glycoprotein IIb–IIIa (Figure 3).
Interestingly, both clones expressed a CD3 component of the chicken T-cell receptor (Figure 3; Chen et al., 1986), but were negative for avian CD4 and CD8 as well as for the T-cell receptors 1, 2 and 3 (data not shown). Since our cells expressed at least three isoforms of Ikaros (chIk-1, chIk-2 and chIk-4) by RT–PCR (Liippo and Lassila, 1997; data not shown), the aberrant CD3 expression in the absence of the T-cell receptor may be due to expression of chicken Ikaros, known to upregulate CD3γ (Cortes et al., 1999). None of the MLL–ENL-transformed cultures expressed the Bu-1 antigen, expressed early during B-cell ontogeny (Rothwell et al., 1996). However, MLL–ENL clones reacted with the LT7 monoclonal antibody, which recognizes an antigen selectively expressed on bursal and post-bursal B-cells (Benatar et al., 1991). Cells positive for CD3 and LT7 also express the CB3 glycoprotein (Figure 3), characteristic of bursal and post-bursal B-cells and associated with β2-microglobulin (Pickel et al., 1990). These antibodies are most likely specific for immature T- or B-cells, since they stained the early T-cell line MSB1 (CD3) or the RP9 and RPL12 pre-B-leukaemic cell lines (LT7; CB3), while being negative on various immature erythroid or myeloid control cells [E26-transformed myeloblasts, the HD11 macrophage cell line or the erythroblast cell line HD3 (data not shown)].
Finally, the MLL–ENL clones D8 and D11 were also stained by the MY174 antibody (data not shown), recognizing a core protein of an avian embryonic mesenchymal chondroitin sulfate proteoglycan (Fernandez et al., 1991). This protein corresponds to human NG2, closely associated with MLL rearrangement in ALL (Behm et al., 1996) and AML (Mauvieux et al., 1999). NG2 has not been detected on normal haematopoietic precursors (Smith et al., 1996) and NG2-positive leukaemias without MLL involvement are very rare (Wuchter et al., 2000), suggesting that MLL fusion proteins might cause NG2 expression or modification during leukaemogenesis.
Commitment and differentiation of MLL–ENL-infected cells
Next, we examined the commitment/differentiation potential of the MLL–ENL-transformed multipotent progenitors. When exposed directly to differentiation-inducing cytokines in the absence of SCF, most cells died, with only a minority of cells undergoing complete maturation. Since v-Ski multipotent cells had shown a similar behaviour, we employed a ‘two-step induction protocol’ developed for differentiation induction in this system (Beug et al., 1995a). For this, the MLL–ENL clones were first exposed to SCF-containing ‘commitment media’ for 5 days (see Materials and methods) and subsequently treated with cytokines promoting terminal erythroid or myeloid differentiation in the absence of SCF.
During the 5-day commitment phase, the cells grew exponentially and no significant apoptosis was observed (not shown). Subsequently, the cells were cultivated for 5 days in erythroid or myeloid differentiation media, containing anaemic serum plus insulin or cMGF, relCM and IGF-I, respectively. Following myeloid commitment and differentiation, the verified MLL–ENL clones B8 and IIC10 differentiated into benzidine-negative cells (Figure 4, right panels) with a monocyte/macrophage or neutrophil morphology (see insets). Importantly, the same clones differentiated into mature erythrocytes plus a few immature erythroid cells, when exposed to erythroid commitment and differentiation conditions (Figure 4, left panels, see also insets). A few myeloid, mainly granulocytic cells (insets) persisted under these conditions, probably kept alive by non-erythroid cytokines present in the anaemic serum used as a source of erythropoietin. Similar results were obtained for the other two clones (D8 and D11). We did not attempt to induce lymphoid differentiation, since the required avian cytokines and culture conditions are still badly defined (Vainio et al., 1990).
Fig. 4. MLL–ENL-transformed multipotent clones can be induced to differentiate along erythroid and myeloid lineages. Cells from MLL–ENL clones B8 (top panels) and IIC10 (bottom panels) were cytocentrifuged on to slides after cultivation in multipotent cell medium (left panels) or after induction of terminal erythroid (middle panels) or myeloid differentiation (right panels), employing a two-step commitment/differentiation protocol (Beug et al., 1995a; Materials and methods). Photographs of representative cytospins are shown after staining with acid benzidine (haemoglobin, yellow-brown) plus Wright–Giemsa (Beug et al., 1995b). Insets: cells at higher magnification.
In conclusion, renewing multipotent progenitors induced by MLL–ENL plus c-Kit retain the ability to undergo commitment and terminal differentiation, if exposed to proper commitment and differentiation signals.
