Abstract
RNA interference (RNAi) is a powerful tool for identifying gene function in Trypanosoma brucei. We generated an RNAi library, the first of its kind in any organism, by ligation of genomic fragments into the vector pZJMβ. After transfection at ∼5-fold genome coverage, trypanosomes were induced to express double-stranded RNA and screened for reduced con canavalin A (conA) binding. Since this lectin binds the surface glycoprotein EP-procyclin, we predicted that cells would lose affinity to conA if RNAi silenced genes affecting EP-procyclin expression or modification. We found a cell line in which RNAi switches expression from glycosylated EP-procyclins to the unglycosylated GPEET-procyclin. This switch results from silencing a hexokinase gene. The relationship between procyclin expression and glycolysis was supported by silencing other genes in the glycolytic pathway, and confirmed by observation of a similar upregulation of GPEET- procyclin when parental cells were grown in medium depleted of glucose. These data suggest that T.brucei ‘senses’ changes in glucose level and modulates procyclin expression accordingly.
Keywords: concanavalin A/hexokinase/MALDI-TOF mass spectrometry/RNA interference library
Introduction
Trypanosoma brucei is the protozoan parasite that causes sleeping sickness. Trypanosomes inhabit the bloodstream of their mammalian host and are transmitted by a tsetse fly vector. In a mammalian infection, trypanosomes proliferate within the blood as long slender forms and then differentiate into short stumpy forms that are pre-adapted for life in the insect. When the insect ingests short stumpy trypanosomes during a blood meal, the parasites differentiate into procyclic forms within the fly midgut (Vickerman, 1985). After ∼3 weeks, they are found in the salivary glands, where they develop into metacyclic forms that are infectious to mammals.
Numerous developmental changes occur during the differentiation of bloodstream forms to procyclic forms, including replacement of the variant surface glycoprotein (VSG) surface coat with a coat consisting of members of the procyclin family of glycoproteins (Roditi et al., 1989; Ziegelbauer et al., 1990). The GPI-anchored procyclins are divided into two classes, EP-procyclins and GPEET- procyclin, and they are named for the type of amino acid repeats in their C-terminal regions. EP-procyclins have 22–30 Glu-Pro repeats, whereas GPEET-procyclin has five or six Gly-Pro-Glu-Glu-Thr repeats followed by three EP repeats (Mowatt and Clayton, 1987; Roditi et al., 1987; Richardson et al., 1988; Mowatt et al., 1989). In the trypanosome strain 427, the EP-procyclins are encoded by three genes (EP1, EP2 and EP3), while GPEET-procyclin is encoded by a single gene (GPEET) (Roditi and Clayton, 1999). EP1-1, EP1-2 and EP3 are N-glycosylated with a homogeneous Man5GlcNAc2, whereas EP2 and GPEET are unglycosylated (Treumann et al., 1997; Acosta-Serrano et al., 1999; Hwa et al., 1999). The expression pattern of procyclins varies between parasite strains and can change after long-term in vitro culturing (Butikofer et al., 1997; Treumann et al., 1997). Con tinuous GPEET expression can be achieved in vitro by addition of glycerol to the culture medium, suggesting that external signals may influence procyclin expression (Vassella et al., 2000). During the first few hours of differentiation of bloodstream forms to procyclic forms in vitro, all procyclin isoforms are expressed at comparable levels (Vassella et al., 2001). Within 24 h, GPEET becomes predominant, to be replaced by glycosylated EP-procyclins later in development (Vassella et al., 2001). During development in the fly vector, procyclins could not be detected until day 3 (because insufficient parasites were available for analysis), when GPEET was detected. By day 7, GPEET had been replaced by glycosylated EP-procyclins (Vassella et al., 2000; Acosta-Serrano et al., 2001).
The lectin concanavalin A (conA) binds the N-glycan on EP-procyclins and induces cell death (Welburn et al., 1996; Hwa et al., 1999; Pearson et al., 2000). Parasites lacking EP-procyclins because of genetic knockout (Ruepp et al., 1997) are resistant to conA killing even though they continue to express the naturally unglycosylated GPEET-procyclin (Pearson et al., 2000). Similarly, chemically generated mutants with altered N-glycans are resistant to conA (Hwa et al., 1999; Acosta-Serrano et al., 2000).
RNA interference (RNAi) is a powerful tool for analysis of T.brucei gene function (Ngo et al., 1998; Wang and Englund, 2001). We developed an RNAi vector, pZJM, for silencing trypanosome genes (Wang et al., 2000). We now describe insertion of genomic fragments into pZJM, yielding an RNAi-based library that provides a bona fide forward genetic method for these diploid organisms. We have used this library for the identification of genes playing a role in conA sensitivity. We hypothesized that cells would become conA resistant by silencing genes involved in procyclin expression, GPI biosynthesis, N-glycan biosynthesis or cell death. Unexpectedly, the first gene we discovered by this approach was hexokinase, a glycolytic enzyme. We found that silencing of hexokinase alters procyclin expression, with GPEET (a naturally unglycosylated procyclin that does not bind conA) expression completely replacing that of EP. We also found that silencing other genes involved in glucose transport or glycolysis led to the same switch in procyclin expression. Furthermore, using an approach totally independent of RNAi, we observed that growth of trypanosomes in glucose-depleted medium caused a similar rise in GPEET expression. Given that GPEET expression is upregulated relative to that of EP-procyclins during the natural differentiation process, we suggest that glycolytic flux or changes in levels of glycolytic metabolites, which in turn are sensitive to the glucose levels within the fly midgut, may be a factor influencing procyclin expression.
Results
Identification of a gene conferring conA sensitivity
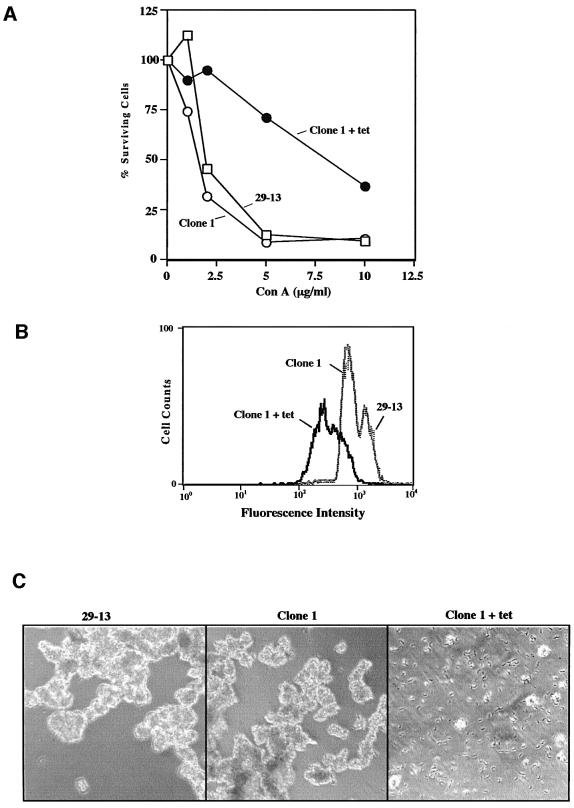
To identify genes involved in procyclin expression or modification, we transfected trypanosomes with an RNAi genomic library in the vector pZJMβ. After selecting transfected cells, we induced RNAi for 7 days. We then used six rounds of conA–Sepharose chromatography and two fluorescence-activated cell sorting (FACS) enrichments to isolate a single cell line, clone 1, which shows RNAi-dependent resistance to conA. Following induction of RNAi for 2 weeks, clone 1 had an EC50 of ∼8 µg/ml conA compared with ∼2 µg/ml for uninduced clone 1 or parental cells (Figure 1A). Analysis of conA binding by flow cytometry confirmed that clone 1 had reduced affinity for conA when compared with either uninduced or 29-13 cells (Figure 1B). Uninduced and parental cells grown in 5 µg/ml conA for 24 h agglutinated, whereas RNAi cells appeared normal (Figure 1C).
