Abstract
Proper placement of the bacterial cell division site requires the site-specific inactivation of other potential division sites. In Escherichia coli, selection of the correct mid-cell site is mediated by the MinC, MinD and MinE proteins. To clarify the functional role of the bacterial cell division inhibitor MinD, which is a membrane-associated ATPase that works as an activator of MinC, we determined the crystal structure of a Pyrococcus furiosus MinD homologue complexed with a substrate analogue, AMPPCP, and with the product ADP at resolutions of 2.7 and 2.0 Å, respectively. The structure reveals general similarities to the nitrogenase iron protein, the H-Ras p21 and the RecA-like ATPase domain. Alanine scanning mutational analyses of E.coli MinD were also performed in vivo. The results suggest that the residues around the ATP-binding site are required for the direct interaction with MinC, and that ATP binding and hydrolysis play a role as a molecular switch to control the mechanisms of MinCDE-dependent bacterial cell division.
Keywords: ATPase/crystal structure/filaments/MinD/protein interaction
Introduction
The mechanisms of bacterial cell division are poorly understood, although many genes responsible for the division process have been characterized. In Escherichia coli, cell division is initiated by the localization of the tubulin-like GTPase FtsZ to the future division site. FtsZ assembles into a ring, and other proteins are recruited to form the septal ring organelle (Rothfield and Justice, 1997; Margolin, 1998). It was suggested that the formation of this FtsZ assembly, which is a key regulation step in the cell division phase, occurs through a self-organization reaction involving a GTP cycle (Lutkenhaus, 1993). The formation of the FtsZ ring itself is not specific for the mid-cell site and is restricted by several inhibitors of the cell division reaction, such as SulA and MinCDE (Bi and Lutkenhaus, 1993; Justice et al., 2000).
The division septum is normally placed at the mid-point of the cell, but potential division sites (PDSs) also exist near each of the cell poles (Alder et al., 1967; Teather et al., 1974). Restriction of the division site at the mid-point is governed by the products of the three genes of the minB operon, minC, minD and minE (Cook et al., 1989; de Boer et al., 1990a). MinC inhibits the division at all of the PDSs but normally requires the activity of peripheral membrane ATPase MinD for its function (de Boer et al., 1991). At normal levels of minB gene expression, MinC and MinD act cooperatively to form a non-specific division inhibitor complex (de Boer et al., 1990b, 1992; Mulder et al., 1992). MinE is thought to have two functions: it suppresses the MinCD-mediated division inhibition and recognizes the mid-cell points from the cell poles. In the absence of MinE, the cells form filaments and the other two Min proteins are distributed randomly along the cell membrane (de Boer et al., 1989). This observation led to the conclusion that MinE plays two roles in the cell: as an antagonist of the MinCD division inhibitor and as a topological specificity factor. In other words, MinE promotes mid-cell division by excluding MinCD from the mid-cell site. A recent study has revealed that MinE is required for MinD segregation/oscillation (Raskin and de Boer, 1999a). Moreover, the MinC oscillation requires MinD, but not vice versa. It is suggested that MinD may play a role as the oscillation motor, while MinC is a cargo (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999b). Although these three proteins are interdependent, as described above, precise information about their roles remains elusive.
The role of MinD in the cell division stage is not as clearly defined as those of the other minB products. What roles do nucleotide binding and hydrolysis play? How does MinD stimulate the MinC function? The MinD functions were defined previously as follows: first, MinD interacts directly with MinC in the yeast two-hybrid system (Huang et al., 1996) and the ATPase activity of MinD is required for MinC activation (de Boer et al., 1991). Secondly, MinD is required for the site-specific modulation of division inhibition by MinE. Thirdly, MinD is required to localize MinE at the mid-cell site (Raskin and de Boer, 1997). The structural analysis of E.coli MinD has not been successful yet, because of difficulty in crystallization. There exist three MinD homologues in the hyperthermophilic archaeon Pyrococcus furiosus (Bernander, 1998; Gerard et al., 1998), and we could crystallize one of the P.furiosus MinD homologues. Two distinct crystal structures, the complexes with ADP-Mg2+ and with AMPPCP-Mg2+, were determined at 2.0 and 2.7 Å resolution, respectively. Based on the crystal structures, we also performed mutational analyses in vivo to gain functional insight into the specific interactions of E.coli MinD with MinC. The results indicate that MinC activation in vivo depends on the residues around the nucleotide-binding site in MinD.
Results
Structure of the MinD–nucleotide complexes
The structures of P.furiosus MinD complexed with ADP and AMPPCP were determined by multiple isomorphous replacement (MIR) followed by density modification procedures (Table I). Both of the final models consist of residues 1–237, one nucleotide, one Mg2+ and solvent molecules. The C-terminal region of MinD was structurally disordered.
Table I. Crystallographic statistics.