MLL–ENL cooperates with the ts-v-sea oncogene to generate multilineage leukaemia in vivo
Finally, we asked whether MLL–ENL was able to induce leukaemia in chicks and whether or not the potential leukaemic cells would represent multipotent progenitors. However, injection of MLL–ENL virus-producing fibroblasts failed to generate any leukaemia (Table I). A similar situation was observed for the v-ski oncogene, which cooperates with c-Kit to transform multipotent progenitors in vitro, but fails to induce leukaemia on its own. v-Ski, however, did cause multipotential leukaemia in combination with a temperature-sensitive v-sea erythroblastosis virus (Larsen et al., 1992), which on its own causes anaemia, but not leukaemia (Knight et al., 1988). Accordingly, we tested whether or not ts-v-Sea and MLL–ENL might cooperate in vivo in a similar fashion. Eight-day-old chicks were injected with MLL–ENL- and ts-v-Sea-producing fibroblasts. A disease consistent with leukaemia developed in all of these chicks (Table I). Bone marrow cells of three chicks sacrificed just before leukaemic death (leukaemia 2, day 32; leukaemias 6 and 7, day 49) appeared as pale cell pellets and contained elevated numbers of immature or partially mature cells. Southern blot analysis of retroviral integration sites in genomic DNA extracted from these three leukaemias revealed that leukaemia 2 and leukaemia 7 appeared to be biclonal and polyclonal, respectively, whereas leukaemia 6 seemed to be monoclonal (Figure 5A). Analysis with restriction enzymes releasing the complete MLL–ENL sequence revealed a truncation of the 6 kb MLL–ENL cDNA to a size of ∼4 kb (Figure 5A). The nature of this truncation was determined by nested PCR analysis of leukaemias 2 and 6 and sequencing of PCR products (excluding leukaemia 7 from analysis because of its polyclonality). In both leukaemias, an in-frame deletion had occurred, linking nucleotide 705 to nucleotide 2710 of MLL, thus removing the third AT-hook and one of the two trithorax-related sequences known to confer nuclear punctate distribution (Yano et al., 1997) (Figure 5B). This deleted region of MLL has been shown to be dispensable for MLL–ENL transforming activity in a myeloid cell transformation assay (Slany et al., 1998) and could not be detected by either Southern blot or PCR in the virus-producing fibroblasts used for injection.
Table I. In vivo pathology of MLL–ENL and ts-v-Sea coinfected chicken.
| Virus | Disease | Number | Latency (days) |
|---|---|---|---|
| ts-v-Sea | Anaemia | 1/4 | 22 |
| MLL–ENL | None | 0/11 | NA |
| ts-v-Sea/MLL–ENL | Leukaemia | 8/8 | 31–49 |
Eight-day-old chicks were coinjected with fibroblasts producing MLL–ENL and/or ts-v-Sea virus and monitored for 3 months. NA, not applicable.

Fig. 5. Molecular and phenotypic analysis of MLL–ENL/ts-v-Sea- induced leukaemias. (A) Southern blot of genomic DNA extracted from bone marrow of leukaemic chickens. Digestion with EcoRI reveals a truncated MLL–ENL cDNA of ∼4 kb in all leukaemias analysed (see legend to Figure 2). As a control, the MLL–ENL clone D11 containing the 6 kb full-length MLL–ENL cDNA and the chicken T-cell line MSB1 are shown. (B) Schematic representation of MLL–ENL-specific sequences detected in leukaemias 2 and 6, revealing an in-frame deletion. See Results and Supplementary data for a detailed description of the respective nested PCR analyses. (C) Analysis of ts-v-Sea in leukaemias 2, 6 and 7 by PCR analysis generated the expected 1.4 kb product, encompassing the S13 viral env sequence plus the transmembrane and complete tyrosine kinase domains of v-Sea. Positive control, in vitro-transformed ts-v-Sea/MLL–ENL clone; negative control, MLL–ENL clone D8; W, water control. (D) Cultured cells from leukaemia 2 (two fields, top) and leukaemia 6 (bottom) are shown after cytocentrifugation on to slides and staining with acid benzidine and Wright–Giemsa.
Next, we tested the same leukaemias for the presence of the ts-v-sea oncogene by PCR. Only leukaemia 2 was positive for retroviral ts-v-Sea (Figure 5C), while this sequence could not be detected in leukaemias 6 and 7. In line with this, we were unable to expand the leukaemic cells from the latter two leukaemias, using media which support proliferation of ts-v-Sea-transformed erythroblasts (see Materials and methods). Instead, these leukaemic cells persisted in the cultures as partially mature myeloid cells for ∼7 days (Figure 5D, bottom panel; data not shown). In contrast, bone marrow cells from leukaemia 2 could be expanded in the absence of SCF and maintained for 2 months as proliferating, immature cells without visible signs of senescence, far exceeding the much shorter in vitro lifespan of ts-v-Sea-transformed erythroblasts. Cytospin analysis revealed that cultured cells from leukaemia 2 contained blast-like progenitors plus immature erythroid and myeloid cells (Figure 5D, top panels). Furthermore, FACS analysis showed that leukaemia 2 cells expressed a similar set of multipotent, erythroid and myeloid lineage marker antigens to the in vitro-transformed bone marrow MLL–ENL clones (data not shown). In conclusion, v-Sea and MLL–ENL cooperate in causing multilineage leukaemia in vivo.
