Abstract
RNA silencing is a eukaryotic genome defence system that involves processing of double-stranded RNA (dsRNA) into 21–26 nt, short interfering RNA (siRNA). The siRNA mediates suppression of genes corresponding to the dsRNA through targeted RNA degradation. In some plant systems there are additional silencing processes, involving systemic spread of silencing and RNA-directed methylation/transcriptional suppression of homologous genomic DNA. We show here that siRNAs produced in plants from a green fluorescent protein (GFP) transgene are in short (21–22 nt) and long (24–26 nt) size classes, whereas those from endogenous retroelements are only in the long class. Viral suppressors of RNA silencing and mutations in Arabidopsis indicate that these classes of siRNA have different roles. The long siRNA is dispensable for sequence-specific mRNA degradation, but correlates with systemic silencing and methylation of homologous DNA. Conversely, the short siRNA class correlates with mRNA degradation but not with systemic signalling or methylation. These findings reveal an unexpected level of complexity in the RNA silencing pathway in plants that may also apply in animals.
Keywords: DNA methylation/double-stranded RNA/retroelements/RNA silencing/silencing suppressor/systemic signalling
Introduction
In eukaryotic cells, gene silencing operating at the RNA level has roles in adaptive protection against viruses (Voinnet, 2001), in genome defense against mobile DNA elements (Ketting et al., 1999; Wu-Scharf et al., 2000) and in developmental regulation of gene expression (Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001). Many of these silencing systems involve double-stranded RNA (dsRNA) that is generated by host- or virus-encoded RNA-dependent RNA polymerases (Dalmay et al., 2000; Mourrain et al., 2000; Sijen et al., 2001a), by transcription either through inverted repeats (Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001) or from converging promoters (Aravin et al., 2001). Various terms including RNA interference, post-transcriptional gene silencing and quelling have been used to refer to these examples of gene silencing. However, based on genetic and molecular analyses, it seems that their mechanisms share similarities and here we use the generic term ‘RNA silencing’.
A second component of RNA silencing, in addition to dsRNA, is a 21–26 nt RNA known as short interfering RNA (siRNA) (Hamilton and Baulcombe, 1999; Elbashir et al., 2001a). In Drosophila, the siRNA is derived from dsRNA (Zamore et al., 2000) by the action of an RNaseIII-like enzyme named Dicer (Bernstein et al., 2001). The siRNA guides a multi-subunit ribonuclease, referred to as RNA-induced silencing complex (RISC) (Hammond et al., 2000, 2001a; Elbashir et al., 2001a; Nykanen et al., 2001), and ensures that it specifically degrades RNAs that share sequence similarity with the dsRNA. It is thought that the specificity is mediated by base pairing of the siRNA and the target.
In addition to mRNA degradation, RNA silencing in plants acts at several other levels, including DNA methylation and transcriptional suppression (Wassenegger and Pelissier, 1998; Mette et al., 2000; Jones et al., 2001), pre-mRNA processing (Mishra and Handa, 1998) and translation (VanHoudt et al., 1997). Aspects of RNA silencing have also been implicated in translational control in other eukaryotes (Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001). In many of these examples the target nucleic acids share nucleotide sequence similarity with the initial dsRNA trigger, and it is likely therefore that siRNAs are the specificity determinants. However, it is not known whether RISC is involved and it remains possible that different siRNA-containing complexes act at various levels of gene expression.
As well as intracellular RNA silencing, there is also transmission of the silencing state between cells (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Voinnet et al., 1998). In plants, a signal of silencing moves from cell to cell through plasmodesmata and for greater distances through the vascular system. The signal is likely to incorporate a nucleic acid because it mediates a nucleotide sequence-specific effect. A similar signal may exist in Caenorhabditis elegans, where RNA silencing is also non-cell autonomous (Fire et al., 1998; Winston et al., 2002).
One possible role of the extracellular signal of silencing in plants is anti-viral. The signal would move together with, or in advance of the virus, and mediate silencing of the viral RNA in the newly infected cells. Consequently the infection would progress slowly or would be arrested (Voinnet et al., 2000). Many plant viruses produce proteins that suppress RNA silencing in order to counteract this defence mechanism (Anandalakshmi et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998). These proteins share no obvious common structural motifs and appear to act against different stages of the RNA silencing mechanism (Voinnet et al., 1999; Anandalakshmi et al., 2000; Llave et al., 2000; Mallory et al., 2001), including synthesis or movement of the systemic signal (Voinnet et al., 2000; Guo and Ding, 2002).
Here, we characterize siRNAs produced from a transgene and from several endogenous retroelements, and we investigate the effects of viral suppressors of silencing. We show that there are two size classes of siRNAs associated with RNA silencing of a green fluorescent protein (GFP) transgene and that these siRNAs are differentially affected by the viral suppressors of RNA silencing. In contrast, siRNA from retroelements is composed of only the long class. If the abundance of the long siRNA was reduced by viral suppressors or by mutation of the SDE4 Arabidopsis gene, systemic silencing and methylation of genomic DNA were prevented or reduced, whereas degradation of the target RNA was unaffected. Conversely, if the short class was reduced in abundance or was absent, there was no degradation of target RNAs corresponding to the siRNA. Based on these results, we propose that the short siRNA is incorporated into RISC and is involved in degradation of the target RNA. We further propose that the long siRNA plays a separate role that is associated with systemic signalling of RNA silencing and RNA-directed DNA methylation.
Results
Agrobacterium-mediated silencing of GFP in plants
When a liquid culture of Agrobacterium tumefaciens is pressure-injected into leaves, the transferred (T)-DNA of the bacterial Ti plasmid is transferred into plant cells, where transient expression of the T-DNA-encoded genes procedes. Thus, in Nicotiana benthamiana, the ‘agro-infiltration’ of a GFP transgene coupled to a cauliflower mosaic virus 35S promoter (35S) results in strong green fluorescence in the infiltrated zone that contrasts with the surrounding red fluorescence from chlorophyll. GFP fluorescence and GFP mRNA reach peak levels after 2–3 days in the infiltrated patch (Voinnet and Baulcombe, 1997) and then decline as a consequence of RNA silencing activation (Johansen and Carrington, 2001; O.Voinnet, S.Rivas, P.Mestre and D.C.Baulcombe, manuscript submitted). We refer to this phenomenon as ‘local silencing’ of GFP.
