Abstract
The mammalian SIN3 complex consists of histone deacetylases (HDAC1, HDAC2), several known proteins (SAP30, N-CoR) and as yet unidentified proteins. Here we show that the mouse tetradecanoyl phorbol acetate induced sequence 7 (TIS7) protein is a novel transcriptional co-repressor that can associate with the SIN3 complex. We have identified tis7 as a gene that is up-regulated upon loss of polarity in a mouse mammary gland epithelial cell line expressing an estrogen-inducible c-JunER fusion protein. In unpolarized cells, TIS7 protein levels increase and TIS7 translocates into the nucleus. Overexpression of tis7 causes loss of polarity and represses a set of genes, as revealed by cDNA microarray analysis. We have shown that TIS7 protein interacts with several proteins of the SIN3 complex (mSin3B, HDAC1, N-CoR and SAP30) by yeast two-hybrid screening and co-immunoprecipitations. TIS7 co-immunoprecipitated HDAC complex is enzymatically active and represses a GAL4-dependent reporter transcription. The transcriptional repression of endogenous genes by tis7 overexpression is HDAC dependent. Thus, we propose TIS7 as a transcriptional co-repressor affecting the expression of specific genes in a HDAC activity-dependent manner during cell fate decisions, e.g. scattering.
Keywords: HDAC/SIN3/TIS7/transcription
Introduction
The complex regulation of gene expression at the transcriptional level is controlled by the combinatorial action of sequence-specific DNA-binding proteins, transcriptional co-regulators and components of the basal transcriptional machinery (Chi et al., 1995; Goodrich et al., 1996; Mannervik et al., 1999). Acetylation of internal lysine residues of core histone N-terminal domains by histone acetyltransferases (HATs) and deacetylation by histone deacetylases (HDACs) are associated with transcriptional regulation (Hebbes et al., 1988; Braunstein et al., 1993). Several HATs and HDACs have been shown to also acetylate and deacetylate non-histone protein substrates (e.g. transcription factors), implicating internal lysine acetylation as a possible mechanism of rapid and reversible regulation (Kouzarides, 2000; Humphrey et al., 2001). The two major class I-containing histone deacetylation complexes are NuRD (nucleosomal remodelling and histone deacetylation) and SIN3 (reviewed in Knoepfler and Eisenman, 1999; Ahringer, 2000). The SIN3 complex shares four core proteins with NuRD (HDAC1, HDAC2, RbAp46 and RbAp48) and additionally contains Sin3, SAP18 and SAP30 (Ayer, 1999). RbAp46 and RbAp48 proteins interact with histones and histone deacetylases HDAC1 and HDAC2. The SIN3 complex does not bind to DNA directly, but through sequence-specific DNA-binding proteins: passive repressors, e.g. nuclear hormone receptors (Nagy et al., 1997). Sin3 proteins, mSin3A and mSin3B, act as adaptor molecules linking repressive transcription factors to accessory proteins. This facilitates targeting of the HDAC complexes (Hassig et al., 1997; Laherty et al., 1997; Knoepfler and Eisenman, 1999; Kyrylenko et al., 2000). However, SIN3-mediated repression is not exclusively HDAC dependent, suggesting that the SIN3 complex either contains an intrinsic repressor activity or it associates with other co-repressors (Laherty et al., 1997; Wong and Privalsky, 1998).
To study transcriptional regulation during cell fate decisions, we have used the previously established c-JunER mammary epithelial system (Fialka et al., 1996). Activation of the estrogen-responsive c-JunER fusion protein results in reduced transepithelial resistance, disruption of intercellular junctions, their barrier function and in the formation of irregular multilayers. The reversible loss of epithelial polarity resembles a scattering phenotype with redistributed apical and basolateral proteins to the entire plasma membrane. Activation of c-JunER mimics the c-Jun activation in vivo and transcriptionally regulates a variety of AP-1 target genes (Fialka et al., 1996).
In this study we identified, by RT–PCR-based differential display (DD RT–PCR), the TPA induced sequence 7 (tis7) gene (Varnum et al., 1989a) to be up-regulated upon the loss of epithelial polarity in c-JunER cells. TIS7, also known as PC4 (Tirone and Shooter, 1989) or IFRD1 (interferon related developmental regulator 1) (Buanne et al., 1998), was described previously as an immediate early gene (Tirone and Shooter, 1989; Varnum et al., 1989b; Guardavaccaro et al., 1994, 1995) that is possibly involved in the differentiation of various cell types (Tirone and Shooter, 1989; Guardavaccaro et al., 1994, 1995). However, the molecular function of TIS7 has not yet been elucidated.
Here we present evidence that TIS7, up-regulated after c-Jun activation, translocates into the nucleus and acts as a transcriptional co-repressor. We identified by cDNA microarray analysis several genes specifically repressed by overexpressed TIS7. TIS7 can associate with the mammalian SIN3 complex and HDACs, and thereby affect the expression of specific genes.
Results
TIS7 is up-regulated and translocates into the nucleus of c-JunER cells losing epithelial polarity
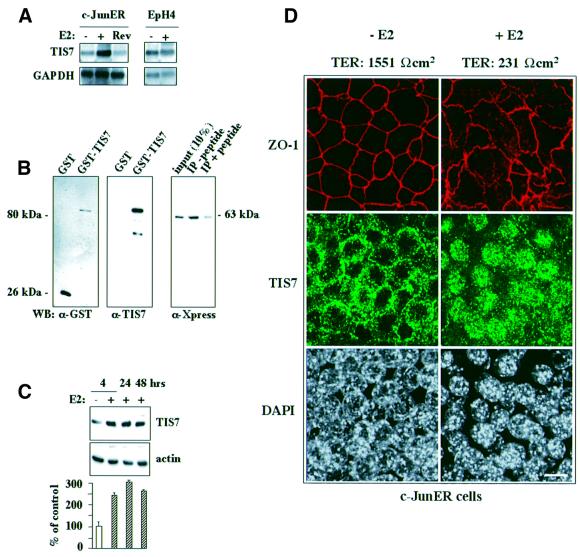
We were interested in the identification of genes whose expression changed as a result of activation of c-JunER. In the present study we performed DD RT–PCR with RNA samples isolated from untreated and 48 h β-estradiol (E2)-induced c-JunER cells. One of the several differentially expressed genes was tis7. A full-length TIS7 cDNA was subsequently cloned, which was identical to the one described in Varnum et al. (1989a). This nucleotide sequence encodes a protein of 449 amino acids that migrates on SDS–PAGE at ∼53 kDa. After 48 h of E2 treatment, the tis7 gene was up-regulated 5- to 10-fold (Figure 1A). Upon removal of E2, as the cells regain their epithelial polarity, the tis7 mRNA expression returned back to basal levels. Since estrogen hormones are potent mitogens for mammary epithelial cells, EpH4 parental cells of the c-JunER cell line were treated with an identical concentration of E2 as a control. The mRNA levels of the tis7 gene were not affected (Figure 1A, right panel).
