Significance
The sequence of amino acids within a protein dictates its structure and function. Protein engineering campaigns seek to discover protein sequences with desired functions. Data-driven models of the sequence–function relationship can be used to guide and accelerate this process. In this work, we combine two paradigms in deep generative modeling—transformer-based protein language models and variational autoencoders—to introduce the Protein Transformer Variational AutoEncoder (ProT-VAE) as an accurate, generative, fast, and transferable model for data-driven protein design. In experimental testing of the ProT-VAE designs, we identify a phenylalanine hydroxylase enzyme with a 2.5× elevation in catalytic activity and a γ-carbonic anhydrase enzyme with a 61 °C elevation in melting temperature.
Keywords: protein design, transformers, protein language models, variational autoencoders, generative modeling
Abstract
Deep generative models have demonstrated success in learning the protein sequence to function relationship and designing synthetic sequences with engineered functionality. We introduce the Protein Transformer Variational AutoEncoder (ProT-VAE) as an accurate, generative, fast, and transferable model for data-driven protein design that blends the merits of variational autoencoders to learn interpretable, low-dimensional latent embeddings for conditional sequence design with the expressive, alignment-free featurization offered by transformer-based protein language models. We implement the model using NVIDIA’s BioNeMo framework and validate its performance in retrospective functional prediction and prospective functional design. The model identifies a phenylalanine hydroxylase enzyme with 2.5× catalytic activity over wild-type, and a γ-carbonic anhydrase enzyme with a melting temperature elevation of ΔTm = +61 °C relative to the most thermostable sequence reported to date and activity in 23% v/v methyl diethanolamine at pH 11.25 and 93°C corresponding to industrially relevant conditions for enzymatic carbon capture technologies. The ProT-VAE model presents a powerful and experimentally validated platform for machine learning-guided directed evolution campaigns to discover synthetic proteins with engineered function.
Proteins are molecular machines that are the workhorses of biology. The ability to design synthetic sequences with engineered functionalities is a long-standing goal of synthetic biology with enormous potential in multiple fields including medicine, public health, biochemical engineering, and clean energy. Rational design of protein sequences with programmed function requires models of the sequence–function (i.e., genotype–phenotype) relationship as a means to guide generation of candidate sequences with the desired functionality for experimental synthesis and testing (1, 2). Historically, the sequence–structure relationship has frequently been adopted as a proxy for the sequence–function relationship (3), and a number of powerful approaches exploiting modern tools such as equivariant neural networks and diffusion models have been deployed to engineer desired three-dimensional protein structures (2, 4, 5). More recently, approaches employing techniques such as recurrent neural networks (6, 7), variational autoencoders (8–14), generative adversarial networks (15), reinforcement learning (16), and transformers (17–24) have been developed to learn the sequence–function relationship and have demonstrated remarkable performance in functional prediction tasks such as fluorescence, stability, and epistasis (17, 25).
A protein design model should possess four key characteristics: i) accurate learning of the sequence–function relationship, ii) generative design of sequences under this learned mapping, iii) fast and transferable model training, and iv) the capacity for unsupervised training over unlabeled sequence data and semisupervised retraining over labeled data. The first and second properties require a sufficiently expressive and powerful model to learn the correlated patterns of amino acid mutations (i.e. the “syntax”) underpinning the sequence–function relationship and permit design of synthetic sequences consistent with these learned patterns. The third property is important to enable efficient training of large and expensive neural network models and amortization of training costs via transferability to multiple protein families. The fourth property is germane to protein engineering applications where the vast size of protein sequence space and time and labor costs of experimental assays mean that labeled data points (i.e., sequences annotated with experimental measurements) tend to be eclipsed by unlabeled data. Based on these desiderata, a number of deep generative model architectures have been employed for data-driven protein design. Two approaches in particular have received substantial attention: variational autoencoders (VAEs) (8, 9, 13, 14, 26) and transformer-based models (17–24, 27). VAEs comprise encoding and decoding neural networks that compress and decompress the high-dimensional sequence data into a low-dimensional latent space exposing ancestral and functional relationships and can be used to guide conditional generative sequence generation (8, 13, 26, 28–33). VAEs typically necessitate that sequences be provided as fixed length vectors within multiple sequence alignments (MSAs) (34) that can be laborious to construct, introduce bias, and limit applications to homologous families. This limitation can be alleviated through the use of convolutional or recurrent layers (10, 26), but this can present challenges in learning long-range mutational correlations. Transformers use the attention mechanism to learn many-body and long-range correlated patterns by self-supervised training (35) and underpin a number of protein language models (pLMs) for protein functional prediction and design (18, 20, 22, 23, 27, 36). The ability to train over variable length sequences has enabled training of generic pLMs over millions or billions of nonhomologous protein sequences residing in large public databases such as UniProt (37) and BFD (38, 39). Sequence generation can be conditioned on control characters or partial sequences to guide synthetic protein design (20, 36). The high dimensionality of the fixed-length latent space of typical transformer-based pLMs, however, sacrifices easy interpretability of phylogeny and functional patterns and frustrates conditional generative design of synthetic proteins with tightly controlled functionality.
In this work, we introduce the Protein Transformer Variational AutoEncoder (ProT-VAE) as a model that blends the relative merits of VAEs and transformers to achieve all four of the desired criteria above by sandwiching a VAE between the encoder and decoder stacks of a transformer pLM in order to compress and decompress the fixed-length internal representation of the transformer into a low-dimensional latent space. We construct and train the ProT-VAE model using the NVIDIA BioNeMo framework and demonstrate its capacity as a powerful, extensible, and lightweight model for data-driven protein design in retrospective computational prediction tasks and prospective design of synthetic functional proteins for experimental synthesis and testing.
Results
Protein Transformer Variational AutoEncoder (ProT-VAE).
Prior art.
Wang and Wan developed the Transformer-Based Conditioned Variational Autoencoder wherein a VAE was used to learn a distribution over story plots and serve as a conditioning variable for the transformer decoder in story completion tasks (40). Jiang et al. developed the Transformed Variational AutoEncoder by combining the Music Transformer and Deep Music Analogy to develop a model capable of learning long-range dependencies within musical melodies, furnish interpretable embeddings via a disentangling conditional VAE, and a means to transfer melody and rhythm between contexts (41). Li et al. and Park and Lee employed, respectively, pretraining and fine-tuning approaches to mitigate posterior collapse in a transformer VAE model for text (42, 43). Arroyo, Postels, and Tombari proposed the Variational Transformer Network as a synthesis of self-attention encoders and decoders within a VAE architecture for layout detection and generation (44). Henderson and Fehr employed VAEs as an information bottleneck regularizer for transformer embeddings and used this model to embed and generate text within a nonparametric space of mixture distributions (45). The present work is most closely related to the recent work of Castro et al., who introduced the Regularized Latent Space Optimization (ReLSO) approach for data-driven protein engineering (32). The jointly trained autoencoder architecture underpinning this approach comprises a transformer encoder, low-dimensional projection into a latent space bottleneck, 1D convolutional neural network decoder, and fully connected network to predict function from the latent space embedding. The ProT-VAE shares similarities with the ReLSO approach in the use of a transformer-based featurization and subsequent compression of this encoding into a low-dimensional latent space but is distinguished by its use of an attention-based decoder stack for sequence generation to efficiently learn long-range correlations, its use of a transferable encoder and decoder such that the lightweight VAE is the only model component that requires retraining for each protein engineering task, the capacity for unsupervised/semisupervised training that does not require all sequences to have attendant experimental measurements, and validation in wet lab testing of synthetic protein sequences.