When a 35S–GFP transgene is agro-infiltrated into plants that are already transformed with a GFP transgene, the infiltrated patch appears bright green due to the transient GFP expression superimposed on fainter green fluorescence from the resident transgene (Voinnet et al., 1998). As on the non-transformed plants, transient expression of GFP peaks after 2–3 days and then declines. At later times, the tissue becomes uniformly red fluorescent (Voinnet et al., 1998), indicating that the newly infiltrated transgenes and the resident GFP transgene have both become locally silenced. This local silencing precedes ‘systemic silencing’, in which GFP expression is suppressed in newly emerging, non-infiltrated leaves of the GFP transgenic plants (Voinnet et al., 1998).
Two classes of siRNA
Local and systemic silencing of GFP in N.benthamiana is associated with two size classes of 21–25 nt GFP siRNA (Figure 1), corresponding to both sense and antisense strands. In the extracts of local silencing tissue, the longer siRNAs (25 nt) are as abundant as the shorter species (Figure 1B and C, lanes 4 and 8, respectively) whereas in the systemic silencing tissue the shorter siRNAs (21–22 nt) are by far the more abundant species (Figure 1B and C, lanes 3 and 7, respectively). We can rule out that the different size classes of siRNA are artefacts caused by contaminants in the RNA preparations because labelled marker RNA had the same electrophoretic mobility, irrespective of whether it was analysed alone or after mixing with the plant RNA (data not shown).

Fig. 1. Differential accumulation of long and short siRNA in local and systemic GFP silencing. (A) Local silencing (LS) of GFP (left) was induced in leaves of wild-type (WT) N.benthamiana by agro-infiltration of the 35S–GFP construct. Local silencing also occurs in GFP transgenic N.benthamiana (right) and precedes systemic silencing (SS), in which transgene expression is suppressed in new, emerging, non-infiltrated tissues. (B–C) Low molecular weight RNA was hybridized with GFP antisense-specific (B) or sense-specific (C) probes. Lanes ‘SS’ were from systemically silenced upper leaves of GFP transgenic N.benthamiana. ‘5X’ indicates that the amount of RNA loaded in lanes 3 and 7 was 5-fold higher than the amount loaded in lanes 4 and 8. The samples in lanes ‘LS’ were from leaves of wild-type N.benthamiana exhibiting local silencing following infiltration with the 35S–GFP strain of A.tumefaciens. Control samples (lanes ‘NS’) were from non-infiltrated leaves of wild-type N.benthamiana. Both GFP siRNA classes accumulate to similar, high levels in GFP transgenic and wild-type plants (data not shown).
The effect of viral suppressors on local silencing
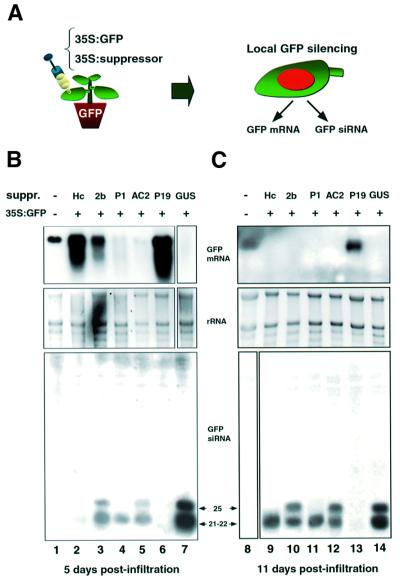
The onset of GFP silencing after agro-infiltration provides a convenient system for induction of siRNA. Furthermore, by infiltrating mixed A.tumefaciens cultures, the effect of viral suppressors on GFP siRNA, mRNA and systemic silencing can be assessed. In such an experiment one of the T-DNA constructs carries the 35S–GFP transgene as an initiator of silencing and the second encodes a viral suppressor (Figure 2A). The cultures are infiltrated into the leaves of GFP transgenic N.benthamiana and the plants are monitored for local and systemic GFP silencing. This approach has been used previously to investigate suppressors of silencing encoded in several viruses (Voinnet et al., 2000; Johansen and Carrington, 2001; Dunoyer et al., 2002; Guo and Ding, 2002).
Fig. 2. The effects of viral suppressors on local RNA silencing. (A) Local silencing was induced in leaves of GFP transgenic N.benthamiana by infiltration of the 35S–GFP strain of Agrobacterium together with a second strain designed to express a viral suppressor of silencing. The onset of local silencing was then monitored and samples were collected for GFP mRNA and siRNA analysis. RNA was extracted from the infiltrated leaves after 5 (B) and 11 days (C). GFP mRNA was detected by hybridization with 32P-labelled GFP cDNA. GFP siRNA was detected by hybridization with labelled GFP sense RNA. The length of the siRNA is indicated as 21–22 nt (short class) and 25 nt (long class). Lanes 2–6 and 9–12 are from plants in which the GFP A.tumefaciens was mixed 1:1 with a second culture expressing a viral suppressor. The suppressors were Hc-Pro (HC, lanes 2 and 9), 2b (lanes 3 and 10), P1 (lanes 4 and 11), AC2 (lanes 5 and 12) and P19 (lanes 6 and 13). Lanes 7 and 14 are from plants in which the second A.tumefaciens culture carried the 35S–GUS transgene construct. Lanes 1 and 8 correspond to non-infiltrated control leaves.
The viral suppressors tested include the P1 protein of rice yellow mottle virus (RYMV), the P19 protein of tomato bushy stunt virus (TBSV), the helper component protease (Hc-Pro) of potato virus Y (PVY), the 2b protein of cucumber mosaic virus (CMV) and the AC2 protein of African cassava mosaic virus (ACMV) (Voinnet et al., 1999). We used an Agrobacterium culture carrying a 35S-β glucuronidase (GUS) transgene as a non-suppressor control. Both GFP mRNA and GFP siRNA levels were monitored at 5 and 11 days post-infiltration (d.p.i.) in three separate experiments.
Figure 2B and C illustrates that both GFP siRNA size classes accumulated in tissues infiltrated with a GFP Agrobacterium strain in the absence of a viral suppressor (lanes 7 and 14, respectively). In contrast, in all GFP combinations with viral suppressor constructs, the accumulation of siRNA was reduced, although to different extents (lanes 2–6 and 8–12). The strongest effect, with P19, resulted in suppressed accumulation of both long and short siRNAs for at least 11 days (lanes 6 and 13). Hc-Pro suppressed both siRNA classes at 5 d.p.i., but by 11 d.p.i. there was accumulation of the smaller class only (lanes 2 and 9). P1 suppressed accumulation of the longer class of siRNA throughout the 11-day duration of the experiments, but caused only a moderate reduction in the shorter class (lanes 4 and 11). AC2 and 2b were the weakest suppressors and caused a similar, moderate reduction of both siRNA classes (lanes 5 and 12 and lanes 3 and 10, respectively).