Fig. 1. Clone 1 is resistant to conA killing and has reduced affinity for conA following RNAi induction. (A) Trypanosome strain 29-13, uninduced clone 1 and tetracycline-induced clone 1 (for 14 days) were grown for 24 h in the presence of increasing concentrations of conA. Cell densities were determined using a Coulter counter (model Z1) and cell numbers plotted as a percentage of control untreated cells. The Coulter counter was set to exclude cell aggregates larger than 7.65 µm. This method may not discriminate between living and some dead cells, leading to a conservative estimation of the EC50. The data presented here are representative of four independent experiments in which the EC50 for induced cells was consistently 4- to 6-fold greater than that of parental 29-13 cells. (B) Living trypanosomes were incubated with 10 µg/ml fluorescein-conjugated conA for 15 min at room temperature in cytoM and then analyzed by flow cytometry (10 000 cells/assay). Cell line 29-13, uninduced clone 1 and induced clone 1 (14 days) were analyzed. The two peaks seen in both 29-13 and uninduced clone 1 could be due to the presence of both normal cells and large cells about to divide. Laser intensity was adjusted to yield autofluorescence from unstained cells of ∼10 fluorescence intensity units. (C) 29-13 cells, uninduced clone 1 and tetracycline-induced (14 days) clone 1 were incubated with 5 µg/ml conA for 24 h and then visualized by phase microscopy.
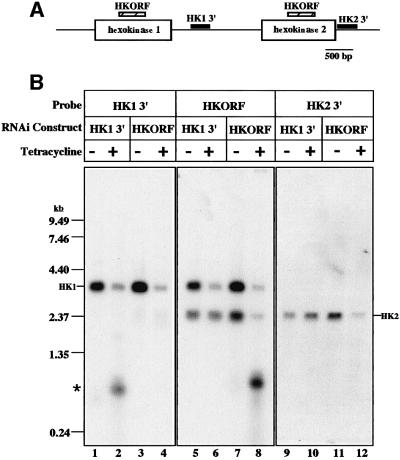
To identify the silenced gene in clone 1, we amplified the genomic DNA fragment within pZJMβ. Searching the T.brucei sequence database, we identified the insert (341 bp) as a sequence starting 397 bp downstream of the hexokinase 1 gene (DDBJ/EMBL/GenBank accession No. AJ345044) on chromosome 10. The sequence contains stop codons in all frames and is likely to be in the 3′ untranslated region. A second hexokinase gene (hk2) is arrayed in tandem to hexokinase 1 (see the map in Figure 2A). Although hexokinases 1 and 2 are virtually identical, they have distinct 3′ UTRs, with the largest region of identity being only five nucleotides. Therefore, hexokinase 2 should not be targeted by RNAi using the 341 bp insert.
Fig. 2. The hexokinase 1 gene is silenced by RNAi using a construct targeting either the 3′ UTR or the open reading frame. (A) The hexokinase 1 and 2 genomic locus showing DNA fragments (HKORF, HK1 3′ and HK2 3′) used to silence hexokinase expression by RNAi and as probes. (B) Northern analysis of total RNA from uninduced (–) and tetracycline-induced (+) (for 40 h) cells containing the constructs pZJM(HK1 3′) or pZJM(HKORF). Total RNA was purified from 5 × 107 parasites and electrophoresed on a formaldehyde–1.5% agarose gel. rRNA levels were estimated by ethidium bromide staining to ensure equal loading of RNA. The asterisk indicates dsRNA expressed from the pZJM insert.
To confirm that silencing of the hexokinase 1 gene was responsible for conA resistance, we cloned the same 3′ UTR sequence (HK1 3′), as well as 502 bp of hexokinase 1 open reading frame (HKORF, which targets both hexokinase genes), into pZJM followed by transfection into 29-13 cells. After RNAi induction, we found that hexokinase 1 mRNA (4 kb) was significantly reduced (Figure 2B, lanes 1 and 2) in the pZJM(HK1 3′) cells. In the pZJM(HKORF) cells, we also found reduction not only of the 4 kb transcript, but also of one of 2.5 kb (Figure 2B, lanes 7 and 8).
The 2.5 kb transcript corresponds to the HK2 gene. Using a probe specific to the 3′ UTR of HK2 (HK2 3′), we found that silencing of HK1 (using the HK1 3′ UTR) did not cause silencing of the HK2 transcript (compare lanes 9 and 10), and in fact it correlated with an apparent (and not understood) increase in abundance of this transcript. Targeting the HKORF, however, led to a significant reduction in the HK2 2.5 kb transcript (lanes 11 and 12). Hybridization of the same northern with an HKORF-specific probe (lanes 5–8) showed that RNAi of the HK1 3′ UTR caused no significant change in the 2.5 kb transcript (lanes 5 and 6).
RNAi of the hexokinase genes dramatically affects hexokinase enzyme activity in cell lysates. Silencing hexokinase 1 [using pZJM(HK1 3′)] reduces activity 79%: from 0.36 ± 0.01 µmol/min/mg in parental 29-13 cell lysates to 0.077 ± 0.005 µmol/min/mg in lysates from cells induced for 21 days. Targeting both genes using pZJM(HKORF) had an even greater effect, reducing activity by 95% in the 21 day induced cell lysate (to 0.020 ± 0.001 µmol/min/mg).
