Abstract
During evolution, placental mammals appear to have lost cyclobutane pyrimidine dimer (CPD) photolyase, an enzyme that efficiently removes UV-induced CPDs from DNA in a light-dependent manner. As a consequence, they have to rely solely on the more complex, and for this lesion less efficient, nucleotide excision repair pathway. To assess the contribution of poor repair of CPDs to various biological effects of UV, we generated mice expressing a marsupial CPD photolyase transgene. Expression from the ubiquitous β-actin promoter allowed rapid repair of CPDs in epidermis and dermis. UV-exposed cultured dermal fibroblasts from these mice displayed superior survival when treated with photoreactivating light. Moreover, photoreactivation of CPDs in intact skin dramatically reduced acute UV effects like erythema (sunburn), hyperplasia and apoptosis. Mice expressing the photolyase from keratin 14 promoter photo reactivate CPDs in basal and early differentiating keratinocytes only. Strikingly, in these animals, the anti-apoptotic effect appears to extend to other skin compartments, suggesting the presence of intercellular apoptotic signals. Thus, providing mice with CPD photolyase significantly improves repair and uncovers the biological effects of CPD lesions.
Keywords: cyclobutane pyrimidine dimers/photolyase transgenic mice/photoreactivation/UV sensitivity
Introduction
Absorption of UV light energy by DNA induces various types of lesions. Although single- and double-strand breaks, as well as DNA–protein cross-links, can occur, >99% of the UV-induced damage consists of chemical base modifications, with cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6–4) pyrimidone photoproducts (6–4PPs) as the most frequent types of photolesions (Mitchell, 1988). CPDs and 6–4PPs occur at two adjacent pyrimidines in the DNA and affect proper base pairing, which results in interference with key cellular processes like transcription and replication. Lesions in the DNA can lead to reduction of RNA synthesis, arrest of cell cycle progression, and induction of apoptosis. Moreover, persisting DNA damage can give rise to gene mutations that may allow cells to escape from controlled growth, which may ultimately lead to cancer (Friedberg et al., 1995).
To counteract the deleterious effects of DNA damage produced by endogenous and environmental genotoxic agents (including UV-induced lesions), all organisms have developed a complex network of repair systems with complementary substrate specificity that keeps the DNA under continuous surveillance (Friedberg et al., 1995; Hoeijmakers, 2001). Removal of photolesions from the DNA is performed by the versatile and evolutionary highly conserved nucleotide excision repair (NER) pathway. NER is a complex multi-step process, and involves the concerted action of 30 or so proteins to sequentially execute damage recognition, chromatin remodeling, excision of a small oligonucleotide containing the damage, and gap-filling DNA synthesis followed by strand ligation (Friedberg et al., 1995; Wood, 1996; de Laat et al., 1999; Hoeijmakers 2001). The NER system is comprised of two subpathways. Global genome NER (GG-NER) operates genome-wide, but has the disadvantage that certain types of damage (like UV-induced CPDs) are less well recognized and accordingly less efficiently repaired. To prevent such lesions hampering transcription too long by stalling RNA polymerase II, a distinct NER subpathway has evolved, called transcription-coupled repair (TC- NER). This process directs the repair machinery, preferentially to the blocked polymerase on the template strand of actively transcribed DNA, and operates as a selective backup system for lesions that are slowly, or not at all, repaired by GG-NER.
Inherited defects in NER genes cause photosensitive disorders such as xeroderma pigmentosum (XP, subdivided over seven complementation groups, XP-A through to -G) and Cockayne syndrome (CS, complementation groups A and B; for a review, see Bootsma et al., 2001). All XP patients are deficient in GG-NER and, with the exception of XP-C and -E, also in TC-NER. A specific defect in TC-NER is encountered in CS (Venema et al., 1990; van Hoffen et al., 1993). XP and CS patients display an increased sensitivity of the skin to UV light. In addition, XP patients have a highly elevated risk of developing UV-induced skin cancer. Obviously, the repair of UV-induced DNA damage strongly reduces the many undesirable consequences of UV exposure.
It has proven difficult to disentangle the individual contribution of different types of UV lesions (i.e. CPDs and 6–4PPs) to the range of deleterious effects exerted by UV irradiation. CPDs are known to be repaired much slower by NER than 6–4PPs (Mitchell, 1988). In NER-proficient human fibroblasts, repair of most of the CPD lesions introduced by a modest dose of UV light takes >24 h, while 6–4PPs are removed within a few hours (Bohr et al., 1985). The GG-NER system is responsible for CPD removal anywhere in the genome, whereas CPDs that stall elongating RNA polymerase are selectively eliminated by TC-NER (Bohr et al., 1985; Mellon et al., 1987; van Hoffen et al., 1993). Recognition and repair of CPDs by GG-NER requires p53-dependent upregulation of the p48 subunit of the XP-E p48-p125 UV-DDB heterodimer (Hwang et al., 1999). Importantly, the p48 gene is not induced in rodent cells (Hwang et al., 1998); therefore, repair of CPDs is limited to the transcribed strand of active genes by TC-NER (Bohr et al., 1985; van der Horst et al., 1997). In contrast to CPD repair, both human and rodent cells efficiently remove 6–4PPs from their genome (Mitchell, 1988; van Hoffen et al., 1995), mainly performed by GG-NER. In the epidermis of humans and mice, keratinocytes remove CPDs and 6–4PPs in the same fashion as cultured fibroblasts (Hanawalt et al., 1981; Mitchell et al., 1990; Ruven et al., 1994; Qin et al., 1995). Some studies have pointed to the CPDs as the most cytotoxic and carcinogenic lesions (Broughton et al., 1990; Nishigaki et al., 1998; Chiganças et al., 2000), but, to date, the actual magnitude of the biological effects provoked by the different types of UV-induced DNA lesions has remained unclear.
Many organisms of all kingdoms mount an additional repair system to remove UV lesions called photoreactivation. In contrast to the complex NER pathway, photoreactivation is performed by photolyases that rapidly convert UV lesions back to the original undamaged bases in a simple enzymatic reaction using visible light as an energy source. To perform this reaction, photolyases are equipped with two different chromophoric co-factors. Depending on the photolyase, either 5,10-methyl tetrahydrofolate (MTHF) or 8-hydroxy-5-deazaflavin (8-HDF) serve as light-harvesting antennas that pass on energy to reduced FAD, the chromophore that acts as the reaction center in dimer splitting (for a review, see Yasui and Eker, 1997). Notably, photolyases show substrate specificity for either CPDs (CPD photolyase) or 6–4PPs (6–4PP photolyase; for a review, see Yasui and Eker, 1997). Photolyases occur in bacteria, lower eukaryotes, plants and many animals including marsupials. Remarkably, despite the strong conservation of photoreactivation, this repair mechanism is absent in placental mammals (e.g. humans and mice), implying that photolyase genes have been lost during evolution of this subclass. The inability to photoreactivate CPDs and 6–4PPs leaves placental mammals with only the NER system for removal of these photolesions. Here, we report the generation and characterization of mice that express the CPD photolyase transgene from the marsupial Potorous tridactylis either in all tissues or specifically in the basal keratinocytes of the epidermis.
Results
Generation of β-actin promoter-driven CPD photolyase transgenic mouse and cell lines
To generate mice ubiquitously expressing a CPD photolyase transgene, we assembled a construct containing the P.tridactylis CPD photolyase cDNA (Yasui et al., 1994), preceded by a cytomegalovirus (CMV) enhancer containing the chicken β-actin promoter and hooked up to human genomic β-globin sequences, including exons 2 and 3, intron 2, the 3′ untranslated region and the polyadenylation signal (Figure 1A). After oocyte injections, we obtained two independent β-actin promoter-driven photolyase mouse lines, designated β-act-CPD-1 and β-act-CPD-2 (carrying two and three copies of the transgene, respectively; data not shown). FISH analysis of metaphase chromosome spreads of mouse dermal fibroblasts (MDFs) isolated from β-act-CPD-1 and β-act-CPD-2 animals revealed integration of the photolyase transgene on chromosome 3C and 15A2, respectively (data not shown).