ts-v-Sea can replace requirement for SCF in vitro
Finally, we addressed the question of whether ts-v-Sea could substitute for SCF-activated c-Kit in the cooperation with MLL–ENL to cause multipotent cell transformation in vitro. Chick bone marrow cells were coinfected with the two retroviruses and MLL–ENL/ts-v-Sea-transformed colonies selected in methocel, in the presence of G418 and in the absence of SCF. Several clones carrying the complete MLL–ENL cDNA and expressing ts-v-Sea and MLL–ENL as detected by RT–PCR (data not shown) were obtained. As expected from the results with leukaemia 2, ts-v-Sea/MLL–ENL-expressing clones no longer required SCF for continuous proliferation at 37°C (Figure 6, triangles). This SCF dependence could be restored after complete inactivation of ts-v-Sea (42°C plus the glycosylation inhibitor castanospermine; Knight et al., 1988). In the presence of exogenous SCF, the temperature-shifted and castanospermine-treated cells proliferated for >6 days (Figure 6, squares), while they ceased to grow after 4–5 days in the absence of SCF (Figure 6, circles). In conclusion, ts-v-Sea signals can substitute for those emanating from activated c-Kit, while complete inhibition of ts-v-Sea restores SCF requirement in the MLL–ENL-transformed cells.
Fig. 6. ts-v-Sea can substitute for SCF-induced c-Kit signalling required for leukaemic transformation by MLL–ENL. In vitro-transformed MLL–ENL/ts-v-Sea clones C7, D6 and G8 were cultivated in S13 medium without additions at 37°C (triangles) or at 42°C in the same medium plus 300 µM of the glycosylation inhibitor castanospermine, in the presence (squares) or absence (circles) of SCF. Cumulative cell numbers were determined daily using an electronic cell counter.
Discussion
Here, we demonstrate that primary, avian bone marrow cells expressing MLL–ENL show sustained renewal, display a multipotent progenitor phenotype and have a strongly enhanced in vitro lifespan. Both survival and renewal of these multipotent progenitors as immature cells required activation of endogenous c-Kit by its ligand SCF, replaceable by signals from constitutively active receptor tyrosine kinase oncoproteins such as v-Sea.
MLL–ENL-transformed bone marrow clones expressed several multipotential cell markers such as 11C3 and thrombomucin (MEP21). Although these markers are also present on thrombocytes, the MLL–ENL-transformed clones could be induced to undergo either myeloid or erythroid differentiation in response to different cytokine combinations, confirming their multipotential nature. Similarly, expression of three markers for immature T- or B-cells suggested that MLL–ENL progenitors represented lymphohematopoietic cells, a notion supported by our finding that MLL–ENL-transformed clones also expressed the chicken homologue of NG2, a marker frequently detected in infant ALL with MLL involvement (Behm et al., 1996). Since induction of lymphoid differentiation of chicken cells in vitro is still a very inefficient process, we cannot formally prove that expression of lymphoid markers on these cells does in fact indicate lymphoid lineage potential. Alternatively, immature MLL–ENL-transformed progenitors may be primed to simultaneously express multilineage-associated markers (Hu et al., 1997), a possibility that might explain the seemingly aberrant expression of CD3 in the absence of the T-cell receptor. Renewal of multipotent progenitors in vitro could not be induced by MLL–ENL alone, but required cooperation with ligand-activated c-Kit. Similar cooperations between receptor tyrosine kinases (c-Kit/v-Sea/v-ErbB) and oncogenes derived from transcription factors or chromatin regulators, e.g. v-ErbA (Bauer et al., 2001) and v-Ski (Beug et al., 1995a) were required for induction of progenitor renewal in other avian leukaemia models. One physiological in vivo role of such cooperations between tyrosine kinases and transcriptional regulators is the enhancement of erythroid progenitor renewal during stress erythropoiesis (Bauer et al., 1999).
Despite detailed molecular studies involving a variety of MLL fusion proteins, it has been difficult to directly assess their biological function. A prevalent feature of MLL-associated leukaemias is the coexpression of myeloid and lymphoid antigens in the leukaemic blasts [e.g. CD15 in ALL with t(4,11); Pui et al., 1991]. In rare cases, they present as biphenotypic leukaemias (Carbonell et al., 1996) supporting a stem cell origin of this type of leukaemia. In addition, recent evidence from microarray analysis of leukaemias harbouring MLL translocations display gene expression profiles distinctly different from other B-cell ALLs (Armstrong et al., 2002). In human leukaemias expressing the MLL–ENL fusion protein, over half of the leukaemias are characterized as lymphoid (Moorman et al., 1998).
In contrast, none of these phenotypes were displayed in murine models, where expression of MLL–ENL almost exclusively caused leukaemic transformation of myeloid cells in vitro and in vivo. Our studies identify a possible reason for this discrepancy. Since transformation of multipotent cells by MLL–ENL requires concomitant activation of c-Kit, it is likely that additional alterations occurred in the MLL–ENL-induced methocel colonies during their establishment into respective myeloid cell lines (Lavau et al., 1997). A similar need for secondary events may be responsible for the fact that chimaeric mice expressing MLL–AF9 progressed to ALL only rarely and after long latency periods (>11 months) (Dobson et al., 1999). The strong dependence of MLL–ENL on cooperating signals during transformation of multipotent progenitors is also indicated by the fact that the differentiation arrest induced by MLL–ENL plus c-Kit was neither absolute nor irreversible. Rather, the transformed multipotent cells could be committed to erythroid or myeloid progenitors and then induced to terminally differentiate by a two-step procedure involving respective, lineage-specific cytokines combined with the controlled withdrawal of SCF.