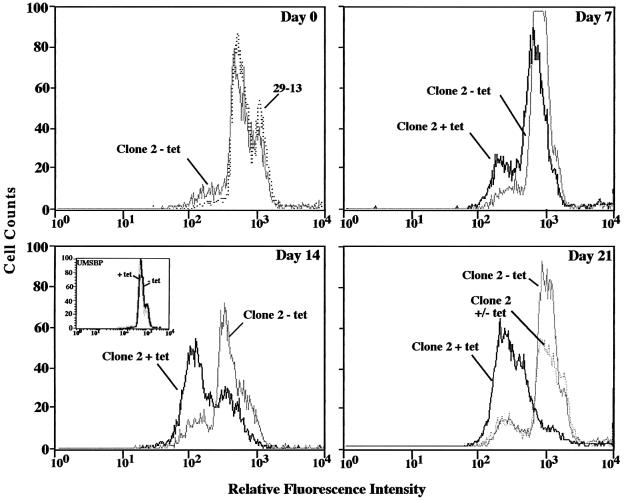
We next induced RNAi in pZJM(HKORF) cells (clone 2, isolated by limiting dilution) to confirm that RNAi of hexokinase was responsible for the observed phenotype. We found that RNAi for 16 days conferred conA resistance, with an EC50 of ∼6.5 µg/ml conA (data not shown). Furthermore, we used flow cytometry to show that RNAi caused a loss of conA binding (Figure 3). Before induction of RNAi (day 0), ∼5% of uninduced pZJM(HKORF) cells showed decreased conA binding not seen in parental cells, possibly due to incomplete transcriptional repression from pZJM in a small population of cells, as seen previously (Wang et al., 2000). After 7 days of tetracycline induction, 26% of the cells had a lower affinity for conA, while only 7% of uninduced cells displayed this phenotype (Figure 3). After 14 days of tetracycline induction, 60% of the population had reduced conA affinity, whereas 10% of the uninduced cells were deficient (Figure 3). After 21 days of RNAi, ∼85% bound reduced levels of conA (Figure 3). To confirm that this phenotype was due to RNAi, pZJM(HKORF) cells that had been induced for 14 days were washed free of tetracycline. We found that, after 7 days of growth in the absence of tetracycline (day 21), ∼70% of the population reverted to parental cell levels of conA binding (Figure 3). To prove that reduction of conA binding was not a general consequence of RNAi, cells expressing double-stranded (ds) RNA for 14 days to a homolog of the Crithidia fasciculata UMSBP (Tzfati et al., 1992; Wang et al., 2000) were found to bind conA at a level comparable to that of parental 29-13 cells (Figure 3, day 14, inset).
Fig. 3. Cells expressing dsRNA corresponding to the open reading frame of hexokinase have reduced ConA–fluorescein affinity. Trypanosomes (clone 2) were grown in the presence of tetracycline and analyzed by flow cytometry as described in Figure 1B. The inset in the day 14 panel shows cells harboring pZJM(UMSBP) induced for 14 days. The day 21 panel also shows cells induced for 14 days and then washed free of tetracycline for 7 days (Clone 2 +/– tet).
Effects of silencing of the hexokinase 1 gene on procyclin expression
ConA kills trypanosomes by binding to glycosylated EP-procyclins (Pearson et al., 2000). Mutant trypanosomes are resistant to conA-induced cell death if they express unglycosylated EP-procyclins, have altered N-glycans, or have had their EP-procyclin genes deleted (in the latter case they continued to express GPEET- procyclin, an unglycosylated species) (Ruepp et al., 1997; Hwa et al., 1999; Acosta-Serrano et al., 2000; Pearson et al., 2000).
To clarify the biochemical basis for conA resistance in induced pZJM(HK1 3′) cells, we studied whether RNAi affected the biosynthesis of either GPI or N-glycan precursors. We used a cell-free system labeled with GDP-[3H]mannose (in the presence and absence of tunicamycin) that allows evaluation of biosynthetic intermediates (Masterson et al., 1989). Analysis of the products by TLC showed no detectable difference in the synthesis of either type of precursor in induced cells (14 days) when compared with uninduced or parental cells (data not shown).
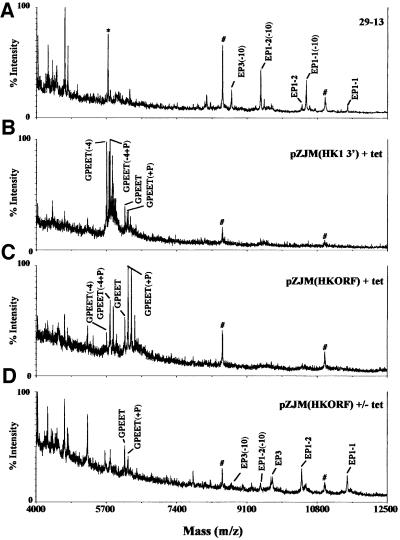
We then analyzed the parasite’s procyclin repertoire by purification of these proteins followed by removal of their GPI anchors and MALDI-TOF mass spectrometry (MS). This method identifies which procyclins are expressed, as well as their glycosylation state. We found that parental 29-13 and uninduced pZJM(HK1 3′) express a variety of procyclins, including EP1-1, EP1-2 and EP3, which all have N-glycans (Figure 4A; data not shown). In both cases, however, there was virtually no detectable GPEET- procyclin. Upon tetracycline induction for 21 days, the parasites no longer synthesized detectable EP-procyclins and instead had switched expression to the naturally unglycosylated GPEET-procyclin (Figure 4B). We confirmed assignment of procyclin species by MALDI-TOF analyses after mild acid hydrolysis, which cleaves EP- procyclins at Asp-Pro bonds (data not shown) (Acosta-Serrano et al., 1999). Analysis of pZJM(HKORF) cells following 21 days of induction confirmed that hexokinase silencing led to a change in procyclin expression from EP to GPEET (Figure 4C). Furthermore, removal of tetracycline at day 14 led to a decrease in GPEET expression concomitant with return of the wild-type EP-procyclin repertoire (Figure 4D).
Fig. 4. Trypanosomes with silenced hexokinase express GPEET- procyclin. Procyclins were purified and prepared for MALDI-TOF MS. An internal standard (insulin, 5733 Da, indicated by an asterisk) was added to some samples. Identical spectra were obtained when insulin was omitted. [M-H]– ions were identified using the assignments in Acosta-Serrano et al. (1999). EP1-1, EP1-2 and EP3 had m/z of 11 531, 10 430 and 9723, respectively. Full-length GPEET and phosphorylated GPEET (+P) had m/z of 6142 and 6222, respectively. Species indicated as (-10) lack 10 amino acids from the N-terminus, a cleavage occurring during HF treatment (Acosta-Serrano et al., 1999). GPEET species identified as (-4) lack the four N-terminal amino acids. GPEET can also be phosphorylated, with the predominant form being phosphorylated once (+P). Other species correspond to multiply phosphorylated forms (Mehlert et al., 1999). The ions at m/z 8504 and 10 987 (indicated with a hash) are not procyclins; the latter is probably KMP-11 (Acosta-Serrano et al., 1999). (A) Parental 29-13 cells. (B) Cells containing pZJM(HK1 3′) induced for 21 days. (C) Cells containing pZJM (HKORF) induced for 21 days. (D) Cells containing pZJM (HKORF) induced for 14 days and then washed free of tetracycline for 7 days. Prior to washing out tetracycline, cells harboring pZJM(HKORF) (and induced for 14 days) were virtually indistinguishable from cells induced for 21 days, with the exception being that EP1-1(-10) and EP3(-10) could be detected at very low levels. These procyclins are from the cells examined by flow cytometry in Figure 3.
Glycolysis and procyclin expression
To further investigate the relationship between glucose metabolism and procyclin expression, we used RNAi to silence expression of the trypanosome hexose transporter gene (THT) family. RNAi using a sequence fragment common to all THT family members silenced all THT transcripts (as determined by northern analysis, data not shown). Furthermore, after induction of RNAi for 14 days, [3H]2-deoxyglucose uptake assays indicated that glucose transport was reduced by >90% (data not shown). Interestingly, these parasites also became deficient in conA binding, as measured by flow cytometry (data not shown), similar to that in hexokinase-deficient cells. MALDI-TOF MS analysis confirmed a switch in procyclin expression from EP-procyclins to GPEET-procyclins, although detectable EP species were still present, perhaps a result of incomplete silencing of the hexose transporters (data not shown).