Fig. 1. TIS7 mRNA and protein levels are up-regulated and TIS7 protein translocates into the nucleus of E2-activated c-JunER cells. (A) Northern blot analysis of total RNA isolated from c-JunER cells hybridized with a TIS7 cDNA probe. Control cells (–), 48 h E2-treated cells (+), reverted cells (Rev = cells grown for 48 h with E2 and then cultured for two additional passages in the absence of E2). Northern blot analysis of EpH4 parental cell line. One representative northern blot from five (c-JunER cells) or seven (EpH4 cells) independent experiments is shown. (B) Specificity of the affinity-purified TIS7 rabbit polyclonal antibodies. Left panel: GST–TIS7 fusion proteins detected by western blotting using α-GST antibodies. Middle panel: same proteins analysed using the affinity-purified α-TIS7 antibodies. Right panel: immunoprecipitation experiment using same the α-TIS7 antibodies without (–peptide) or after pre-incubation with the peptide used for immunization. HeLa cells were transfected with Xpr–TIS7 vector construct and equal amounts of cell lysate were used for immunoprecipitation. Immunoprecipitated protein Xpr–TIS7 was detected by western blotting using α-Xpress monoclonal antibodies. (C) Western blot analysis of TIS7 protein in control (–E2) and E2-treated (+E2) c-JunER cells, respectively. Equal loading is documented by the actin immunostaining (bottom). Western blot images were scanned, quantified and are presented as a ratio between the intensity of the specific TIS7 and actin input control signals (mean ± SD, n = 3). (D) Laser scanning confocal immunofluorescence analysis of TIS7 and ZO-1 localization in c-JunER polarized cells (–E2; left panel) and cells treated with 10–6 M E2 for 4 days (right panels). Loss of polarity of the epithelial cells was followed by measuring the transepithelial resistance (TER values). Size bar = 10 µm.
We generated a rabbit polyclonal antiserum against a peptide comprising 18 N-terminal amino acid residues of TIS7. The TIS7 antiserum recognized the GST–TIS7 protein with the predicted size of 80 kDa on western blots (Figure 1B, middle panel). It did not cross-react with GST protein loaded in comparable amounts, as documented by the α-GST antibody blotting (Figure 1B, left panel). Next, we tested the affinity-purified α-TIS7 antibody in immunoprecipitations of lysates from HeLa cells transfected with the His6Xpr-TIS7 expression construct. The α-TIS7 antibody specifically immunoprecipitated the 63 kDa His6Xpr-TIS7 band recognized by the α-Xpress monoclonal antibody. The specificity of the immunoprecipitation was confirmed by its peptide inhibition (Figure 1B, right panel).
In accordance with the DD RT–PCR results, TIS7 protein levels in total cell lysates were also up-regulated upon c-JunER activation (up to 3-fold) (Figure 1C, graph). TIS7 protein levels were induced 4 h following the onset of the E2 treatment and remained up-regulated for >48 h (Figure 1C). Effects of c-JunER activation on the subcellular distribution of TIS7 were analysed by confocal laser scanning immunofluorescence microscopy. In fully polarized cells [4 days in culture; epithelial polarity documented by the ring-like staining of the tight junction protein ZO-1 and high transepithelial electric resistance (TER) values], the majority of TIS7 localized in the vicinity of the lateral plasma membrane (Figure 1D, –E2). Upon c-JunER activation, cells lost epithelial polarity (as documented by the disrupted ZO-1 staining and low TER values), the lateral staining of TIS7 was reduced and the TIS7 protein accumulated in the nuclei (Figure 1D, +E2), similar to sparsely seeded undifferentiated cultures (data not shown).
In summary, the loss of polarity caused by the activation of the c-JunER fusion protein in mouse mammary epithelial cells was accompanied by an increase in both tis7 mRNA and protein levels, and by translocation of the TIS7 protein into the nucleus.
TIS7 overexpression changes the polarity of epithelial cells
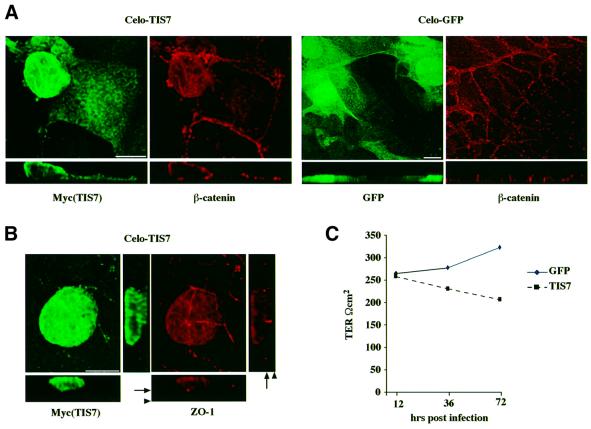
Next we examined whether overexpression of TIS7 protein itself would affect the polarity of the mammary epithelial cells. We constructed a recombinant TIS7 chicken adenovirus (CELO) and expressed TIS7 by infection of fully polarized monolayers of c-JunER cells. Virus expressing eGFP was used as a control. Immunofluorescence analysis revealed that the majority of TIS7-overexpressing cells lost the polarized epithelial phenotype, as assessed by the distribution of junctional proteins β-catenin and ZO-1, respectively (Figure 2A and B). These cells were higher, rounded up and protruded from the monolayer (Figure 2A and B, x–z section; note the difference in the thickness of the x–z section, reflecting the number of cell layers). The cumulative effect of prolonged TIS7 overexpression on the epithelial cell polarity also reflected the fall in the transepithelial resistance, as shown in the graph (Figure 2C). Although eGFP was overexpressed in similar amounts and also accumulated in the nuclei of infected cells, it did not affect epithelial cell polarity. We conclude that overexpression of TIS7 may have interfered with the polarized epithelial phenotype since contact with substratum and adhesion to neighboring cells were impaired. The nuclear localization of TIS7 in unpolarized cells suggested a role in the regulation of gene expression.
Fig. 2. TIS7-overexpressing cells lose epithelial polarity. (A) Cells were infected with CELO-recombinant viruses. Overexpressed TIS7 was detected by laser scanning confocal immunofluorescence microscopy using α-Myc antibody, endogenous β-catenin with the monoclonal antibody and eGFP directly. Top panels show sections through the x–y planes and underneath the x–z side view is shown. Bar = 10 µm. (B) TIS7-overexpressing cell grows on the top of the monolayer of otherwise polarized epithelial cells (arrowhead marks the filter insert). In polarized cells, tight junctional protein ZO-1 forms a typical ring-like structure (arrow). (C) TIS7 overexpression caused a significant decrease in the transepithelial resistance.
Analysis of TIS7 effects on gene transcription using cDNA microarray analysis
In order to find out whether nuclear TIS7 had the capacity to affect transcription, we performed a cDNA microarray analysis. The experimental set-up of this analysis is explained in detail in the Supplementary data (available at The EMBO Journal Online). The expression data were ranked using a web-based software tool developed in our laboratory to query a relational database (MicroarrayViewer; see Supplementary data). Ninety-three percent of genes were specifically down-regulated by TIS7 overexpression, while only two genes were up-regulated. The expression differences corresponded to an average expression ratio of 2.2. Transcriptional effects of TIS7 were independently, quantitatively, reconfirmed for several of the candidate genes using Real-Time PCR (Light Cycler™) (Table I). Western blot analysis also showed that protein levels of two candidate genes (CRABP II and OPN) were down-regulated in cells overexpressing TIS7 (Supplementary data).