The GFP mRNA levels were inversely related to the abundance of the short siRNAs. Thus, in the samples without a viral suppressor, agro-infiltration of 35S-GFP induced a high level of short siRNAs, and the GFP mRNA from the stable integrated transgene and from the transiently expressed DNA was below the limit of detection (Figure 2B and C, lanes 7 and 14, respectively). In samples with intermediate levels of the short siRNA due to the 2b, P1 and AC2 suppressors (Figure 2B, lanes 3–5), the GFP mRNA at 5 d.p.i. was at approximately the same level or was less abundant than in the non-silenced controls (Figure 2B, lane 1). However, by 11 d.p.i., with these intermediate suppressors, the GFP mRNA silencing was as strong as in the absence of viral suppressors (Figure 2C, lanes 10–12). The 11 d.p.i. sample with Hc-Pro also represented the intermediate situation in which the short siRNA class was moderately abundant and the GFP mRNA was markedly reduced (Figure 2C, lane 9). The extreme situation, in which short siRNA was reduced to levels that were at or close to the detection limit, was with Hc-Pro at 5 d.p.i. and with p19 at both time points. In these samples (Figure 2B, lanes 2 and 6, and Figure 2C, lane 13), there was strong suppression of silencing and the GFP mRNA was more abundant than in the non-silenced plants (Figure 2B and C, lanes 1 and 8, respectively).
In contrast, there was no obvious relationship between the accumulation of the long siRNA and GFP mRNA levels. Thus, GFP mRNA was suppressed if the long siRNAs were detectable, as in the 2b and AC2 samples (Figure 2B, lanes 3 and 5, and Figure 2C, lanes 10 and 12). The GFP mRNA was also suppressed if the longer siRNAs were not detected, as in the Hc-Pro (11 d.p.i.) and P1 samples (Figure 2B, lane 2, and Figure 2C, lanes 9 and 11). Taken together, these RNA analyses indicate that the short siRNA but not the long siRNA has a role in local silencing.
The effect of viral suppressors on systemic silencing
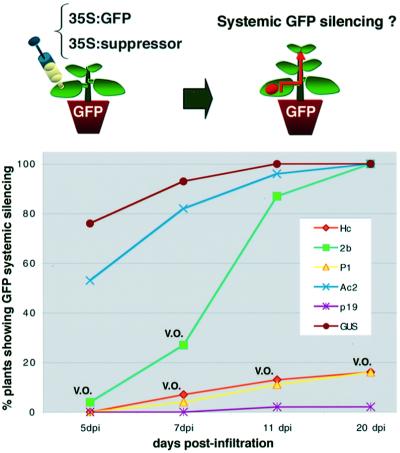
Systemic silencing of GFP in the upper leaves of the infiltrated plants (Figure 3) was strongly inhibited when the 35S–GFP trigger of local silencing was co-expressed with P1, Hc-Pro or P19. It was striking that the suppressors were those affecting production of the longer siRNA (Figure 2B, lane 2, 4 and 6, and Figure 2C, lanes 9, 11 and 13). Conversely, the suppressors that had either a slight (AC2) or moderate (2b) effect on systemic silencing (Figure 3) in our experimental conditions were those with only a slight effect on the level of the long siRNA (Figure 2B, lanes 3 and 5, and Figure 2C, lanes 10 and 12). These results were consistently reproduced in three independent experiments involving 10 plants each. They suggested a correlation between the production of the long GFP siRNA species in the infiltrated patch and the subsequent onset of systemic silencing.
Fig. 3. The effects of viral suppressors on systemic RNA silencing. GFP transgenic N.benthamiana plants were infiltrated with mixed A.tumefaciens cultures as described in Figure 2, and were monitored for the onset of systemic silencing at 5, 7, 11 and 20 d.p.i. For each experimental treatment, a total of 30 individual seedlings (three separate experiments with 10 plants each) were tested. Abbreviations for the viral suppressors are as shown in the legend to Figure 2. V.O., veins only: this indicates that systemic silencing was incomplete and limited to the veins of a few leaves.
To investigate this possible correlation between the longer siRNA and systemic silencing further, we exploited mutant forms of PVX incorporating a GFP gene (PVX–GFP) (Figure 4A). Replication-competent forms of PVX–GFP are able to initiate local silencing of the GFP mRNA and siRNA production in GFP transgenic N.benthamiana. However, only mutant forms of PVX–GFP in which the P25 movement protein gene had been deleted are able to initiate systemic silencing, indicating that P25 is a suppressor of the systemic signal but not of the local silencing triggered by virus replication (Voinnet et al., 2000).
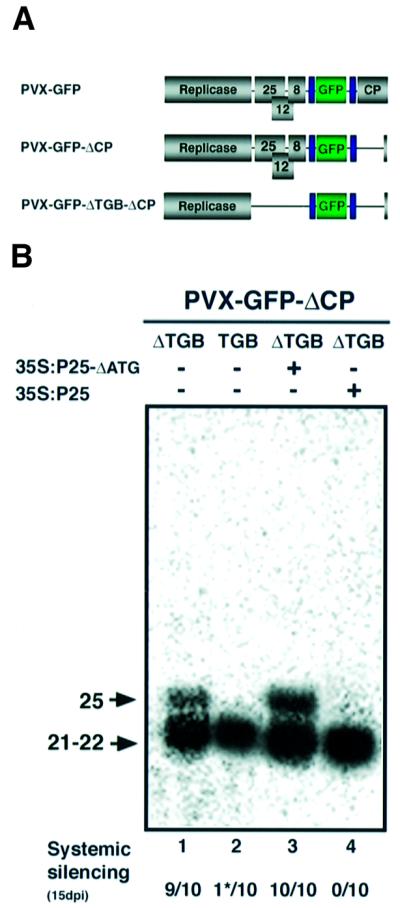
Fig. 4. The effects of PVX P25 on siRNA accumulation and systemic RNA silencing. (A) PVX–GFP has been described previously (Ruiz et al., 1998; Voinnet et al., 2000). Expression of the inserts in the PVX vector is controlled by a duplicated coat protein (CP) promoter, as indicated in blue. The replicase ORF is essential for viral replication; the 25, 12 and 8 kDa proteins are all strictly required for viral cell-to-cell movement and are collectively referred to as triple gene block (TGB) proteins. The CP is essential for encapsidation as well as cell-to-cell and systemic movement. PVX–GFP–ΔCP carries a deletion span ning the entire CP ORF. PVX–GFP–ΔTGB–ΔCP is based on PVX–GFP–ΔCP and carries a deletion spanning the entire TGB (Voinnet et al., 2000). These PVX–GFP mutants were inserted between the 35S promoter and terminator of the pBin61 T-DNA, and viral inocula were provided to plants by Agrobacterium-mediated transient expression of the above-mentioned T-DNAs. (B) Four-week-old seedlings of GFP transgenic N.benthamiana were inoculated with PVX–GFP-based replicons. After 5 days, RNA was extracted from the inoculated leaf and antisense GFP siRNA was assayed as described in Figure 2. The onset and progression of systemic GFP silencing was monitored in 10 plants for each inoculum and the number of plants showing systemic silencing is indicated. The asterisk at the bottom of lane 2 indicates partial systemic silencing that was only restricted to a few leaf veins in this particular plant. The viral inocula were either PVX–GFP–ΔTGB–ΔCP (lanes 1, 3 and 4) or PVX–GFP–ΔCP (lane 2). The 35S–P25–ΔATG (lane 3) and 35S–P25 (lane 4) constructs were transiently co-expressed in the inoculated tissue, as described in Figure 2.