We next assessed the overall role of the glycolytic pathway in procyclin regulation, using RNAi to target the two genes for pyruvate kinase (PK; the last enzyme in the glycolytic pathway). Unlike the case with hexokinase or the hexose transporters, RNAi silencing of the PK genes was lethal after 7 days of induction, with inhibition of cell growth by day 4 (Figure 5A, upper panel). However, if we adapted the parasites to grow in low glucose medium, a condition in which they downregulate glycolysis and use amino acids as a carbon source (ter Kuile, 1997), induction of PK RNAi has only a minimal effect on cell growth (Figure 5A, lower panel). Analysis of the PK-deficient cells grown in low glucose indicates that they have reduced conA binding (by flow cytometry) similar to cells with silenced hexokinase (Figure 5B, lower panel). Furthermore, MALDI-TOF MS confirms that this loss of binding is due to a complete switch in procyclin expression from EP to GPEET (data not shown).
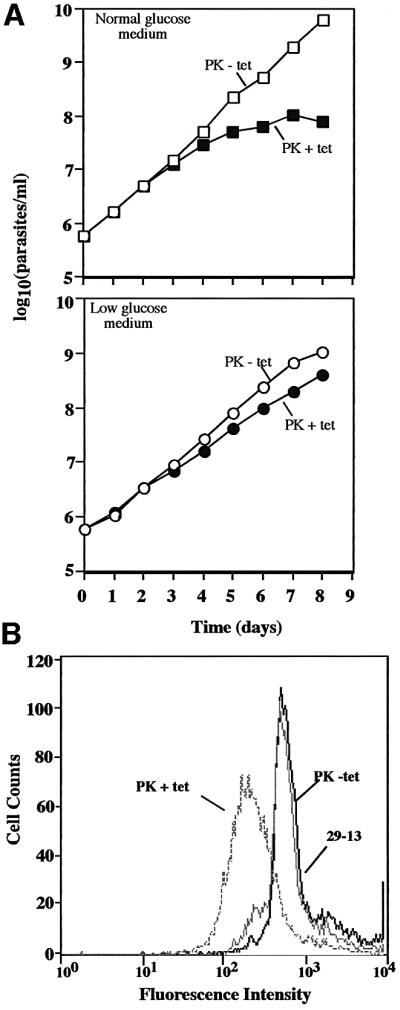
Fig. 5. Effect of silencing PKs on cell growth and conA binding. (A) Trypanosomes grown in triplicate in normal SDM-79 medium (upper panel) or low glucose SDM-79 (lower panel) were induced to express PK dsRNA as indicated. Cell densities were determined with a Coulter counter. Cells were diluted 10-fold when the density reached 5 × 106/ml, and densities shown in the graph are the products of cell density and the dilution factor. Error bars are too small to be visible. (B) Trypanosomes were grown in low glucose SDM-79 and then incubated with 10 µg/ml fluorescein-conjugated conA and analyzed by flow cytometry as described in Figure 1. Cells were induced for 5 days prior to analysis.
Effect of glucose levels on procyclin expression
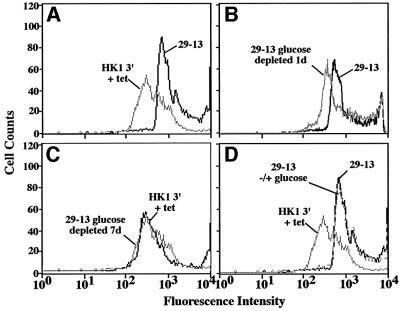
Growth of 29-13 procyclics under low (0.5 mM) glucose conditions does not alter conA binding or procyclin expression (Figure 5B; data not shown). To determine the effect of growing parasites in glucose-depleted medium, we shifted 29-13 cells from growth in low glucose medium to a glucose-depleted medium (∼0.03 mM glucose, based on the manufacturer’s analysis of the dialyzed serum). After 1 day of growth in a glucose-depleted environment, cells exhibited a loss of conA–FITC binding (Figure 6B). To determine whether glucose could reverse the loss of lectin binding, the culture (after growing for 5 days in glucose-depleted medium) was divided and half was supplemented with glucose (5 mM) for 2 days. Cells maintained in the glucose-depleted environment for 7 days were essentially unchanged from day 5 (data not shown), with a substantial loss of conA binding almost identical to that seen in pZJM(HK1 3′) cells induced for 10 days (Figure 6C). However, cells supplemented with glucose on day 5 and analyzed on day 7 are nearly indistinguishable by flow cytometry from cells grown in normal medium (Figure 6D), indicating that loss of conA binding is reversible.
Fig. 6. Growth in glucose-depleted medium leads to loss of conA binding in parental 29-13 cells. Trypanosomes (29-13) grown in normal SDM-79 were seeded into low glucose medium, grown for 4 days and then passed (1:10) into glucose-depleted medium when they reached a density of ∼5 × 106 cell/ml. After 5 days growth in glucose-depleted medium, glucose (5 mM) was added back to a portion of the culture (29-13 –/+ glucose). Cells were analyzed by flow cytometry on day 7 using conA–FITC as described in Figure 1. As a control, cells harboring pZJM(HK1 3′) were induced for 10 days.
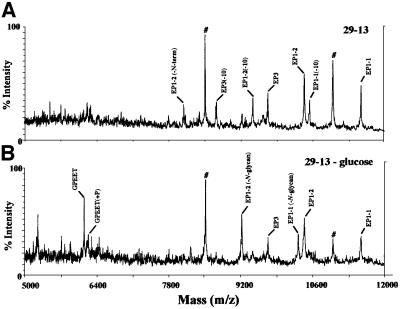
MALDI-TOF MS confirmed that cells grown in glucose-depleted medium for 4 days had significantly changed procyclin expression. Consistent with the flow cytometry (Figure 6), these cells expressed abundant GPEET-procyclin (Figure 7B). Interestingly, they also expressed unglycosylated EP species, as well as normally glycosylated EP1-1 and EP1-2.
Fig. 7. Trypanosoma brucei grown in glucose-depleted medium upregulates GPEET-procyclin. 29-13 cells adapted to low glucose medium were grown for 4 days in glucose-depleted medium prior to procyclin isolation and analysis as described in Figure 4. EP1-2(-N-term), the ion at m/z 8089, probably results from non-specific cleavage of 24 residues from the N-terminus of EP1-2 (Acosta-Serrano, 1999). See Figure 4 for discussion of ions at m/z 8504 and 10 987 (indicated with a hash). (A) Parental 29-13 cells cultured in SDM-79 medium. (B) 29-13 cells grown for 4 days in glucose-depleted medium.
Discussion
RNAi is a powerful technique for analysis of gene function in many organisms, including T.brucei. Here we advanced the use of this method by developing an RNAi-based library, allowing use of RNAi for forward genetic screens. Forward genetic approaches had previously been limited by the diploidy of this organism. This genomic RNAi library is, to our knowledge, the first developed for any organism.