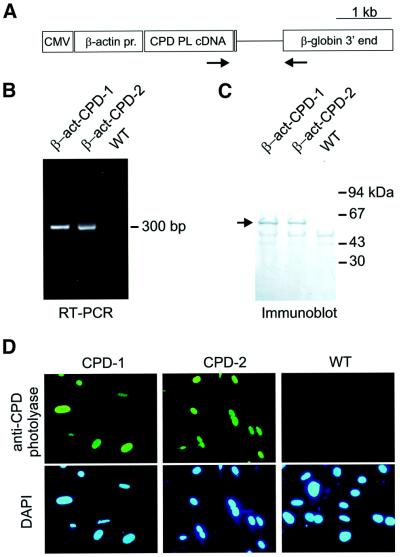
Fig. 1. Expression of the β-actin CPD photolyase gene in transgenic mice. (A) Expression construct for the generation of β-act-CPD photolyase transgenic mice, containing the CMV enhancer and chicken β-actin promoter, the P.tridactylis CPD photolyase cDNA, and human genomic β-globin sequences, including exons 2 and 3, intron 2, the 3′ untranslated region and the polyadenylation signal. Arrows indicate the position of the primers used for the RT–PCR experiment. (B) RT–PCR on skin extracts of β-act-CPD photolyase transgenic mice results in a 300 bp band. (C) Immunoblot analysis of protein extracts (30 µg per lane) from cultured fibroblasts obtained from β-act-CPD photolyase transgenic mice. The arrow indicates the position of the 62 kDa photolyase protein. (D) Immunocytochemical detection of CPD photolyase in cultured mouse dermal fibroblasts, using an affinity-purified anti-CPD photolyase. Nuclei are visualized by DAPI staining.
RT–PCR analysis of total skin RNA showed expression and correct splicing of the CPD photolyase mRNA (Figure 1B). In addition, immunoblot analysis of protein extracts from wild-type, β-act-CPD-1 and β-act-CPD-2 MDFs, using a polyclonal antibody raised against the Potorous CPD photolyase, showed a band of the expected size (61 kDa), indicating expression of the full-length protein (Figure 1C). Immunocytochemical analysis of these cultured MDFs revealed a fluorescent signal in the nuclei of transgenic cell lines only, indicating correct nuclear localization of the CPD photolyase protein, probably due to a putative nuclear localization signal sequence (24-ARKKRK-29; Yasui et al., 1994) in the N-terminus of the protein (Figure 1D). Taken together, these data demonstrate that the two independent CPD photolyase mouse lines properly express the transgene.
To facilitate genotyping, to avoid the potential risk of adverse side-effects originating from inactivation of loci at the transgene integration site, and to obtain transgenic and wild-type animals at equal amounts, mouse strains were kept in a heterozygous state. Importantly, β-act-CPD photolyase transgenic mice are born at a Mendelian ratio are fertile and, up to the age of 1 year, do not show any overt phenotype. This indicates that insertion of the CPD photolyase transgene does not interfere with development, viability and fertility. Moreover, β-act-CPD-1 and β-act- CPD-2 cells and animals are indistinguishable for all the parameters tested.
Photoreactivation of CPDs in cultured CPD photolyase fibroblasts
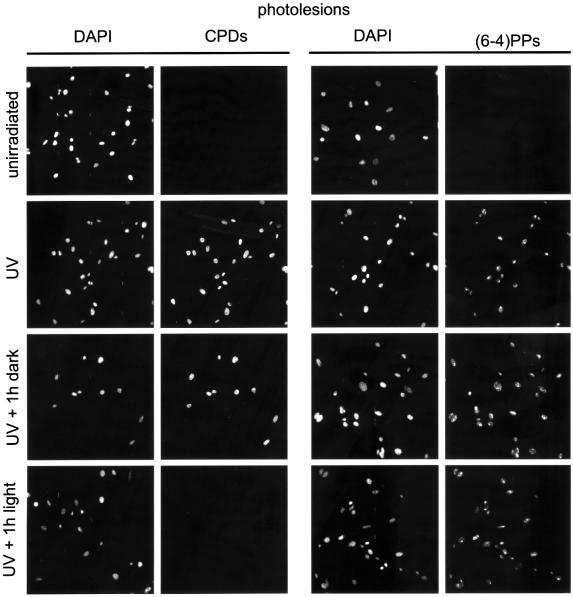
The transgenically expressed CPD photolyase is expected to allow lesion-specific repair of CPDs upon exposure to photoreactivating light. To investigate light-dependent removal of CPDs in MDFs, we have applied an immunocytochemical assay using antibodies specific for either CPDs or 6–4PPs (Mori et al., 1991). Cells were irradiated with 20 J/m2 of UV-C light and, as expected for both antibodies, a bright immunofluorescent signal could be detected in the nucleus of UV-treated cells, while non-irradiated cells remained unlabeled. This confirms the presence of both CPDs and 6–4PPs immediately after UV exposure (Figure 2). When cells were subsequently kept in the dark for 1 h (thus withholding the photolyase from the energy source required for enzymatic activity), CPD and 6–4PP staining remained present. In line with the known repair rates for both photolesions in rodents (Bohr et al, 1985; Mitchell, 1988), this indicates that, in the first hour after UV exposure, TC-NER can only remove a small fraction of the induced photolesions. In marked contrast, however, after exposure of UV-irradiated cells to 1 h of photoreactivating light, CPD labeling was hardly detectable, while the signal for 6–4PPs was comparable with that observed in cells kept in the dark (Figure 2). The lesion-specific and light-dependent removal of CPDs clearly demonstrates that the marsupial CPD photolyase is enzymatically active in mouse cells.
Fig. 2. Photoreactivation of CPDs in cultured CPD photolyase transgenic fibroblasts. Induction of CPD and 6–4PP lesions in cultured MDFs from β-act-CPD-1 photolyase mice by 20 J/m2 of UV-C light and the effect of subsequent exposure of cells photoreactivating light for 1 h. Photolesions were detected by immunofluorescent labeling, using CPD- or 6–4PP-specific antibodies and FITC-conjugated goat anti-mouse antibodies. Nuclei were visualized by DAPI staining.
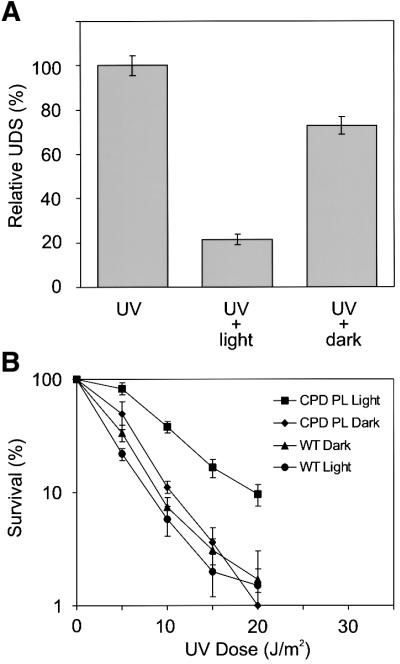
In repair-proficient mammalian cells, UV-induced DNA lesions are exclusively repaired by NER. From the previous experiment, it is evident that CPDs in UV-C exposed transgenic MDFs are rapidly removed by the CPD photolyase. Accordingly, it is expected that cells show reduced UV-induced NER activity after photoreactivation of CPD lesions. NER activity can be measured in cultured cells by the unscheduled DNA synthesis (UDS) assay, which quantifies the incorporation of radiolabeled thymidine in newly synthesized DNA in repair patches. As shown in Figure 3A, the relative UDS level in β-act- CPD-1 MDFs that were exposed to UV light and subsequently kept in the dark for 1 h drops to 75% of the UDS activity measured immediately after exposure to UV light. This difference reflects the NER activity during the first hour following UV exposure. As predicted, relative UDS levels decreased even further when UV-exposed cells were given 1 h of photoreactivating light. The reduction in NER activity confirms the effectiveness of the CPD photolyase and reveals the fraction of UDS derived from CPD repair.
Fig. 3. Effect of photoreactivation of CPDs in MDFs on NER activity and cellular survival. (A) NER activity in cultured MDFs from β-act-CPD-1 photolyase mice exposed to 16 J/m2 of UV-C light and the effect of subsequent exposure of cells to 1 h of photoreactivating light. UDS was measured by the incorporation of [3H]thymidine, followed by counting of grains above nuclei of non-dividing cells. UDS levels are expressed as the percentage of activity observed immediately after UV exposure. (B) UV survival of primary β-act-CPD-1 MDFs after exposure to increasing doses of UV-C light, with or without 1 h exposure to photoreactivating light. Error bars indicate the standard error of the mean. Similar results were obtained with β-act-CPD-2 MDFs.