We have now extended these analyses using conditions that, unlike those used previously (Voinnet et al., 2000), allow resolution of long and short siRNAs. We inoculated GFP transgenic N.benthamiana with forms of PVX–GFP that either did or did not encode P25, and monitored production of siRNAs and systemic silencing in the infected plants. The PVX constructs were incapable of moving out of the inoculated cells because they were defective for coat protein (Figure 4A) (Voinnet et al., 2000). Consequently, any systemic silencing would be due to spread of the silencing signal rather than the virus.
The results with these constructs show that there was systemic silencing and long siRNA only if the PVX–GFP had a deletion that included the P25 gene (Figure 4B, lanes 1 and 3). If the P25 gene was included as part of the PVX–GFP genome (Figure 4B, lane 2) or was provided in trans, there was suppression of systemic silencing and the longer siRNA was absent (Figure 4B, lane 4). These data therefore reinforce the association of the long siRNA with systemic silencing. They further suggest that P25 blocks systemic silencing by interfering with production of this long siRNA.
Retrotransposon siRNA
We also investigated the size distribution of endogenous plant siRNAs. Although there are no known natural targets of RNA silencing in plants, it was originally suggested (Flavell, 1994) that the mechanism is a natural defence against transposable elements in plants. This idea is supported by recent molecular and genetic evidence from animals and lower plants (Ketting et al., 1999; Tabara et al., 1999; Wu-Scharf et al., 2000; Aravin et al., 2001). To find out whether plants also have transposon siRNA-like species, we probed extracts of Arabidopsis thaliana and Nicotiana species for siRNAs corresponding to three different retroelements. In each instance, as shown below, we detected RNA of both sense and antisense polarities and of a size similar to transgene siRNA (Figures 5 and 6). However, unlike transgene siRNA, in each case the small RNA corresponded only to the longer size class.
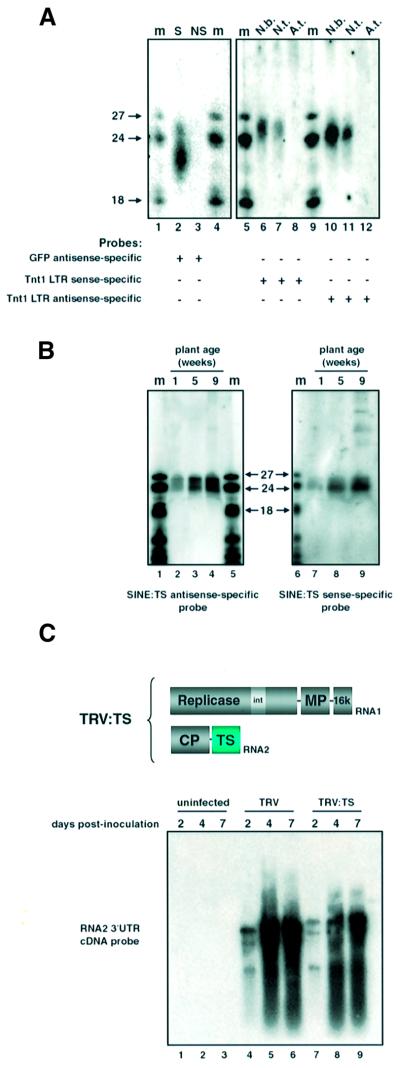
Fig. 5. Retroelement siRNA in Nicotiana sp. (A) Low molecular weight RNA from N.benthamiana (lanes ‘N.b.’); 5-week-old N.tabacum var. Petite Havana (lanes ‘N.t.’) and 6-week-old A.thaliana landrace C24 (lanes ‘A.t.’) were hybridized with 32P-labelled Tnt1 LTR sense or antisense RNA. Samples in lanes ‘S’ and ‘NS’ were as described in Figure 1 and the siRNA was detected by hybridization with a GFP antisense-specific probe. (B) RNA was extracted from the leaves of 1-, 5- and 9-week-old N.tabacum var. Petite Havana. Low molecular weight RNA was hybridized with a 32P-labelled sense or antisense RNA corresponding to a cloned TS SINE. The two panels show hybridization to the same filter stripped of probe between hybridizations. (C) Leaves of N.benthamiana were inoculated with TRV:00 or TRV:TS and total RNA was extracted from the inoculated leaves after 2, 4 and 7 days. RNA was hybridized with a 32P-labelled, 617-bp DNA fragment of TRV RNA2 generated by EcoRI digestion of vector pTV00. This fragment corresponds to the 3′ UTR of RNA2 and thus cross-hybridizes with RNA1. The multiple RNA species hybridizing to the probe are genomic and subgenomic RNAs that are typically associated with TRV infection (Ratcliff et al., 2001). The size shift of the TRV:TS RNA species (lanes 7–9) confirms detection of the recombinant virus.
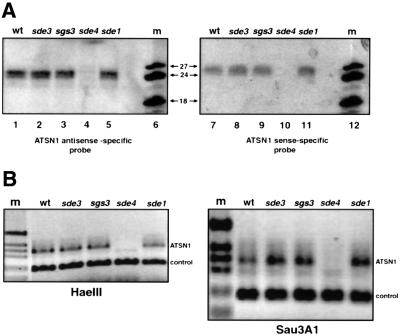
Fig. 6. Retroelement siRNA and DNA methylation in Arabidopsis. (A) RNA was extracted from wild-type and silencing-defective mutants of 6-week-old A.thaliana (landrace C24) seedlings. The mutations were at the sde1/sgs2, sgs3, sde3 and sde4 loci (Dalmay et al., 2000, 2001; Mourrain et al., 2000). AtSN1 siRNA was detected by hybridization with sense RNA transcribed from the cloned AtSN1 sequence. After probe stripping, the filter was reprobed with antisense RNA from the cloned AtSN1 sequence. (B) DNA was extracted from the lines of A.thaliana as in (A) and digested with the methylation-sensitive enzymes HaeIII or Sau3AI. Similar extent of digestion was confirmed by electrophoresis and ethidium staining of digested DNA. Each sample of digested DNA was used as template for PCR with AtSN-specific primers and primers that would amplify an unrelated sequence (to control for variations in efficiency of individual PCR). The AtSN primers would only amplify AtSN DNA if it had remained undigested.