The selection and screening for RNAi-generated conA resistance were challenging. In our initial experiment, we attempted a selection in which we cultured cells in the presence of a lethal concentration of conA. Resistant cells emerged, but they proved to be spontaneously generated mutants as their conA-resistant phenotype did not depend on RNAi. As an alternative approach, we screened for cells with reduced conA binding using conA–Sepharose chromatography and FACS. With this approach we isolated a single clone (clone 1) that contained a 341 bp fragment of the 3′ UTR of the hexokinase 1 gene within pZJMβ (see below for a description of an improved screening procedure).
The EC50 for conA resistance of clone 1 was ∼8 µg/ml (Figure 1A), a value lower than those reported previously for chemically generated conA mutants 1-1 and 4-1 (with EC50s of 25 and 100 µg/ml, respectively) (Hwa et al., 1999). One possible reason for this difference is that the Coulter counter might not accurately discriminate living cells from dead ones, leading to a conservative estimation of the EC50. Another possible explanation is that RNAi does not cause a complete knockdown of EP-procyclin expression. Flow cytometry of this population supports this suggestion, with some remaining trypanosomes that bind parental levels of conA (Figure 1B).
Northern analysis indicates that a single transcript containing the 3′ UTR of hexokinase 1 is expressed, and this transcript is silenced following RNAi (Figure 2B, lanes 1 and 2). A second hexokinase gene (with a distinct 3′ UTR) resides on chromosome 10 in tandem to the hexokinase gene described here. Since the same phenotype is seen whether hexokinase 1 is silenced alone [using the pZJM(HK1 3′) construct] or both hexokinase genes are targeted simultaneously [using pZJM(HKORF)], hexokinase 2 may not be involved in this conA resistance, or is at least epistatic to hexokinase 1. Targeting both genes using pZJM(HKORF) leads to greater loss (95%) of hexokinase activity than targeting hexokinase 1 alone (79%), probably because both gene products contribute to the total hexokinase activity. However, we cannot rule out the possibility that pZJM(HKORF) targets hexokinase 1 more efficiently than pZJM(HK1 3′), which would be consistent with hexokinase 1 being responsible for all of the observed activity.
How does silencing of hexokinase 1, the glucose transporters or PKs lead to reduced conA binding? We considered the possibility that RNAi of these proteins could disrupt sugar metabolism and that this disruption, in turn, could affect procyclin N-glycosylation or GPI anchoring. Since glucose starvation in mammalian cells leads to altered N-glycan biosynthesis (Rearick et al., 1981), we were surprised to find no difference between the levels of mature GPI and N-glycan precursors in parental 29-13 cells and in cells induced for RNAi. While incomplete RNAi silencing might allow a sufficient amount of glucose to be metabolized and utilized for sugar nucleotide biosynthesis, we considered the possibility that trypanosomes could employ a portion of the gluconeogenic pathway to synthesize sugars for sugar nucleotides. Trypanosoma brucei glycosomes contain phosphoenolpyruvate carboxykinase (PEPCK) (Hart et al., 1984; Hunt and Kohler, 1995) and we have identified a candidate gene for fructose-1,6-bisphosphatase from the T.brucei genome sequence database. Both enzymes would be required for conversion of oxaloacetate (derived from proline via α-ketoglutarate) to glucose-6-phosphate, which could ultimately form sugar nucleotides. Some of these activities may be expressed only under very low glucose concentrations, as procyclic trypanosomes grown in normal glucose medium lack detectable fructose-1,6-bisphosphatase activity (Cronin et al., 1989). Our data on cells grown under glucose-depleted conditions (∼0.03 mM glucose) support the possibility of gluconeogenesis, as these cells still express some glycosylated EP-procyclins with GPI anchors (Figure 7). However, the concomitant expression of unglycosylated EP-procyclins raises the possibility that gluconeogenesis may not be sufficient for synthesis of enough N-glycan for all EP-procyclins. Alternatively, the very low sugar concentration in these cultures may be efficiently funneled to the N-glycan and GPI biosynthetic pathways, yielding some glycosylated EP-procyclins. Nevertheless, since synthesis of the N-glycan and GPI precursors is not altered in hexokinase-deficient cells, there must be another reason for loss of conA binding.
We found, using MS, that RNAi silencing of hexokinase caused a dramatic switch from nearly exclusively EP-procyclins to the normally unglycosylated GPEET- procyclin. Our finding that RNAi-induced silencing of the hexose transporters and PKs also upregulated GPEET- procyclin indicates that a reduction in flux through the glycolytic pathway may affect procyclin expression. It is notable that glycolytic flux serves as a glucose sensor in other cells. For example, secretion of insulin by pancreatic β cells is regulated by blood glucose levels, which the cells monitor by a measurement of the rate of glycolysis (reviewed in Ashcroft and Gribble, 1999).
The molecular link(s) between glucose sensing and procyclin expression remains to be determined. The effect could be direct or indirect, and glycolysis could be only one of several factors influencing procyclin expression. However, the effect of glucose metabolism on procyclin expression could have biological significance, as the parasite naturally undergoes changes in surface coat expression at a time of dramatic changes in availability of metabolic fuels. When the tsetse fly takes a blood meal from the mammalian host, the parasites enter the insect midgut, where they differentiate to procyclic forms. An early event in differentiation is a switch in surface coats, from VSG to procyclin, a process that can be initiated in vitro by growth of bloodstream forms in low glucose medium (Milne et al., 1998). In the early stages of differentiation in the fly midgut, the parasite has access to high glucose levels derived from the mammalian blood, but subsequently the glucose concentration is rapidly reduced (C.M.Turner, personal communication). At later stages of development, the major carbon and energy sources for procyclic trypanosomes are amino acids, predominantly proline. The changes in concentration of glucose (and possibly other metabolites) could be factors controlling the switches in procyclin expression.
At the earliest stages of differentiation in vitro, all procyclin species are expressed (Vassella et al., 2001). This is followed, in 24 h, by a change in expression to predominantly GPEET-procyclin. It is this switch that could be controlled by glycolytic flux or metabolite levels. Ultimately, at 72 h, some of the EP-procyclins become more abundant (Vassella et al., 2001). Attempts to analyze procyclin expression throughout the parasite’s development in the tsetse fly were limited due to inadequate numbers of parasites in the earliest stages of development (Acosta-Serrano et al., 2001). However, the earliest stages that could be analyzed expressed exclusively GPEET- procyclin. [See Acosta-Serrano et al. (2001) and Vassella et al. (2001) for speculations on the significance of these shifts in procyclin expression.]
While we could modulate procyclin expression by silencing the glycolytic pathway, we were unable to trigger GPEET expression by growing 29-13 cells in medium containing low glucose (0.5 mM), probably because 0.5 mM glucose is not low enough. However, in the near absence of glucose (using glucose-depleted medium with dialyzed FBS, which yielded a final glucose concentration of ∼0.03 mM), cells rapidly (<1 day) began to change the procyclin expression profiles, as determined by flow cytometry (Figure 6). After 4 days, as shown by MALDI-TOF MS, these cells had a marked increase in GPEET expression (Figure 7B). Growth under glucose-depleted conditions did not eliminate EP-procyclin expression, but rather led to expression of both non-glycosylated and glycosylated EP species (Figure 7B). The glucose, which is present at very low levels, may be low enough to trigger increased GPEET expression but high enough to sustain EP expression. Alternatively, the mechanism that triggers GPEET upregulation may be directly linked to glucose levels in the medium; suppression of EP may require additional signals that were effectively generated in the hexokinase-deficient cells but not recapitulated in this experiment.