| Data collection | ADP form | ATP form | |||
|---|---|---|---|---|---|
| Space group | P213 | P213 | |||
| Unit cell dimensions (Å) | a = 96.04 | a = 98.64 | |||
| Data set | Native | MeHgCl | NaOsCl6 | K2Pt(SCN)4 | |
| X-ray source | BL41 | BL41 | CuKα (Dip2030) | CuKα (Dip2030) | CuKα (Dip2030) |
| Wavelength (Å) | 0.710 | 1.0083 | 1.542 | 1.542 | 1.542 |
| Data range (Å) | 39.2–2.0 | 39.2–2.0 | 20.0–3.0 | 15.0–3.0 | 20.0–2.7 |
| Unique reflections | 20 215 | 20 048 | 5669 | 5904 | 8972 |
| Completeness (%)a | 100 (100) | 100 (99.9) | 91.0 (91.1) | 96.4 (96.2) | 99.4 (95.8) |
| Rmergea,b | 0.078 (0.240) | 0.081 (0.279) | 0.041 (0.086) | 0.038 (0.087) | 0.038 (0.217) |
| Risoa,c | 0.267 (0.315) | 0.201 (0.241) | 0.151 (0.180) | ||
| Phasing power (centric/acentric)d | 0.72/0.82 | 0.78/0.99 | 0.06/0.08 | ||
| Overall figure of merit (centric/acentric) | 0.268/0.387 | ||||
| Refinement | |||||
| Resolution range (Å) | 19.6–2.0 | 19.7–2.7 | |||
| No. of reflections in working set | 19 195 | 8478 | |||
| Rcryst (Rfree)e | 0.213 (0.236) | 0.209 (0.251) | |||
| R.m.s.d. bond length (Å) | 0.0048 | 0.0065 | |||
| R.m.s.d. bond angles (°) | 1.33 | 1.35 | |||
| No. of protein atoms | 1774 | 1774 | |||
| No. of solvent atoms | 122 | 71 | |||
| No. of nucleotide atoms | 27 | 31 | |||
| No. of Mg2+ ions | 1 | 1 | |||
aNumbers in parentheses refer to statistics for the highest shell of data.
bRmerge = Σ|Iobs – <I>|/ΣIobs, where Iobs is the intensity measurement and <I> is the mean intensity for multiply recorded reflections (Otwinowski and Minor, 1997).
cRiso = Σ||Fp| – |Fph||/Σ|Fp|, where Fph and Fp are the derivative and native structure factors, respectively.
dPhasing power = <Fh>/E, where <Fh> is the root-mean-square heavy atom structure factor and E is the residual lack of closure error.
eRcryst and Rfree = Σ||Fobs = |Fcalc||/|Fobs| for reflections in the working and test sets, respectively.
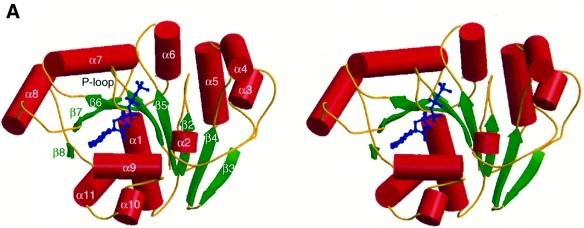
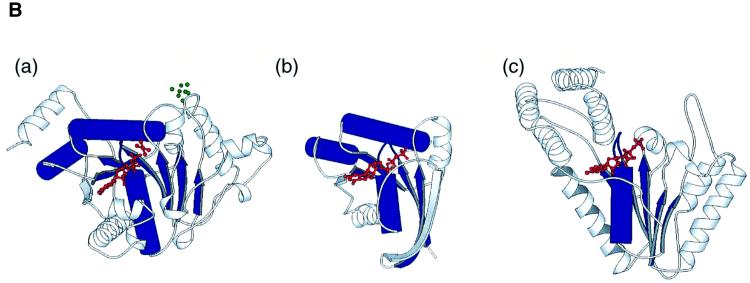
The polypeptide of MinD folds in an α–β–α layered structure. The central eight-stranded β-sheet, which contains seven parallel strands, is buttressed by several α-helices (Figure 1A). The adenine nucleotides lie on the C-terminal side of the parallel β-sheets and are recognized by the nucleotide-binding motifs, such as the P-loop or the Walker A motif (Walker et al., 1982; Saraste et al., 1990). An architectural comparison, using DALI (Holm and Sander, 1993), shows that MinD is similar to the nitrogenase iron protein (NIP; Z score = 24.0; Georgiadis et al., 1992). The major differences are found in α4, α5, β5 (MinD numbering) and at the extra helical C-terminus of NIP. The 4Fe:4S cluster of NIP, which is important for dinitrogen reduction, is located at two cysteine residues in α4 and β5, which are not conserved in MinD (Figure 1B, a). As reported in the structural analyses of NIP (Georgiadis et al., 1992), both proteins show structural similarities to the H-Ras G-protein and the RecA-like ATPase domain. The superposition of the H-Ras GTPase domain (Pai et al., 1990) onto MinD shows a high similarity in the foldings between the two proteins (four parallel β-strands: β7, β6, β1, β5; and α7, α8, the P-loop and the following α1), with a root mean square deviation (r.m.s.d.) of 1.2 Å over 60 Cα atoms (Figure 1B, b). In the RecA-like ATPase family, the five parallel β-strands, the P-loop and the following α-helix show structural similarity to the MinD framework, although the connectivity of the inter-β-strands differs. For example, the five β-strands (β6, β1, β5, β2 and β4), the P-loop and α1 of MinD superimpose onto the equivalent residues in the RecA-like ATP-binding domain of the N-ethylmaleimide-sensitive fusion protein (NSF; Lenzen et al., 1998) with an r.m.s.d. of 1.1 Å for 52 Cα atoms (Figure 1B, c). Furthermore, NIP and MinD exhibit similar architectural regions in these ATPase and GTPase domains (β7, β6, β1, β5, β2, β4, β3, the P-loop, α1, α7 and α8) shown in Figure 1B, a (1.2 Å r.m.s.d. value for 81 Cα atoms).