Next, we studied whether the fast removal of CPDs renders MDFs supplemented with a CPD photoreactiva ting system less sensitive to UV-C light. As shown in Figure 3B, photoreactivation of the photolyase MDFs gave rise to a higher percentage of surviving cells over the whole range of UV doses tested when compared with non-photoreactivated transgenic and wild-type cells. The dose-reducing effect of the CPD photolyase is ∼2-fold, meaning that photoreactivation increases the UV survival to the level normally observed at half the UV dose. Thus, NER-proficient mouse cells significantly benefit from CPD photoreactivation.
Photoreactivation of CPDs in the mouse skin
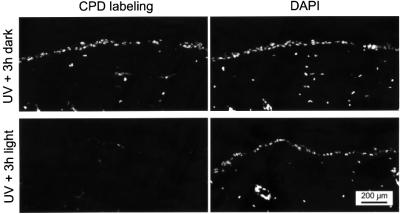
To investigate whether the skin, the prime target of UV light in the intact animal, also profits from the addition of the expressed CPD photolyase transgene, we analyzed the repair of CPDs in the dermis and epidermis of β-actin promoter-driven CPD photolyase transgenic mice. To this end, the immunofluorescent assay used to visualize photolesions in cultured MDFs was adapted for use on skin sections (see Materials and methods). One-third of a depilated area on the back of photolyase mice was covered, while the remaining part was exposed to 1 minimal erythemal dose (MED; the UV dose at which erythema starts to appear) of UV-B light. Next, half of the UV-exposed area, as well as the non-UV-exposed area, was covered, while the remaining part of the skin was exposed to photoreactivating light for 3 h. As expected, the non-UV-exposed part of the skin did not show any CPD labeling (data not shown), whereas the UV-irradiated/non-photoreactivated part of the skin contained clear nuclear labeling of CPDs in the epidermis and the upper part of the dermis (Figure 4, top panels). Importantly, exposure of the UV-irradiated skin to photoreactivating light resulted in a strong decrease in CPD labeling in epidermis and dermis. In contrast, labeling of 6–4PPs was not reduced detectably upon photoreactivation (data not shown). These data clearly demonstrate that the ubiquitously expressed CPD photolyase is active in all epidermal and dermal cells and that the enzyme can specifically photoreactivate the majority of CPDs in these cells within 3 h following UV exposure, leaving the 6–4PPs unaffected. In addition, these data show that both UV-B light and photoreactiva ting light do penetrate the epidermis and reach into the dermis.
Fig. 4. Photoreactivation of CPDs in the skin of β-act-CPD photolyase transgenic mice. Induction of CPD lesions in the depilated dorsal skin of β-act-CPD-1 photolyase mice by 1 MED of UV-B light and the effect of subsequent exposure to photoreactivating light for 3 h. Photolesions were detected by immunofluorescent labeling, using CPD-specific antibodies and FITC-conjugated goat anti-mouse antibodies. Nuclei are visualized by DAPI staining.
Reduction of acute skin effects by photoreactivation of CPDs
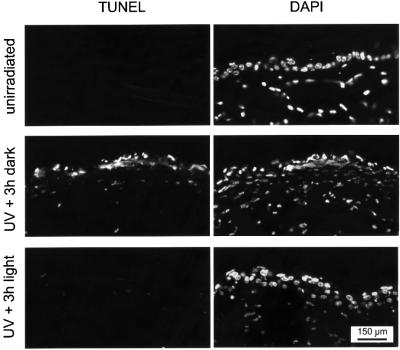
Exposure of the skin to (repeated) UV light induces apoptosis in the epidermis, accompanied by redness (erythema, commonly known as sunburn), swelling (edema), and followed several days later by thickening of the epidermis (hyperplasia). To investigate the effect of CPD photoreactivation on induction of apoptosis, the depilated dorsal skin of CPD photolyase mice was exposed to a single dose of 1 MED of UV-B light and partly exposed to photoreactivating light for 3 h, as described in the previous section. Next, animals were kept in the dark and, after 40 h, skin samples were processed for analysis of apoptosis by a TUNEL assay. In the absence of photoreactivating light, we observed a strong induction of apoptosis in the epidermis of UV-exposed skin with few apoptotic dermal cells (Figure 5, middle panels), a staining pattern very similar to that observed in UV-exposed wild-type animals (data not shown). The non-UV-exposed area of the skin remained unstained (Figure 5, top panels), indicating that apoptosis was specifically induced by exposure to UV light. In marked contrast, the photoreactivated UV-exposed skin showed little or no apoptotic signal (Figure 5, bottom panels). Similar results were obtained when animals were exposed to 1.5 MED of UV-B, or when the TUNEL assay was performed 24 h after UV irradiation (data not shown). Thus, photoreactivation of CPDs in CPD photolyase transgenic mice clearly reduces the apoptotic response.
Fig. 5. Effect of CPD photoreactivation on UV-induced apoptosis in the skin of β-act-CPD photolyase transgenic mice. Induction of apoptosis in the depilated dorsal skin of β-act-CPD-1 mice exposed to 1 MED of UV-B light, without (middle panels) or with (bottom panels) subsequent treatment of animals with photoreactivating light for 3 h. Non-UV-exposed animals were used as a control (upper panels). Except for the photoreactivation step, animals were kept in the dark immediately after UV treatment. Apoptosis was measured 40 h after UV exposure by a TUNEL assay and nuclei were visualized by DAPI staining.
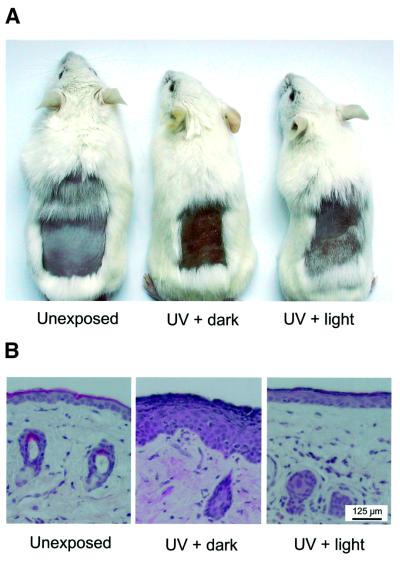
To study the induction of erythema and hyperplasia, we exposed the depilated dorsal skin of CPD photolyase mice to 1.5 MED of UV-B light per day for four consecutive days. Animals were continuously kept in the dark, except for animals that received 3 h of photoreactivating light immediately after each UV exposure. Three days after the last UV exposure, a clear redness and swelling was observed on the back of UV-treated animals that had not been exposed to photoreactivating light (Figure 6A). In marked contrast, photoreactivated animals did not display detectable swelling and only showed slight discoloration of the skin when compared with the unirradiated transgenic mice used as a control (Figure 6A). Histological examination of skin sections from these mice revealed that photoreactivation strongly reduced formation of epidermal hyperplasia, as clearly visible in samples from the non-photoreactivated UV-exposed skin (Figure 6B).
Fig. 6. Effect of CPD photoreactivation on UV-induced erythema and hyperplasia in the skin of β-act-CPD photolyase transgenic mice. The depilated back of β-act-CPD-1 transgenic mice was exposed to UV-B light for four consecutive days (1.5 MED per day) and were either given 3 h of photoreactivating light or kept in the dark. As a control, non-UV-exposed animals were used. Animals were killed 3 days after the last exposure and, except for the photoreactivation step, had been kept in the dark throughout the experiment. (A) Appearance of the dorsal skin of non-UV-exposed (left), UV-exposed (middle) and UV-exposed/photoreactivated animals (right), showing clear erythema when photoreactivation of UV-exposed animals is omitted. (B) Repre sentative examples of hematoxilin/eosin stained sections of the dorsal skin of the mice shown in (A). Note the thick epidermal layer in UV-exposed skin that had not received photoreactivating light, indicative of the induction of hyperplasia.
Taken together, these data demonstrate that fast removal of CPDs by photoreactivation has a clear protective effect on the skin of the CPD photolyase transgenic mice.