ProT-VAE.
A schematic of the ProT-VAE architecture is presented in Fig. 1. The architecture comprises three nested components. The exterior component is a pretrained ProtT5nv transformer-based T5 encoder and decoder available within the NVIDIA BioNeMo framework that is currently generally available and planned for future open source release (https://www.nvidia.com/en-us/clara/bionemo/) (46, 47). The intermediate block is a compression/decompression block that compresses the ∼300,000-dimensional ProtT5nv hidden state into a more parsimonious 32,768-dimensional intermediate-level representation. Similar to the ProT5nv block, these intermediate layers are also pretrained on large protein databases. The compression block comprises 1 × 1 convolutions, LayerNorm, and GeLU activations over three layers with filter sizes of 512, 256, and 64. The decompression block mirrors the compression block but with filters of size 256, 512, and 768. Empirically, we find reductions of 16× or more in the size of the ProtT5nv hidden state are possible without any noticeable degradation of reconstruction quality. The innermost block is a three layer fully connected maximum mean discrepancy variational autoencoder (MMD-VAE) employing ReLU activations (48) that takes the flattened output of the compression block and further compresses it into a protein family-specific, low-dimensional latent space before decompressing it and passing its output to the decompression block. The VAE is a lightweight network that is trained anew for each particular homologous protein family. The dimensionality of the latent space is a key hyperparameter of the model. Training of the VAE is fast and is the only nontransferable component of the architecture. We note that the model takes as input only protein sequence data and is trained in an unsupervised manner that does not appeal to any labels on the sequence data (e.g., functional activity, environmental conditions). This is advantageous in scaling training to large unlabeled sequence databases but does necessitate that generative sequence design is limited to producing sequences similar to the training data without the provision for more specific guided design toward specific structure, function, or environmental conditions. Full details of model architecture and training are provided in Materials and Methods.
Fig. 1.

Overview of the ProT-VAE model architecture comprising three nested components and how the model can be used for embedding and generation. (A) A pretrained ProtT5nv transformer-based T5 encoder and decoder defines the exterior component. This is followed by a pretrained compression/decompression block that compresses the ∼300,000-dimensional ProtT5nv hidden state into a 32,768-dimensional intermediate-level representation using a series of stacked 1 × 1 convolutions. The innermost component is a protein family-specific VAE that reduces the intermediate-level representation into a low-dimensional (typically <10-dimensional) information bottleneck. The ProtT5nv and convolutional compression/decompression components are trained on large libraries comprising millions of protein sequences whereas the VAE is trained on specific protein families for each particular design task. After pretraining, the T5 encoder/decoder blocks are frozen during the training of the global dimensionality reduction and family-specific encoder–decoders. (B) The outputs of the family specific encoder define an interpretable latent space suitable for extraction of comprehensible patterns of phylogeny and function for conditioning the generative decoding of synthetic protein sequences. (C) In turn, the VAEs furnish low-dimensional latent embeddings to guide generative sequence design and which can be quickly and iteratively retrained in an unsupervised or semisupervised fashion. Essentially, the ProT-VAE combines the properties of transformers as generic, transferable, and powerful featurizers capable of learning long-range correlations and operating on variable length sequence data, with the capacity of VAEs to furnish low-dimensional latent embeddings to guide generative sequence design.
Validation systems.
We test the capabilities of ProT-VAE in applications to three proteins. We first conduct retrospective functional and phylogenetic analyses of the learned latent space of the Src homology 3 (SH3) protein family involved in diverse signaling functions. We then perform prospective design of two synthetic enzymes: phenylalanine hydroxylase (PAH), which catalyzes conversion of phenylalanine to tyrosine, and β- and γ-carbonic anhydrases (CA), which catalyze the conversion of carbon dioxide and water into carbonic acid. In each case, we test the capability of the ProT-VAE model to learn meaningful and interpretable latent spaces organizing protein sequences by ancestry and function, make accurate predictions of protein function from the learned latent space, and, for PAH and CA, prospectively design and experimentally test synthetic sequences with measured function commensurate or superior to natural sequences.
SH3.
The SH3 is a family of small β folds that mediate protein signaling within cells by binding to type II poly-proline peptides with sequences N-R/KXXPXXP-C or N-XPXXPXR/K-C (49, 50). SH3 domains have evolved to perform a variety of functions within various organisms by evolving differential binding specificities, resulting in a number of distinct paralogs (i.e., homologous proteins performing different functions within the same species) within the SH3 family. Recent work by Lian et al. trained VAEs over an MSA of ∼5,300 SH3 homologs to develop a deep generative model for synthetic SH3 design (13). The VAE learned an unsupervised 3D latent space embedding in which the natural sequences demonstrated an emergent hierarchical clustering by phylogeny and function. The Sho1SH3 domain in Saccharomyces cerevisiae (baker’s yeast) mediates transduction of an osmotic stress signal by binding a Pbs2 ligand that activates a homeostatic response to balance the osmotic pressure by intracellular production of glycerol (51). A high-throughput in vivo osmosensing assay was developed to measure the relative enrichment of deep sequencing counts of S. cerevisiae Sho1SH3 knockouts into which mutant SH3 genes designed by the VAE were transformed. The normalized relative enrichment (r.e.) score has been shown to quantitatively report on the binding free energy of the mutant SH3 with the Pbs2 ligand. The assay demonstrated that natural Sho1SH3 orthologs reside within a localized cluster within the VAE latent space and generative design of mutant sequences in the vicinity of this cluster conferred equal or superior high osmolarity protection to wild type Sho1SH3, which, by construction, possesses a normalized r.e. score of unity.