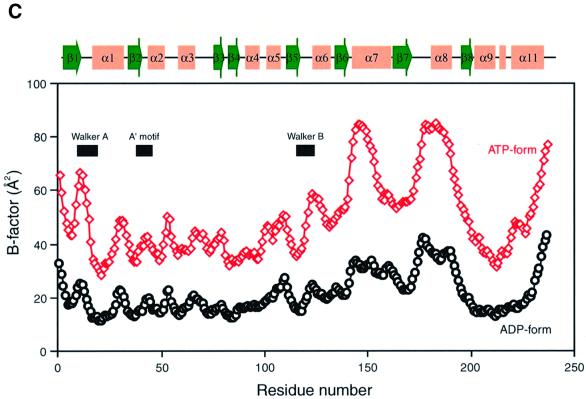
Fig. 1. Structure of P.furiosus MinD. (A) Stereo diagram of MinD in complex with AMPPCP. The helices are shown as red cylinders, β-strands as green arrows. The bound nucleotide is shown with thickened blue bonds. (B) Ribbon diagram of NIP (a), H-Ras (b) and NSF (c), shown in the same orientation as in (A). Structural elements compared with MinD are coloured in blue (see text). The bound nucleotides, ADP-AlF4– (a), GTP (b) and ATP (c) are shown in red; the 4Fe:4S of NIP is in green. (C) A plot of the average main chain temperature factors for MinD–nucleotide complexes (red line for the ATP form and black for the ADP form). Secondary structural elements are indicated on the top.
The temperature factors (B-factors) of the MinD backbone atoms show that the residues around the adenine nucleotide are relatively mobile: the Walker A and B motifs (residues 7–18 and 118–121, respectively), α7 and α8 (Figure 1C). In ADP-bound MinD, most of the B-factors vary between 10 and 30 Å2, with 22.6 Å2 on average. AMPPCP-bound MinD has B-factors that are twice as high as those of the ADP-bound form, but the B-factor profiles correlate well between the two forms. In particular, the Walker A and B motifs and the α7 helix are strikingly mobile in ATP-bound MinD, as revealed by the higher B-values. These regions are presumed to sense the dissociation of the γ-phosphate. In contrast, the central β-sheet exhibits lower temperature factors in both AMPPCP- and ADP-bound MinD, indicating the formation of a common rigid core in ATP/ADP-binding proteins. Interestingly, the B-factor profile observed in MinD was never found in NIP (Schlessman et al., 1998), although their architectures resemble each other. The NIP protein forms a homodimer, whose interface contains both of the Walker A and B motifs and α7. It is thus likely that the interface of MinD, which recognizes a counterpart in an ATP-dependent manner, also involves the mobile α7.
The nucleotide-binding site
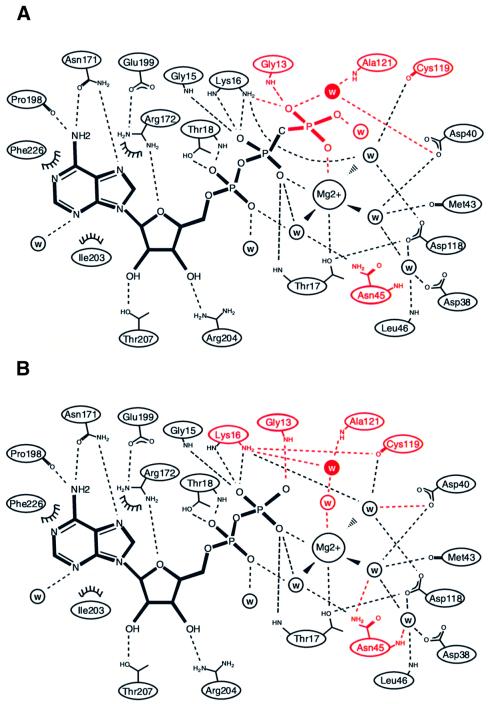
The previous report (de Boer et al., 1991) showed that ADP and dATP inhibit the ATPase activity of E.coli MinD, but AMP and the nucleotide triphosphates with other bases cannot. This implies that the protein would recognize at least the adenine base and the β- and γ-phosphates of the ATP nucleotide. In agreement, it was observed that the ATP analogue, AMPPCP, and the ADP nucleotide were bound to MinD through many hydrogen bonding interactions in the crystal structures (Figure 2A and B). Unlike other nucleotide-binding proteins, no large conformational change were observed between the AMPPCP-bound and ADP-bound MinD structures, as revealed by an r.m.s.d. value of 0.38 Å for all Cα atoms.
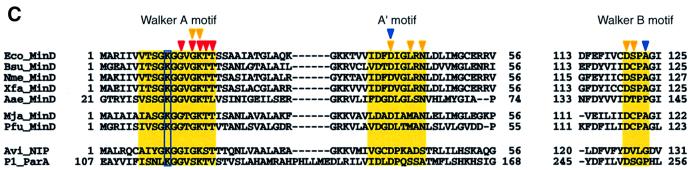
Fig. 2. Nucleotide binding of P.furiosus MinD. (A and B) Scheme showing the interactions between P.furiosus MinD and the nucleotide [AMPPCP (A) and ADP (B)]. Dashed lines indicate hydrogen bonds (<3.5 Å). Water molecules are indicated by ‘w’; van der Waals contacts are indicated by arcs. The differences between AMPPCP- and ADP-bound MinD are highlighted in red. The attacking water molecule is shown in white inside the closed circle. (C) Walker motif sequence conservation in MinD family proteins. Bacterial MinD homologues (Eco, E.coli; Bsu, Bacillus subtilis; Nme, Neisseria meningitidis; Xfa, Xylella fastidiosa; Aae, Aquifex aeolicus), archaeal MinD homologues (Mja, Methanococcus jannaschii; Pfu, P.furiosus) and MinD-related proteins (Avi, Azotobacter vinelandii) were aligned with CLUSTAL_W (Thompson et al., 1994) and edited manually. Key motifs are highlighted in yellow. The Lys11 residue is boxed. Triangles indicate residues involved in the specific binding of phosphate (red), magnesium (yellow) and attacking water (blue).
The adenine base with an anti conformation is strictly recognized by the conserved residues in the MinD homologues. The side chain of Asn171 makes two hydrogen bonds with the adenine base: an oxygen interacts with the exocyclic amine N6, whereas another amine group binds to the endocyclic nitrogen N1. N6 of adenine also forms a hydrogen bond with the main chain carbonyl of Pro198. Additional non-polar interactions are observed: the Ile203 side chain extends vertically to the adenine ring to make a hydrophobic contact and the aliphatic atoms of the Arg172 side chain make non-polar contacts with the same adenine on the opposite side.