Photoreactivation of CPDs in basal keratinocytes
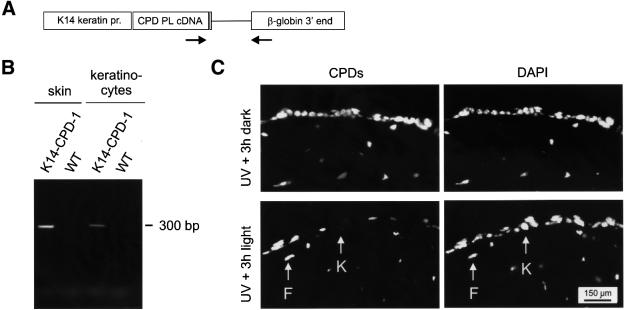
The ubiquitous expression of the CPD photolyase from the β-actin promoter results in the photoreactivating light-dependent removal of CPDs from epidermis and upper dermis (Figure 4). Since basal keratinocytes are the prime target for UV-induced carcinogenesis, we generated a second expression construct in which the β-actin promoter and CMV enhancer were replaced by the human K14 promoter (Figure 7A), which, in the skin, has been reported to be only active in basal keratinocytes of the epidermis (Vassar et al., 1989). Using this construct, we obtained a transgenic mouse line (K14-CPD-1) containing ∼25 copies of the photolyase transgene, as determined by Southern blot analysis of mouse genomic DNA (data not shown). RT–PCR analysis of total skin RNA, using the same primer set previously applied for detection of the β-actin promoter-derived CPD photolyase transcript, revealed that the transgene is transcribed and properly spliced (Figure 7B). A similar result was obtained when cultured basal keratinocytes from this transgenic mouse line were used (Figure 7B). This shows that the CPD photolyase transgene is expressed in basal keratinocytes. Heterozygous transgenic animals were born at the expected Mendelian frequency, were fertile and did not show an aberrant phenotype up to the age of 1 year.
Fig. 7. Expression of the K14-CPD photolyase gene in transgenic mice. (A) Expression construct for the generation of K14-CPD photolyase transgenic mice, containing the human K14 promoter, the P.tridactylis CPD photolyase cDNA, and human genomic β-globin sequences, including exons 2 and 3, intron 2, the 3′ untranslated region and the polyadenylation signal. Arrows indicate the position of the primers used for the RT–PCR experiment. (B) RT–PCR on skin extracts and primary cultured keratinocytes of K14-CPD photolyase transgenic mice results in a 300 bp band. (C) CPD lesions in the depilated dorsal skin of K14-CPD-1 photolyase mice following exposure to 1 MED of UV-B light and without (top panels) or with (bottom panels) subsequent exposure to photoreactivating light for 3 h. Photolesions were detected by immunofluorescent labeling, using CPD-specific antibodies and FITC-conjugated goat anti-mouse antibodies. Note the specific absence of labeling in basal keratinocytes (K) and persistence of CPDs in dermal fibroblasts (F) after exposure of UV-exposed mice to photoreactivating light. Nuclei were visualized by DAPI staining.
To investigate whether the K14 promoter-driven transgene is active, we performed photoreactivation ex periments on UV-B-exposed K14-CPD photolyase mice using the immunocytochemical labeling assay for CPD lesions in skin sections. Similar to the experiment with the β-actin promoter-driven CPD photolyase transgenic animals (Figure 4, upper panels), the non-photoreactivated UV-exposed part of the skin of K14-CPD photolyase transgenic animals displayed a clear labeling of CPDs in the epidermis and the upper part of the dermis (Figure 7C). However, light-dependent removal of CPDs was only observed in the basal region of the epidermis, while CPDs persisted in the dermis and the upper layers of the epidermis (Figure 7C). This fits with the notion that the K14 promoter is not active in the dermis and is switched off in the epidermis once basal keratinocytes start to differentiate (Vassar et al., 1989). With respect to the latter, the turnover of the photolyase protein is apparently sufficiently high to deplete terminally differentiated keratinocytes from photoreactivating capacity. In conclusion, this result shows that it is possible to photoreactivate CPDs in a specific skin compartment by using a cell type-specific promoter.
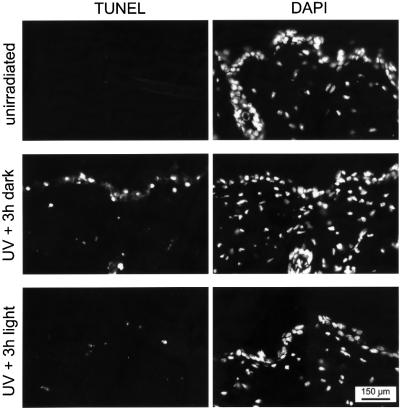
The K14-CPD photolyase transgene now allows removal of CPDs from proliferating basal and early differentiating keratinocytes in the UV-exposed epidermis, which appears to be potentially the most dangerous cell type in the development of skin cancer. To study the protective effect of CPD photoreactivation in these cells on the induction of apoptosis, we subjected K14-CPD photolyase mice to UV-B light (with and without subsequent photoreactivation) and performed a TUNEL assay on skin sections, following the same protocol as used for β-act-CPD photolyase mice. Light-mediated removal of CPDs from the basal keratinocytes almost completely reduced the apoptotic response in the epidermis (Figure 8). Strikingly, not only did we see a reduction in the apoptotic response throughout the epidermis, but a reduction was also seen in the dermis. This suggests that CPDs in basal keratinocytes are an important trigger for apoptosis in the whole skin and thus exert a systemic effect.
Fig. 8. Effect of CPD photoreactivation in basal keratinocytes on UV-induced apoptosis in the skin of K14-CPD photolyase transgenic mice. Induction of apoptosis in the depilated dorsal skin of K14-CPD-1 mice exposed to 1 MED of UV-B light, without (middle panels) or with (bottom panels) subsequent treatment of animals with photoreactivating light for 3 h. Non-UV-exposed animals were used as a control (upper panels). Except for the photoreactivation step, animals were kept in the dark immediately after UV treatment. Apoptosis was measured 40 h after UV exposure by a TUNEL assay and nuclei are visualized by DAPI staining.
Discussion
Photolyase transgenic mice
We have generated transgenic mouse lines that either ubi quitously express the CPD photolyase from P.tridactylis in all tissues (using the β-actin promoter), or specifically express the enzyme in the basal region of the epidermis (using the K14 promoter). Whereas photolyase genes from numerous species have been identified, we opted for the Potorous gene as, from an evolutionary point of view, marsupials are most closely related to placental mammals. Interestingly, although we observed a high copy number of the CPD photolyase transgene in the K14-CPD photolyase mouse line, we could not obtain β-act-CPD photolyase transgenic mouse lines at high copy number. This suggests that, at least in some tissues, ubiquitous overexpression of the CPD photolyase might not be favorable for proper embryonic development. As photolyases have some affinity for unirradiated DNA, high levels of this enzyme in the cell might interfere with key cellular processes, such as transcription and replication. Importantly, we have shown that the CPD photolyase transgenic mice can specifically and rapidly remove UV-induced CPDs by photoreactivation. This allowed the light-dependent and lesion-specific removal of CPDs from all parts of the skin or specifically from basal keratinocytes in the epidermis. The proper functioning of the heterologous photolyase in the mouse implies that the chromophoric co-factors necessary for light-dependent catalysis of dimer splitting (FAD and possibly MTHF; Yasui et al., 1994) are available in mouse cells and are properly incorporated in the enzyme. With these mouse models, we set out to determine how CPDs and 6–4PPs, the two main types of UV lesions, contribute to the acute deleterious effects of UV exposure.
Photoreactivation of CPDs increases UV resistance of cultured mouse cells
Using cultured MDFs from β-act-CPD photolyase transgenic mice, we have shown that photoreactivation occurs fast, in a lesion-specific manner. Using up to 20 J/m2 of UV-C, a dose sufficient to kill >95% of the cells, the majority of CPD lesions could be removed by photoreactivation within 1 h, leaving the 6–4PPs unaffected. Within this period, the slower endogenous NER system is only capable of dealing with a fraction of the photolesions, as evident from the amount of CPDs and 6–4PPs left in the cells when photoreactivating light is omitted. Moreover, in rodents, NER of CPD lesions is restricted to the transcribed strand of active genes. Yet, we have shown that rapid photoreactivation of CPDs in β-act-CPD photolyase transgenic mouse cells has a pronounced effect on NER, as evident from the ∼3-fold (light-dependent) reduction in UDS. A comparable reduction in UDS has been observed after microinjection of Anacystis nidulans CPD photolyase into cultured human fibroblasts (Zwetsloot et al., 1985, 1986), which is somewhat surprising as rodent cells, in contrast to human cells, hardly remove CPD lesions by GG-NER. Apparently, TC-NER of CPD lesions constitutes a prominent portion of NER activity in the first hour after UV exposure.