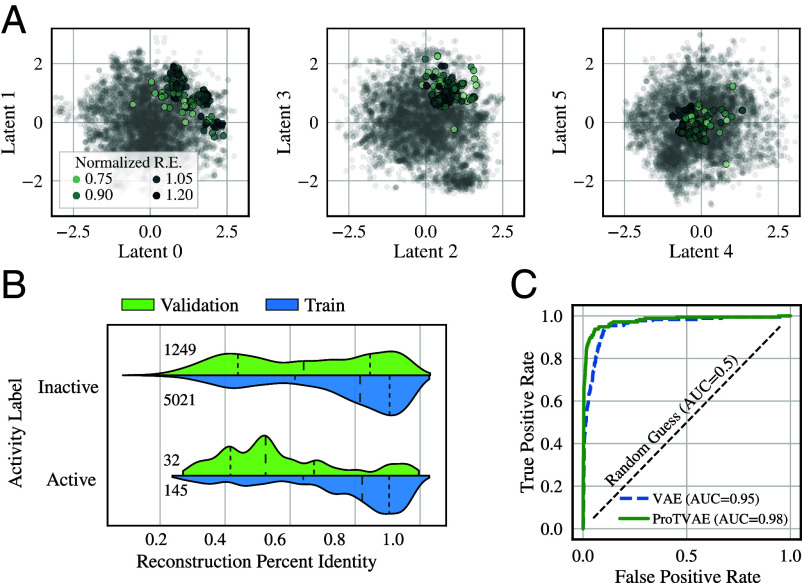
We first test the capacity of the ProT-VAE model to learn a latent space representation capable of accurate prediction of the experimental measurements of SH3 activity reported in ref. 13. To do so, we fine-tuned the inner two VAE layers of the ProT-VAE model on this SH3 dataset employing a 6D latent space. We present in Fig. 2A a series of 2D projections of the latent space highlighting those sequences with high normalized r.e. scores (i.e., high osmosensing activity). In Fig. 2B, we test the generative capacity of the trained model by passing each training and validation sequence through the model and calculating the percentage identity of each decoded sequence with the original sequence. For each of the three proteins considered in this work, we apply a similarity cutoff range of 20% to 80% to all sequences recovered during an iterative BLAST search. We then perform an 85–15% random split into training and validation sets. This approach ensures that each split contains unique, diverse sequences, allowing for proper model evaluation during training. The reconstruction accuracy, particularly of the validation set, is relatively poor, but we suggest that this may be attributable to high variability in the training data in amino acid positions not relevant to osmosensing function. A more important test of the model for the purposes of data-driven protein engineering is its ability to predict functional activity based on location in the latent space. To assess this predictive capacity, we stratified sequences into a binary dataset of high (normalized r.e. > 0.6) and low activity, and trained a logistic classifier to predict these binary labels based on location in the 6D latent space. The predictions of the trained classifier over the validation partition are presented in Fig. 2C, where we present the receiver operating characteristic (ROC) curve. The ProT-VAE slightly outperforms the MSA-based VAE model in predicting functional performance with an area under the ROC curve (AUC) of 0.98 compared to 0.95. The high predictive accuracy of the trained ProT-VAE model demonstrates that protein activity is strongly localized within the latent space such that the trained model can accurately predict activity based on latent space location despite a relatively poor reconstruction accuracy and, contrary to the VAE, can do so without the requirement for an MSA that can be laborious to construct, introduce bias, and limit applications to homologous families.
Fig. 2.
ProT-VAE organizes the SH3 family by functional activity without an MSA. (A) Two-dimensional projections of the latent space are shown colored by normalized relative enrichment (r.e.), where darker points correspond to more active sequences. Note the clustering of these sequences in all three projections. (B) Reconstruction percent identity of SH3 sequences stratified by high (normalized r.e. > 0.6) and low activity. The distributions for each classification are represented by violin plots separated into those in the training set (blue) and validation set (green). The number of sequences within each split are printed on the Left of the plot. Dashed lines correspond to the 25%, median, and 75% quartile ranges. (C) Receiver operating characteristic (ROC) curves for a logistic classifier predictions of the binary activity labels as a function of the 6D latent space coordinates trained on the network training data via fivefold cross validation and evaluated on the validation data. The green solid line corresponds to the ProT-VAE model, the blue dashed line corresponds to the previous MSA-based VAE model, and the black dashed line is the null hypothesis. In the legend, we report the area under the ROC curve (AUC) scores for each classifier.
We hypothesized that the observed localization of osmosensing function may be due to a learned clustering of the various SH3 paralog groups within the latent space. Accordingly, we next assess the degree to which the trained ProT-VAE model learns to separate sequences according to phylogenetic ancestry within the learned latent space (8, 13). Lian et al. previously observed a hierarchical nesting of phylogeny and function for the SH3 family within the latent space of their MSA-based VAE: the latent space first separates by paralog group (i.e., function), and then by phylogeny within each paralog cluster (13). We seek to determine whether we also observe these trends within the trained ProT-VAE model. In Fig. 3A, we demonstrate that the ProT-VAE model is unable to visually separate phylogeny between Ascomycota and Basidiomycota in any latent dimension. In Fig. 3B, we show that there is clear clustering of the paralog groups of the SH3 family according to the four annotated paralog groups Abp1, Rvs167, Sho1, and Bzz1. In Fig. 3C, we illustrate that we now do observe improved separation between Ascomycota and Basidiomycota within the Sho1 paralog cluster. In SI Appendix, Fig. S1, we show that the model achieves similar phylogenetic separations for the other three paralog groups. The capacity of ProT-VAE to learn functional and phylogenetic separation within the latent space without the need for MSAs demonstrates its capacity to learn the underlying correlated patterns of amino acid mutations and is a prerequisite for MSA-free generative design of synthetic proteins.
Fig. 3.
The ProT-VAE latent space hierarchically organizes the SH3 family first by paralog group and then by phylogeny. (A) Two-dimensional projections of the latent space are shown colored by two phylogenetic groups, Ascomycota (blue) and Basidiomycota (orange) with no apparent organization. (B) Two-dimensional projections of the latent space colored by paralog groups—Abp1 in blue, Rvs167 in orange, Sho1 in green, and Bzz1 in yellow—exhibit strong clustering. (C) Considering the Sho1 paralog group (green), we now additionally distinguish by phylogeny—Ascomycota as circles and Basidomycota as triangles—and observe a hierarchically nested separation according to phylogeny within this paralog cluster. Analogous plots for the other three paralog clusters are presented in SI Appendix, Fig. S1.
PAH.
PAH is a member of the family of aromatic amino acid hydroxylases (AAAH). Human PAH (hPAH) is an enzyme that catalyzes the catabolism of one amino acid, phenylalanine, into another, tyrosine, by hydroxylation of the Phe side chain (52). This reaction is critical in eliminating surplus phenylalanine and producing tyrosine as an essential precursor for the biosynthesis of hormones, neurotransmitters, and pigments. Starting from a human PAH variant, 2PAH (53), we conducted a psiBLAST search over the NCBI nr database (54) to collate a dataset of 20,000 homologous sequences for training of the inner VAE of the ProT-VAE with a 6D latent space.
We present our analysis of the trained ProT-VAE model in Fig. 4. Inspection of the learned latent space reveals good separation and clustering of the primary functional substrates of the AAAH family (Fig. 4A). Again, the inference of phylogenetic and functional relationships within a learned latent space demonstrates that the model is learning the correlated patterns of amino acid mutations underpinning the sequence–function relationship as a prerequisite to subsequent data-driven functional protein design. To test the generative capacity of the model, we passed the training and validation sequences through the trained model and calculated the percentage identity of each decoded sequence with the original sequence (Fig. 4C), and, in this case, we observe high median reconstruction accuracies across all four substrates. Excellent parity between the distributions for the training and validation indicate that the model is not overfit. A k = 5-nearest neighbors classifier trained to predict substrate specificity as a function of location of a sequence within the 6D latent space is capable of highly accurate predictions (Fig. 4E).
Fig. 4.

ProT-VAE organizes the AAAH family by substrate specificity and phylogeny within the latent space. Two-dimensional projections of the latent space colored by (A) substrate specificity and (B) phylogeny. Reconstruction accuracy of AAAH family sequences stratified by (C) substrate specificity and (D) phylogeny. The distributions for each classification are represented by violin plots separated into those in the training set (blue) and validation set (green). The number of sequences within each split are printed in the Left of the plot. Dashed lines correspond to the 25%, median, and 75% quartile ranges. (E) Prediction of functionality and (F) phylogeny via latent space trained classification model. A k = 5-nearest neighbors classifier was trained on the training data and evaluated in predicting the labels for the validation set based on the 6D latent space coordinates. The confusion matrices demonstrate that substrate specificity and phylogeny are localized in the latent space. (G) Two-dimensional projections of spherical linear interpolation (SLERP) paths in the latent space to test its smoothness and suitability for optimization and design. A substrate specificity path (black) traverses between a human PAH (hPAH) represented by a white star and a human tyrosine hydroxylase (hTyrH) represented by a black star, and a phylogeny path (red) traverses between the same hPAH and a bacterial PAH (bacPAH) represented by a red star. Illustration of the fractional similarity to the hPAH starting sequence for (H) the substrate specificity path traversing between hPAH and hTyrH and (I) the phylogeny path traversing between hPAH and bacPAH. The nodes of the paths are not constructed to coincide with the embedding of any existing sequences in the latent space and so typically decode to non-natural sequences. We present AlphaFold (4) predicted structures for the sequence residing at the approximate inflection point of the path (purple) aligned to the structure of the hPAH at the beginning of the path (blue) and hTyrH or bacPAH at the end of the path (red).