The ribose ring adopts the typical C2′-endo conformation. Both hydroxyl groups at the 2′ and 3′ positions are recognized by the side chains of Thr207 and Arg204, respectively. The O4′ atom is bound to the amino group of Arg172, which forms a hydrogen bond to the carboxylate group of Glu199.
The phosphate-binding region of MinD reveals high similarity to those of other proteins with Walker motifs. The P-loop (Walker A motif; residues 7–18), connecting β1 and α1, has well conserved sequences, and its amino acid side chains exhibit the same conformations among all other proteins with the GXXXXGKT motif. The side chain of Lys16 forms hydrogen bonds to the β- and γ-phosphate oxygens, and that of Thr17 interacts with a magnesium ion. The main chain imino groups of Gly13 and Gly15 interact with the β- or γ-phosphate oxygens. Thr18 in the Walker A motif recognizes the α-phosphate oxygen through the hydroxyl side chain and the main chain amide group.
Other conserved motifs are also observed in MinD. The Walker B motif (residues 118–121) is located near the terminus of β5, and has the modified sequence, Asp-X-Pro-Ala, differing from those of NIP and the Ras protein (Asp-X-X-Gly; Schindelin et al., 1997). The first residue of the B motif is the conserved aspartate, which is identical to those previously observed in the P-loop protein-Mg2+-nucleotide structure, and is suggested to be important for ATP hydrolysis (Walker et al., 1982). The conserved Asp118 is involved in the nucleotide-binding site, where it makes hydrogen bonds with two water molecules that interact with the Mg2+ and the hydroxyl group of Thr17. The carbonyl group of Cys119 interacts with the same coordinated water molecule. The A′ motif (residues 37–45), which is found in members of the MinD family (Koonin, 1993; Vitale et al., 1996), lies adjacent to the Walker A motif in the sequence alignment (Figure 2C). This motif is located at the end of β2, and the highly conserved Asp40 and Asn45 side chains form hydrogen bonds to the oxygen atoms liganded to Mg2+. Furthermore, the conserved Asp38 in the A′ motif interacts indirectly with Asp118 of the Walker B motif through a water molecule. As reported for other nucleotide-binding proteins, such as RecA, the A′ motif seems to play a role as the sensor of ATP binding/hydrolysis (Story and Steitz, 1992).
A possible mechanism for ATP hydrolysis
In the MinD-AMPPCP complex, we have observed a water molecule, which is aligned for an in-line nucleophilic attack of the γ-phosphate (Figure 2A) and occupies the same position as the γ-phosphate in the MinD-ADP complex (Figure 2B). This water molecule is hydrogen bonded to the main chain imino group of Ala121 and to the carboxyl group of Asp40. In MinD homologues, Asp40 is highly conserved (Figure 2C) and, notably, this residue functions as a general base for ATP hydrolysis in NIP (Schindelin et al., 1997).
As reported previously, E.coli MinD possesses a relatively weak ATPase activity (de Boer et al., 1991). The same tendency was also observed in P.furiosus MinD (I.Hayashi, unpublished data) and a MinD-related protein, ParA (Bouet and Funnell, 1999; Figure 2C). This weak activity may be explained from the two crystal structures of P.furiosus MinD, which is assumed to be a functional homologue of E.coli MinD. First of all, we could observe no structural differences in the environment around the γ-phosphate of the adenine nucleotide between AMPPCP-bound and ADP-bound MinD. Particularly, the numbers of hydrogen bonds between the protein and the two nucleotides are almost identical in the two states of MinD. This suggests that MinD binds ADP as strongly as ATP, and that the nucleotide release may not be induced by ATP hydrolysis. The electrostatic aspect of the nucleotide-binding site may also be related to the poor ATPase activity of MinD. The importance of basic residues near the γ-phosphate oxygen, which would stabilize the negative charge in a transition state, has been mentioned in the case of F1 ATPase (Abrahams et al., 1994) and many other nucleotide hydrolases (Coleman et al., 1994; Sondek et al., 1994). In F1 ATPase, the basic residue, which stabilizes the pentacoordinate transition state, is supplied by the adjacent subunit. In contrast, MinD lacks the corresponding basic residue, and this might account for its low ATPase activity. A similar feature has also been reported in the structure of the ATP-binding domain of NSF, which displays little ATPase activity (Lenzen et al., 1998). Provided that either ATP hydrolysis or turnover dominates the rate-limiting steps, the low catalytic activity of MinD may be related to the features of the ATP-binding site.
Structure-based site-directed mutagenesis
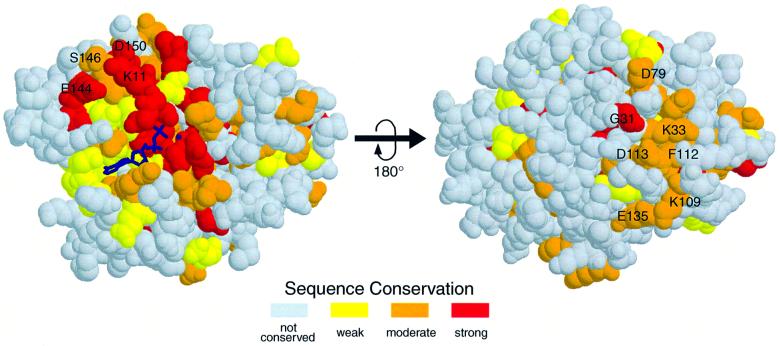
We have addressed the question of what functional significance could be drawn from the two MinD crystal structures. A possible counterpart (i.e. MinC in E.coli) may induce the ATPase activity of MinD, like GAPs that accelerate GTP hydrolysis of G-proteins (Gamblin and Smerdon, 1998). In order to examine the ATP-dependent effectors of MinD, we have performed mutational analyses of MinD, using the E.coli MinCD cell division system. Figure 3 shows the sequence conservation map on the molecular surface of P.furiosus MinD. Expectedly, many highly conserved residues are located in the nucleotide-binding region and the central β-sheets. As shown in Figure 3 in the right panel, these conserved residues in the central β-sheets line the surface and participate in specific intramolecular interactions. In another conserved region, Lys11 of MinD, which stabilizes the ATP leaving group in the NIP structure (Schindelin et al., 1997), interacts with Glu144, Ser146 and Asp150 in α7.