The MLL–ENL fusion protein also contributed to leukaemia induction in chicks. Interestingly, all three characterized leukaemias contained a similar deletion in the MLL component. Previous evidence suggests that loss of these MLL sequences does not impair myeloid transformation (Slany et al., 1998). Since the length of the MLL–ENL retrovirus approaches the upper size limit for packaging into viral particles, the most likely reason for generation of these truncations is in vivo selection for a smaller transforming retrovirus, as demonstrated by others (Vennstrom et al., 1994).
Clearly, full leukaemic transformation of multipotent cells in vivo (leukaemia 2) required cooperation of MLL fusion proteins with the ts-v-Sea receptor tyrosine kinase, similar to the situation in leukaemias generated by v-Ski. Interestingly, two leukaemias (6 and 7) could not be expanded in vitro. These leukaemias arose with a long latency period (49 days after infection compared with 32 days for leukaemia 2) and expressed MLL–ENL but not v-Sea, when analysed after in vitro culture. Since none of the chicks injected with MLL–ENL-producing fibroblasts alone ever developed leukaemia, we have to assume that v-Sea plays a role early on during leukaemia induction, but may not be required during later stages. This agrees with our earlier observation that rare clones of multipotent v-Ski/ts-v-Sea-transformed bone marrow cells were observed to lose v-Sea, resulting in loss of renewal potential and differentiation into partially mature myeloid cells (Larsen et al., 1993).
Do receptor tyrosine kinases such as c-Kit or RON, the human homologue of Sea (Ronsin et al., 1993) play a role in human leukaemias harbouring MLL fusion proteins? No proof for such a concept has been obtained to date but some circumstantial evidence exists. Recently, enhanced activity of c-Kit due to mutations was found in human leukaemias with a variable incidence from extremely rare to 50% of AML with inv(16) (Gari et al., 1999; Beghini et al., 2000). In addition, expression of c-Kit is enhanced in myeloid leukaemia and T-ALL (Legitimo et al., 1999; Ashman et al., 2000). Another member of the stem cell tyrosine kinase family, FLT3, was identified as a relatively common leukaemogenic target in AML (Yamamoto et al., 2001) and found as a highly upregulated gene in microarray analysis of MLL rearranged B-precursor ALL (Armstrong et al., 2002). Finally, oncogenic mutants of RON induced transformation of NIH 3T3 cells, by causing accumulation of c-myc transcripts via activation of the Tcf/LEF transcription factor (Danilkovitch-Miagkova et al., 2001). Since the differentiation arrest of murine MLL–ENL-transformed myelomonocytic progenitors was dependent on Myc (Schreiner et al., 2001), enhanced RON or Sea tyrosine kinase activity might contribute to MLL–ENL transformation via c-Myc induction, at least in our avian model. This raises the possibility that either secondary mutations in Myc or activated RON might cooperate with MLL–ENL in human leukaemia.
This novel possibility raised by our studies, i.e. that leukaemia characterized by MLL fusion proteins is dependent on hyperactive signalling from stem cell receptor tyrosine kinases, could be of crucial importance for the development of treatment. First results from clinical use of the FDA-approved kinase c-Abl inhibitor Gleevec (STI-571) in chronic myelogenous leukaemia (CML) are very promising (Mauro and Druker, 2001). However, this drug is not specific for c-Abl, inhibiting also the tyrosine kinase activity of c-Kit (Buchdunger et al., 2000) and PDGFR, suggesting that it may be useful for treatment of other leukaemias including those involving MLL fusion proteins.
Materials and methods
Cloning of retroviral constructs and establishment of producer cell lines
A 1 kb fragment containing the 3′ end of AF9 and encompassing the fusion point with MLL was cloned by RT–PCR from total RNA isolated from the MonoMac6 cell using a primer from exon 5 in MLL and a C-terminal AF9 primer incorporating an additional sequence encoding a Myc-tag epitope and an EcoRI site. A 740 bp PflMI–EcoRI fragment was then cloned into the corresponding sites of the pBEXd11t construct containing the MLL–ENL cDNA (a gift from Dr M.Cleary). The resulting MLL–AF9 cDNA fuses MLL exon 8 in frame with a central exon of AF9 (nucleotide 1321 in DDBJ/EMBL/GenBank accession No. D16688). The 5 kb MLL–AF9 cDNA was then cloned into the pCRNCM retroviral vector (Steinlein et al., 1994). A C-terminal Myc tag was also introduced into the MLL–ENL cDNA by a similar procedure and subcloned into the CRNCM vector. All PCR products were cloned and the sequence confirmed. Such Myc-tagged MLL fusion proteins were shown earlier to retain normal transforming activity.
Retroviral constructs were cotransfected in a 10:1 ratio with the RCAS helper virus (Hughes et al., 1987) into chicken embryo fibroblasts (CEFs) and cells were selected in 0.8 mg/ml G418. Correct size and expression of both constructs was tested by northern blot and immunofluorescence cell staining after transient expression in CEFs (data not shown).