Annotation of the PAH latent space by phylum demonstrates that the learned latent space is also well clustered by sequence phylogeny and, similar to the substrate specificity results, we observe good clustering of the top phylum labels (Fig. 4B) and high reconstruction accuracy over all classes, despite a substantial imbalance across class labels (Fig. 4D). A second k = 5-nearest neighbors classifier trained over the phylum labels also achieves high prediction accuracy (Fig. 4F).
A hallmark of a latent space suitable for optimization and conditional generative design is sufficient smoothness for interpolative sequence design. We present in Fig. 4G an illustration of two interpolative pathways: one between sequences possessing similar phylogeny but different substrates, and one between different phylogenies acting on the same substrate. The substrate path was traversed by interpolating 50 points using spherical linear interpolation (SLERP) (55) between the 2PAH human PAH (hPAH) and a human tyrosine hydroxylase (hTyrH), while the phylogeny path was interpolated between the same hPAH and a flavobacteriaceae PAH sequence (bacPAH). By decoding the sequences at each node in the pathway, we find that each path executes a smooth transition covering a range of ∼40% in the fractional sequence similarity for the substrate path (Fig. 4H) and ∼80% for the phylogeny path (Fig. 4I).
Taken together, these results indicate that the ProT-VAE has learned a smooth and interpolatable latent space embedding that organizes and localizes sequences by both substrate specificity and phylogeny, and suggests that the latent space can be used to condition generation of synthetic sequences with desirable functional properties.
We now experimentally test the PAH sequences generatively designed by the trained ProT-VAE. In Fig. 5A, we present PAH latent space embeddings annotated by experimentally measured fold-over-wild-type (FOWT) activities relative to hPAH generatively designed sequences in the vicinity of hPAH in the latent space (SI Appendix). Of the 190 proteins tested, 69 (36%) showed activity and 19 (10%) were more active than hPAH, with a maximum measured activity of 2.5× that of hPAH. In Fig. 5B, we illustrate the FOWT activity of the designed sequences as a function of their mutational distance from hPAH, and in SI Appendix, Fig. S2, we present an analogous plot as a function of their mutations from the nearest natural sequence. The ProT-VAE model has designed a number of highly active sequences relatively similar to the hPAH (<30/333 mutations) but also a number of highly mutated sequences (>100/333 mutations) with measurable activity. AlphaFold (4) predicted structures for our highest activity design (2.5× FOWT, 19 mutations) and the most sequence divergent design (0.2× FOWT, 130 mutations) suggest that the ProT-VAE model has learned to preserve the near native fold of the wild-type hPAH despite not being furnished any structural information.
Fig. 5.
Latent-conditioned generative design of synthetic PAH sequences. (A) Latent space projections of the ∼20,000 training sequences (gray) and hPAH (gold star). The colored points represent the 190 latent vectors sampled in the vicinity of hPAH and passed for generative decoding into synthetic PAH sequences for gene assembly and experimental testing. The color of the points indicates the measured fold-over-wild-type (FOWT) activity. Black points are inactive sequences. (B) Activity distribution of the designed sequences as a function of mutational distance from hPAH. The wild-type hPAH contains 333 amino acid positions. We present AlphaFold (4) predicted structures for the (C) highest activity design (2.5× FOWT, 19 mutations from hPAH; tan) and the (D) most sequence divergent functional design (0.2× FOWT, 130 mutations from hPAH; green) both alone and aligned to the crystal structure of the wild-type hPAH (blue).
CA.
We next consider the design of synthetic CA enzymes that reversibly catalyze the interconversion of carbon dioxide to bicarbonate. Five nonhomologous CA families—α, β, γ, δ, and ζ—have emerged through convergent evolution (56–58) and are the subject of industrial interest for biochemical carbon capture and storage (CCS) technologies (59). The design challenge is to engineer high thermal and chemical stability required for the enzyme to tolerate CCS operating conditions without compromising activity. Commencing from a natural β-CA [PDB: 1G5C (60)], and γ-CA [PDB: 1THJ (61)], we conducted a psiBLAST search over the NCBI nr database (54) to collect, respectively, 73,458 and 73,045 homologous training sequences. These training data were used to train β-CA and γ-CA-specific interior VAE models employing 4D latent spaces for each model.
We present in Fig. 6 the learned ProT-VAE latent space embeddings for β and γ-CA. In each case, we observe good clustering with respect to phylogeny, with the latent space showing a clear localization of the four leading phyla (Fig. 6 A and B). As was the case for PAH, the sequence identity of the decoded sequences shows good correspondence between the training and validation datasets (Fig. 6 C and D). For β-CA, a k = 5-nearest neighbor classifier exhibits high predictive accuracy in phylum prediction indicating good localization and organization within the learned latent space (Fig. 6E). In γ-CA, a catalytic triad of histidines separated by approximately 20 amino acids in primary structure is required for function (62). A random forest model exhibits high accuracy prediction (AUC = 0.98) of the presence or absence of this triad as a function of location in the learned latent space (Fig. 6F). This not only allows discrimination of sequences possessing the motif from those that do not, but it also enables us to condition generative sequence design from regions where we have very high confidence that the motif will be included in the artificial sequence and elevate our anticipated yield of functional sequences.
Fig. 6.
ProT-VAE organizes the β and γ-CA families in latent space. Two-dimensional projections of the (A) β-CA and (B) γ-CA latent spaces colored by phylogenetic labels. (C) β-CA and (D) γ-CA model sequence reconstructions by phylum. The distributions for each classification are represented by violin plots separated into those in the training set (blue) and validation set (green). The number of sequences within each classification and dataset split are printed in the Left of the plot. Dashed lines correspond to the 25%, median, and 75% quartile ranges moving from Left to Right. (E) Confusion matrix predictions of β-CA phylogenetic labels based on the 4D latent space coordinates under a k = 5-nearest neighbors classifier trained on the network training data and evaluated on network validation data. (F) ROC curve for a random forest classifier predictions of the presence or absence of a γ-CA histidine catalytic triad as a function of the 4D latent space coordinates trained on the network training data and evaluated on network validation data. The training set contained 58,762 sequences with the triad and 3,326 sequences without; the validation set contained 10,392 sequences with the triad and 565 sequences without. The trained classifier possesses an AUC = 0.98.