Fig. 3. Sequence conservation for MinD homologous proteins mapped on the P.furiosus structure. The alignment analysis was done with the seven MinD homologous proteins described in Figure 2C. Variable regions are coloured in white, and increasing conservation is indicated with deepening red colour. Two views of the protein are shown, including the bound nucleotide represented by thickened blue bonds. The conserved residues indicated in the right panel are located mostly at the N-termini of the β-strands.
We are interested in the molecular mechanism of MinD coupled with nucleotide binding and hydrolysis, and hence in the gene products that are activated by this ATPase. Archaea, including P.furiosus, lack the genes of the E.coli minB operon, except minD (Bernander, 1998; Gerard et al., 1998). Sequence comparison between E.coli and P.furiosus MinD reveals substantial conservation over the full length, with 31% identity and 73% similarity without large gaps (data not shown).
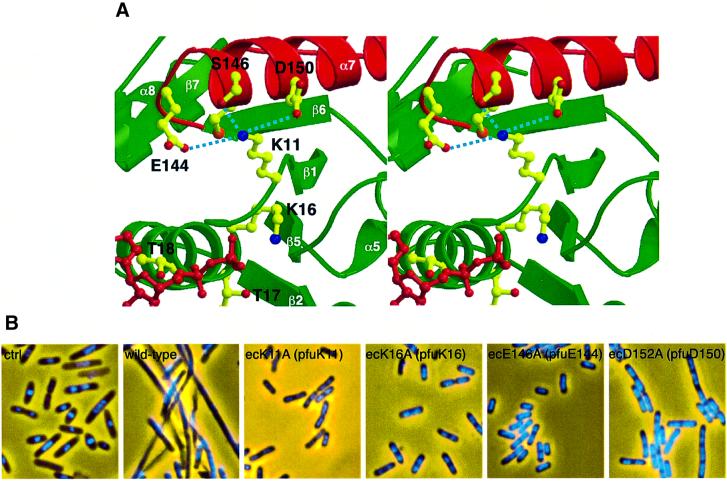
We constructed several E.coli minD mutant alleles, in which highly conserved residues were replaced by alanine. It was reported that MinC interacts directly with MinD in the two-hybrid system (Huang et al., 1996), and that the nucleotide-binding motif of MinD is important for MinC activation in vivo (de Boer et al., 1991). However, it remains unknown how MinD activates MinC, depending upon ATP hydrolysis. We applied the two-hybrid system to examine the effect of the MinD mutation. After the co-transformation of plasmids into the Saccharomyces cerevisiae Y187 strain carrying the LacZ gene, transformants were assayed for β-galactosidase activity. The results are summarized in Table II. A signal for strong interaction was obtained for the MinC–MinD combination. The E.coli K16A mutant [ecK16A, corresponding to P.furiosus K16 (pfuK16)], which shows a drastic reduction of ATPase activity in other ATPases (Rao et al., 1988; Hishida et al., 1999) and results in the inability of MinC activation (de Boer et al., 1991), does not interact with MinC. The ecK11A (pfuK11) mutant also completely abolishes binding to MinC. These results imply that MinC activation depends upon the interaction with MinD. Furthermore, the ecE146A (pfuE144) and ecD152A (pfuD150) mutations were shown to reduce the ability to bind to MinC, suggesting the importance of the two acidic residues in α7 (Figure 4A).
Table II. Interaction between E.coli MinC and MinD derivatives.
| Fusion to BD | Fusion to AD | P.furiosus residue | X-gal | β-galactosidase activity (U) |
|---|---|---|---|---|
| MinC | – | white | <1 | |
| – | MinDwt | white | <1 | |
| MinC | MinDwt | dark blue | 320 | |
| MinC | MinDK11A | K11 | white | <1 |
| MinC | MinDK16A | K16 | white | <1 |
| MinC | MinDE146A | E144 | very pale blue | 9 |
| MinC | MinDD152A | D150 | pale blue | 73 |
| – | P.furiosus MinD | white | <1 | |
| MinC | P.furiosus MinD | very pale blue | 3 |
Fig. 4. Effects of the local conformation around the K11 residue. (A) Stereo diagram of the interface between the Walker A motif region (green) and α7 (red). K11, ADP and the coordinating side chains are shown as ball-and-stick, with ADP and oxygens in red, and nitrogens in blue. (B) Phenotypes of mutations at residues around E.coli K11 (ecK11). Fixed cells were stained with DAPI to observe the nucleoids and photographed. From left to right: BL21(DE3) [minCDE+] with control pET21a, pEMDwt [PT7::minD+], pEMD11 [PT7::minDecK11A], pEMD16 [PT7::minDecK16A], pEMD146 [PT7::minDecE146A] and pEMD152 [PT7::minDecD152A]. The amino acids in P.furiosus (pfu) are also indicated in parentheses.