Retroviral infection, methocel cloning and maintenance of cells
Freshly isolated bone marrow from 3- to 7-day-old chicks was infected with retroviral constructs as described previously (Beug et al., 1995b). Bone marrow cells were cultured in CFU-E medium (S13) (Hayman et al., 1993) supplemented with 200 ng/ml recombinant avian SCF, 10–6 M estradiol, IGF-1 (40 ng/ml) and 2–10% conditioned medium from c-rel-transformed CEFs (relCM), containing various avian cytokines including chicken myeloid growth factor (cMGF; for details see Beug et al., 1995b). Infected cultures were maintained at an optimal density of 4 × 106 cells/ml by daily partial medium changes plus addition of fresh growth factors. Infected bone marrow cells or uninfected control cells were plated in methocel cultures at various cell densities (5 × 105, 1 × 106 and 2 × 106 cells/plate) in the presence of SCF, relCM, estradiol, IGF-1 and selected with G418. For coinfection of bone marrow cells with MLL–ENL and ts-v-Sea, the bone marrow was infected as described above, mixing CEFs producing MLL–ENL and ts-v-Sea retroviruses at a ratio of 10:1.
Induction of lineage commitment and terminal differentiation
To analyse differentiation of MLL–ENL cultures, a two-step protocol developed previously for v-Ski multipotent progenitors was employed (Beug et al., 1995a). To induce erythroid commitment, cells were seeded at 4 × 106 cells/ml in Epotest medium (lacking chicken serum) containing SCF, anaemic serum as a source of chicken erythropoietin, the glucocorticoid receptor agonist dexamethasone and IGF-I. Myeloid commitment was induced in the same medium containing SCF, relCM, cMGF, dexamethasone, the estrogen receptor antagonist ICI164384) and IGF-I. After continuous proliferation in these commitment media for 5–7 days, terminal erythroid differentiation was induced in the cells by switching them to S13 medium containing high-titre anaemic serum plus insulin, while terminal myeloid differentiation was induced in Epotest medium containing cMGF, relCM and IGF-I. After 5 days of terminal differentiation, cells were analysed for their phenotype by cytocentrifugation and subsequent staining with Wright–Giemsa and acid benzidine.
DNA and RNA analysis
Genomic DNA was isolated using the GenomeStar kit (Hybaid) and Southern blot analysis was performed using standard procedures. The DNA was digested with appropriate enzymes to establish the clonal status and size of the MLL fusion cDNA and hybridized with probes as indicated in Results. Labelling was performed using a random primer DNA labelling kit (Boehringer Mannheim).
Total RNA was isolated using the TRIzol Reagent (Gibco BRL) and treated with RNase-free DNase (Boehringer Mannheim). Reverse transcription was performed using oligo(dT) primers and the GeneAmp RNA PCR Kit (Applied Bisosystems). PCR analysis was performed using MLL primer (MLL, DDBJ/EMBL/GenBank accession No. L04284) 5′-CGGTCAATAAGCAGGAGAAT-3′ and ENL primer (ENL, DDBJ/EMBL/GenBank accession No. L04285) 5′-GAGAGTGGAATTGT GGGTAA-3′ generating products spanning the MLL–ENL (720 bp) fusion point. Ikaros expression was determined by RT–PCR using primer pairs chEx4F/chEx6R and chEx2F/chEx7R published previously (Liippo and Lassila, 1997). Ts-v-Sea expression of in vitro-transformed ts-v-Sea/MLL–ENL was determined by RT–PCR with a v-Sea-specific primer pair 5′-CTAACTTGACAACATCACTCCTCG-3′ and 5′-GTTGATGTAGT GCTCACCTTCCAG-3′.
FACS analysis
For FACS analysis, 1 × 106 cells were washed once, resuspended in 100 µl phosphate-buffered saline (PBS) containing 10% fetal calf serum (FCS) and incubated on ice with the primary mouse monoclonal antibody for 1 h. Cells were washed three times and incubated with secondary rabbit anti-mouse immunoglobulin R-phycoerythrin-conjugated antibody (Dako A/S, Denmark). Cells were washed three times with PBS prior to analysis with a FACScalibur (Becton Dickinson). Antibodies used for immunofluorescence staining were the mouse monoclonals anti-c-Kit (kit2c75), MEP21, MEP26, 51-2, LT7, L22 and 5-11G2 recognizing the chicken Bu-1a and Bu-1b antigens respectively, CB3, AV20, 11C3, MY174 (Kimata et al., 1986). The phycoerythrin-conjugated CD3 (CT3), CD4 (CT4), CD8 (CT8), TcR1, TcR2 and TcR3 monoclonal antibodies were obtained from Southern Biotechnology (Birmingham, AL, USA). As isotype control murine IgG1 and IgG2 labelled with R-phycoerythrin were used. For all non-directly labelled mouse monoclonals an anti-chicken mouse monoclonal antibody was used as isotype control.