Table I. Identification of genes that transcription specifically changed following the TIS7 overexpression by cDNA microarray analysis.
| Sequence annotation | UniGene accession No. | Differential expression: TIS7–EGFP | Real-Time PCR |
|---|---|---|---|
| Cellular retinoic acid binding protein II (crabp II)* | W81912 | –4.1 | –7.5 |
| Osteopontin/secreted phosphoprotein I (spp1)* | AA108928 | –2.9 | –2.1 |
| w.s.t. CG5676 gene product # | AA238637 | –2.6 | |
| h.s.t. AF150733_1 AD-014 protein # | W74865 | –2.2 | |
| Unknown EST | 34061 | –2.1 | |
| h.s.t. NADH-ubiquinone oxireductase B14 SU # | 62323 | –2.1 | |
| m.s.t. 60S ribosomal protein L39 # | AA178015 | –2.1 | |
| h.s.t. 60S ribosomal protein L22 # | AA174807 | –2.0 | |
| Unknown EST | W41563 | –2.0 | |
| Myosin heavy polypeptide 8, skeletal muscle, perinatal* | W13528 | –1.9 | |
| Synapsin Ib* | AA033398 | –1.9 | |
| w.s.t. wdnm1 protein precursor # | AA105830 | –1.8 | |
| Feminization-1a homolog (fem-1a)* | AA434902 | –1.8 | |
| h.s.t. cystathione-γ-lyase # | AA245993 | –1.7 | |
| Emopamil binding protein (ebp)* | W08348 | –1.7 | –2.8 |
| Cathepsin Z* | W14289 | –1.7 | |
| Peptidylprolyl-isomerase A* | AA499926 | –1.7 | |
| Fibroblast inducible secreted protein 12 (fisp12)* | W36541 | –1.7 | |
| Pituitary tumor transforming gene protein 1 (pttg1)* | A250500 | –1.6 | |
| Thymidine kinase 1* | AA041834 | –1.6 | |
| Unknown EST | W09957 | –1.6 | |
| Homeobox, msh-like 2* | AA016730 | –1.5 | |
| DNA segment, Chr 15, Wayne State University 77 | AA014127 | –1.5 | |
| Torsin family 2, member A* | AA190292 | –1.5 | |
| Hexokinase 1* | W11571 | –1.5 | |
| h.s.t. SRB7 # | AA067890 | –1.5 | |
| Split hand/foot deleted gene 1* | AA462396 | –1.5 | |
| h.s.t. ubiquinol–cytochrome c reductase complex (7.2 kDa) # | AA087137 | –1.5 | |
| h.s.t. myelin protein zero (p0) precursor # | AA097191 | +1.5 | +3.8 |
| Intracisternal A-particle mRNA | AA404014 | +1.5 |
Using the MicroarrayViewer program (see Supplementary data), genes specifically regulated by ectopic TIS7 could be extracted from the vector associated and from the great majority of unaffected genes. Candidate genes are listed according to their level of differential expression. w.s.t., weakly similar to; m.s.t., moderately similar to; h.s.t., highly similar to; *, functionally annotated TIS7 target genes; #, TIS7 target genes of ‘uncertain’ identity. Confirmation of the cDNA microarray hybridization results by Real-Time PCR analysis. Values of TIS7-regulated gene expressions were normalized and then compared as a ratio of TIS7 sample values and eGFP sample values.
In conclusion, the cDNA microarray experiment revealed that TIS7 overexpression preferentially caused down-regulation of a specific set of genes and this finding, together with TIS7 nuclear localization, suggest a potential role for TIS7 as a transcriptional co-repressor.
TIS7 interacts with the SIN3 HDAC complex
In order to identify the mechanism by which TIS7 affects transcription, we decided to identify proteins interacting with the mouse TIS7 protein. In a yeast two-hybrid screen, we used a mouse embryo cDNA library and the full-length cDNA (amino acids 1–449), as well as N-terminal (amino acids 1–227), C-terminal (amino acids 262–449) and central domains (amino acids 240–280) of TIS7 as baits. We identified mSin3B, a member of the SIN3 HDAC complex (four positive clones), as a protein potentially interacting with the N-terminal domain of the TIS7 protein.
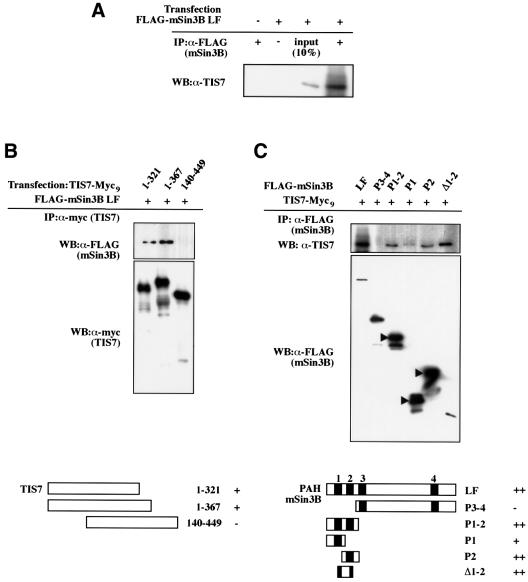
Having established the interaction in yeast two-hybrid screens, we tested whether the TIS7 interaction with mSin3B could also be demonstrated in vivo. First, HeLa cells were transfected with a FLAG-tagged mSin3B long form (FLAG-mSin3B LF) (Naruse et al., 1999) and immunoprecipitated using α-FLAG antibodies. Endo genous TIS7 protein co-immunoprecipitated with FLAG-mSin3B (Figure 3A).
Fig. 3. TIS7 interacts with mSin3B. (A) HeLa cells were transfected with FLAG-mSin3B LF expression plasmid. Cell lysates were immunoprecipitated with α-FLAG tag monoclonal antibody and co-immunoprecipitated endogenous TIS7 was detected by western blot analysis. (B) HeLa cells were co-transfected with N- or C-terminal Myc9-tagged TIS7 truncation mutants and FLAG-tagged mSin3B long-form construct as indicated. Cell lysates were immunoprecipitated with the antibodies against the Myc tag, and co-immunoprecipitated mSin3B was detected by immunoblotting using α-FLAG antibodies. The bottom panel shows TIS7-Myc9 truncation mutants immunoprecipitated with the α-Myc antibodies. (C) HeLa cells were co-transfected with C-terminally Myc9-tagged full-length TIS7 construct and different N-terminally FLAG-tagged mSin3B constructs (LF, long form; P, PAH domain; arrowheads point towards the Sin3 fusion protein bands, smaller bands are degradation products). Protein complexes were immunoprecipitated with α-FLAG tag antibodies. In the lower panel are mSin3B constructs immunoprecipitated with α-FLAG antibodies detected with the same antibody. Schemes under both panels depict expressed parts of the proteins and level of interaction (–, +, ++).
Next, we mapped the mSin3B interaction domains on the TIS7 protein. FLAG-tagged mSin3B LF was transfected into HeLa cells either alone or together with a series of Myc9-tagged TIS7 deletion constructs and immunoprecipitations were performed using α-Myc antibodies. Two TIS7 constructs encompassing the N-terminal amino acids 1–321 and 1–367 co-immunoprecipitated mSin3B. However, a partially overlapping construct expressing the protein encoded by amino acid residues 140–449, lacking the N-terminal domain of the TIS7 protein, was not capable of binding mSin3B (Figure 3B). The results of this domain mapping experiment confirmed our initial finding that TIS7 interacted in the yeast two-hybrid screen with the mSin3B protein through its N-terminal domain. Thus, we concluded that the region necessary and sufficient for the interaction with mSin3B is the TIS7 sequence between the amino acids 1 and 140.