To confirm these results, the wild-type and mutant minD alleles were inserted downstream of the T7 promoter, and the resultant plasmids (pEMDwt for wild-type minD, pEMD11 for minDecK11A, pEMD16 for minDecK16A, pEMD146 for minDecE146A and pEMD152 for minDecD152A) were introduced into the strain BL21(DE3) in which minCDE is constitutively expressed. As de Boer et al. (1991) reported, when the recombinant MinD was expressed in minCDE+ cells, domestic MinC activation led to cell filamentation by functionally expressed MinD, otherwise the defective MinD mutants could not inhibit the cell division process. The results of the two-hybrid assay reflect the transformants’ phenotypes (Figure 4B): the plasmid expressing the wild-type minD product caused filamentation, whereas the plasmids of pEMD11 [PT7::minDecK11A], pEMD16 [PT7::minDecK16A] and pEMD146 [PT7::minDecE146A] did not cause filamentation. The transformants with pEMD152 [PT7::minDecD152A] led to partial filamentation, which suggests that ecD152A is less effective in activating the MinC-dependent division inhibition than wild-type MinD. The P.furiosus crystal structures support these genetic data (Figure 4A): the amino group of Lys11 interacts electrostatically with the carboxyl groups of Glu144 and Asp150 in α7, in addition to its hydrogen bond to the Ser146 side chain. Our results imply that the MinD ATPase activity and the conformation around the ATPase-binding site play an essential role in the E.coli cell division inhibition mechanism. It should be noted that we detected a weak interaction between P.furiosus MinD and E.coli MinC in the two-hybrid assay (Table II). This result supports the two MinD proteins from P.furiosus and E.coli sharing the same structural features, although the introduction of P.furiosus minD into BL21(DE3) did not induce filamentation (data not shown).
Discussion
Previous studies have demonstrated that MinD is a membrane-associated ATPase, which activates MinC in vivo (de Boer et al., 1991). Although interactions between MinC and MinD or between MinD and MinE were suggested by microscopic or two-hybrid analyses (Zhao et al., 1995; Huang et al., 1996), the function of MinD has not yet been defined. Thus far, the molecular biology of MinD has been restricted to the ATPase characterization of MinD reported by de Boer et al. (1991).
We have determined the atomic structure of the P.furiosus MinD homologue. As is the case with some ATPases (Ban et al., 1999; Benz et al., 1999), the ATP hydrolysis by MinD is not directly coupled to its conformational change. We presume that MinD could interact with its putative counterparts, MinC and MinE, depending on ATP hydrolysis. Indeed, our two-hybrid analysis with minDecK16A has shown that the MinD mutant fails to interact with MinC. Furthermore, the MinD-related protein, ParA, was found to change its oligomeric state and ATPase activity in the presence of DNA and ParB (Davis et al., 1992).
The structure-based site-directed mutagenesis revealed that the MinCD interaction depends on the residues around the MinD nucleotide-binding motif, and that a defect in the MinCD interaction leads to MinC inactivation and causes the inability for cell division. The highly conserved residue, K11, is suggested to play a major role in the interaction with MinC. This key residue occupies a central position in the network of polar interactions in the vicinity of the ATP/ADP-binding site. The alterations of this network by the mutation analyses weaken or collapse the MinCD interaction. The residues involved in this network, K11, E144, S146 and D150, are well conserved even in eukaryotic MinD homologues, as described below.
Previous sequence analyses have reported that the genes homologous to MinD are found in all living organisms, including yeast, plants and higher eukaryotes (Shahrestanifar et al., 1994; Vitale et al., 1996; Bernander, 1998). However, in the Gram-positive bacterium Bacillus subtilis, the min locus contains the minC and minD genes, but lacks a minE homologue. Instead, a non-homologous protein, DivIVA, plays the same role as MinE at the cell division stage (Edwards and Errington, 1997). In eukaryotes and archaea, furthermore, no genes corresponding to minC have been observed, and there is no evidence that MinD joins the cell division machinery. However, it should be noted that Arabidopsis thaliana MinD mediates the plastid division placement, although whether or not MinC and MinE exist is not yet known (Colletti et al., 2000). On the other hand, a yeast MinD homologue, NBP35p, was reported to be essential for cell viability. The mutation of the nucleotide-binding motif in this protein also leads to a lethal phenotype, implying that this motif is involved in essential interactions within a certain cellular machinery.
There have been many reports on molecular switches that are coupled to nucleotide binding and hydrolysis: GTPase regulation in signal transduction (Wittinghofer and Nassar, 1996); ATP-regulated switches in DNA mismatch repair (Ban et al., 1999); transposition (Yamauchi and Baker, 1998); and plasmid partition systems (Bouet and Funnell, 1999). These switches are considered to regulate each reaction of such cellular phenomena through an ATP-dependent modulation of the intermolecular interactions. Furthermore, a MinD-related protein, ParA, which belongs to the partitioning ATPase superfamily (Gerdes et al., 2000), has been reported to utilize an ATP-regulated switch in its plasmid partitioning pathway (Bouet and Funnell, 1999). Although it is still unclear how MinD functions in the cell, it is likely that MinD is a cell oscillation motor that recruits and activates MinC, depending on its own ATPase activity.
Materials and methods
Pyrococcus furiosus MinD protein expression and purification
The genomic DNA sequence encoding the P.furiosus MinD protein was amplified by PCR, and cloned into the bacterial expression vector pET28a (Novagen) using the NdeI and XhoI cloning sites. The recombinant DNA analysis indicated that it is free of PCR errors. The His6-tagged MinD protein was expressed in the bacterial strain BL21(DE3). Cells were grown at 30°C in 1.5 l of LB medium, induced at A600 = 0.8 with 0.67 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h, and harvested by centrifugation. The bacterial pellet was lysed by sonication in 50 mM Tris–HCl pH 8.0, 5 mM MgCl2, 0.2 M NaCl, 2 mM dithiothreitol (DTT) and 5% glycerol, and then centrifuged at 30 000 g. The supernatant was heated to 80°C for 20 min, centrifuged again and loaded onto a HiTrap Ni-chelating column (Pharmacia). Thrombin protease (Pharmacia) was added to the eluted protein fraction. The mixture was incubated at room temperature for 16 h. After dialysis against the initial buffer, the protein solution was reloaded onto the Ni-chelating column to remove the non-cleaved MinD protein, and then fractionated on a ResourceQ anion exchange column (Pharmacia). The protein solution was exchanged finally with a buffer containing 10 mM Tris–HCl pH 8.0, 0.2 M NaCl and 1 mM DTT, concentrated to 30 mg/ml and used for crystallization.