Interestingly, the glucose-depleted phenotype was quickly reversed by the addition of glucose to the medium (Figure 6D). The loss of N-glycosylation on some EP-procyclins (which are GPI anchored) suggests that trypanosomes may direct limited sugar resources preferentially to GPI biosynthesis. The finding that use of a glucose-depleted medium upregulates GPEET-procyclin, in an experimental approach totally independent of RNAi, strengthens the connection between glycolysis and procyclin expression.
It was surprising that RNAi silencing of hexokinase and the glucose transporters was not lethal to the trypanosome, whereas silencing of the PKs (in normal glucose medium) caused cell death. The reason for this difference could be that the loss of activity of hexokinase and THT was slow (perhaps due in part to a high copy number or stability of the proteins, or to a slower loss of the mRNA). As shown in Figure 3, it took ∼21 days for the hexokinase phenotype to be fully expressed. This slow loss probably gives the parasites necessary time to adapt to a metabolic state less dependent on glycolysis and more dependent on proline metabolism, as described previously (ter Kuile, 1997). In contrast, RNAi silencing of the PKs led to growth inhibition by day 4, followed by cell death. In this case, death may have occurred before the cells had time to adapt to proline metabolism. Support for this idea comes from our finding that silencing of the PKs had only a minimal effect on cell growth if the parasites had been pre-adapted to low glucose medium prior to RNAi induction. Under those conditions, silencing of the PKs caused the same switch in procyclin expression observed for RNAi-induced silencing of hexokinase and the glucose transporter.
This work proves the feasibility of using the RNA library for forward genetic screens. It is also provides a powerful method for discovering novel biological functions of T.brucei genes. This approach has limitations, however. For example, RNAi phenotypes appear at different rates, based on transcript abundance and protein levels and stability. Furthermore, production of small amounts of dsRNA without tetracycline addition (Wang et al., 2000), probably due to incomplete repression of transcription by the tetracycline repressor (Wirtz et al., 1999), could be a problem with some genes. Also, RNAi using a given gene fragment sometimes does not cause degradation of the cognate mRNA. Finally, cells can become resistant to RNAi, which might explain the small percentage of cloned pZJM(HKORF) cells that do not exhibit reduced conA binding levels after 21 days of RNAi induction (Figure 3). Nevertheless, the prospect of identifying a few genes using this forward genetic approach makes these limitations bearable.
We are currently performing library screens to study other aspects of trypanosome biology. We are also revisiting conA binding in screens using new protocols. Since it is likely that the combination of six rounds of conA–Sepharose chromatography and two separations by FACS led to bias in our initial screen (resulting in isolation of only a single conA-resistant cell), we have recently undertaken a new approach in which we sort fluorescein-labeled conA-stained cells by one round of FACS and then immediately clone cells with reduced lectin binding. We isolated 156 clones in a preliminary screen, of which 16 have confirmed tetracycline-inducible loss of conA binding. The false positives were probably the result of imprecise cell sorting. One of the genes recovered in this screen encoded EP-procyclin, a finding that validates this approach. Studies of other genes discovered in this screen could clarify the link between glycolysis and procyclin gene expression. Alternatively, they could illuminate other aspects of procyclin expression or modification.
Materials and methods
RNAi library construction
The pZJM vector (Wang et al., 2000) was modified to insert a multicloning site. Annealing the oligonucleotides (5′-TCGAGGGCCAGTGAGGCCTCTAGAGGGCCCCATATGGTTAACGGCCACACAGGCCA-3′ and 5′-AGCTTGGCCTGTGTGGCCGTTAACCATAT GGGGCCCTCTAGAGGCCTCACTGGCCC-3′) generated a duplex with XhoI and HindIII cohesive ends. This DNA was ligated into the XhoI and HindIII sites of pZJM, generating pZJMβ, which has XhoI, HindIII, XbaI, NdeI, HpaI and two SfiI cloning sites.
To generate the RNAi library, 100 µg of T.brucei strain 927 DNA were sonicated for 1 min on ice at 50% maximum power using a Branson Model W140 sonicator, generating DNA with an average size of ∼1 kb. The sonicated DNA was incubated with BAL-31 nuclease (New England Biolabs; 2 U in 200 µl) for 5 min at 25°C, followed by extraction with phenol:chloroform:isoamyl alcohol [25:24:1 (v/v)] and precipitation of the DNA. The DNA was then incubated with T4 DNA polymerase (Invitrogen; 5 U in 100 µl) and 0.2 mM dNTPs for 20 min at 37°C, followed by heat inactivation of the enzyme (65°C, 10 min). Adapter oligonucleotides (AGGCCTCGCGA and 5′ phosphorylated TCGCGA GGCCTCAC) were annealed and then ligated to the genomic fragments. Free adapters were removed by passing the ligation mixture twice over a Qiaquick PCR purification column (Qiagen, Valencia, CA).
Inserts were ligated into the SfiI sites of pZJMβ and the plasmid electroporated into Escherichia coli XL-1 blue cells. A total of ∼5 × 105 colonies were plated and pooled, and plasmid DNA was isolated. To assess the quality of the library, E.coli were re-transformed with purified pZJMβ library, plated and plasmids purified from individual colonies. Inserts were found in 92% of plasmids and the average size was 660 bp.
Trypanosome growth
Procyclic 29-13 T.brucei (a gift from Drs Elizabeth Wirtz and George Cross, Rockefeller University), a 427 strain that expresses T7 RNA polymerase and the tetracycline repressor (Wirtz and Clayton, 1995; Wirtz et al., 1998), were grown in SDM-79 as described previously (Wirtz et al., 1999). Low glucose SDM-79 was prepared with glucose-free RPMI 1640 replacing the liquid MEM. Also, additional glucose (normally present in SDM-79 at 1 mg/ml) was omitted. This mixture was supplemented with normal FBS (10%), resulting in a final glucose concentration of ∼0.5 mM. Glucose-depleted medium (final glucose concentration ∼0.03 mM) differed from low glucose medium in that 10% dialyzed FBS (Invitrogen, Carlsbad, CA) was used in place of normal serum. Prior to growth in glucose-depleted medium, parasites were first adapted for 4 days to low glucose medium.
Transfection
To cover the parasite genome five times with the pZJM library (average insert size 660 bp), we needed to generate 2.7 × 105 transfected parasites. To determine the transfection efficiency, we electroporated 1 × 108 parasites with 10 µg of NotI-linearized library and allowed the cells to recover for 24 h. Cells were centrifuged and resuspended in medium containing phleomycin (2.5 µg/ml), and plated in the first row of a 96-well plate at 5 × 106/ml. Subsequent rows of the plates were seeded with 1:1 dilutions of the preceding row. The greatest dilution yielding viable wells was used (along with an observed plating efficiency of 30%) to calculate the transfection efficiency. Two independent experiments yielded values of ∼1 × 10–4. Given this transfection efficiency, we electroporated ∼5 × 109 parasites in 50 transfections (1 × 108 cells/transfection) (Wang et al., 2000).