Some of the specific domains of mSin3B important for its interaction with interacting partners are already well characterized, e.g. interaction of the paired amphipathic helix (PAH)2 domain with the transcription factor MAD (Ayer et al., 1996; Harper et al., 1996; Laherty et al., 1998), or of PAH1 and PAH3 domains with N-CoR (Alland et al., 1997; Heinzel et al., 1997). Therefore, we examined the in vivo interactions between the series of mSin3B deletion constructs and full-length TIS7. Among the series of mSin3B deletion constructs examined (Figure 3C), mSin3B LF as well as constructs containing the PAH2 domain and the intervening region between the PAH1 and PAH2 domains represented significant binding to TIS7. The PAH1 domain alone also bound TIS7, though less efficiently. In contrast, constructs containing the C-terminal half of mSin3B only, encompassing PAH3 through PAH4 domains (mSin3B P3-4), showed no interaction with TIS7 (Figure 3C).
It was shown previously that mSin3 proteins (A and B), as part of the SIN3 complex, recruit HDACs to specific transcription factors (e.g. Mad or NRSF) and thereby repress transcription (Ayer et al., 1995, 1996; Korhonen et al., 1998; Naruse et al., 1999). To test whether TIS7 is associated with HDAC1 in vivo, TIS7 immunoprecipitates were assayed for HDAC1 by western blot analysis. TIS7 antibody specifically co-immunoprecipitated HDAC1 and formation of this precipitate was blocked with the cognate immunogen. Endogenous HDAC1 was co-immunoprecipitated in a dose-dependent manner with increasing amounts of the α-TIS7 antibody (Figure 4A). In the reciprocal experiment, endogenous TIS7 was co-immunoprecipitated with the monoclonal antibody against the endogenous HDAC1 (Figure 4B). To test whether TIS7 has the capacity to associate with other HDACs besides HDAC1, we performed in vitro pull-down assays using GST–TIS7 fusion protein. Radioactively labelled HDAC1, HDAC2 and HDAC3 specifically interacted with the GST–TIS7 protein (Figure 4C). Similarly, GST–TIS7 interacted with N-CoR nuclear receptor co-repressor, a component of HDAC complexes. These interactions were specific since, first, products of in vitro coupled transcription/translation reaction did not interact with GST control protein alone and, secondly, GST–TIS7 did not interact with an unrelated nuclear protein, human cohesin subunit SCC1, synthesized in an identical way (Figure 4C).
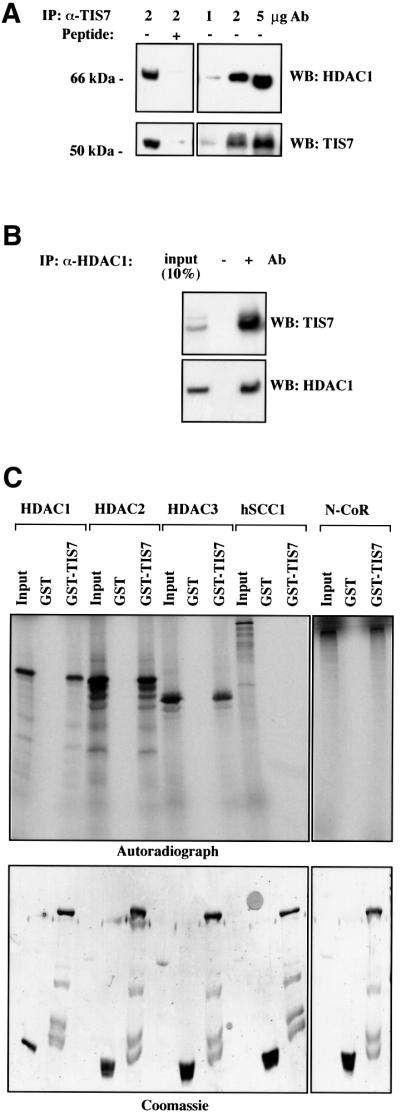
Fig. 4. TIS7 interacts with HDAC1. (A) HeLa cell lysates were immunoprecipitated with the affinity-purified rabbit α-TIS7 antibodies, without (–) or with (+) pre-incubation with the peptide used to raise the TIS7 antibodies (left panel). Co-immunoprecipitated HDAC1 was detected by western blotting. Increasing amounts of the α-TIS7 antibodies co-immunoprecipitated increasing amounts of the endogenous HDAC1. Bottom panels show immunoprecipitated endogenous TIS7 detected by western blotting using the same α-TIS7 antibodies. (B) In a vice versa experiment, HeLa cell lysates were immunoprecipitated with the monoclonal antibodies against HDAC1 and co-immunoprecipitated endogenous TIS7 was detected by western blotting (–Ab, pre-immune serum). The bottom panel documents amounts of immunoprecipitated endogenous HDAC1. (C) GST pull-down analysis using recombinant GST–TIS7 protein. 35S-labelled HDAC1, HDAC2, HDAC3, hSCC1 and N-CoR products of in vitro coupled transcription/translation reaction in rabbit reticulocyte lysate were incubated with GST proteins bound to the glutathione beads, washed and eluted with sample buffer. Bound proteins were analysed by autoradiography. Equal loading of GST and GST–TIS7 proteins was confirmed by Coomassie Blue staining (bottom panel).
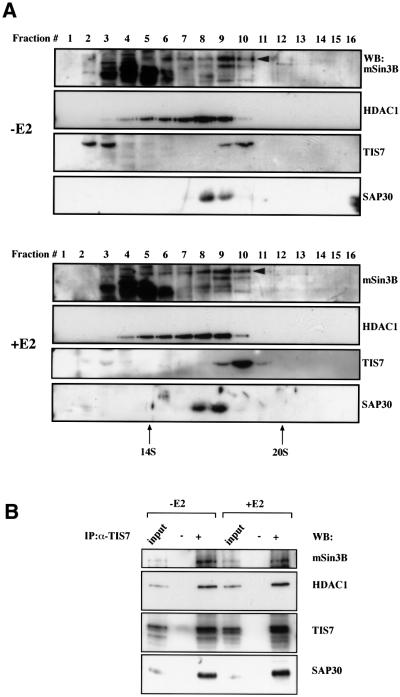
In the next experiment, we tested whether TIS7 may associate with some of the currently known subunits of the SIN3 complex in mouse epithelial c-JunER cells. We analysed c-JunER nuclear extract fractions after separation by continuous sucrose gradients. In E2-untreated cells, part of the TIS7 pool co-sedimented with mSin3B, HDAC1 and SAP30 during sucrose gradient centrifugation (Figure 5A, fraction 9). Another part of the TIS7 pool in these normally polarized cells appeared in lighter fractions from the gradient centrifugation representing monomers. However, following 4 days treatment of c-JunER cells with E2, there was a significant increase in the amount of TIS7 that co-sedimented with mSin3B and HDAC1 during sucrose gradient centrifugation (fraction 10). At the same time, TIS7 sedimenting in the lightest fractions of the sucrose gradient (fractions 2 and 3) disappeared. We have confirmed that TIS7 associates specifically with the SIN3 complex in an independent sucrose gradient separation of a nuclear extract by blotting with antibodies against MTA2, a component of a different HDAC complex. MTA2 fractionated in different fractions than TIS7 (data not shown).