In vivo experiments
Eight-day-old chicks were injected intravenously with 1 × 106 ts-v-Sea producing fibroblasts, 5 × 106 MLL–ENL producing fibroblasts or coinjected with 5 × 106 MLL–ENL and 1 × 106 ts-v-Sea producing fibroblasts (injected volume, 100 µl). The chickens were monitored for 3 months. Genomic DNA was extracted from the bone marrow cells of highly moribund chickens and Southern blot analysis was carried out as described above. Detailed analysis of MLL truncations was carried out by PCR of genomic DNA with appropriate primers (MLL, DDBJ/EMBL/GenBank accession No. L04284, for primer sequences see Supplementary data). The presence of ts-v-Sea was determined by PCR with a v-Sea-specific primer pair 5′-CTAACTTGACAACATCACTCCTCG-3′ and 5′-GTTGATGTAGTGCTCACCTTCCAG-3′ (v-sea, DDBJ/EMBL/GenBank accession No. M25158). FACS analysis was carried out as described above. Trials to cultivate the leukaemic cells were performed in S13 medium (Beug et al., 1995b) supplemented with rel-CM and IGF-1.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors wish to thank Dr Michael Cleary for the MLL–ENL cDNA, Drs Dave Carrino, Chen Lo Chen, Catherine Corbel, Fred Davison, Michael Ratcliffe and Olli Vainio for gifts of antibodies, Olli Vainio for helpful advice, Evi Deiner for expert technical assistance and Mel Greaves for critical reading of the manuscript. This work was supported by a Leukaemia Research Fund Specialist Programme Grant (Grant No. 9845), European Union Network grants Nos ERB-FMRX-CT98-0197 and HPRN-CT-2000-00083 and grants from the Austrian Research funding agency (FWF; SFB 006/612) and the Austrian Industrial Research Promotion Fund (FFF project No 803776).
References
- Armstrong S.A. et al. (2002) MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet., 30, 41–47. [DOI] [PubMed] [Google Scholar]
- Ashman L.K., Ferrao,P., Cole,S.R. and Cambareri,A.C. (2000) Effects of mutant c-Kit in early myeloid cells. Leuk. Lymphoma, 37, 233–243. [DOI] [PubMed] [Google Scholar]
- Ayton P.M. and Cleary,M.L. (2001) Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene, 20, 5695–5707. [DOI] [PubMed] [Google Scholar]
- Bauer A., Tronche,F., Wessely,O., Kellendonk,C., Reichardt,H.M., Steinlein,P., Schutz,G. and Beug,H. (1999) The glucocorticoid receptor is required for stress erythropoiesis. Genes Dev., 13, 2996–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A., Gandrillon,O., Samarut,J. and Beug,H. (2001) Nuclear receptors in hematopoietic development: cooperation with growth factor receptors in regulation of proliferation and differentiation. In Zon,L. (ed.), Hematopoiesis: A Developmental Approach. Oxford University Press, Oxford, pp. 368–390.
- Beghini A., Peterlongo,P., Ripamonti,C.B., Larizza,L., Cairoli,R., Morra,E. and Mecucci,C. (2000) c-Kit mutations in core binding factor leukemias. Blood, 95, 726–727. [PubMed] [Google Scholar]
- Behm F.G., Smith,F.O., Raimondi,S.C., Pui,C.H. and Bernstein,I.D. (1996) Human homolog of the rat chondroitin sulfate proteoglycan, NG2, detected by monoclonal-antibody 7.1, identifies childhood acute lymphoblastic leukemias with t(4;11)(q21;q23) or t(11;19)(q23;p13) and MLL gene rearrangements. Blood, 87, 1134–1139. [PubMed] [Google Scholar]
- Benatar T., Iacampo,S., Tkalec,L. and Ratcliffe,M.J. (1991) Expression of immunoglobulin genes in the avian embryo bone marrow revealed by retroviral transformation. Eur. J. Immunol., 21, 2529–2536. [DOI] [PubMed] [Google Scholar]
- Beug H., Dahl,R., Steinlein,P., Meyer,S., Deiner,E.M. and Hayman,M.J. (1995a) In vitro growth of factor-dependent multipotential hematopoietic cells is induced by the nuclear oncoprotein v-Ski. Oncogene, 11, 59–72. [PubMed] [Google Scholar]
- Beug H., Steinlein,P., Bartunek,P. and Hayman,M.J. (1995b) Avian hematopoietic cell culture: in vitro model systems to study oncogenic transformation of hematopoietic cells. Methods Enzymol., 254, 41–76. [DOI] [PubMed] [Google Scholar]
- Beug H., Metz,T., Mullner,E.W. and Hayman,M.J. (1996) Self-renewal and differentiation in primary avian hematopoietic cells—an alternative to mammalian in vitro models. Curr. Top. Microbiol. Immunol., 211, 29–39. [DOI] [PubMed] [Google Scholar]
- Buchdunger E., Cioffi,C.L., Law,N., Stover,D., Ohno-Jones,S., Druker,B.J. and Lydon,N.B. (2000) Abl protein-tyrosine kinase inhibitor STI571 inhibits in vitro signal transduction mediated by c-kit and platelet-derived growth factor receptors. J. Pharmacol. Exp. Ther., 295, 139–145. [PubMed] [Google Scholar]
- Caldas C., Kim,M.-H., MacGregor,A., Cain,D., Aparicio,S. and Wiedemann,L.M. (1998) Isolation and characterization of a pufferfish MLL (mixed lineage leukemia) like gene (fMLL) reveals evolutionary conservation in vertebrate genes related to Drosophila trithorax. Oncogene, 16, 3233–3241. [DOI] [PubMed] [Google Scholar]