ConA screening
To screen the library-transfected trypanosomes for cells resistant to conA binding, we induced RNAi with 1 µg/ml tetracycline (Wang et al., 2000) for 7 days. The culture (5 ml at 5 × 106 cells/ml) was then passed over a 1 ml conA–Sepharose 4B column (15 mg lectin/ml packed gel; Sigma, St Louis, MO) pre-equilibrated with cytomix (van den Hoff et al., 1992) supplemented with 1 mM MnCl2 (cytoM). Parasites not binding to the column or that washed off in 1 ml of cytoM were recovered by centrifugation and resuspended in fresh medium. The culture was allowed to recover for 5–7 days and the process was repeated five times. This was followed by a round of FACS (see below), another column enrichment, and a final round of FACS.
FACS and flow cytometry
Cells (5 × 107) purified by conA–Sepharose were centrifuged and resuspended in 5 ml of cytoM containing 10 µg/ml fluorescein-labeled conA (Sigma). After 15 min at room temperature, trypanosomes were filtered through a cell strainer (35 µm pore size) to remove large aggregates. FACS was performed on a FACStar Plus (Becton Dickinson Biosciences, Franklin Lakes, NJ) at the Johns Hopkins School of Medicine Flow Cytometry Core Analytic Laboratory using an argon laser with 488 nm emission. Cells incubated without lectin were used to establish the forward scattering gate and as a control for autofluorescence. Cells with reduced fluorescein-labeled conA binding (typically ∼5% of the population) were sorted and, after the second sorting (see previous paragraph), cloned by limiting dilution into uncoated 24-well tissue culture plates (Falcon 351174) and grown under 5% CO2 at 28°C. (This cloning method is a personal communication from Dr W.Gibson, University of Bristol, Bristol, UK.)
Flow cytometry using a FACScan flow cytometer (Becton Dickinson Biosciences) was used to analyze the tetracycline-dependent loss of conA binding. Cells were prepared as described for FACS and 10 000 cells were analyzed per sample.
Identification of the gene responsible for conA resistance
Genomic DNA was isolated from sorted and cloned cells using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN). The insert was amplified using BetaF (CCCCAAGGGGTTATGCTAGT) and BetaR (GATCTAGCCCCTGCAGGAAT) primers, and the resulting PCR product was cloned into pCR2.1-TOPO (Invitrogen), sequenced, and analyzed by a BLAST search of the T.brucei sequence database.
The insert was identified as a region of the 3′ UTR of an annotated hexokinase 1 gene (AJ345044). A second hexokinase gene (see Figure 2A) is not targeted by this construct. We also tested a 502 bp fragment of the open reading frame of the hexokinase 1 gene, which is 98% identical to the same region in the hexokinase 2 gene. We obtained this fragment by PCR using primers HKORFF (CCTTCTTCAGGCTGCTTTTG) and HKORFR (GAACTGAAGTCCGGGCATAC), and ligated it into pZJM. We generated a 342 bp probe for northern analysis of the hexokinase 2 gene using PCR primers HK2F (GCGAACGTAGTGTAATTTTCTG) and HK2R (CTCCCCCGCTCAGTTATAAC).
RNAi of other genes
Other fragments were inserted into pZJM for RNAi experiments. A 350 bp fragment from the open reading frame of the hexose transporter THT1e (Bringaud and Baltz, 1993) was generated using primers THTF (CACATCACCGGGTTTCTTCT) and THTR (TGCACTTCCCGGCTATAATC). In RNAi experiments, this fragment targets all THT1 and THT2 gene family members. A 503 bp fragment targeting expression of both PK genes was generated using primers PKF (CACCCTGGTGGTTTTCAATC) and PKR (TATTGGTATTGCGCTGGACA).
Hexokinase enzyme assays
Assays were performed as described previously (Misset and Opperdoes, 1984). Cells (1 × 107) were washed, lysed in 100 µl of 0.1% Triton X-100, 20 mM Tris–HCl pH 7.4 for 10 min on ice, and assayed in triplicate. Protein concentrations were determined by Bradford assay using BSA as a standard.
Procyclin purification and MALDI-TOF MS
Procyclins were purified from 5 × 107 cells essentially as described previously (Acosta-Serrano et al., 1999). Briefly, parasites were harvested by centrifugation and washed in cold PBS. Procyclins were then extracted with 0.1% Triton X-100 for 15 min at 0°C (modified from Vassella et al., 2001). Samples were centrifuged (14 000 g for 5 min) and the supernatant extracted four times with water-saturated n-butanol. The aqueous phase was dried and procyclin GPI anchors removed by a 16 h incubation in 48% hydrofluoric acid (HF) on ice (Mehlert et al., 1999). Procyclins were purified on a C18 ZipTip (Millipore, Bedford, MA).
Negative-ion mass spectra were acquired using a Voyager-DE STR BioSpectrometry Workstation (PerSeptive Biosystems, Framingham, MA) at the Applied Biosystems Mass Spectrometry Facility at Johns Hopkins. Procyclins and bovine insulin (in some cases) were co-crystallized with sinapinic acid, and masses calibrated against the internal and external standards.
Acknowledgments
Acknowledgements
We appreciate the helpful advice from Elisabetta Ullu in building the RNAi library. We thank Alvaro Acosta-Serano, Isabel Roditi, Erik Vassella, Tina Saxowsky and Soo Hee Lee for helpful comments on this manuscript. Last, we would like to remember Viiu Klein—her skilled technical assistance and kind nature will be sorely missed. This work was supported by NIH grant AI21334.