Fig. 5. TIS7 can associate with the SIN3 complex. (A) mSin3B, HDAC1, TIS7 and SAP30 partially co-fractionate. Nuclear extracts prepared from control (–E2) or 4 day E2-treated c-JunER cells (+E2) were fractionated on a 10–35% continuous sucrose gradient. Aliquots from 0.6 ml fractions were resolved on a 10% SDS–PAGE gel and immunoblotted with the indicated antibodies. In mSin3B western blot, the arrowhead indicates the full-length mSin3B LF band; other bands detected by this antibody, mainly in lighter fractions, represent degradation products. Sedimentation coefficients of 14 and 20S fractions were marked according to the peaks of proteasome and cohesin proteins used as markers. (B) c-JunER cells were either left untreated (–E2) or cultured in the presence of 10–6 M E2 for 4 days. Nuclear extracts were prepared as described in Dignam et al. (1983), and immunoprecipitated with the affinity-purified α-TIS7 antibody. Co-immunoprecipitated endogenous proteins were detected by western blotting with the indicated antibodies. The additional band in the TIS7 western blot is an as yet unidentified post-translational modification present in E2-treated cells.
Performing the immunoprecipitation with the affinity-purified α-TIS7 antibody, we found that besides mSin3B, other endogenous protein components of the SIN3 complex, e.g. HDAC1 and SAP30, also co-immunoprecipitated specifically with the endogenous TIS7 (Figure 5B). These interactions were detectable both in polarized (–E2) as well as unpolarized (+E2) c-JunER cells. Although there was also a minor increase in the amount of immunoprecipitated TIS7 in the +E2 sample, the amounts of co-immunoprecipitated SIN3 complex proteins did not increase. These immunoprecipitation results confirmed our initial two-hybrid screen findings, namely the interaction of TIS7 with the SIN3 complex subunits in vivo.
The TIS7 HDAC complex is enzymatically active
Our data demonstrated that TIS7 can interact with mSin3B and HDAC1, as well as other members of the HDAC complex. To establish that TIS7 associates with a functional HDAC, TIS7 immunoprecipitates from HeLa and c-JunER cells were assayed for HDAC activity. Endogenous TIS7 immunocomplexes contained substantial HDAC activity when compared with control precipitates using an antibody directed against an unrelated epitope (Myc) (Figure 6A). The specificity of the immunoprecipitation was confirmed by the absence of HDAC activity from the immunoprecipitate where the antibody was blocked by the cognate immunogen peptide. Importantly, HDAC activity also co-immunoprecipitated with TIS7 in the c-JunER cell line. Since immunocomplexes with HDAC activity could also contain enzyme isoforms other than HDAC1, we decided to compare TIS7 immunoprecipitates that were derived from wild-type and HDAC1–/– mouse embryonic stem cells. The total HDAC activity in the HDAC1–/– cells was only partially, but not completely, decreased because of the up-regulated activity of the remaining HDAC isoforms (Lagger et al., 2002). The HDAC activity co-immunoprecipitated with TIS7 in the HDAC1–/– sample was significantly decreased (Figure 6B). On the other hand, the HDAC activity was not totally abolished, showing a possible interaction between TIS7 and other HDAC proteins, as suggested by our GST pull-down assays (Figure 4B). Subsequent western blotting confirmed that TIS7 interacted with HDAC2, which is more abundant in HDAC1–/– cells (Figure 6B). These results demonstrate that TIS7 associates with a functional HDAC complex in vivo.

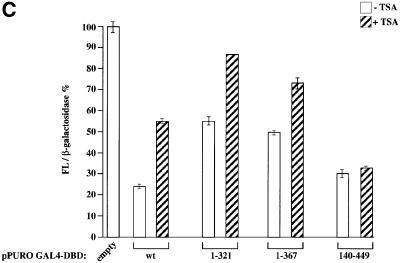
Fig. 6. TIS7 co-immunoprecipitated HDAC is enzymatically active. (A) HeLa and c-JunER cell lysates (as indicated below the graphs) were immunoprecipitated with affinity-purified α-TIS7 antibodies, without (–pept.) or with TIS7 N-terminal peptide pre-incubation (+ pept.), or with unrelated antibodies (α-Myc) as a negative control. HDAC activity was measured as described previously (Doetzlhofer et al., 1999). HDAC1 was detected by western blot analysis with mouse monoclonal α-HDAC1 antibodies. The input lane represents 4% of the total lysate used for the immunoprecipitation and documents the functionality of the enzymatic assay. This experiment was repeated several times with similar results; one representative experiment is shown, values of the bars are mean ± SD, n = 3. (B) TIS7 interacts mainly with HDAC1. The left side of the graph shows the HDAC enzymatic activity of total cell lysates derived from wild-type and homozygous HDAC1–/– embryonic stem cells. The right side of the graph shows the result of the HDAC enzymatic activity measured in the same cell lysates following the immunoprecipitation with affinity-purified α-TIS7 antibodies. The western blot underneath the graph shows HDAC1 and HDAC2 detected using monoclonal antibodies. (C) TIS7 inhibits transcriptional activity. HeLa cells were transiently co-transfected with the pLGCLuc-E351 luciferase reporter construct, β-galactosidase expression plasmid and either GAL4 DNA-binding domain (DBD) alone or GAL4-DBD-TIS7 wild-type, N- (1–321; 1–367) and C-terminal (140–449) TIS7 plasmid, respectively. After 36 h, the transfected cells were treated with 50 ng/ml TSA or left untreated, and 12 h later luciferase and β-galactosidase activities were measured. Luciferase intensities were normalized to β-galactosidase activity. Values of cells transfected with pPuroGAL4-DBD empty vector, reporter construct and not treated with TSA represented 100%.
TIS7 represses transcriptional activity in an HDAC-dependent manner
It was shown previously that targeting HDACs to the vicinity of a promoter resulted in transcriptional repression (Yang et al., 1996; Alland et al., 1997; Hassig et al., 1997; Zhang et al., 1998). Therefore, we tested whether directing TIS7 to a particular promoter would inhibit transcription through recruitment of HDAC activity. HeLa cells were transfected with a construct containing a minimal thymidine kinase–luciferase template regulated by five consensus Gal4-binding sites in the promoter-proximal region (Alkema et al., 1997). Co-transfection of the Gal4-dependent reporter with a mammalian expression vector encoding the Gal4 DNA-binding domain fused to full-length TIS7 sequence resulted in up to 75% repression of the transcriptional activity. Furthermore, the mechanism of this transcriptional repression was HDAC dependent since repression was partially relieved by HDAC inhibitor [trichostatin A (TSA)] treatment of TIS7 co-transfected cells (Figure 6C). Several Gal4-fused TIS7 mutants (expressing the same parts of the TIS7 protein as used for the interaction study in Figure 4A) were generated and their repression activities were analysed in order to determine minimal regions required for the repression activity of TIS7. Both the N-terminal TIS7 constructs 1–321 and 1–367 repressed transcription (Figure 6C), although less strongly than the C-terminal construct 140–449 or wild-type full-length TIS7. Interestingly, transcriptional repression by the N-terminal constructs of TIS7 was almost completely de-repressed by the TSA treatment. In contrast, transcriptional repression by the C-terminal construct (140–449) was not significantly affected by even high TSA concentrations. These results suggest that histone deacetylation is involved in the transcriptional silencing via the N-terminal domain of TIS7. On the other hand, the C-terminus of TIS7 exhibits repressive activity independent of histone deacetylation (Figure 6C).