The Tnt1 element in Nicotiana tabacum is an active retrotransposon (Grandbastien et al., 1989). Sense and antisense specific probes from the long terminal repeat (LTR) of this sequence detected 24–26 nt RNA in N.tabacum (var. Samsun) and N.benthamiana (Figure 5A, lanes 6, 7, 10 and 11) but not in A.thaliana (Figure 5A, lanes 8 and 12), which does not harbour elements of the Tnt1 family. These longer siRNAs corresponded, in equal abundance, to both sense and antisense strands (Figure 5A) and were also detected using probes from the Gag gene of Tnt1 (data not shown). We did not detect Tnt1 siRNA of the small class (21–22 nt).
To determine whether siRNA was produced from other retroelements, we used as a probe the TS SINE element of tobacco (Yoshioka et al., 1993). TS siRNAs in N.benthamiana and N.tabacum were detected with both sense and antisense probes corresponding to the entire length of a cloned TS element (Figure 5B). As with the Tnt1 element, these TS siRNAs corresponded only to the 24–26 nt long class. We infer that the TS siRNAs are abundant because they were detected with the same short fluorographic exposure times used to detect GFP siRNA in systemically silenced leaves (Figure 5B).
The siRNAs associated with transgene silencing mediate resistance against virus infection. Thus, N.benthamiana plants exhibiting virus-induced and transgene-induced systemic silencing of a GFP transgene are resistant against recombinant viruses carrying a GFP insert (Voinnet and Baulcombe, 1997; Ruiz et al., 1998). However, the TS siRNAs did not confer resistance to N.benthamiana against recombinant tobacco rattle virus carrying a TS insert (TRV:TS), because this vector was able to accumulate as rapidly and as extensively as the recombinant TRV without an insert (Figure 5C, lanes 4–6 compared with lanes 7–9).
To test whether abundant siRNA production is a general feature of SINEs, we used the AtSN1 of Arabidopsis as a probe. As with the TS SINE, both sense and antisense siRNA (Figure 6A) were found but detection required much longer exposure time than for the tobacco TS element, suggesting that they are less abundant. The strong siRNA signal obtained with the TS probe is more likely a reflection of the high copy number of the TS element [50 000 copies per genome (Yoshioka et al., 1993) compared with 70 copies of the AtSN1 (Myouga et al., 2001)]. RNA blot analysis indicated the presence of sense and antisense AtSN1 siRNAs (Figure 6A) which, as with the Tnt1 and TS elements, were of the longer class. These AtSN1 siRNAs were present in wild-type plants and in sde1/sgs2, sgs3 and sde3 mutants that are defective for transgene silencing (Elmayan et al., 1998; Dalmay et al., 2000) (Figure 6A). However, in an sde4 mutant background, the AtSN1 siRNAs were absent (Figure 6A, lanes 4 and 10). Based on these results it seemed likely that retroelement siRNAs are produced by mechanisms that are similar but not identical to those involved in transgene RNA silencing.
RNA silencing of transgenes and retroelements in plants is often associated with sequence-specific methylation (Ingelbrecht et al., 1994; English et al., 1996; Jones et al., 1999; Mette et al., 2000; Morel et al., 2000; Miura et al., 2001). Because siRNAs are strong candidates for the molecules that direct methylation, we tested whether methylation of an AtSN1 element was affected in the sde4 mutant. The methylation analysis of AtSN1 DNA was carried out by PCR of genomic DNA from wild-type and silencing-defective mutants of A.thaliana. The primers were based on the A.thaliana (landrace Columbia) DNA that flanked an AtSN1 insertion and the PCR was carried out on DNA that had been digested independently with two separate methylation-sensitive enzymes, Sau3A1 and HaeIII. These enzymes cut within the AtSN1 sequence so that if the template DNA was unmethylated, the amount of PCR product would be less than with methylated DNA. As an internal control, the PCR also included a pair of primers for a DNA fragment that did not contain Sau3A1 or HaeIII sites. Figure 6B shows the results of this analysis and confirms that in the sde4 genotype, the AtSN1 DNA was less methylated than in wild-type plants or any of the other silencing-defective mutants tested. Thus, based on this sde4 phenotype, there is a correlation of long AtSN1 siRNA and methylation of the corresponding DNA.
Discussion
Two classes of siRNA in plants
In this paper we demonstrate that the short RNAs associated with transgene RNA silencing are heterogenous in both size and function. Thus, these RNAs are not a single class of ∼25 nt, as reported previously (Hamilton and Baulcombe, 1999), but instead are two distinct species, co-migrating with 21–22 nt and 25 nt markers. The discrepancy between the present and the previous analysis is most likely due to improved electrophoretic resolution, shorter exposure times and the nature of the size markers. We previously used DNA oligonucleotide markers migrating ∼10% faster than the RNA markers used here (Sambrook et al., 1989). We refer to the two classes as short and long siRNAs. However, until we have sequenced these molecules and determined their length, we acknowledge that the apparent size difference may be due to RNA modifications affecting electrophoretic mobility.
In the presence of viral suppressors, the short GFP siRNAs correlated with degradation of the target GFP mRNA (Figure 2). We propose, therefore, that this 21–22 nt siRNA represents the siRNA that guides the RISC ribonuclease to the target of RNA silencing (Zamore et al., 2000; Elbashir et al., 2001a). The long class of siRNA is similar to the short class in that it corresponds to both RNA strands (Figure 1) and, therefore, is probably derived from dsRNA. However, it is unlikely that the two classes of siRNA have the same function because they accumulate differentially in locally and systemically silenced tissue or in the presence of viral suppressor proteins (Figures 1–3). In addition, there are two lines of evidence indicating that the long siRNA is not the guide for RISC. First, in our assay of the P1 and Hc-Pro suppressors, there was silencing of a GFP target RNA in the absence of long GFP siRNA (Figure 2B and C). The second line of evidence is based upon experiments with TRV:TS showing that targeted RNA degradation does not occur in the presence of abundant, long TS siRNA (Figure 5C). Consistent with the idea that the long siRNA is not a guide for RISC, synthetic siRNAs are inactive in an RISC assay if they are longer than 23 nt (Elbashir et al., 2001b).