We experimentally verified the activity and stability of synthetic β- and γ-CA sequences designed by the trained ProT-VAE models (SI Appendix). Of 88 synthetic β-CA sequences, 35 (40%) expressed in our Escherichia coli expression system, and 20 (23%) exhibited measurable catalytic activity. Of 88 synthetic γ-CA sequences, 22 (25%) expressed in our E. coli expression system, and all of these demonstrated measurable activity, which we attribute to our targeted latent space design strategy that conditions presence of the catalytic motif. A number of these designed β- and γ-CA sequences have remarkably high thermostability, possessing melting temperatures of 101 °C for the most stable synthetic β-CA and 116 °C for the most stable synthetic γ-CA (Fig. 7), while maintaining activity comparable to bovine α-CA (SI Appendix, Fig. S3). The most thermostable β- and γ-CA sequences reported in literature possess melting temperatures of 89 °C (63) and 55 °C (64) respectively. Our top performing designs represent thermostability improvements of ΔTm = +12 °C and ΔTm = +61 °C over the current best.
Fig. 7.

Synthetic β-CA and γ-CA designed using ProT-VAE yields high thermal stability sequences. (A) Experimentally measured melting points (Tm) of designed β-CA and γ-CA sequences as a function of their mutational distance from their nearest naturally occurring sequence. The melting temperatures of the most thermostable β-CA [89 °C (63)] and γ-CA [55 °C (64)] reported in literature are indicated by horizontal dashed lines. (B) AlphaFold (4) predicted structures for the highest stability γ-CA (Tm = 116 °C, ΔTm = +61 °C; tan) presented alone and aligned to the AlphaFold of the highest similarity natural γ-CA (blue). (C) AlphaFold predicted structures for the highest stability β-CA (Tm = 101 °C, ΔTm = +12 °C; green) presented alone and aligned to the highest similarity natural β-CA (blue).
We further characterize the highest-stability γ-CA sequence to quantify its enzymatic efficiency and activity under industrial conditions. The synthetic γ-CA is 0.8× active as bovine α-CA in normal conditions and is stable up to 93 °C in 23% v/v methyl diethanolamine (MDEA) at pH 11.25, representing industrially relevant conditions for carbon capture technologies (65) (SI Appendix, Fig. S4).
Discussion
In this work, we introduce ProT-VAE, an accurate, generative, fast, and transferable model of the sequence–function relationship for data-driven protein engineering. By blending the desirable features of transformers and VAEs, the model admits alignment-free training in an unsupervised or semisupervised fashion and furnishes interpretable low-dimensional latent spaces that facilitate understanding and generative design of functional synthetic sequences. The model comprises a VAE to distill task-specific information from generally pretrained, attention-based transformer encoder and decoder stacks with the aid of intermediate compression/decompression blocks. We validate ProT-VAE in applications to three different protein families: SH3, PAH, and CA. We show that the learned latent spaces are organized by phylogeny and function and can be used for the conditional generative design of synthetic proteins with desired properties. Experimental gene synthesis and assays demonstrate the generative design of a synthetic PAH sequence possessing 19/333 mutations relative to the wild-type human PAH and 2.5× elevated catalytic activity, and a synthetic γ-CA possessing 35 mutations relative to the highest similarity natural γ-CA, a melting temperature of Tm = 116 °C representing a ΔTm = +61 °C elevation relative to the most thermostable sequence reported to date, and stability up to 93 °C in 23% v/v MDEA at pH 11.25 corresponding to industrially relevant conditions for enzymatic carbon capture. The ProT-VAE model represents an experimentally demonstrated deep generative model for data-driven protein design that can be generically applied to other machine learning-guided directed evolution campaigns to iteratively identify novel proteins with elevated function by semisupervised retraining of the VAE blocks on the synthetic sequences and their attendant functional assays (1, 27, 55, 66–73). Compared to other deep generative protein design models ProT-VAE offers advantages in learning the sequence–function mapping to enable direct optimization of protein function in the absence of structure, which opens the door to both larger training data [there are known sequences compared to only solved structures (74, 75)] and the capacity to engineer proteins for which the structural determinants of function are poorly characterized or even unknown. Furthermore, by integrating an exterior transformer with an interior VAE, ProT-VAE eliminates the need to construct a MSA—thereby avoiding the bias inherent in alignment construction and enabling training over large databases of nonhomologous proteins to realize benefits in transfer learning across protein families—while still furnishing an interpretable low-dimensional latent space to expose functional and phylogenetic patterns and guide conditional generative design of synthetic proteins with engineered functionality.
In future work, we envisage a number of avenues for innovations and improvements to the model. First, it is of interest to provision the model with the ability for conditional sequence generation based on desired functional characteristics, environmental conditions, or even natural language prompts (2, 20, 76). This would require elaborating the architecture to introduce conditioning variables and the curation of training data for which these metadata are available. Second, we would like to explore the transferability of not just the exterior encoder/decoder blocks but also the VAE latent space across protein families. Currently, we retrain the VAE for each new protein family of interest, but it would be of interest to explore the degree to which a single latent space may be transferable across multiple related protein families to amortize the training costs and realize potential advantages in elevated data volume through transfer learning (77). Third, it would be interesting to explore the use of the model in scaffolding and in-painting tasks wherein a portion of the protein sequence is defined (e.g., a known binding site, catalytic site, allosteric positions) and the model is used to generatively design the remainder of the sequence (78, 79). Fourth, we are interested to explore the incorporation of physical priors, which may be particularly valuable in regularizing the model in low-data regimes, and exploring multimodal learning paradigms incorporating information on not just sequence but also structure, functional annotations, and dynamics (66, 80).
Materials and Methods
ProT-VAE Architecture and Training.
Exterior ProtT5nv block.
The ProtT5nv block is the first block in the three block architecture. ProtT5nv is a pretrained transformer-based T5 encoder and decoder model trained over approximately 46M unique protein sequences within the UniRef50 (release 05/2022) database after clustering, truncation, and splitting. This model is made available within the NVIDIA BioNeMo framework that is currently generally available and planned for future open source release (https://www.nvidia.com/en-us/clara/bionemo/) (46, 47). The model has 12 layers, 12 attention heads, a hidden dimension of 768, and 198M parameters. Pre-LN layer normalization and GeLU activation are used throughout the model. Additionally, encoder embeddings and decoder projections to logits are shared in this architecture. The model was trained with a maximum input sequence length of 512 and a masking probability of 15%. Unsupervised mask prediction was used as a training objective. Dropout was set to 0.1 during training.
ProtT5nv was trained starting from a T5 model pretrained using a natural language processing (NLP) paradigm. Parameters of all layers from the NLP-pretrained T5 model, except for encoder and decoder embeddings, were used to initialize the ProtT5nv model weights. The original NLP-pretrained T5 model had a dictionary of 29,184 tokens, while the ProtT5nv model only required 128 tokens, including 96 sentinel tokens. ProtT5nv encoder embeddings were therefore initialized with 128 first encoder embedding vectors from the NLP-pretrained T5 model. Then, decoder projections to logits were tied to encoder embeddings.
After initialization, the model was further trained with protein sequences from UniRef50, release 05/2022 (81). Protein sequences longer than 512 amino acids were removed, resulting in approximately 46M samples. The sequences were randomly split with 4.35K in validation, 875K in test, and the remaining in train. ProtT5nv model was trained using data parallelism on 224 V100 GPUs for 58 epochs (approximately 1M iterations) using a microbatch size of 12 protein sequences per GPU. Inverse square root annealing was used as a learning rate scheduler, with a minimum learning rate of 0.0 and 10,000 warmup steps. Fused Adam optimization was used with parameters β1 = 0.9, β2 = 0.999, and weight decay = 0.01.
Intermediate compression/decompression block.