Importantly, we have shown that photoreactivation of CPDs makes the mouse cells more resistant to the killing effects of UV exposure. This observation is in full agreement with previous studies on cultured cells from organisms with a natural photolyase gene (Rosenstein and Setlow, 1980; van Zeeland et al., 1980; Wade and Trosko, 1983) or photolyase-transfected HeLa cells (Chiganças et al., 2000). The photoreactivating light-mediated reduction in UV sensitivity of transgenic CPD photolyase mouse cells might be explained by a genome-wide reduction in CPD damage load by photoreactivation, and/or an increased clearance of CPDs from the transcribed strand of active genes. Several observations support the latter option. First, overexpression of the p48 gene in rodent cells markedly stimulates GG-NER- mediated removal of CPDs from the genome, but hardly improves UV survival (Tang et al., 2000). Furthermore, TC-NER-deficient cells from patients and rodents with the repair disorder CS are very sensitive to UV. Moreover, UV-induced apoptosis is primarily triggered by RNA polymerase II getting stalled at lesions in the template strand of transcribed genes (Yamaizumi et al., 1994; Ljungman and Zhang, 1996; Conforti et al., 2000). Thus, enhanced removal of CPDs from the transcribed strand of active genes appears to be the major determinant in the improved UV resistance of CPD photolyase mouse cells. Yet, we cannot exclude that the genome-wide photoreactivation of CPDs also, to some extent, contributes to enhanced UV resistance. After removal of CPDs by photoreactivation, the GG-NER and TC-NER pathways can fully concentrate on the repair of 6–4PPs, which therefore may be more efficiently removed from transcribed DNA.
CPD photoreactivation reduces acute skin effects
In this study, we have focused on the role of CPDs in the induction of acute effects in the UV-exposed skin: apoptosis, erythema and hyperplasia. Although some studies point to 6–4PPs as the most cytotoxic lesions (Mitchell, 1988), CPDs cause the most abundant damage in the DNA after UV exposure, and have been implicated as the cause of UV-induced cell death (Broughton et al., 1990; Nishigaki et al., 1998; Chiganças et al., 2000). Analogous to our results obtained with MDFs, the intact skin of β-actin promoter-driven CPD photolyase transgenic mice shows an efficient light-dependent removal of CPDs by the photolyase, resulting in a reduction of cell death via apoptosis. It must therefore be concluded that the presence of CPDs prominently contributes to the triggering of apoptosis in the skin. Photoreactivation of CPDs in epidermis and dermis also reduces other acute skin effects of (repeated) exposure to UV light, such as erythema, edema and hyperplasia. UV-mediated induction of the apoptotic pathway in the intact mouse skin, similar to cultured cells, appears to originate from RNA polymerase II blocking lesions in the transcribed strand. GG-NER-deficient XP-C mice can accumulate photolesions in their genome without inducing apoptosis and hyperplasia, whereas TC-NER-deficient CS-B mice (as well as XP-A mice with a complete NER defect) already display these acute effects at low UV doses (van Oosten et al., 2000; Brash et al., 2001).
The effect of photoreactivation of CPDs in β-act-CPD photolyase transgenic mice matches well with previous studies involving animals with an endogenous CPD photo lyase. In the South American opossum Monodelphis domestica, photoreactivation has been shown to clearly protect the skin and eye from aforementioned acute effects (Applegate et al., 1985; Ley, 1985, 1991; Ley et al., 1991). Recently, topical application of CPD photolyase-containing liposomes has been shown to add a higher level of protection to the UV-exposed human skin (Stege et al., 2000). Although, on average, less than half of the CPDs could be repaired by this approach, several immunological and cytotoxic effects of UV light, such as suppression of ICAM-I, immunosuppression, as well as erythema and sunburn cell formation, could be ablated.
Surprisingly, experiments involving K14-CPD photolyase mice revealed that the fast removal of CPDs from basal and early differentiating keratinocytes was sufficient to prevent the induction of apoptosis in both the epidermis and the dermis. This suggests that UV-induced apoptosis in the skin is not just a cell-autonomous effect, but may act in a systemic manner. The Fas–Fas ligand (Fas/FasL) system has been implicated to play an important role in intercellular signaling between keratinocytes during UV-induced apoptosis in the skin (Hill et al., 1999; Wehrli et al., 2000; and references therein). Apparently, as evident from this study, signaling from keratinocytes extends to other cell types, such as dermal fibroblasts.
The potential of photolyase mice
In this study, we have focused on the consequences of photoreactivation of CPDs on, and thus the contribution of, 6–4PPs and other (non-CPD) types of lesions to acute effects in the mouse skin. However, it is obvious that an important application of the β-act-CPD and K14-CPD photolyase transgenic mouse models involves a study of the specific contribution of CPDs and 6–4PPs, as well as their position in the genome (i.e. transcribed versus non-transcribed DNA), to other short- and long-term effects of UV exposure like immune suppression, interference with cell cycle progression, mutagenesis and, importantly, carcinogenesis. A recent study by You and co-workers (You et al., 2001) has shown that, at least in vitro, photoreactivation of CPDs rather than 6–4PPs reduces the mutation frequency in NER-proficient cultured mouse embryonic fibroblasts carrying the BigBlue® mutation reporter system. Currently, we are in the process of crossing the CPD photolyase mice with totally NER-deficient XP-A, GG-NER-deficient XP-C and TC-NER-deficient CS-B animals (all in a hairless genetic background), proven to develop skin cancer upon chronic exposure to UV light (de Vries et al., 1995; Nakane et al., 1995; Cheo et al., 1996; van der Horst et al., 1997). The possibility to investigate the effects of different UV lesions by photoreactivation of a specific type of damage is also becoming increasingly valuable now the mechanisms of damage tolerance, including translesion synthesis and recombination repair, are being unraveled (for a review, see Lehmann, 2000). Many DNA polymerases have been discovered, each with their own accuracy (and presumably specificity) and ability to perform translesion DNA synthesis on different DNA lesions. Once mutant mouse models for these polymerases become available, they can be crossed with photolyase mice. Detailed analysis of such mice may shed new light on the mutagenic and carcinogenic properties of different types of UV-induced damage in the skin and the role of translesion synthesis therein.
In conclusion, we have complemented a placental mammal with a photoreactivation repair system for light-mediated damage reversal of UV-induced CPD lesions. We have shown that mice expressing a CPD photolyase tolerate higher doses of UV irradiation before developing erythema, sunburn and other short-term effects of UV light. Obviously, it is important to gain complete insight into the fate of the various photolesions, not only the efficiency of repair by TC- and GG-NER, but also the way they are dealt with by the various error-free and error-prone DNA translesion synthesizing polymerases.
Finally, the above objectives clearly highlight the need for the counterpart of our CPD photolyase mice, i.e. animals that express a 6–4PP photolyase and can specifically remove this lesion in a light-dependent manner. Recently, we have generated transgenic mice that carry the Arabidopsis thaliana 6–4PP photolyase and specifically remove 6–4PPs from their genome in a light-dependent manner. We are currently comparing the properties of CPD photolyase animals with those of 6–4PP photolyase and CPD photolyase/6–4PP photolyase (double) transgenic mice.