Our finding that long siRNA is associated with both SDE4-mediated DNA methylation (Figure 6B) and systemic silencing (Figures 3 and 4) could mean that this class of siRNA is directly involved in these processes. For example, it could be that long siRNA is the systemic signal of silencing. This long siRNA molecule could also mediate the nucleotide sequence-specific methylation of DNA often associated with systemic RNA silencing in plants. However, our data could also accommodate the possibility that the long siRNA is either a derivative or precursor of the RNAs species that are directly involved in systemic silencing and RNA-directed DNA methylation.
To assess the precise function of the different classes of siRNA in plants, it will be necessary to establish the ways in which these RNAs differ physically and whether they are produced by the same Dicer activity. The Arabidopsis genome encodes at least four proteins with the helicase, PAZ and dsRNA binding domains that are characteristic of the Drosophila Dicer (Y.Klaue and D.Baulcombe, unpublished data), and one possibility is that different members of this protein family produce the short and long siRNAs. Loss for each of these proteins will allow us to assess this scenario. Loss of the true Dicer function would lead to loss of the primary RNA silencing mechanism. Suppression of Dicer paralogues would leave the primary silencing mechanism intact but may prevent production of the long siRNA. If systemic signalling and DNA methylation are dependent upon long siRNA, the loss of Dicer paralogue function would lead to suppression of these aspects of RNA silencing. Other RNA silencing processes such as amplification and transitivity (Sijen et al., 2001a; Vaistij et al., 2002) may also be dependent on the long siRNA.
RNA silencing and the control of retroelements
Mutations in C.elegans, Drosophila and Chlamydomonas that impair RNA silencing of transgenes or host regulatory genes also result in elevated activity of transposable elements (Ketting et al., 1999; Wu-Scharf et al., 2000; Aravin et al., 2001). Based on these observations, there is now widespread agreement that an important natural function of RNA silencing is to restrict transposons (Plasterk and Ketting, 2000; Sharp and Zamore, 2000; Hammond et al., 2001b). It is somewhat ironic that although RNA silencing was discovered in higher plants, (Napoli et al., 1990), there has been no direct evidence of such a function. The data presented here provide this evidence by demonstrating the presence of siRNA corresponding to three different retrotransposons in Nicotiana spp. and A.thaliana (Figures 5 and 6). It will be interesting to determine whether the sde4 mutant, which does not accumulate siRNA of at least one type of retroelement, has a mutator phenotype like mut7 and other silencing-defective lines of C.elegans.
In C.elegans, there is only partial overlap in the RNA silencing-related mechanisms that control retroelement activity and mediate dsRNA interference. Some mutations suppress both processes whereas others affect only one (Ketting et al., 1999; Tabara et al., 1999). Clearly there are degrees of complexity in RNA silencing that are not apparent from the well studied dsRNA/Dicer/RISC-based process. In plants there is probably a similar degree of complexity because both classes of siRNA are associated with transgene silencing, whereas retrotransposon siRNA belongs only to the long class. Similarly, of the four loci identified in a mutant screen as being required for transgene-induced RNA silencing, only sde4 has any effect on AtSN1 siRNA. Moreover, sgs2 and sde1, -3 and -4 interfered with transgene methylation (Dalmay et al., 2000), but only sde4 interfered with methylation of AtSN1 DNA (Figure 6). Presumably, the classes of siRNA or RNA silencing genes are not involved equally in different branches of the silencing pathway. Some of the complexity may simply be a result of compartmentalization with, for example, RNA turnover degradation in the cytoplasm and DNA methylation in the nucleus (Mette et al., 2001).
Retrotransposon DNA in plants is often methylated and transcriptionally silent (Hirochika et al., 2000; Miura et al., 2001). To account for this situation it is likely, from the data here and from prior analyses of RNA-mediated transcriptional silencing (Mette et al., 2000; Sijen et al., 2001b), that the transcriptional inactivation involves RNA silencing. A double-stranded form of the retroelement RNA would be processed into siRNA and there would be RNA-mediated methylation of the corresponding DNA. There is little information about both the transcriptional status and genomic organization of the three elements we tested, and we cannot predict how the putative dsRNA precursor of the siRNAs arose in each case. One possibility is that an RNA-dependent RNA polymerase converted retrotransposon RNA into double-stranded form. The dsRNA derived in this way would include promoter sequences of the retroelement and could target transcription.
A second model to account for retroelement dsRNA invokes transcription through inverted repeat retroelements. In this second model, transcription would be initiated from either an adjacent host promoter or within the retroelement, and the dsRNA would be produced independently of an RdRP. However, irrespective of the mechanism for retroelement dsRNA production, our data show that RNA silencing has the potential to protect plant genomes against foreign DNAs. Given the abundance of retroelements in many plant genomes, the activity of RNA silencing is likely to have played a major role in determining plant genome size and structure.
Materials and methods
Plant and Agrobacterium material, and growth conditions
Wild-type N.tabacum (var. Samsun and Petite Havana), N.benthamiana and the GFP transgenic 16c line of N.benthamiana were grown as described previously (Marano and Baulcombe, 1998; Voinnet et al., 1998). Arabidopsis thaliana landrace C24 and transgenic and mutant lines derived from this were grown as described previously (Dalmay et al., 2000). The previously described sde2 mutant is allelic with sgs3 (Mourrain et al., 2000) (A.Herr, personal communication). GFP fluorescence was monitored visually under ultra-violet epi-illumination (Voinnet et al., 1998). Recombinant A.tumefaciens strain C58C1 was grown to stationary phase at 29°C in L-broth with 50 µg/ml kanamycin and 5 µg/ml tetracycline, collected by centrifugation (5000 g for 15 min at 20°C) and re-suspended in 10 mM MgCl2 and 150 µg/ml acetosyringone. Cells were left in this medium for 3 h and then infiltrated into the abaxial airspaces of 2- to 4-week-old N.benthamiana plants. All the Agrobacterium strains used harboured the pCH32 helper plasmid (Hamilton et al., 1996) and were used undiluted in the experiments described here, except for the P25 strain which was maintained at an optical density (OD600) of 1.0 to avoid toxicity (Voinnet et al., 2000).