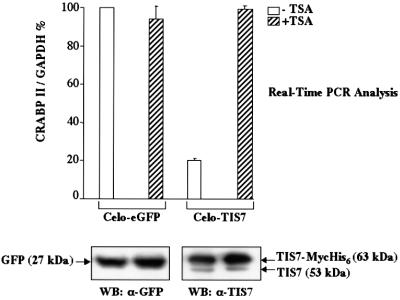
Finally, we asked whether the transcriptional regulation of genes regulated by TIS7 overexpression, identified by the cDNA microarray analysis, was HDAC dependent. c-JunER cells were infected with CELO-eGFP and CELO-TIS7, respectively. TSA treatment substantially reverted the TIS7-mediated repression of one tested gene: crabp II (Figure 7). Importantly, TSA treatment did not have an effect on the crabp II RNA levels in the control, eGFP-overexpressing cells or on the expression of the house-keeping gene gapdh. These data suggest that TIS7 specifically down-regulated the expression of crabp II via a HDAC activity-dependent mechanism.
Fig. 7. TIS7-repressed gene expression is HDAC dependent. c-JunER cells were infected with CELO-eGFP or CELO-TIS7. After 36 h, cells were treated with 10 ng/ml TSA (+TSA) or left untreated (–TSA). After an additional 12 h, total RNAs were isolated and expression of crabp II was measured by Real-Time PCR. Expression of crabp II was normalized to the expression of gapdh. Western blots at the bottom equal expression of eGFP or TIS7 protein in both samples.
Discussion
According to studies in several different biological systems, TIS7 has been postulated to be involved in the regulation of various cell differentiation processes (Tirone and Shooter, 1989; Guardavaccaro et al., 1995; Rubin et al., 1998). Our data provide a molecular mechanism by which TIS7 can regulate gene expression and might be involved in differentiation processes. Here we show that, under specific conditions, TIS7 associates with the SIN3 repressor complex possessing HDAC activity. This interaction suggests a possibility that TIS7 may repress transcription of specific genes via a HDAC activity-dependent mechanism. The Supplementary data for this study provide a list of genes specifically regulated following the overexpression of TIS7, accompanied by online links leading to their characteristics (http://www.imp.univie.ac.at/lh/chip_10_5_01/index.html).
TIS7 shuttles between cytoplasm and nucleus
It was previously shown that TIS7 translocates between the cytoplasm and nucleus (Kujubu et al., 1987; Tirone and Shooter, 1989; Guardavaccaro et al., 1994). Similarly, in sparsely seeded and unpolarized c-JunER cells, TIS7 localized preferentially in the nucleus. As the density of the cultures increased, TIS7 redistributed to the cytoplasm and in fully polarized and contact-inhibited cells localized mainly in the vicinity of the plasma membrane (data not shown). Upon the loss of epithelial polarity, the TIS7 protein translocated from the plasma membrane into the nucleus. Our data suggest that the nuclear localization of TIS7 is essential for transcriptional regulation.
TIS7 is a transcriptional co-repressor
We have demonstrated that the ability of TIS7 to attenuate transcriptional activity is associated with HDAC activity. Our co-immunoprecipitation data suggested that TIS7 can act as a co-repressor that can associate with the SIN3 complex. According to sequence analysis, TIS7 does not contain a DNA-binding domain and, therefore, we do not expect TIS7 to directly bind to DNA, thereby tethering the HDAC activity-containing complex to a particular promoter on its own. However, via its interactions with other components of a multi-molecular complex, TIS7 may interact with DNA-binding proteins and thereby co-repress expression of genes. Since our GST pull-down assays were performed using rabbit reticulocyte lysates without purification of the in vitro transcribed/translated, labelled proteins, one can not rule out the possibility that interactions between GST–TIS7 and HDACs are indirect, through additional proteins in the complex. Our data show that TIS7 has the capacity to interact with several members of the HDAC class I. The experiment showing that TIS7 co-immunoprecipitated HDAC activity in cells originating from HDAC1–/– ES cells opened the possibility that TIS7 interacts with several different HDAC activities, which was confirmed by the co-immunoprecipitation of HDAC2.
By mapping experiments, we identified two distinct domains at the N- and C-terminal ends of the TIS7 molecule exhibiting repressive effects in the Gal4–TK-luciferase reporter assay. Importantly, the only TIS7 domain necessary for the direct interaction with mSin3B resided at its N-terminus. We also showed that the transcriptional repression caused by the overexpression of the N-terminal TIS7 constructs could be relieved by TSA treatment. However, the repression of the C-terminal construct was not TSA sensitive, suggesting that this part of the TIS7 molecule participates in the HDAC-independent transcriptional repression mechanism. This result is analogous to the effects of the neural restrictive silencer factor (NRSF) where the N-terminal repression domain, but not the second, C-terminal domain, is also TSA sensitive (Naruse et al., 1999). We show here that TIS7 interacts with the intervening region between PAH domains 1 and 2 of mSin3B. This binding site was also similar to a domain encompassing the region of PAH1 and PAH2 domains of NRSF, which was shown to have the strongest interaction with the mSin3B SF (Naruse et al., 1999).
Role of TIS7 in HDAC activities in vivo
In summary, the studies presented here demonstrate that TIS7 represents a novel transcriptional co-repressor interacting with the functional HDAC complex in vivo and in vitro. We found expression of a TIS7-regulated gene to be de-repressed by TSA treatment. However, because of a very general HDAC inhibitory effect of TSA, one can not test whether HDAC inhibition would revert loss of epithelial polarity caused by TIS7 overexpression. We propose a model for TIS7 function in which an increase in the amount of nuclear TIS7 protein (e.g. as a result of transfection or loss of epithelial polarity) is capable of transcriptional repression of particular genes in a HDAC-dependent manner. Our future interests remain in confirming the role of TIS7 as a specific transcriptional co-repressor in vivo at the level of the whole organism.
Materials and methods
Cell culture
EpH4 and c-JunER cells were cultured on permeable filter culture inserts, as described in Fialka et al. (1996). HeLa cells were grown in DMEM supplemented with 5% FCS (Gibco BRL), 10 mM HEPES pH 7.3, 1% penicillin/streptomycin (Gibco BRL) and 1% l-glutamine.
Plasmids
Full-length mTIS7 cDNA was amplified by PCR and cloned into pcDNA3.1/HisC, pcDNA3.1(–)/MycHis (Invitrogen), pcDNA3.1(–) with a C-terminal Myc9 tag and in a pGEX2 GST vector (Pharmacia). For the Gal4 transcriptional activity assays, mTIS7 fragments were PCR amplified and cloned into the pPuroGal4-DBD vector (Alkema et al., 1997). The pLGCLuc-E351 luciferase reporter construct was described previously in Braselmann et al. (1992); pCIneoHDAC1-Myc plasmids in Bartl et al. (1997) and Doetzlhofer et al. (1999); and mSin3B constructs in Naruse et al. (1999). The hSCC1 product of in vitro coupled transcription/translation reaction was previously characterized in Waizenegger et al. (2000).
Northern blot analysis
Northern blots were performed as described previously (Vietor et al., 2001). Hybridizations were normalized to gapdh expression on the same blot. Radioactivity of specific bands was measured and evaluated by the ImageQuant software (Molecular Dynamics).
Differential display
Total RNA samples isolated from the controls and E2-treated c-JunER cells were analysed with the Differential Display Kit (Display Systems) according to the manufacturer’s recommendations. Analyses were repeated five times and sequences analysed by the Blast search program at the NCBI home page. Differential regulation of identified genes was confirmed by northern blot analysis.