The dimensionality reduction block serves to efficiently compress and decompress the ∼300,000-dimensional ProtT5 latent space into a more parsimonious intermediate-level representation that serves a fixed-length input to a task-specific VAE. This block was first pretrained on UniRef50, release 05/2022 (81) using a mean squared error (MSE) reconstruction objective. The block consists of three layers, where each layer comprises i) 1 × 1 convolutions, ii) LayerNorm, iii) GeLU activations, and the filter size is incremented at each step. In the results presented here, three layers were used with filter sizes of 512, 256, and 64 in the encoding side and 256, 512, and 768 in the decoding side. In the current implementation, the full output of the transformer hidden state, including the positions corresponding to padding tokens, is compressed to create a resultant 32,768-dimensional intermediate representation that is fed to the family-specific VAE layers.
Interior family-specific VAE block.
The ProtT5nv and compression/decompression stacks are transferable and generic models that need only be trained once over large libraries of diverse protein sequences and can be conceived of as furnishing expressive fixed-length featurizations of arbitrary proteins from unaligned sequences. Only the interior lightweight VAE requires training anew for each protein engineering task. The primary role of the VAE stack is to furnish a smooth, low-dimensional latent space that furnishes interpretable understanding and a springboard for conditional generation of synthetic protein sequences with engineered function. For all models, the inner VAE consisted of three fully connected layers with GELU activations and batchnorm. Layer sizes were for all models were 2,560, 1,280, and N dimensions where N is the latent space size for that model. Latent space size was N = 6 for the PAH and SH3 models, and N = 4 for the β-CA and γ-CA models. To find optimal latent space size, we run a sweep of 3 to 10 dimensions and identify the latent space dimension residing at a knee in the reconstruction loss on the validation set, beyond which only marginal improvements in reconstruction loss are realized with increased latent dimensionality. We apply the learnings from Zhao et al. (30) and set lambda = 1.0 and alpha = 1.0 within the MMD-VAE loss function to maximize the information between the input data and the embedded latent space and construct smoother latent spaces than an evidence lower bound (ELBO) loss alone.
Model training.
A pretrained ProtT5 was obtained from the previously described BioNeMo checkpoint. The intermediate dimensionality reduction layers were pretrained on UniProt using the Adam optimizer with a learning rate of 0.0001 on an MSE objective on reconstruction of the full T5 hidden state. T5 layers were frozen during this pretraining. Family-specific VAE layers were trained on full, unaligned sequences belonging to a single homologous protein family. Sequences were fed in with 15% of positions randomly masked and 20% of masks corrupted to different amino acids. The training objective was the reconstruction of the original sequence, optimized using Adam with a learning rate of 0.0001. The sequence data used to train the SH3, PAH, and CA models are freely available from the publicly accessible Joint Genome Institute (JGI) (https://jgi.doe.gov), Protein Families (Pfam) (https://pfam.xfam.org), and National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov) databases.
PAH Synthetic Sequence Design.
We generated latent space locations for generative decoding by fitting a multidimensional Gaussian with 0.1 variance centered around the location of the hPAH in the 6D latent space and sampling 6D latent vectors. We filtered out sequences that were duplicates of natural ones and imposed a maximum similarity of any generated sequence to any natural sequence in the training data of 99%. To localize the generated sequences around the hPAH in both latent space and sequence space, we also placed a cap on the maximum number of mutations (i.e., substitutions, insertions, deletions) away from hPAH at 140 of the 333 wild-type positions, corresponding to a minimum sequence similarity of 58%. Under these criteria, 190 sequences containing a maximum of 133 mutations away from hPAH and spanning a range of lengths of 223-339 residues were chosen for experimental synthesis and testing. The designed protein sequences assayed in 96-well plates using a Biotek plate reader to evaluate the fluorescence of tyrosine produced over a defined time period, and activities reported relative to hPAH as FOWT activity. Full details of the experimental procedures are provided in SI Appendix.
CA Synthetic Sequence Design.
For both β-CA and γ-CA generation, we sampled sequences from latent space by randomly sampling from a multidimensional Gaussian distribution which was fitted over the latent coordinates of known functional natural sequences. Additionally, for γ-CA, each generated sequence’s latent vectors were passed through the latent space binary classifier for presence of the catalytic triad to gate through only those with 99% binary classifier confidence. In each case, we also limited the degree of exploration away from the training data by stipulating that the generatively decoded proteins must be within 99% to 70% sequence similarity from the nearest training sequence, normalized to the reference natural. We used these strategies to design 88 synthetic β-CA and 88 synthetic γ-CA sequences that were subjected to experimental gene synthesis and measurement of catalytic activity and thermostability. Full details of the experimental procedures are provided in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. Vedant Sachdeva and Dr. Anisha Zaveri for assistance in preparing the AlphaFold structural predictions of the synthetic proteins.
Author contributions
R.R., A.B.C., and A.L.F. designed research; E.S., J. Moller, A.L., J.P., S.Q., J. Mayer, P.S., S.G., D.H., C.D., C.B., T.S., M.K., M.L., and M.G. performed research; E.S., J. Moller, A.L., J.P., S.Q., J. Mayer, P.S., S.G., D.H., C.D., C.B., T.S., M.K., M.L., and M.G. analyzed data; and E.S., J. Moller, A.L., J.P., S.Q., J. Mayer, P.S., S.G., D.H., C.D., C.B., T.S., M.K., M.L., and M.G. wrote the paper.
Competing interests
E.S., J. Moller, A.L., J.P., S.Q., J. Mayer, P.S., S.G., D.H., C.D., C.B., and T.S. are or were employees of Evozyne, Inc. where this work was conducted and who may have the opportunity for stock ownership in the company. M.K., M.L., M.G., and A.B.C. are employees of NVIDIA who collaborated in this work and who may have the opportunity for stock ownership in the company. R.R. and A.L.F. are co-founders of Evozyne, Inc. and possess stock ownership in the company. R.R. and A.L.F. are co-authors of US Provisional Patent Applications 62/900,420 and 63/669,836, US Patent Application 17/642,582, and International Patent Application PCT/US2020/050466. E.S., J. Moller, A.L., and A.L.F. are co-authors of US Provisional Patent Application 63/479,378 and International Patent Application PCT/US24/10805. A.L.F. is co-author of US Provisional Patent Application 63/314,898. R.R. and A.L.F. are faculty members at the University of Chicago.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Anthony B. Costa, Email: acosta@nvidia.com.
Andrew L. Ferguson, Email: andrew.ferguson@evozyne.com.