Constructs for transient expression, viral constructs and viral inoculation procedures
35S–GFP, 35S–25K, 35S–25KΔATG, PVX–GFP–ΔCP and PVX–GFP– ΔTGB–ΔCP constructs were described previously (Voinnet et al., 2000). The other suppressor cDNAs were amplified with primer sequences (available on request) from previously described plasmids (Brigneti et al., 1998; Voinnet et al., 1999) and inserted into the SmaI site of pBin61 (Bendahmane et al., 2000), and confirmed by sequencing. The control 35S–GUS construct was from Jonathan Jones (Sainsbury Laboratory). TRV:TS is based on recombinant, bipartite tobacco rattle virus, (Ratcliff et al., 2001). RNA1 encodes the viral replicase, a movement protein (MP) and a 16 kDa protein of unknown function. The replicase open reading frame (ORF) contains an intron (int). RNA2 encodes the viral coat protein (CP) and was modified to carry the TS SINE insert as a 3′ transcriptional fusion. RNA1 and RNA2 share nucleotide sequence homology in their 3′ UTRs. The cloned RNA1 of TRV:TS was inserted between the 35S promoter and the terminator of the pBin61 T-DNA. The cloned TRV:TS RNA2 was inserted between the 35S promoter and the Nos terminator of the pGreen binary vector T-DNA (Hellens et al., 2000). Viral inocula were provided to plants by Agrobacterium-mediated transient co-expression of the above-mentioned T-DNAs.
RNA isolation
Leaves were harvested and frozen in liquid nitrogen, ground to a fine powder and mixed with RNA extraction buffer (4 M guanidine isothiocyanate, 25 mM Na Citrate pH 7, 0.1% Sarkosyl) (Figures 1, 5 and 6). The resulting slurries were extracted twice with phenol/chloroform/isoamylalcohol (equilibrated at pH 8; Sigma). Alternatively (Figures 2 and 4), the frozen leaf powder was mixed with Tri-reagent (Sigma), extracted once with 1/5 volume of chloroform and then once with an equal volume of phenol/chloroform/isoamylalcohol. RNA was precipitated with the addition of an equal volume of isopropanol and incubated at –20°C for 30 min. mRNA and rRNA (high molecular weight RNAs) were precipitated with 10% polyethylene glycol (mol. wt 8000), 0.5 M NaCl (4°C for 30 min) and redissolved in 50% formamide. Low molecular weight RNAs including siRNA were precipitated from the PEG/NaCl supernatant (3 vol. ethanol at –20°C for 2 h) and redissolved in 50% formamide. Relative quantification of the low molecular weight fraction was by ethidium bromide staining.
RNA analysis
High molecular weight RNA was analysed as described previously (Figures 1, 2 and 4). For the experiments shown in Figure 5, high molecular weight RNA in 50% formamide was denatured with 3.7% formaldehyde in 1× TAE and separated by electrophoresis in a 0.8% agarose/1× TAE gel run at 50°C. Analysis of siRNA was as described previously (Hamilton and Baulcombe, 1999). Low molecular weight RNA markers were produced by RNase T1 digestion of 32P-labelled GFP sense in vitro transcripts yielding fragments of 27 nt, 24 nt (×2), 18 nt and 15 nt. Strand-specific hybridization controls were oligonucleotides spotted onto hybridization membranes and hybridized under the same conditions as described for northern blotting. All hybridization signals were detected by phosphorimaging.
Retroelements
Retroelements corresponding to Tnt1 LTR from N.tabacum (var. Samsun), the TS element (Yoshioka et al., 1993) from N.tabacum (var. Samsun) and the AtSN1 sequence of A.thaliana were PCR-amplified and cloned into plasmid vectors. Direct sequence analysis showed that the TntI sequence corresponded to DDBJ/EMBL/GenBank accession No. X13777, that the TS clone corresponded to the TSa subfamily of SINEs (Yoshioka et al., 1993) and that the AtSN1 sequence corresponded most closely to genomic sequence on BAC clone T15B3.
DNA methylation analysis
Genomic DNA from 5-week-old plants (DNeasy plant DNA extraction kit; Qiagen) was digested with HaeIII or Sau3AI (New England Biolabs). The sequence flanking the cloned AtSN1 insertion was amplified by PCR (30 cycles at 95°C for 30 s, at 55°C for 15 s and at 72°C for 30 s) with primers (ACTTAATTAGCACTCAAATTAAACAAAATAAGT; TTTAAACATAARAARAARTTCCTTTTTCATCTAC) from ∼400 ng of this DNA. Internal PCR control primers based on the Arabidopsis (landrace Columbia) sequence At2g19920 (A.Herr, personal communication) span a region lacking HaeIII or Sau3AI sites.
Acknowledgments
Acknowledgements
We thank members of our laboratory for providing valuable comments on aspects of this manuscript, and Alan Herr and Yvonne Klaue for unpublished data. We also thank Mike Hill and his team for excellent plant care. This work was funded by the Gatsby Charitable Foundation, the Biotechnology and Biological Sciences Research Council and from the Royal Society (Dorothy Hodgkin Fellowship to O.V.). The use of genetically modified plant viruses was licensed by DEFRA [license No. PHL 24B/3654 (3/2001)].