Materials and methods
Generation of CPD photolyase transgenic mice
Constructs for the generation of CPD photolyase transgenic mice were cloned in the vector pSP72 (Promega) and contained either the chicken β-actin promoter with the CMV enhancer (from pCY4B, kindly provided by Dr J.Miyazaki, Osaka, Japan) or the human K14 promoter (2.3 kb PCR fragment, generated using forward primer 5′-AAGCTTATATTCCAT GCTAG-3′ and reverse primer 5′-GGATCCTGAGTGAAGAGAAGG-3′), followed by the P.tridactylis CPD photolyase cDNA. At the 3′ end, we inserted exon 2 (the last 22 bp), intron 2, exon 3 and 3′ untranslated region (including the polyadenylation signal) of the human β-globin gene. The expression constructs were excised from the plasmid using appropriate restriction enzymes, separated from the vector DNA by agarose gel electrophoresis, isolated from the gel with the GeneClean II kit (Bio101), and further purified using elutip-D-mini columns (Schleicher and Schuell, Dassel, Germany). The fragment was dissolved in injection buffer (10 mM Tris–HCl pH 7.5, 0.08 mM EDTA) and injected in the pronucleus of fertilized eggs derived from FVB/N intercrosses as described previously (Hogan et al., 1994).
Transgenic animals were identified by Southern blot analysis of genomic tail DNA, using the CPD photolyase cDNA as a probe. To estimate the number of integrated copies, equal amounts of genomic DNA from transgenic animals were subjected to Southern blot analysis. As a standard, we used equal amounts of wild-type genomic DNA supplemented with 0, 10, 30 or 100 pg of the corresponding CPD photolyase expression construct. The hybridization signal obtained with the CPD photolyase cDNA probe was quantified using a Molecular Dynamics PhosphorImager and ImageQuant software. After comparison of signal intensities, the transgene copy number could be estimated using the supplemented reference samples. The site of chromosomal integration was determined by FISH analysis of metaphase chromosome spreads of transgenic mouse cell lines as described by Arnoldus et al. (1990), using the CPD photolyase cDNA probe.
Routine genotyping of mice was performed by PCR analysis. Primer set 5′-TGAGACTCATCTCCCAGGAC-3′ (forward primer) and 5′-CAC CAATGCCATGTGTTTGC-3′ (reverse primer) was used to amplify a 321 bp fragment of the CPD photolyase coding region.
RNA isolation and RT–PCR
Mouse skin RNA was isolated from a 0.5–1.0 cm2 skin section, after hairs had been plucked from the back of the killed mouse. After addition of 2–3 ml of an ice-cold 3 M LiCl–6 M urea solution, the skin sample was immediately homogenized on ice using a Heidolph Diax 600 homogenizer set at 22 000 r.p.m. The homogenate was left at 4°C overnight, and after centrifugation at 113 000 g for 20 min, the pellet was re suspended in 0.5 ml of 10 mM Tris–HCl, 0.1 mM EDTA, 0.5% SDS and 0.1 mg/ml proteinase K, and incubated at 37°C for 30 min. Total RNA was obtained by subsequent phenol extraction, centrifugation and ethanol precipitation of the supernatant. The pellet was dissolved in 10 mM Tris–HCl and 0.1 mM EDTA. cDNA synthesis was performed using Superscript II RNaseH reverse transcriptase (RT; Life Technologies) according to the protocol of the supplier. A standard PCR reaction was performed on the cDNA using a forward primer in the photolyase transgene (5′-GATCTTCGGAAAGATCCGC-3′) and a reverse primer in exon 3 of the β-globin gene (5′-TGGACAGCAAGAAAGCGAG-3′). The presence of introns in the photolyase transgene allowed us to discriminate the cDNA-derived PCR product from possible genomic DNA contaminations in the isolated RNA.
Isolation and culturing of dermal fibroblasts and keratinocytes
Primary MDFs were isolated from the skin according to the protocol of Mémet et al. (1997). In short, after removal of hairs from a small area on the back of an anesthetized mouse, a small piece of skin (∼0.3 cm2) was removed. The tissue was minced, transferred to a phosphate-buffered saline (PBS) solution containing 0.125% trypsin, 1 mg/ml collagenase type V and 0.3 mg/ml, and incubated at 37°C for 15 min, after which the cell suspension was collected. A fresh enzyme solution was added to the remaining pieces of skin and the procedure was repeated twice more. Cell suspensions from one mouse line were pooled and centrifuged, after which the pellet was resuspended in medium [40% Dulbecco’s modified Eagle’s medium (DMEM), 40% HAM-F10, 20% fetal calf serum (FCS), 1% penicillin + streptomycin]. Cells were transferred to 10 cm dishes and cultured at 37°C in 5% CO2.
Primary keratinocytes from 2-day-old mice were isolated as described previously (Dlugosz et al., 1995). Following genotyping by PCR analyis of tail DNA, selected keratinocyte lines were cultured on collagen– fibronectin-coated dishes in low calcium (0.05 mM) Eagle’s minimal essential medium (EMEM) (BioWhittaker), supplemented with 8% FCS [treated with chelex 100 (Bio-Rad) to remove Ca2+ ions], 1 ng/ml keratinocyte growth factor (R&D systems) and 50 µg/ml penicillin and streptomycin (Gibco).
Immunological procedures
A fusion protein of P.tridactylis CPD photolyase and glutathione S-transferase was overexpressed in Escherichia coli, purified (Yasui et al., 1994) and used for the immunization of two rabbits. The serum was affinity purified against immobilized CPD photolyase.
Western blot analysis. Protein extracts were prepared by harvesting MDFs in sample buffer (2% SDS, 0.1 M Tris pH 6.8, 4% β-mercapto ethanol, 15% glycerol) containing 1 mM PMSF. The protein extract was heat denatured, subjected to SDS–PAGE on a 10% gel and transferred to nitrocellulose. The nitrocellulose blot was incubated for 1 h in non-fat milk, washed with PBGTNa [PBS containing 0.5% bovine serum albumin (BSA; Sigma), 0.05% gelatin, 0.05% Tween-20 and 300 mM NaCl], incubated overnight with the anti-CPD photolyase polyclonal antibody diluted in PBGTNa, washed for 5 min in PBGTNa four times, incubated for 2 h with the secondary antibody goat anti-rabbit coupled to alkaline phosphatase and washed for 5 min with PBGTNa twice and for 5 min with PBS twice. Blots were stained using the BCIP/NBT method (Bio-Rad).
Immunofluorescent detection of CPD photolyase. Coverslips with attached MDFs were rinsed once in PBS and fixed with 2% paraformaldehyde in PBS for 15 min. After fixation, cells were washed twice with PBS and permeabilized with 0.5% Triton X-100 (Sigma) in PBS for 5 min. Cells were subsequently washed twice in PBS, incubated in PBS containing 100 mM glycine (Sigma) for 10 min, incubated in PBG [PBS containing 0.5% BSA (Sigma) and 0.05% gelatin] for 10 min and incubated overnight at 4°C with the anti-CPD photolyase polyclonal antibody diluted in PBG. The coverslips were washed for 5 min with PBG four times and incubated with goat anti-rabbit coupled to FITC for 2 h and washed for 5 min with PBG twice and for 5 min with PBS twice. Finally, coverslips were incubated for 5 min with DAPI (0.2 µg/ml) in PBS, washed with PBS, and mounted in Vectashield (Vector Laboratories, Burlingame, CA). All steps were performed at room temperature, unless stated otherwise.
DNA repair assays
Photoreactivation in MDFs. Cells grown on coverslips were washed with PBS, exposed to 20 J/m2 of UV-C (Philips TUV germicidal lamp), and subsequently kept in Hank’s buffer (137 mM NaCl, 5.4 mM KCl, 4.4 mM KH2PO4, 0.33 mM Na2HPO4, 1.3 mM CaCl2, 0.81 mM MgSO4, 4.2 mM NaHCO3, 1 g/l glucose pH 7.4). Photoreactivation was performed by exposing cells for 1 h to light from four white fluorescent tubes (Philips TLD 18W/54) at a distance of 15 cm and shielded by a 5 mm glass filter. Non-photoreactivated cells were given the same treatment, except that dishes were covered with two layers of aluminum foil and put under the same fluorescent lamps. Next, cells were fixed in a mixture of 75% methanol and 25% acetic acid for 1 h, subsequently incubated in 70% ethanol (1 h) and 70% ethanol containing 0.07 N NaOH (4 min), and washed for 3 × 5 min with TBS (20 mM Tris–HCl, 150 mM NaCl pH 7.4). Next, cells were treated with 1 µg/ml proteinase K in 20 mM Tris–HCl and 2 mM CaCl2 at pH 7.4 for 10 min, incubated for 30 min with TBSF (TBS containing 1% FCS), and incubated overnight at 4°C with an antibody raised against CPD or 6–4PP photolesions (antibody TDM2 and 64M2, respectively; Mori et al., 1991) diluted in TBSF. Coverslips were washed for 5 min with TBSF four times, incubated for 2 h with goat anti-mouse conjugated to FITC, washed for 5 min with TBSF twice and for 5 min with PBS twice. Finally, coverslips were incubated for 5 min with DAPI (0.2 µg/ml) in PBS, washed with PBS and mounted in Vectashield. Cells were analyzed using a Leica DMRBE epifluorescence microscope.