The structure determination revealed that the protein contained ADP. For the preparation of the MinD AMPPCP form, the cell lysate was precipitated with ammonium sulfate and dissolved in 6 M guanidium hydrochloride (GdnHCl), 1 mM DTT and 10 mM EDTA in 50 mM Tris pH 8.0. After dialysis with the buffer containing 6 M GdnHCl, 1 mM DTT and 50 mM Tris, the protein solution was applied to a 10 ml HiTrap Ni-chelating column. MinD was refolded on the column with a 200 ml gradient against 6.0–0 M GdnHCl, 0–0.5 mM AMPPCP, 50 mM Tris pH 8.0, 2 mM MgCl2, 0.5 M NaCl, 1 mM DTT and 5% glycerol. The eluted MinD was loaded onto the ResourceQ column, concentrated and used for crystallization. The bound nucleotides in the crystals were confirmed using a photodiode array detector.
Crystallization and data collection
Crystals were grown at 20°C using the hanging drop mode of the vapour diffusion method. Crystals of the ADP-bound form were obtained by mixing 3 µl of the crystallization buffer, containing 8% 2-methyl-2,4-pentanediol, 0.2 M MgCl2 and 0.1 M sodium citrate pH 5.5, with 3 µl of protein solution. They contain one MinD molecule per asymmetric unit with a solvent content of 56%. The AMPPCP-bound protein was crystallized as described above, except that 0.1 M of sodium cacodylate pH 6.0 was used in place of sodium citrate. Both crystal forms belong to the same cubic space group P213, but have slightly different unit cell dimensions: a = 96.04 Å for the ADP form and a = 98.64 Å for the AMPPCP form.
Prior to data collection, all of the crystals were flash cooled under a dry nitrogen stream at 100 K. Crystals of the ADP form were soaked into cryosolvent containing 0.1 M NaCl and 25% ethylene glycol in the crystallization buffer. For the AMPPCP form crystals, 20% (v/v) 2,3-butanediol was used as the cryoprotectant. Three derivatives were obtained by soaking in the cryosolvent with 1 mM MeHgCl for 3 h, 1 mM NaOsCl6 for 12 h and K2Pt(SCN)4 for 3 h. The data sets for the native high resolution and the mercury derivative crystals of the ADP form were collected at the Spring-8 BeamLine41 in Harima, Japan, using a Mar CCD detector. These data were processed with MOSFLM (Leslie, 1991) and SCALA (CCP4, 1994). Other diffraction data for the AMPPCP-bound crystal and the other two ADP-bound derivatives were collected, using a MAC science imaging plate diffractmeter DIP2030 with a CuKα rotation anode, and processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997).
Phasing, model building and refinement
Experimental phases were obtained by the MIR method. Heavy atom sites in the three derivatives were located on difference Fourier maps or difference Patterson maps, and then MIR phases were calculated using SHARP (de la Fortelle and Bricogne, 1997). The initial phases were improved further by density modification with SOLOMON (Abrahams and Leslie, 1996). The electron density map at 2.0 Å clearly showed the entire structure of MinD, except for the C-terminal region (residues 238–245). The MinD model was built into the electron density map using QUANTA98 (Molecular Simulations Inc.), and refined using CNS (Brünger et al., 1998), in which simulated annealing and subsequent individual B-factor refinement were applied. Several rounds of refinement were repeated with manual rebuilding on QUANTA98.
For the structure determination of AMPPCP-bound MinD, the rotation function and the translation function were calculated with CNS, using the structure of ADP-bound MinD. The bound AMPPCP was defined in an Fo – Fc electron density map, and then the structure of the AMPPCP form was refined.
The Ramachandran plot of ADP-bound MinD by PROCHECK revealed that 94.1% of the residues have backbone angles in the most favoured region, with the remaining 5.9% assigned to the additional allowed region. Likewise, 90.1% of residues in the AMPPCP form were assigned to the most favoured region, and the remaining 9.9% to the additional allowed region.
Plasmids for E.coli MinC, MinD and its mutants
Escherichia coli minC, minD and its mutant genes were amplified by PCR and inserted into the vector pET28a using the same construction method as for the P.furiosus minD homologous gene. The mutants of E.coli minD were prepared using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). The mutants were confirmed by sequencing. Then, the His6 tag-fused genes were inserted in-frame into the vectors of pET21a (Novagen), pGAD424 and pGBT9 (Clontech) vectors. The pET21a-derived vectors carrying minD and its mutants were named pEMDwt, pEMD11, pEMD16, pEMD146 and pEMD152, for wild-type, E.coli K11A (ecK11A), ecK16A, ecE146A and ecD152A, respectively.
The yeast two-hybrid system
The recombinant plasmids with the E.coli minC and minD genes, derived from pGAD424 and pGBT9, were transformed into the yeast strain Y187 (MATα ura3-52 his3 ade2-101 trp1-901 leu2-3 URA3::GAL1-lacZ). Double transformants were selected on dropout media lacking tryptophan and leucine, and assayed for β-galactosidase activity by both the filter assay and the liquid assay. The protocols were described by Clontech and Huang et al. (1996).