References
- Anandalakshmi R., Pruss,G.J., Ge,X., Marathe,R., Smith,T.H. and Vance,V.B. (1998) A viral suppressor of gene silencing in plants. Proc. Natl Acad. Sci. USA, 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi R., Marathe,R., Ge,X., Herr,J.M., Mau,C., Mallory,A., Pruss,G., Bowman,L. and Vance,V.B. (2000) A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science, 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Naumova,N.M., Tulin,A.V., Vagin,V.V., Rozovsky,Y.M. and Gvozdev,V.A. (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol., 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Querci,M., Kanyuka,K. and Baulcombe,D.C. (2000) Agrobacterium transient expression system as a tool for isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J., 21, 73–81. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Brigneti G., Voinnet,O., Li,W.X., Ji,L.H., Ding,S.W. and Baulcombe,D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J., 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dalmay T., Hamilton,A.J., Rudd,S., Angell,S. and Baulcombe,D.C. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay T.D., Horsefield,R., Braunstein,T.H. and Baulcombe,D.C. (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J., 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Pfeffer,S., Fritsch,C., Hemmer,O., Voinnet,O. and Richards,K.E. (2002) Identification, subcellular localization and some properties of a cysteine-rich suppressor of gene silencing encoded by peanut clump virus. Plant J., 29, 555–567. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001a) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001b) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T. et al. (1998) Arabidopsis mutants impaired in cosuppression. Plant Cell, 10, 1747–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J.J., Mueller,E. and Baulcombe,D.C. (1996) Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell, 8, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Flavell R.B. (1994) Inactivation of gene expression in plants as a consequence of specific sequence duplication. Proc. Natl Acad. Sci. USA, 91, 3490–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien M.A., Spielmann,A. and Caboche,M. (1989) TNT1, a mobile retroviral-like transposable element of tobacco isolated by plant-cell genetics. Nature, 337, 376–380. [DOI] [PubMed] [Google Scholar]
- Grishok A. et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Guo H.S. and Ding,S.W. (2002) A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J., 21, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe,D.C. (1999) A novel species of small antisense RNA in post-transcriptional gene silencing. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hamilton C.M., Frary,A., Lewis,C. and Tanksley,S.D. (1996) Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc. Natl Acad. Sci. USA, 93, 9975–9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cell extracts. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001a) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Caudy,A.A. and Hannon,G.J. (2001b) Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet., 2, 110–119. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards,E.A., Leyland,N.R., Bean,S. and Mullineaux,P.M. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacteruim-mediated plant transformation. Plant Mol. Biol., 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Hirochika H., Okamoto,H. and Kakutani,T. (2000) Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell, 12, 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan,J., Pasquinelli,A.E., Balint,E., Tuschl,T. and Zamore,P.D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Ingelbrecht I., Van Houdt,H., Van Montagu,M. and Depicker,A. (1994) Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc. Natl Acad. Sci. USA, 91, 10502–10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.K. and Carrington,J.C. (2001) Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol., 126, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Hamilton,A.J., Voinnet,O., Thomas,C.L., Maule,A.J. and Baulcombe,D.C. (1999) RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell, 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Ratcliff,F. and Baulcombe,D.C. (2001) RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol., 11, 747–757. [DOI] [PubMed] [Google Scholar]
- Kasschau K.D. and Carrington,J.C. (1998) A counterdefensive strategy of plant viruses: suppression of post-transcriptional gene silencing. Cell, 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Ketting R., Haverkamp,T., van Luenen,H. and Plasterk,R. (1999) mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Fischer,S.E.J., Bernstein,E., Sijen,T., Hannon,G.J. and Plasterk,R.H.A. (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C.elegans. Genes Dev., 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Kasschau,K.D. and Carrington,J.C. (2000) Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl Acad. Sci. USA, 97, 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C. et al. (2001) HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell, 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marano M.R. and Baulcombe,D. (1998) Pathogen-derived resistance targeted against the negative-strand RNA of tobacco mosaic virus: RNA strand-specific gene silencing? Plant J., 13, 537–546. [Google Scholar]
- Mette M.F., Aufsatz,W., van der Winden,J., Matzke,M.A. and Matzke,A.J.M. (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J., 19, 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette M.F., Matzke,A.J.M. and Matzke,M.A. (2001) Resistance of RNA-mediated TGS to HC-Pro, a viral suppressor of PTGS, suggests alternative pathways for dsRNA processing. Curr. Biol., 11, 1119–1123. [DOI] [PubMed] [Google Scholar]
- Mishra K.K. and Handa,A.K. (1998) Post-transcriptional silencing of pectin methylesterase gene in transgenic tomato fruits results from impaired pre-mRNA processing. Plant J., 14, 583–592. [Google Scholar]
- Miura A., Yonebayashi,S., Watanabe,K., Toyama,T., Shimada,H. and Kakutani,T. (2001) Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature, 411, 212–214. [DOI] [PubMed] [Google Scholar]
- Morel J.B., Mourrain,P., Beclin,C. and Vaucheret,H. (2000) DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol., 10, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Myouga F., Tsuchimoto,S., Noma,K., Ohtsubo,H. and Ohtsubo,E. (2001) Identification and structural analysis of SINE elements in the Arabidopsis thaliana genome. Genes Genet. Syst., 76, 169–179. [DOI] [PubMed] [Google Scholar]
- Napoli C., Lemieux,C. and Jorgensen,R.A. (1990) Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell, 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A., Haley,B. and Zamore,P.D. (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell, 107, 309–321. [DOI] [PubMed] [Google Scholar]
- Palauqui J.-C., Elmayan,T., Pollien,J.-M. and Vaucheret,H. (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J., 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R.H.A. and Ketting,R.F. (2000) The silence of the genes. Curr. Opin. Genet. Dev., 10, 562–567. [DOI] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez,A.M. and Baulcombe,D.C. (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J., 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Ruiz M.T., Voinnet,O. and Baulcombe,D.C. (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell, 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sharp P.A. and Zamore,P.D. (2000) RNA interference. Science, 287, 2431–2433. [DOI] [PubMed] [Google Scholar]
- Sijen T., Fleenor,J., Simmer,F., Thijssen,K.L., Parrish,S., Timmons,L., Plasterk,R.H.A. and Fire,A. (2001a) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell, 107, 465–476. [DOI] [PubMed] [Google Scholar]
- Sijen T., Vijn,I., Rebocho,A., van Blokland,R., Roelofs,D., Mol,J.N.M. and Kooter,J.M. (2001b) Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol., 11, 436–440. [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian,M., Kelly,W.G., Fleenor,J., Grishok,A., Timmons,L., Fire,A. and Mello,C.C. (1999) The rde-1 gene, RNA interference and transposon silencing in C. elegans. Cell, 99, 123–132. [DOI] [PubMed] [Google Scholar]
- Vaistij F.E., Jones,L. and Baulcombe,D.C. (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell, 14, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanHoudt H., Ingelbrecht,I., VanMontagu,M. and Depicker,A. (1997) Post-transcriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation of 3′ flanking regions. Plant J., 12, 379–392. [Google Scholar]
- Voinnet O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet., 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet O. and Baulcombe,D.C. (1997) Systemic signalling in gene silencing. Nature, 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Vain,P., Angell,S. and Baulcombe,D.C. (1998) Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto,Y.M. and Baulcombe,D.C. (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses. Proc. Natl Acad. Sci. USA, 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Lederer,C. and Baulcombe,D.C. (2000) A viral movement protein prevents systemic spread of the gene silencing signal in Nicotiana benthamiana. Cell, 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Wassenegger M. and Pelissier,T. (1998) A model for RNA-mediated gene silencing in higher plants. Plant Mol. Biol., 37, 349–362. [DOI] [PubMed] [Google Scholar]
- Winston W.M., Molodowitch,C. and Hunter,C.P. (2002) Systemic RNA in C.elegans requires the putative transmembrane protein SID-1. Science, 295, 2456–2459. [DOI] [PubMed] [Google Scholar]
- Wu-Scharf D., Jeong,B.-R., Zhang,C. and Cerutti,H. (2000) Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-Box RNA helicase. Science, 290, 1159–1162. [DOI] [PubMed] [Google Scholar]
- Yoshioka Y., Matsumoto,S., Kojima,S., Ohshima,K., Okada,N. and Machida,Y. (1993) Molecular characterisation of a short interspersed repetitive element from tobacco that exhibits sequence homology to specific transfer-RNAS. Proc. Natl Acad. Sci. USA, 90, 6562–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]