UV survival. UV radiation sensitivity was determined as described previously (Sijbers et al., 1996), except that a photoreactivation step was incorporated in the assay. MDFs were grown in 6-well culture plates (Costar), washed with PBS, exposed to different doses of UV-C light (Philips TUV germicidal lamp) and subsequently kept in Hank’s buffer. Next, cells were either exposed to photoreactivating light or kept in the dark for 1 h as described above, after which cell culturing was continued in the dark. After 2 days, the number of proliferating cells was determined from the amount of radioactivity incorporated during a 3 h pulse with [3H]thymidine. Cell survival is expressed as the percentage of radioactivity in exposed cells relative to that in untreated cells.
Unscheduled DNA synthesis. UV-induced global genome repair was assayed using UDS as described previously (Vermeulen et al., 1994). Cells were exposed to 16 J/m2 of UV-C light and labeled for 3 h with [methyl-3H]thymidine (10 µM, specific activity 40–60 Ci/mmol), followed by processing for autoradiography. Repair capacity was quantified by grain counting after autoradiographic exposure.
Photoreactivation in the mouse skin. Mice were anesthetized and hairs were removed from a small area on the back of the animal. One-third of the hairless area was covered with black non-adhesive tape and the remaining area was exposed to the light of two Philips TL-12 (40 W) tubes emitting UV-B light. Typically, 1 MED was obtained with an exposure of 2 min, at a distance of 21 cm. Subsequently, half of the UV-exposed area was covered with tape, and the mice were exposed for 3 h to the light of four cool white fluorescent tubes (GE Lightning Polylux XL F36W/840 lamps) filtered through 5 mm of glass. Skin samples were taken from the unexposed area, the UV-irradiated area that was covered, and the UV-irradiated area that was exposed to photoreactivating light. The skin samples were fixed for 1 h in a mixture of 75% methanol and 25% acetic acid, incubated in 70% ethanol for 1 h, followed by 5% sucrose (1 h), and snap-frozen in TissueTek (Sakura) by immersion in liquid nitrogen. Cryosections of 8 µm were made with a Leica microtome (Jung CM3000) and deposited on SuperFrost (Menzel-Glazer) glass slides. Sections on slides were incubated in 70% ethanol containing 0.07 N NaOH for 4 min and subsequently washed, incubated and labeled like MDFs as described above.
Apoptosis assay
For detection of apoptotic cells in the skin, we used a TUNEL assay (Fluorescein Apoptosis Detection System, Promega). Mice were exposed to UV and photoreactivating light as described above, and subsequently kept in the dark. Skin samples, taken 40 h after UV exposure, were fixed overnight in 4% paraformaldehyde, washed in PBS and embedded in paraffin. Skin sections (5 µm) were deparafinized and incubated as described by the manufacturer.
Erythema and hyperplasia assays
Mice were anesthetized and an area on the back was depilated by plucking. After recovery, animals were either kept in the dark or exposed to UV-B (1.5 MED per day), immediately followed by photoreactivating light (3 h) for four consecutive days. Except for the above treatment, animals were continuously kept in the dark. Seven days after the start of the experiment, mice were physically examined for the formation of erythema. For analysis of epidermal hyperplasia, skin biopsies were taken and processed for routine histology.
Acknowledgments
Acknowledgements
We thank Dr Jun-Ichi Miyazaki (Osaka University Medical School, Osaka, Japan) for providing us with the pCY4B vector. This work was supported by the Dutch Cancer Foundation (EUR 98-1774, EMCR 2002-2701), the Association for International Cancer Research (AICR Grant 98-259) and the Japanese Ministry of Education, Science and Culture (MONBUSHO Grant 10044231).
References
- Applegate L.A., Stuart,T.D. and Ley,R.D. (1985) Ultraviolet radiation-induced histopathological changes in the skin of the marsupial Monodelphis domestica. I. The effects of acute and chronic exposure and of photoreactivation treatment. Br. J. Dermatol., 113, 219–227. [DOI] [PubMed] [Google Scholar]
- Arnoldus E.P., Wiegant,J., Noordermeer,I.A., Wessels,J.W., Beverstock, G.C., Grosveld,G.C., Van der Ploeg,M. and Raap,A.K. (1990) Detection of the Philadelphia chromosome in interphase nuclei. Cytogenet. Cell Genet., 54, 108–111. [DOI] [PubMed] [Google Scholar]
- Bohr V.A., Smith,C.A., Okumoto,D.S. and Hanawalt,P.C. (1985) DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell, 40, 359–369. [DOI] [PubMed] [Google Scholar]
- Bootsma D., Kraemer,K.H., Cleaver,J.E. and Hoeijmakers,J.H.J. (2001) Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. In Scriver,C.R., Beaudet,A.L., Sly,W.S. and Valle,D. (eds), The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York, NY, pp. 677–703.
- Brash D.E., Wikonkal,N.M., Rmenyik,E., van der Horst,G.T.J., Friedberg,E.C., Cheo,D.L., van Steeg,H., Westerman,A. and van Kranen,H.J. (2001) The DNA damage signal for Mdm2 regulation, Trp53 induction and sunburn cell formation in vivo originates from actively transcribed genes. J. Invest. Dermatol., 117, 1234–1240. [DOI] [PubMed] [Google Scholar]
- Broughton B.C., Lehmann,A.R., Harcourt,S.A., Arlett,C.F., Sarasin,A., Kleijer,W.J., Beemer,F.A., Nairn,R. and Mitchell,D.L. (1990) Relationship between pyrimidine dimers, 6–4 photoproducts, repair synthesis and cell survival: studies using cells from patients with trichothiodystrophy. Mutat. Res., 235, 33–40. [DOI] [PubMed] [Google Scholar]
- Cheo D.L., Meira,L.B., Hammer,R.E., Burns,D.K., Doughty,A.T.B. and Friedberg,E.C. (1996) Synergistic interactions between XPC and p53 mutations in double-mutant mice: neural tube abnormalities and accelerated UV-radiation-induced skin cancer. Curr. Biol., 6, 1691–1694. [DOI] [PubMed] [Google Scholar]
- Chiganças V., Miyaji,E.N., Muotri,A.R., de Fátima Jacysyn,J., Amarante-Mendes,G.P., Yasui,A. and Menck,C.F.M. (2000) Photorepair prevents ultraviolet-induced apoptosis in human cells expressing the marsupial photolyase gene. Cancer Res., 60, 2458–2463. [PubMed] [Google Scholar]
- Conforti G., Nardo,T., D’Incalci,M. and Stefanini,M. (2000) Proneness to UV-induced apoptosis in human fibroblasts defective in transcription coupled repair is associated with the lack of Mdm2 transactivation. Oncogene, 19, 2714–2720. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers,N.G. and Hoeijmakers,J.H.J. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- de Vries A. et al. (1995) Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature, 377, 169–173. [DOI] [PubMed] [Google Scholar]
- Dlugosz A.A., Glick,A.B., Tennenbaum,T., Weinberg,W.C. and Yuspa,S.H. (1995) Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol., 254, 3–20. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (eds) (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Hanawalt P.C., Liu,S.C. and Parsons,S. (1981) DNA repair responses in human skin cells. J. Invest. Dermatol., 77, 86–90. [DOI] [PubMed] [Google Scholar]
- Hill L.L., Ouhtit,A., Loughlin,S.M., Kripke,M.L., Ananthaswamy,H.N. and Owen-Schaub,L.B. (1999) Fas ligand: a sensor for DNA damage critical in skin cancer etiology. Science, 285, 898–900. [DOI] [PubMed] [Google Scholar]
- Hogan B., Beddington,R., Costantini,F. and Lacy,E. (1994) Manipulating the Mouse Embryo Section E: Production of Transgenic Mice. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 217–252. [Google Scholar]