Data, Materials, and Software Availability
Some study data are available: The sequence data used to train the SH3, PAH, and CA models are freely available from the publicly accessible JGI (https://genome.jgi.doe.gov/portal/) (82), Pfam (https://www.ebi.ac.uk/interpro/) (83), and NCBI (https://www.ncbi.nlm.nih.gov/protein/) (84) databases. The BioNeMo framework used to construct the machine learning models is freely available from https://github.com/NVIDIA/bionemo-framework/ (85). The trained machine learning models and their outputs cannot be shared due to commercial and legal constraints. However, by using the two components above (i.e., the training data and the BioNeMo framework) together with the detailed description of the model architecture and training protocol provided in Materials and Methods, an interested reader could reconstruct these models. All other data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Yang K. K., Wu Z., Arnold F. H., Machine-learning-guided directed evolution for protein engineering. Nat. Methods 16, 687–694 (2019). [DOI] [PubMed] [Google Scholar]
- 2.J. Ingraham et al. , Illuminating protein space with a programmable generative model. bioRxiv [Preprint] (2022). 10.1101/2022.12.01.518682 (Accessed 15 February 2025). [DOI] [PMC free article] [PubMed]
- 3.Leman J. K., et al. , Macromolecular modeling and design in Rosetta: Recent methods and frameworks. Nat. Methods 17, 665–680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anishchenko I., et al. , Protein tertiary structure prediction and refinement using deep learning and Rosetta in CASP14. Proteins Struct. Funct. Bioinf. 89, 1722–1733 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin J. E., et al. , Protein design and variant prediction using autoregressive generative models. Nat. Commun. 12, 2403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alley E. C., Khimulya G., Biswas S., AlQuraishi M., Church G. M., Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 16, 1315–1322 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding X., Zou Z., Brooks C. L. III, Deciphering protein evolution and fitness landscapes with latent space models. Nat. Commun. 10, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riesselman A. J., Ingraham J. B., Marks D. S., Deep generative models of genetic variation capture the effects of mutations. Nat. Methods 15, 816–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Z. Costello, H. G. Martin, How to hallucinate functional proteins. arXiv [Preprint] (2019). http://arxiv.org/abs/1903.00458. Accessed: 15 Feb 2025.
- 11.Greener J. G., Moffat L., Jones D. T., Design of metalloproteins and novel protein folds using variational autoencoders. Sci. Rep. 8, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. Sinai, E. Kelsic, G. M. Church, M. A. Nowak, Variational auto-encoding of protein sequences. arXiv [Preprint] (2017). http://arxiv.org/abs/1712.03346 (Accessed 15 February 2025).
- 13.Lian X., et al. , Deep learning-based design of synthetic orthologs of a signaling protein. Cell Syst. 15, P725–737.E5 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giessel A., et al. , Therapeutic enzyme engineering using a generative neural network. Sci. Rep. 12, 1–17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Repecka D., et al. , Expanding functional protein sequence spaces using generative adversarial networks. Nat. Mach. Intell. 3, 324–333 (2021). [Google Scholar]
- 16.C. Angermueller et al. , “Model-based reinforcement learning for biological sequence design” in International Conference on Learning Representations 2020 (International Conference on Learning Representations, 2020), pp. 1–23.
- 17.Rao R., et al. , Evaluating protein transfer learning with TAPE. Adv. Neural Inf. Process. Syst. 32, 9689–9701 (2019). [PMC free article] [PubMed] [Google Scholar]
- 18.Rives A., et al. , Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. U.S.A. 118, e2016239118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. Madani et al. , Progen: Language modeling for protein generation. arXiv [Preprint] (2020). http://arxiv.org/abs/2004.03497 (Accessed 15 February 2025).
- 20.Madani A., et al. , Large language models generate functional protein sequences across diverse families. Nat. Biotechnol. 41, 1099–1106 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.P. Notin et al. , “Tranception: Protein fitness prediction with autoregressive transformers and inference-time retrieval” in International Conference on Machine Learning (Proceedings of Machine Learning Research, 2022), pp. 16990–17017.
- 22.Elnaggar A., et al. , ProtTrans: Toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Ferruz N., Höcker B., Controllable protein design with language models. Nat. Mach. Intell. 4, 521–532 (2022). [Google Scholar]
- 24.Luo Y., et al. , ECNet is an evolutionary context-integrated deep learning framework for protein engineering. Nat. Commun. 12, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.C. Dallago et al. , Benchmark tasks in fitness landscape inference for proteins. bioRxiv [Preprint] (2021). 10.1101/2021.11.09.467890 (Accessed 15 February 2025). [DOI]
- 26.Praljak N., Lian X., Ranganathan R., Ferguson A. L., ProtWave-VAE: Integrating autoregressive sampling with latent-based inference for data-driven protein design. ACS Synth. Biol. 12, 3544–3561 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bepler T., Berger B., Learning the protein language: Evolution, structure, and function. Cell Syst. 12, 654–669 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D. P. Kingma, M. Welling, Auto-encoding variational Bayes. arXiv [Preprint] (2013). http://arxiv.org/abs/1312.6114 (Accessed 15 February 2025).
- 29.Kingma D. P., et al. , An introduction to variational autoencoders. Found. Trends Mach. Learn. 12, 307–392 (2019). [Google Scholar]
- 30.S. Zhao, J. Song, S. Ermon, “Infovae: Balancing learning and inference in variational autoencoders” in Proceedings of the AAAI Conference on Artificial Intelligence (Association for the Advancement of Artificial Intelligence, 2019), vol. 33, pp. 5885–5892.
- 31.Gómez-Bombarelli R., et al. , Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castro E., et al. , Transformer-based protein generation with regularized latent space optimization. Nat. Mach. Intell. 4, 840–851 (2022). [Google Scholar]
- 33.Frassek M., Arjun A., Bolhuis P., An extended autoencoder model for reaction coordinate discovery in rare event molecular dynamics datasets. J. Chem. Phys. 155, 064103 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Chatzou M., et al. , Multiple sequence alignment modeling: Methods and applications. Brief. Bioinform. 17, 1009–1023 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Vaswani A., et al. , Attention is all you need. Adv. Neural Inf. Process. Syst. 30, 6000–6010 (2017). [Google Scholar]
- 36.N. S. Keskar, B. McCann, L. R. Varshney, C. Xiong, R. Socher, Ctrl: A conditional transformer language model for controllable generation. arXiv [Preprint] (2019). http://arxiv.org/abs/1909.05858 (Accessed 15 February 2025).
- 37.Uniprot: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed]
- 38.Steinegger M., Mirdita M., Söding J., Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nat. Methods 16, 603–606 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Steinegger M., Söding J., Clustering huge protein sequence sets in linear time. Nat. Commun. 9, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.T. Wang, X. Wan, “T-cvae: Transformer-based conditioned variational autoencoder for story completion” in Proceedings of the Twenty-Eighth International Joint Conference on Artificial Intelligence (IJCAI-19) (2019), pp. 5233–5239.
- 41.J. Jiang, G. G. Xia, D. B. Carlton, C. N. Anderson, R. H. Miyakawa, “Transformer VAE: A hierarchical model for structure-aware and interpretable music representation learning” in ICASSP 2020–2020 IEEE International Conference on Acoustics Speech and Signal Processing (ICASSP) (Institute of Electrical and Electronics Engineers, 2020), pp. 516–520.
- 42.C. Li et al. , Optimus: Organizing sentences via pre-trained modeling of a latent space. arXiv [Preprint] (2020). http://arxiv.org/abs/2004.04092 (Accessed 15 February 2025).
- 43.S. Park, J. Lee, Finetuning pretrained transformers into variational autoencoders. arXiv [Preprint] (2021). http://arxiv.org/abs/2108.02446 (Accessed 15 February 2025).
- 44.D. M. Arroyo, J. Postels, F. Tombari, “Variational transformer networks for layout generation” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Institute of Electrical and Electronics Engineers, 2021), pp. 13642–13652.
- 45.J. Henderson, F. Fehr, A variational autoencoder for transformers with nonparametric variational information bottleneck. arXiv [Preprint] (2022). http://arxiv.org/abs/2207.13529 (Accessed 15 February 2025).
- 46.NVIDIA BioNeMo cloud service: An end-to-end AI-powered drug discovery pipelines (2024). https://www.nvidia.com/en-us/gpu-cloud/bionemo/.
- 47.NVIDIA Clara Discovery (2024). https://www.nvidia.com/en-us/clara/drug-discovery/.