- Carbonell F., Swansbury,J., Min,T., Matutes,E., Farahat,N., Buccheri,V., Morilla,R., Secker-Walker,L. and Catovsky,D. (1996) Cytogenetic findings in acute biphenotypic leukaemia. Leukemia, 10, 1283–1287. [PubMed] [Google Scholar]
- Chen C.L., Ager,L.L., Gartland,G.L. and Cooper,M.D. (1986) Identification of a T3/T-cell receptor complex in chickens. J. Exp. Med., 164, 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral J. et al. (1996) An Mll–AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell, 85, 853–861. [DOI] [PubMed] [Google Scholar]
- Cortes M., Wong,E., Koipally,J. and Georgopoulos,K. (1999) Control of lymphocyte development by the Ikaros gene family. Curr. Opin. Immunol., 11, 157–171. [DOI] [PubMed] [Google Scholar]
- Danilkovitch-Miagkova A., Miagkov,A., Skeel,A., Nakaigawa,N., Zbar,B. and Leonard,E.J. (2001) Oncogenic mutants of RON and MET receptor tyrosine kinases cause activation of the β-catenin pathway. Mol. Cell. Biol., 21, 5857–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C.L., Warren,A.J., Pannell,R., Forster,A., Lavenir,I., Corral,J., Smith,A.J. and Rabbitts,T.H. (1999) The mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J., 18, 3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez M.S., Dennis,J.E., Drushel,R.F., Carrino,D.A., Kimata,K., Yamagata,M. and Caplan,A.I. (1991) The dynamics of compartmentalization of embryonic muscle by extracellular matrix molecules. Dev. Biol., 147, 46–61. [DOI] [PubMed] [Google Scholar]
- Gari M., Goodeve,A., Wilson,G., Winship,P., Langabeer,S., Linch,D., Vandenberghe,E., Peake,I. and Reilly,J. (1999) c-kit proto-oncogene exon 8 in-frame deletion plus insertion mutations in acute myeloid leukaemia. Br. J. Haematol., 105, 894–900. [DOI] [PubMed] [Google Scholar]
- Graf T., McNagny,K., Brady,G. and Frampton,J. (1992) Chicken ‘erythroid’ cells transformed by the Gag-Myb-Ets-encoding E26 leukemia virus are multipotent. Cell, 70, 201–213. [DOI] [PubMed] [Google Scholar]
- Greaves M.F. (1997) Aetiology of acute leukaemia. Lancet, 349, 344–349. [DOI] [PubMed] [Google Scholar]
- Hayman M.J., Beug,H. and Savin,K.W. (1982) Changes in the expression of membrane antigens during the differentiation of chicken erythroblasts. J. Cell Biochem., 18, 351–362. [DOI] [PubMed] [Google Scholar]
- Hayman M.J., Meyer,S., Martin,F., Steinlein,P. and Beug,H. (1993) Self-renewal and differentiation of normal avian erythroid progenitor cells: regulatory roles of the TGF α/c-ErbB and SCF/c-kit receptors. Cell, 74, 157–169. [DOI] [PubMed] [Google Scholar]
- Hu M., Krause,D., Greaves,M., Sharkis,S., Dexter,M., Heyworth,C. and Enver,T. (1997) Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev., 11, 774–785. [DOI] [PubMed] [Google Scholar]
- Hughes S.H., Greenhouse,J.J., Petropoulos,C.J. and Sutrave,P. (1987) Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol., 61, 3004–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata K., Oike,Y., Tani,K., Shinomura,T., Yamagata,M., Uritani,M. and Suzuki,S. (1986) A large chondroitin sulfate proteoglycan (PG-M) synthesized before chondrogenesis in the limb bud of chick embryo. J. Biol. Chem., 261, 13517–13525. [PubMed] [Google Scholar]
- Knight J., Beug,H., Marshall,J. and Hayman,M.J. (1988) Abnormal glycosylation of the env-sea oncogene product inhibits its proteolytic cleavage and blocks its transforming ability. Oncogene, 2, 317–326. [PubMed] [Google Scholar]
- Larsen J., Beug,H. and Hayman,M.J. (1992) The v-ski oncogene cooperates with the v-sea oncogene in erythroid transformation by blocking erythroid differentiation. Oncogene, 7, 1903–1911. [PubMed] [Google Scholar]
- Larsen J., Meyer,S., Steinlein,P., Beug,H. and Hayman,M.J. (1993) Transformation of chicken bone marrow cells by v-ski oncogene. Oncogene, 8, 3221–3228. [PubMed] [Google Scholar]
- Lavau C., Szilvassy,S.J., Slany,R. and Cleary,M.L. (1997) Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX–ENL. EMBO J., 16, 4226–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legitimo A., Consolini,R., Cocito,M.G., Buffoni,R., Basso,G. and Macchia,P. (1999) The c-kit receptor and its ligand stem cell factor in childhood malignant lymphoid precursors. J. Interferon Cytokine Res., 19, 981–987. [DOI] [PubMed] [Google Scholar]
- Liippo J. and Lassila,O. (1997) Avian Ikaros gene is expressed early in embryogenesis. Eur. J. Immunol., 27, 1853–1857. [DOI] [PubMed] [Google Scholar]
- Mauro M.J. and Druker,B.J. (2001) STI571: targeting BCR-ABL as therapy for CML. Oncologist, 6, 233–238. [DOI] [PubMed] [Google Scholar]