References
- Acosta-Serrano A., Cole,R.N., Mehlert,A., Lee,M.G., Ferguson,M.A. and Englund,P.T. (1999) The procyclin repertoire of Trypanosoma brucei. Identification and structural characterization of the Glu-Pro-rich polypeptides. J. Biol. Chem., 274, 29763–29771. [DOI] [PubMed] [Google Scholar]
- Acosta-Serrano A., Cole,R.N. and Englund,P.T. (2000) Killing of Trypanosoma brucei by concanavalin A: structural basis of resistance in glycosylation mutants. J. Mol. Biol., 304, 633–644. [DOI] [PubMed] [Google Scholar]
- Acosta-Serrano A., Vassella,E., Liniger,M., Kunz Renggli,C., Brun,R., Roditi,I. and Englund,P.T. (2001) The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc. Natl Acad. Sci. USA, 98, 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft F.M. and Gribble,F.M. (1999) ATP-sensitive K+ channels and insulin secretion: their role in health and disease. Diabetologia, 42, 903–919. [DOI] [PubMed] [Google Scholar]
- Bringaud F. and Baltz,T. (1993) Differential regulation of two distinct families of glucose transporter genes in Trypanosoma brucei. Mol. Cell. Biol., 13, 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butikofer P., Ruepp,S., Boschung,M. and Roditi,I. (1997) ‘GPEET’ procyclin is the major surface protein of procyclic culture forms of Trypanosoma brucei brucei strain 427. Biochem. J., 326, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin C.N., Nolan,D.P. and Voorheis,H.P. (1989) The enzymes of the classical pentose phosphate pathway display differential activities in procyclic and bloodstream forms of Trypanosoma brucei. FEBS Lett., 244, 26–30. [DOI] [PubMed] [Google Scholar]
- Hart D.T., Misset,O., Edwards,S.W. and Opperdoes,F.R. (1984) A comparison of the glycosomes (microbodies) isolated from Trypanosoma brucei bloodstream form and cultured procyclic trypomastigotes. Mol. Biochem. Parasitol., 12, 25–35. [DOI] [PubMed] [Google Scholar]
- Hunt M. and Kohler,P. (1995) Purification and characterization of phospho enol pyruvate carboxykinase from Trypanosoma brucei. Biochim. Biophys. Acta, 1249, 15–22. [DOI] [PubMed] [Google Scholar]
- Hwa K.Y., Acosta-Serrano,A., Khoo,K.H., Pearson,T. and Englund,P.T. (1999) Protein glycosylation mutants of procyclic Trypanosoma brucei: defects in the asparagine-glycosylation pathway. Glycobiology, 9, 181–190. [DOI] [PubMed] [Google Scholar]
- Masterson W.J., Doering,T.L., Hart,G.W. and Englund,P.T. (1989) A novel pathway for glycan assembly: biosynthesis of the glycosyl-phosphatidylinositol anchor of the trypanosome variant surface glycoprotein. Cell, 56, 793–800. [DOI] [PubMed] [Google Scholar]
- Mehlert A., Treumann,A. and Ferguson,M.A. (1999) Trypanosoma brucei GPEET-PARP is phosphorylated on six out of seven threonine residues. Mol. Biochem. Parasitol., 98, 291–296. [DOI] [PubMed] [Google Scholar]
- Milne K.G., Prescott,A.R. and Ferguson,M.A. (1998) Transformation of monomorphic Trypanosoma brucei bloodstream form trypomastigotes into procyclic forms at 37°C by removing glucose from the culture medium. Mol. Biochem. Parasitol., 94, 99–112. [DOI] [PubMed] [Google Scholar]
- Misset O. and Opperdoes,F.R. (1984) Simultaneous purification of hexokinase, class-I fructose-bisphosphate aldolase, triosephosphate isomerase and phosphoglycerate kinase from Trypanosoma brucei. Eur. J. Biochem., 144, 475–483. [DOI] [PubMed] [Google Scholar]
- Mowatt M.R. and Clayton,C.E. (1987) Developmental regulation of a novel repetitive protein of Trypanosoma brucei. Mol. Cell. Biol., 7, 2838–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowatt M.R., Wisdom,G.S. and Clayton,C.E. (1989) Variation of tandem repeats in the developmentally regulated procyclic acidic repetitive proteins of Trypanosoma brucei. Mol. Cell. Biol., 9, 1332–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H., Tschudi,C., Gull,K. and Ullu,E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T.W., Beecroft,R.P., Welburn,S.C., Ruepp,S., Roditi,I., Hwa,K.Y., Englund,P.T., Wells,C.W. and Murphy,N.B. (2000) The major cell surface glycoprotein procyclin is a receptor for induction of a novel form of cell death in African trypanosomes in vitro. Mol. Biochem. Parasitol., 111, 333–349. [DOI] [PubMed] [Google Scholar]
- Rearick J.I., Chapman,A. and Kornfeld,S. (1981) Glucose starvation alters lipid-linked oligosaccharide biosynthesis in Chinese hamster ovary cells. J. Biol. Chem., 256, 6255–6261. [PubMed] [Google Scholar]
- Richardson J.P., Beecroft,R.P., Tolson,D.L., Liu,M.K. and Pearson,T.W. (1988) Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol. Biochem. Parasitol., 31, 203–216. [DOI] [PubMed] [Google Scholar]
- Roditi I. and Clayton,C. (1999) An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol. Biochem. Parasitol., 103, 99–100. [DOI] [PubMed] [Google Scholar]
- Roditi I., Carrington,M. and Turner,M. (1987) Expression of a polypeptide containing a dipeptide repeat is confined to the insect stage of Trypanosoma brucei. Nature, 325, 272–274. [DOI] [PubMed] [Google Scholar]
- Roditi I. et al. (1989) Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J. Cell Biol., 108, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp S., Furger,A., Kurath,U., Renggli,C.K., Hemphill,A., Brun,R. and Roditi,I. (1997) Survival of Trypanosoma brucei in the tsetse fly is enhanced by the expression of specific forms of procyclin. J. Cell Biol., 137, 1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Kuile B.H. (1997) Adaptation of metabolic enzyme activities of Trypanosoma brucei promastigotes to growth rate and carbon regimen. J. Bacteriol., 179, 4699–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treumann A., Zitzmann,N., Hulsmeier,A., Prescott,A.R., Almond,A., Sheehan,J. and Ferguson,M.A. (1997) Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J. Mol. Biol., 269, 529–547. [DOI] [PubMed] [Google Scholar]
- Tzfati Y., Abeliovich,H., Kapeller,I. and Shlomai,J. (1992) A single-stranded DNA-binding protein from Crithidia fasciculata recognizes the nucleotide sequence at the origin of replication of kinetoplast DNA minicircles. Proc. Natl Acad. Sci. USA, 89, 6891–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoff M.J., Moorman,A.F. and Lamers,W.H. (1992) Electro poration in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res., 20, 2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassella E., Den Abbeele,J.V., Butikofer,P., Renggli,C.K., Furger,A., Brun,R. and Roditi,I. (2000) A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev., 14, 615–626. [PMC free article] [PubMed] [Google Scholar]
- Vassella E., Acosta-Serrano,A., Studer,E., Lee,S.H., Englund,P.T. and Roditi,I. (2001) Multiple procyclin isoforms are expressed differentially during the development of insect forms of Trypanosoma brucei. J. Mol. Biol., 312, 597–607. [DOI] [PubMed] [Google Scholar]
- Vickerman K. (1985) Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull., 41, 105–114. [DOI] [PubMed] [Google Scholar]
- Wang Z. and Englund,P.T. (2001) RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J., 20, 4674–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Morris,J.C., Drew,M.E. and Englund,P.T. (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem., 275, 40174–40179. [DOI] [PubMed] [Google Scholar]
- Welburn S.C., Dale,C., Ellis,D., Beecroft,R. and Pearson,T.W. (1996) Apoptosis in procyclic Trypanosoma brucei rhodesiense in vitro. Cell Death Differ., 3, 229–236. [PubMed] [Google Scholar]
- Wirtz E. and Clayton,C. (1995) Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science, 268, 1179–1183. [DOI] [PubMed] [Google Scholar]
- Wirtz E., Hoek,M. and Cross,G.A. (1998) Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res., 26, 4626–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E., Leal,S., Ochatt,C. and Cross,G.A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol., 99, 89–101. [DOI] [PubMed] [Google Scholar]
- Ziegelbauer K., Quinten,M., Schwarz,H., Pearson,T.W. and Overath,P. (1990) Synchronous differentiation of Trypanosoma brucei from bloodstream to procyclic forms in vitro. Eur. J. Biochem., 192, 373–378. [DOI] [PubMed] [Google Scholar]