Yeast two-hybrid analysis
The two-hybrid screen was performed as described in the Clontech MATCHMAKER LexA manual, using full-length, N-terminal (amino acid 1–227), C-terminal (amino acid 262–449) and central domain (amino acid 240–280) constructs of TIS7 obtained by PCR. The LexA DBD-containing pBTM116-TIS7 vector was co-transformed with a mouse embryo cDNA library fused to the activation domain of Gal4 of the pACT2 vector into a yeast strain L40. Positive clones were re-analysed in a β-galactosidase (β-gal) overlay assay and prey clones were directly PCR amplified and sequenced.
Real-Time PCR
The primer sequences and protocol of these analyses are available in the Supplementary data.
Construction of recombinant CELO adenovirus vector
A recombinant CELO virus expressing the TIS7 gene was prepared as a derivative of CELO AIM46, previously described in Michou et al. (1999). Full-length TIS7 cDNA, tagged at the 3′ end with Myc epitope and His6 tag sequences, was cloned into the plasmid pPM7 flanked upstream by the CMV IE promoter/enhancer and downstream by a rabbit β-globin intron/polyadenylation sequence. The CMV/TIS7/β-globin cassette was excised and recombined into pAIM46, thus replacing the luciferase cDNA. Construction of a recombinant CELO-eGFP virus (CELO AIM53) was described previously (Michou et al., 1999).
Antibodies
The rabbit antiserum to TIS7 was raised against a peptide comprising 18 N-terminal amino acid residues (TIS7; DDBJ/EMBL/GenBank accession No. X17400; Swissprot P19182; AA 30–47; GGPHRTVQPFSDEHA SIC) and affinity purified. Blocking of the antibody with the peptide used for immunization was performed by 60 min incubation at room temperature with 1 µg of peptide/ml of antibody solution. Rabbit polyclonal affinity-purified α-Myc antibodies were from Gramsch Laboratories (Schwabhausen, Germany); mSin3B (E-20), HDAC1 (C-19) from Santa Cruz and HDAC2 from Upstate Biotechnology; α-GST from Chemicon; α-Flag antibodies (M1) (Eastman Kodak) from Sigma and Alexa 488 goat α-mouse IgG-conjugated secondary antibodies from Molecular Probes. Mouse monoclonal antibodies against Xpress epitope were from Invitrogen, ZO-1 and actin from Chemicon, β-catenin from Transduction Laboratories and GFP from Roche Molecular Biochemicals. Mouse monoclonal α-HDAC1 antibodies were prepared in the laboratory of C.Seiser. Rabbit polyclonal antibodies against mouse osteopontin were a generous gift from Toshimitsu Uede, Hokkaido University, Sapporo; rabbit anti-cellular retinoic acid binding protein II antibodies were from P.Chambon, CNRS Strasbourg. H68.4 mouse monoclonal antibodies against α-transferrin receptor were kindly provided by I.S.Trowbridge.
Immunoprecipitation and immunoblotting
Cells were lysed on ice in buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 0.5 mM EDTA, 0.5 mM EGTA, 50 mM NaF, 5 mM NaPi, 0.2 mM Na3VO4 and a cocktail of protease inhibitors. Lysates were sonicated and centrifuged in order to separate insoluble proteins. Equal amounts of soluble proteins were immunoprecipitated with 5 µg of the respective antibodies followed by 20 µl of (1:1) protein A– or protein G–agarose slurry (Ultralink; Pierce). Following four washes with the lysis buffer, immunoprecipitated proteins were eluted with the sample buffer and boiling. Western blots were performed as described previously (Vietor et al., 2001).
Cell extracts and fractionation
For sedimentation analysis, HeLa cells were harvested in exponential phase. The nuclear extract was prepared according to Dignam et al. (1983), snap-frozen in liquid nitrogen and stored at –80°C. The final buffer composition was 20 mM HEPES, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF and 20% glycerol. Nuclear extract of HeLa cells containing 0.5 mg of protein was centrifuged for 18 h at 30 000 r.p.m. and 4°C through a 10–35% continuous sucrose gradient (in a buffer containing 150 mM NaCl, 20 mM Tris pH 7.4, 0.2% Triton X-100 and 0.5 mM DTT) using a SW40 rotor (Beckman), as described previously (Waizenegger et al., 2000).
Immunofluorescence
Preparation of samples and laser confocal scanning immunofluorescence analysis were performed as described previously (Vietor et al., 2001).
Transient transfections and luciferase assay
Transient transfections were performed using the Lipofectamine Plus reagent (Life Technologies). In luciferase assays, pCMV-β-Gal plasmid was used to normalize for transfection efficiency. The total amount of transfected DNA was equalized by addition of empty vector DNA.
HDAC activity assay
HDAC activity assays were carried out as described previously (Lechner et al., 1996; Bartl et al., 1997; Doetzlhofer et al., 1999). To measure HDAC enzymatic activity, equal amounts (4% of whole-cell lysate) of immunoprecipitate obtained from 375 µl of whole-cell lysate were used.
GST pull-down assay
In vitro expression of radiolabelled proteins was performed in a TNT-coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine. GST–TIS7 recombinant protein was expressed in Escherichia coli BL21. GST pull-down assays with glutathione– Sepharose 4B beads (Pharmacia) coated with GST fusion proteins (2 µg) were performed as described previously (Karlseder et al., 1996; Doetzlhofer et al., 1999). Bound proteins were eluted with loading buffer, resolved by SDS–PAGE and visualized by autoradiography. In order to confirm equal loading, gels were stained with Coomassie Blue.
cDNA microarray experiment
Technical details and the exact protocol concerning this experiment are described in Supplementary data.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank M.Busslinger for helpful comments and discussion on the manuscript. We would like to thank C.K.Glass (UCSD) for providing us with the N-CoR expression plasmid. We would also like to thank C.Pasquali, R.Kurzbauer, M.Bister, C.Gieffers and K.Paiha for excellent technical assistance. This work was supported by Boehringer Ingelheim and by the Austrian Science Foundation grant (FWF, P13577-GEN).