- Mauvieux L., Delabesse,E., Bourquelot,P., Radford-Weiss,I., Bennaceur, A., Flandrin,G., Valensi,F. and MacIntyre,E.A. (1999) NG2 expression in MLL rearranged acute myeloid leukaemia is restricted to monoblastic cases. Br. J. Haematol., 107, 674–676. [DOI] [PubMed] [Google Scholar]
- McNagny K.M., Pettersson,I., Rossi,F., Flamme,I., Shevchenko,A., Mann,M. and Graf,T. (1997) Thrombomucin, a novel cell surface protein that defines thrombocytes and multipotent hematopoietic progenitors. J. Cell Biol., 138, 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman A.V., Hagemeijer,A., Charrin,C., Rieder,H. and Secker-Walker,L.M. (1998) The translocations, t(11;19)(q23;p13.1) and t(11;19)(q23;p13.3): a cytogenetic and clinical profile of 53 patients. European 11q23 Workshop participants. Leukemia, 12, 805–810. [DOI] [PubMed] [Google Scholar]
- Ody C., Vaigot,P., Quere,P., Imhof,B.A. and Corbel,C. (1999) Glyco protein IIb–IIIa is expressed on avian multilineage hematopoietic progenitor cells. Blood, 93, 2898–2906. [PubMed] [Google Scholar]
- Pickel J.M., Chen,C.L. and Cooper,M.D. (1990) An avian B-lymphocyte protein associated with β2-microglobulin. Immunogenetics, 32, 1–7. [DOI] [PubMed] [Google Scholar]
- Pui C.-H. et al. (1991) Clinical characteristics and treatment outcome of childhood acute lymphoblastic leukemia with the t(4;11)(q21;q23): a collaborative study of 40 cases. Blood, 77, 440–447. [PubMed] [Google Scholar]
- Ronsin C., Muscatelli,F., Mattei,M.G. and Breathnach,R. (1993) A novel putative receptor protein tyrosine kinase of the met family. Oncogene, 8, 1195–1202. [PubMed] [Google Scholar]
- Rothwell C.J., Vervelde,L. and Davison,T.F. (1996) Identification of chicken Bu-1 alloantigens using the monoclonal antibody AV20. Vet. Immunol. Immunopathol., 55, 225–234. [DOI] [PubMed] [Google Scholar]
- Schreiner S., Birke,M., Garcia-Cuellar,M.P., Zilles,O., Greil,J. and Slany,R.K. (2001) MLL–ENL causes a reversible and myc-dependent block of myelomonocytic cell differentiation. Cancer Res., 61, 6480–6486. [PubMed] [Google Scholar]
- Slany R.K., Lavau,C. and Cleary,M.L. (1998) The oncogenic capacity of HRX–ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol. Cell. Biol., 18, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F.O. et al. (1996) The human homolog of rat NG2, a chondroitin sulfate proteoglycan, is not expressed on the cell-surface of normal hematopoietic cells but is expressed by acute myeloid-leukemia blasts from poor-prognosis patients with abnormalities of chromosome band 11q23. Blood, 87, 1123–1133. [PubMed] [Google Scholar]
- Steinlein P., Deiner,E., Leutz,A. and Beug,H. (1994) Recombinant murine erythropoietin receptor expressed in avian erythroid progenitors mediates terminal erythroid differentiation in vitro. Growth Factors, 10, 1–16. [DOI] [PubMed] [Google Scholar]
- Tran Quang C., Wessely,O., Pironin,M., Beug,H. and Ghysdael,J. (1997) Cooperation of Spi-1/PU.1 with an activated erythropoietin receptor inhibits apoptosis and Epo-dependent differentiation in primary erythroblasts and induces their Kit ligand-dependent proliferation. EMBO J., 16, 5639–5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio O., Mansikka,A. and Lassila,O. (1990) Analysis of avian T and B lymphocytes. In Lefkovits,I. (ed.), Immunological Methods. Academic Press, pp. 265–279.
- Vainio O., Dunon,D., Aissi,F., Dangy,J.P., McNagny,K.M. and Imhof,B.A. (1996) HEMCAM, an adhesion molecule expressed by c-kit+ hemopoietic progenitors. J. Cell Biol., 135, 1655–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennstrom B., Raynoscheck,C., Jansson,L., Doederlein,G., Lhotak,V., Johnsson,A. and Beug,H. (1994) Retroviral capture of c-erbB proto-oncogene sequences: rapid evolution of distinct viral genomes carrying mutant v-erbB genes with different transforming capacities. Oncogene, 9, 1307–1320. [PubMed] [Google Scholar]
- Wuchter C. et al. (2000) Detection of acute leukemia cells with mixed lineage leukemia (MLL) gene rearrangements by flow cytometry using monoclonal antibody 7.1. Leukemia, 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y. et al. (2001) Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood, 97, 2434–2439. [DOI] [PubMed] [Google Scholar]
- Yano T., Nakamura,T., Blechman,J., Sorio,C., Dang,C.V., Geiger,B. and Canaani,E. (1997) Nuclear punctate distribution of ALL-1 is conferred by distinct elements at the N terminus of the protein. Proc. Natl Acad. Sci. USA, 94, 7286–7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B.D., Hess,J.L., Horning,S.E., Brown,G.A.J. and Korsmeyer,S.J. (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature, 378, 505–508. [DOI] [PubMed] [Google Scholar]