- 48.S. Zhao, J. Song, S. Ermon, InfoVAE: Information maximizing variational autoencoders. arXiv [Preprint] (2017). http://arxiv.org/abs/1706.02262 (Accessed 15 February 2025).
- 49.Musacchio A., Noble M., Pauptit R., Wierenga R., Saraste M., Crystal structure of a Src-Homology 3 (SH3) domain. Nature 359, 851–855 (1992). [DOI] [PubMed] [Google Scholar]
- 50.Mayer B. J., SH3 domains: Complexity in moderation. J. Cell Sci. 114, 1253–1263 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Zarrinpar A., Park S. H., Lim W. A., Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426, 676–680 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Flydal M. I., Martinez A., Phenylalanine hydroxylase: Function, structure, and regulation. IUBMB Life 65, 341–349 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Fusetti F., Erlandsen H., Flatmark T., Stevens R. C., Structure of tetrameric human phenylalanine hydroxylase and its implications for phenylketonuria. J. Biol. Chem. 273, 16962–16967 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Pruitt K. D., Tatusova T., Maglott D. R., NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, D501–D504 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Detlefsen N. S., Hauberg S., Boomsma W., Learning meaningful representations of protein sequences. Nat. Commun. 13, 1914 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Supuran C. T., Structure and function of carbonic anhydrases. Biochem. J. 473, 2023–2032 (2016). [DOI] [PubMed] [Google Scholar]
- 57.R. S. Rowlett, Structure and catalytic mechanism of the β-carbonic anhydrases. Biochim. Biophys. Acta Proteins Proteomics 1804, 362–373 (2010). [DOI] [PubMed]
- 58.Ferry J. G., The gamma class of carbonic anhydrases. Biochim. Biophys. Acta Proteins Proteomics 1804, 374–381 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russo M., et al. , Post-combustion carbon capture mediated by carbonic anhydrase. Sep. Purif. Technol. 107, 331–339 (2013). [Google Scholar]
- 60.Strop P., Smith K. S., Iverson T. M., Ferry J. G., Rees D. C., Crystal structure of the “cab”-type β class carbonic anhydrase from the archaeon methanobacterium thermoautotrophicum. J. Biol. Chem. 276, 10299–10305 (2001). [DOI] [PubMed] [Google Scholar]
- 61.Kisker C., Schindelin H., Alber B. E., Ferry J. G., Rees D. C., A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon methanosarcina thermophila. EMBO J. 15, 2323–2330 (1996). [PMC free article] [PubMed] [Google Scholar]
- 62.Iverson T. M., Alber B. E., Kisker C., Ferry J. G., Rees D. C., A closer look at the active site of γ-class carbonic anhydrases: High-resolution crystallographic studies of the carbonic anhydrase from methanosarcina thermophila. Biochemistry 39, 9222–9231 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Alvizo O., et al. , Directed evolution of an ultrastable carbonic anhydrase for highly efficient carbon capture from flue gas. Proc. Natl. Acad. Sci. U.S.A. 111, 16436–16441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alber B. E., Ferry J. G., Characterization of heterologously produced carbonic anhydrase from methanosarcina thermophila. J. Bacteriol. 178, 3270–3274 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hasan S., Abbas A. J., Nasr G. G., Improving the carbon capture efficiency for gas power plants through amine-based absorbents. Sustainability 13, 72 (2021). [Google Scholar]
- 66.Ferguson A. L., Ranganathan R., 100th anniversary of macromolecular science viewpoint: Data-driven protein design. ACS Macro Lett. 10, 327–340 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Freschlin C. R., Fahlberg S. A., Romero P. A., Machine learning to navigate fitness landscapes for protein engineering. Curr. Opin. Biotechnol. 75, 102713 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding W., Nakai K., Gong H., Protein design via deep learning. Brief. Bioinform. 23, bbac102 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mazurenko S., Prokop Z., Damborsky J., Machine learning in enzyme engineering. ACS Catal. 10, 1210–1223 (2019). [Google Scholar]
- 70.Osadchy M., Kolodny R., How deep learning tools can help protein engineers find good sequences. J. Phys. Chem. B 125, 6440–6450 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Wittmann B. J., Johnston K. E., Wu Z., Arnold F. H., Advances in machine learning for directed evolution. Curr. Opin. Struct. Biol. 69, 11–18 (2021). [DOI] [PubMed] [Google Scholar]
- 72.Xu Y., et al. , Deep dive into machine learning models for protein engineering. J. Chem. Inf. Model. 60, 2773–2790 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Frappier V., Keating A. E., Data-driven computational protein design. Curr. Opin. Struct. Biol. 69, 63–69 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.UniProt Consortium, Uniprot: The universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richardson L., et al. , Mgnify: The microbiome sequence data analysis resource in 2023. Nucleic Acids Res. 51, D753–D759 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.S. Liu et al. , A text-guided protein design framework. arXiv [Preprint] (2023). http://arxiv.org/abs/2302.04611 (Accessed 15 February 2025).
- 77.Strokach A., Kim P. M., Deep generative modeling for protein design. Curr. Opin. Struct. Biol. 72, 226–236 (2022). [DOI] [PubMed] [Google Scholar]
- 78.Wang J., et al. , Scaffolding protein functional sites using deep learning. Science 377, 387–394 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.B. Lai, M. McPartlon, J. Xu, End-to-end deep structure generative model for protein design. bioRxiv [Preprint] (2022). 10.1101/2022.07.09.499440 (Accessed 15 February 2025). [DOI]
- 80.Malbranke C., Bikard D., Cocco S., Monasson R., Tubiana J., Machine learning for evolutionary-based and physics-inspired protein design: Current and future synergies. Curr. Opin. Struct. Biol. 80, 102571 (2023). [DOI] [PubMed] [Google Scholar]
- 81.Suzek B. E., Huang H., McGarvey P., Mazumder R., Wu C. H., UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288 (2007). [DOI] [PubMed] [Google Scholar]
- 82.I. Grigoriev et al. , Data from “Genome portal.” Genomes Online Database. https://genome.jgi.doe.gov/portal/. Accessed 25 August 2022.
- 83.S. Chuguransky et al. , Data from “Pfam Legacy”. Protein Families Database. https://www.ebi.ac.uk/interpro/. Accessed 25 August 25 2022.
- 84.M. Romiti, P. Cooper, Data from “RefSeq FTP.” NCBI Protein. https://www.ncbi.nlm.nih.gov/protein/. Accessed 25 August 2022.
- 85.D. Lin et al. , Data from “Bionemo-framework” GitHub. https://github.com/NVIDIA/bionemo-framework/. Accessed 15 March 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Some study data are available: The sequence data used to train the SH3, PAH, and CA models are freely available from the publicly accessible JGI (https://genome.jgi.doe.gov/portal/) (82), Pfam (https://www.ebi.ac.uk/interpro/) (83), and NCBI (https://www.ncbi.nlm.nih.gov/protein/) (84) databases. The BioNeMo framework used to construct the machine learning models is freely available from https://github.com/NVIDIA/bionemo-framework/ (85). The trained machine learning models and their outputs cannot be shared due to commercial and legal constraints. However, by using the two components above (i.e., the training data and the BioNeMo framework) together with the detailed description of the model architecture and training protocol provided in Materials and Methods, an interested reader could reconstruct these models. All other data are included in the manuscript and/or SI Appendix.