References
- Ahringer J. (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet., 16, 351–356. [DOI] [PubMed] [Google Scholar]
- Alkema M.J., Jacobs,J., Voncken,J.W., Jenkins,N.A., Copeland,N.G., Satijn,D.P., Otte,A.P., Berns,A. and van Lohuizen,M. (1997) MPc2, a new murine homolog of the Drosophila polycomb protein is a member of the mouse polycomb transcriptional repressor complex. J. Mol. Biol., 273, 993–1003. [DOI] [PubMed] [Google Scholar]
- Alland L., Muhle,R., Hou,H.,Jr, Potes,J., Chin,L., Schreiber-Agus,N. and DePinho,R.A. (1997) Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature, 387, 49–55. [DOI] [PubMed] [Google Scholar]
- Ayer D.E. (1999) Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol., 9, 193–198. [DOI] [PubMed] [Google Scholar]
- Ayer D.E., Lawrence,Q.A. and Eisenman,R.N. (1995) Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell, 80, 767–776. [DOI] [PubMed] [Google Scholar]
- Ayer D.E., Laherty,C.D., Lawrence,Q.A., Armstrong,A.P. and Eisenman,R.N. (1996) Mad proteins contain a dominant transcription repression domain. Mol. Cell. Biol., 16, 5772–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartl S., Taplick,J., Lagger,G., Khier,H., Kuchler,K. and Seiser,C. (1997) Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol. Cell. Biol., 17, 5033–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braselmann S., Bergers,G., Wrighton,C., Graninger,P., Superti-Furga,G. and Busslinger,M. (1992) Identification of Fos target genes by the use of selective induction systems. J. Cell Sci. Suppl., 16, 97–109. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- Buanne P., Incerti,B., Guardavaccaro,D., Avvantaggiato,V., Simeone,A. and Tirone,F. (1998) Cloning of the human interferon-related developmental regulator (IFRD1) gene coding for the PC4 protein, a member of a novel family of developmentally regulated genes. Genomics, 51, 233–242. [DOI] [PubMed] [Google Scholar]
- Chi T., Lieberman,P., Ellwood,K. and Carey,M. (1995) A general mechanism for transcriptional synergy by eukaryotic activators. Nature, 377, 254–257. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetzlhofer A., Rotheneder,H., Lagger,G., Koranda,M., Kurtev,V., Brosch,G., Wintersberger,E. and Seiser,C. (1999) Histone deacetylase I can repress transcription by binding to Sp1. Mol. Cell. Biol., 19, 5504–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialka I., Schwarz,H., Reichmann,E., Oft,M., Busslinger,M. and Beug,H. (1996) The estrogen-dependent c-JunER protein causes a reversible loss of mammary epithelial cell polarity involving a destabilization of adherens junctions. J. Cell Biol., 132, 1115–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.A., Cutler,G. and Tjian,R. (1996) Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell, 84, 825–830. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D., Montagnoli,A., Ciotti,M.T., Gatti,A., Lotti,L., Di Lazzaro,C., Torrisi,M.R. and Tirone,F. (1994) Nerve growth factor regulates the subcellular localization of the nerve growth factor-inducible protein PC4 in PC12 cells. J. Neurosci. Res., 37, 660–674. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D., Ciotti,M.T., Schafer,B.W., Montagnoli,A. and Tirone,F. (1995) Inhibition of differentiation in myoblasts deprived of the interferon-related protein PC4. Cell Growth Differ., 6, 159–169. [PubMed] [Google Scholar]
- Harper S.E., Qiu,Y. and Sharp,P.A. (1996) Sin3 co-repressor function in Myc-induced transcription and transformation. Proc. Natl Acad. Sci. USA, 93, 8536–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassig C.A., Fleischer,T.C., Billin,A.N., Schreiber,S.L. and Ayer,D.E. (1997) Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell, 89, 341–347. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Thorne,A.W. and Crane-Robinson,C. (1988) A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J., 7, 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T. et al. (1997) A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature, 387, 43–48. [DOI] [PubMed] [Google Scholar]
- Humphrey G.W., Wang,Y., Russanova,V.R., Hirai,T., Qin,J., Nakatani,Y. and Howard,B.H. (2001) Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem., 276, 6817–6824. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Rotheneder,H. and Wintersberger,E. (1996) Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol., 16, 1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler P.S. and Eisenman,R.N. (1999) Sin meets NuRD and other tails of repression. Cell, 99, 447–450. [DOI] [PubMed] [Google Scholar]
- Korhonen P., Tapiola,T., Suuronen,T. and Salminen,A. (1998) Expression of transcriptional repressor protein mSin3A but not mSin3B is induced during neuronal apoptosis. Biochem. Biophys. Res. Commun., 252, 274–277. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujubu D.A., Lim,R.W., Varnum,B.C. and Herschman,H.R. (1987) Induction of transiently expressed genes in PC-12 pheochromo cytoma cells. Oncogene, 1, 257–262. [PubMed] [Google Scholar]
- Kyrylenko S., Korhonen,P., Kyrylenko,O., Roschier,M. and Salminen,A. (2000) Expression of transcriptional repressor proteins mSin3A and 3B during aging and replicative senescence. Biochem. Biophys. Res. Commun., 275, 455–459. [DOI] [PubMed] [Google Scholar]
- Lagger G. et al. (2002) Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J., 21, 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty C.D., Yang,W.M., Sun,J.M., Davie,J.R., Seto,E. and Eisenman,R.N. (1997) Histone deacetylases associated with the mSin3 co-repressor mediate mad transcriptional repression. Cell, 89, 349–356. [DOI] [PubMed] [Google Scholar]
- Laherty C.D. et al. (1998) SAP30, a component of the mSin3 co-repressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell, 2, 33–42. [DOI] [PubMed] [Google Scholar]
- Lechner T., Lusser,A., Brosch,G., Eberharter,A., Goralik-Schramel,M. and Loidl,P. (1996) A comparative study of histone deacetylases of plant, fungal and vertebrate cells. Biochim. Biophys. Acta, 1296, 181–188. [DOI] [PubMed] [Google Scholar]
- Mannervik M., Nibu,Y., Zhang,H. and Levine,M. (1999) Transcriptional coregulators in development. Science, 284, 606–609. [DOI] [PubMed] [Google Scholar]
- Michou A.I., Lehrmann,H., Saltik,M. and Cotten,M. (1999) Mutational analysis of the avian adenovirus CELO, which provides a basis for gene delivery vectors. J. Virol., 73, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L., Kao,H.Y., Chakravarti,D., Lin,R.J., Hassig,C.A., Ayer,D.E., Schreiber,S.L. and Evans,R.M. (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell, 89, 373–380. [DOI] [PubMed] [Google Scholar]
- Naruse Y., Aoki,T., Kojima,T. and Mori,N. (1999) Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl Acad. Sci. USA, 96, 13691–13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R., Townsley,F. and Bienz,M. (2000) The APC tumour suppressor has a nuclear export function. Nature, 406, 1009–1012. [DOI] [PubMed] [Google Scholar]
- Rubin D.C., Swietlicki,E.A., Wang,J.L. and Levin,M.S. (1998) Regulation of PC4/TIS7 expression in adapting remnant intestine after resection. Am. J. Physiol., 275, G506–G513. [DOI] [PubMed] [Google Scholar]
- Tirone F. and Shooter,E.M. (1989) Early gene regulation by nerve growth factor in PC12 cells: induction of an interferon-related gene. Proc. Natl Acad. Sci. USA, 86, 2088–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum B.C., Lim,R.W. and Herschman,H.R. (1989a) Characterization of TIS7, a gene induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene, 4, 1263–1265. [PubMed] [Google Scholar]
- Varnum B.C., Lim,R.W., Kujubu,D.A., Luner,S.J., Kaufman,S.E., Greenberger,J.S., Gasson,J.C. and Herschman,H.R. (1989b) Granulocyte-macrophage colony-stimulating factor and tetradecanoyl phorbol acetate induce a distinct, restricted subset of primary-response TIS genes in both proliferating and terminally differentiated myeloid cells. Mol. Cell. Biol., 9, 3580–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietor I., Bader,T., Paiha,K. and Huber,L.A. (2001) Perturbation of the tight junction permeability barrier by occludin loop peptides activates β-catenin/TCF/LEF-mediated transcription. EMBO rep., 2, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger I.C., Hauf,S., Meinke,A. and Peters,J.M. (2000) Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell, 103, 399–410. [DOI] [PubMed] [Google Scholar]
- Wong C.W. and Privalsky,M.L. (1998) Components of the SMRT co-repressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF–RARα, and BCL-6. J. Biol. Chem., 273, 27695–27702. [DOI] [PubMed] [Google Scholar]
- Yang W.M., Inouye,C., Zeng,Y., Bearss,D. and Seto,E. (1996) Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl Acad. Sci. USA, 93, 12845–12850. [DOI] [PMC free article] [PubMed] [Google Scholar]