- Hoeijmakers J.H.J. (2001) Genome maintenance mechanisms for preventing cancer. Nature, 411, 366–367. [DOI] [PubMed] [Google Scholar]
- Hwang B.J., Toering,S., Francke,U. and Chu,G. (1998) p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol., 18, 4391–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B.J., Ford,J.M., Hanawalt,P.C. and Chu,G. (1999) Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA, 96, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A.R. (2000) Replication of UV-damaged DNA: new insights into links between DNA polymerases, mutagenesis and human disease. Gene, 253, 1–12. [DOI] [PubMed] [Google Scholar]
- Ley R.D. (1985) Photoreactivation of UV-induced pyrimidine dimers and erythema in the marsupial Monodelphis domestica. Proc. Natl Acad. Sci. USA, 82, 2409–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.D., Applegate,L.A., Fry,R.J.M. and Sanchez,A.B. (1991) Photoreactivation of ultraviolet radiation-induced skin and eye tumors of Monodelphis domestica. Cancer Res., 51, 6539–6542. [PubMed] [Google Scholar]
- Ljungman M. and Zhang,F. (1996) Blockage of RNA polymerase as a possible trigger for U.V. light-induced apoptosis. Oncogene, 13, 823–831. [PubMed] [Google Scholar]
- Mellon I., Spivak,G. and Hanawalt,P.C. (1987) Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell, 51, 241–249. [DOI] [PubMed] [Google Scholar]
- Mémet S., Lilienbaum,A. and Israël,A. (1997) Rapid isolation of mouse primary fibroblasts: a tool for the analysis of transgenic mice. Technical Tips Online. http://research.bmn.com/tto
- Mitchell D.L. (1988) The relative cytotoxicity of (6–4) photoproducts and cyclobutane dimers in mammalian cells. Photochem. Photobiol., 48, 51–57. [DOI] [PubMed] [Google Scholar]
- Mitchell D.L., Cleaver,J.E. and Epstein,J.H. (1990) Repair of pyrimidine (6–4) pyrimidone photoproducts in mouse skin. J. Invest. Dermatol., 95, 55–59. [DOI] [PubMed] [Google Scholar]
- Mori T., Nakane,M., Hattori,T., Matsunaga,T., Ihara,M. and Nikaido,O. (1991) Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6–4) photoproducts from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol., 54, 225–232. [DOI] [PubMed] [Google Scholar]
- Nakane H. et al. (1995) High incidence of ultraviolet-B- or chemical-carcinogen-induced skin tumors in mice lacking the xeroderma pigmentosum group A gene. Nature, 377, 165–168. [DOI] [PubMed] [Google Scholar]
- Nishigaki R., Matani,H. and Shima,A. (1998) Evasion of UVC-induced apoptosis by photorepair of cyclobutane pyrimidine dimers. Exp. Cell Res., 244, 43–53. [DOI] [PubMed] [Google Scholar]
- Qin X., Zhang,S., Oda,H., Nakatsuru,Y., Shimizu,S., Yamazaki,Y., Nikaido,O. and Ishikawa,T. (1995) Quantitative detection of ultraviolet light-induced photoproducts in mouse skin by immunohistochemistry. Jpn J. Cancer Res., 86, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein B.S. and Setlow,R.B. (1980) DNA repair after ultraviolet irradiation of ICR 2A frog cells. Biophys. J., 31, 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruven H.J.T., Seelen,M.J., Lohman,P.H.M., van Kranen,H., van Zeeland,A.A. and Mullenders,L.H.F. (1994) Strand-specific removal of cyclobutane pyrimidine dimers from the p53 gene in the epidermis of UVB-irradiated hairless mice. Oncogene, 9, 3427–3434. [PubMed] [Google Scholar]
- Sijbers A.M. et al. (1996) Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell, 86, 811–822. [DOI] [PubMed] [Google Scholar]
- Stege H., Roza,L., Vink,A.A., Grewe,M., Ruzicka,T., Grether-Beck,S. and Krutmann,J. (2000) Enzyme plus light therapy to repair DNA damage in ultraviolet-B-irradiated human skin. Proc. Natl Acad. Sci. USA, 97, 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.Y., Hwang,B.J., Ford,J.M., Hanawalt,P.C. and Chu,G. (2000) Xeroderma pigmentosum p48 gene enchances global genome repair and suppresses UV-induced mutagenesis. Mol. Cell, 5, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst G.T.J. et al. (1997) Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell, 89, 425–435. [DOI] [PubMed] [Google Scholar]
- van Hoffen A., Natarajan,A.T., Mayne,L.V., van Zeeland,A.A., Mullenders,L.H.F. and Venema,J. (1993) Deficient repair of the transcribed strand of active genes in Cockayne’s syndrome cells. Nucleic Acids Res., 21, 5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoffen A., Venema,J., Meschini,R., van Zeeland,A.A. and Mullenders,L.H.F. (1995) Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6–4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. EMBO J., 14, 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten M. et al. (2000) Differential role of transcription-coupled repair in UVB-induced G2 arrest and apoptosis in mouse epidermis. Proc. Natl Acad. Sci. USA, 97, 11268–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeeland A.A., Natarajan,A.T., Verdegaal-Immerzeel,E.A.M. and Filon,A.R. (1980) Photoreactivation of UV induced cell killing, chromosome aberrations, sister chromatid exchanges, mutations and pyrimidine dimers in Xenopus laevis fibroblasts. Mol. Gen. Genet., 180, 495–500. [DOI] [PubMed] [Google Scholar]
- Vassar R., Rosenberg,M., Ross,S., Tyner,A. and Fuchs,E. (1989) Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc. Natl Acad. Sci. USA, 86, 1563–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J., Mullenders,L.H.F., Natarajan,A.T., van Zeeland,A.A. and Mayne,L.V. (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl Acad. Sci. USA, 87, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen W. et al. (1994) Clinical heterogeneity within xeroderma pigmentosum associated with mutations in the DNA repair and transcription gene ERCC3. Am. J. Hum. Genet., 54, 191–200. [PMC free article] [PubMed] [Google Scholar]
- Wade M.H. and Trosko,J.E. (1983) Enhanced survival and decreased mutation frequency after photoreactivation of UV damage in rat kangaroo cells. Mutat. Res., 112, 231–243. [DOI] [PubMed] [Google Scholar]
- Wehrli P., Viard,I., Bullani,R., Tschopp,J. and French,L.E. (2000) Death receptors in cutaneous biology and disease. J. Invest. Dermatol., 115, 141–148. [DOI] [PubMed] [Google Scholar]
- Wood R.D. (1996) DNA repair in eukaryotes. Annu. Rev. Biochem., 65, 135–167. [DOI] [PubMed] [Google Scholar]
- Yamaizumi M., Sugano,T. (1994) U.V.-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene, 9, 2775–2784. [PubMed] [Google Scholar]
- Yasui A. and Eker,A.P.M. (1997) DNA photolyases. In Nickoloff,J.A. and Hoekstra,M.F. (eds) DNA Damage and Repair: Biochemistry, Genetics and Cell Biology. Vol. 2. Humana Press, Totowa, NJ, pp. 9–32.
- Yasui A., Eker,A.P., Yasuhira,S., Yajima,H., Kobayashi,T., Takao,M. and Oikawa,A. (1994) A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J., 15, 6143–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y.H., Lee,D.H., Yoon,J.H., Nakajima,S., Yasui,A. and Pfeifer,G.P. (2001) Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem., 276, 44688–44694. [DOI] [PubMed] [Google Scholar]
- Zwetsloot J.C.M., Vermeulen,W., Hoeijmakers,J.H.J., Yasui,A., Eker,A.P.M. and Bootsma,D. (1985) Microinjected photoreactivating enzymes from Anacystis and Saccharomyces monomerize dimers in chromatin of human cells. Mutat. Res., 146, 71–77. [DOI] [PubMed] [Google Scholar]
- Zwetsloot J.C.M., Hoeymakers,J.H.J., Vermeulen,W., Eker,A.P.M. and Bootsma,D. (1986) Unscheduled DNA synthesis in xeroderma pigmentosum cells after microinjection of yeast photoreactivating enzyme. Mutat. Res., 165, 109–115. [DOI] [PubMed] [Google Scholar]