Escherichia coli cell microscopy
Escherichia coli BL21(DE3) [F′ ompT hsdSB (rB– mB–) gal (λcI857 ind1 Sam7 nin5 lacUV5-T7gene1) dcm (DE3)] cells, carrying vectors expressing E.coli minD or its mutant alleles, were grown overnight at 30°C in LB medium, supplemented with 100 µg/ml ampicillin and 1% glucose. The cultures were diluted 100-fold in fresh LB medium with appropriate supplements and 20 µM IPTG. The expression of each recombinant protein was checked by SDS–PAGE of cell lysates, which allowed the detection of the full-length proteins. Cells were examined by fluorescence microscopy after cultivation for 3 h at 30°C, chemical fixation and staining with 4′,6-diamidino-2-phenylindole (DAPI; Ishioka et al., 1997).
Figure preparation
Figures 1A and B, 3 and 4B were made with MOLSCRIPT (Kraulis, 1991) and Raster3D (Merrit and Bacon, 1997).
Protein Data Bank ID codes
The coordinates have been deposited in the Protein Data Bank. The accession codes are 1G3R for AMPPCP-MinD and 1G3Q for ADP-MinD, respectively.
Acknowledgments
Acknowledgements
We thank M.Kawamoto of Spring-8 for help in data collection. We also acknowledge D.Tsuchiya and M.Ishikawa for data collection and helpful discussions, H.Toh for the alignments, S.Iwai for the nucleotide analysis, and Y.W.Han, H.Iwasaki and H.Shinagawa for the microscopic observations of E.coli cells.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 31–42. [DOI] [PubMed] [Google Scholar]
- Abrahams J.P., Leslie,A.G.W., Lutter,L.R. and Walker,J.E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature, 370, 621–628. [DOI] [PubMed] [Google Scholar]
- Alder H.I., Fisher,W.D., Cohen,A. and Hardigree,A.A. (1967) Miniature Escherichia coli cells deficient in DNA. Proc. Natl Acad. Sci. USA, 57, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C., Junop,M. and Yang,W. (1999) Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell, 97, 85–97. [DOI] [PubMed] [Google Scholar]
- Benz J., Trachsel,H. and Baumann,U. (1999) Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae—the prototype of the DEAD box protein family. Struct. Fold. Des., 7, 671–679. [DOI] [PubMed] [Google Scholar]
- Bernander R. (1998) Archaea and the cell cycle. Trends Microbiol., 29, 955–961. [DOI] [PubMed] [Google Scholar]
- Bi E. and Lutkenhaus,J. (1993) Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol., 175, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet J.Y. and Funnell,B.E. (1999) P1 ParA interacts with the P1 partition complex at parS and an ATP–ADP switch controls ParA activities. EMBO J., 18, 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. et al. (1998) Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Coleman D.E., Berghuis,A.M., Lee,E., Linder,M.E., Gilman,A.G. and Sprang,S.R. (1994) Structures of active conformations of Gi α1 and the mechanism of GTP hydrolysis. Science, 265, 1405–1412. [DOI] [PubMed] [Google Scholar]
- Colletti K.S., Tattersall,E.A., Pyke,K.A., Froelich,J.E., Stokes,K.D. and Osteryoung,K.W. (2000) A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol., 10, 507–516. [DOI] [PubMed] [Google Scholar]
- Cook W.R., de Boer,P.A.J. and Rothfield,L.I. (1989) Differentiation of the bacterial cell division site. Int. Rev. Cytol., 118, 1–31. [DOI] [PubMed] [Google Scholar]
- Davis M.A., Martin,K.A. and Austin,S.J. (1992) Biochemical activities of the parA partition protein of the P1 plasmid. Mol. Microbiol., 6, 1141–1147. [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E. and Rothfield,L.I. (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E.coli. Cell, 56, 641–649. [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Cook,W.R. and Rothfield,L.I. (1990a) Bacterial cell division. Annu. Rev. Genet., 24, 249–274. [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E. and Rothfield,L.I. (1990b) Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc. Natl Acad. Sci. USA, 87, 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E., Hand,A.R. and Rothfield,L.I. (1991) The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J., 10, 4371–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E. and Rothfield,L.I. (1992) Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol., 174, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomorous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Edwards D.H. and Errington,J. (1997) The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol., 24, 905–915. [DOI] [PubMed] [Google Scholar]
- Gamblin S.J. and Smerdon,S.J. (1998) GTPase-activating proteins and their complexes. Curr. Opin. Struct. Biol., 8, 195–201. [DOI] [PubMed] [Google Scholar]
- Georgiadis M.M., Komiya,H., Chakrabarti,P., Woo,D., Kornuc,J.J. and Rees,D.C. (1992) Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science, 257, 1653–1659. [DOI] [PubMed] [Google Scholar]
- Gerard E., Labedan,B. and Forterre,P. (1998) Isolation of a minD-like gene in the hyperthermophilic archaeon Pyrococcus AL585, and phylogenetic characterization of related proteins in the three domains of life. Gene, 222, 99–106. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Moller-Jensen,J. and Jensen,R.B. (2000) Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol., 37, 455–466. [DOI] [PubMed] [Google Scholar]
- Hishida T., Iwasaki,H., Yagi,T. and Shinagawa,H. (1999) Role of Walker motif A of RuvB protein in promoting branch migration of Holliday junctions. Walker motif A mutations affect ATP binding, ATP hydrolyzing, and DNA binding activities of Ruvb. J. Biol. Chem., 274, 25335–25342. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hu Z. and Lutkenhaus,J. (1999) Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol., 34, 82–90. [DOI] [PubMed] [Google Scholar]
- Huang J., Cao,C. and Lutkenhaus,J. (1996) Interaction between FtsZ and inhibitors of cell division. J. Bacteriol., 178, 5080–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishioka K., Iwasaki,H. and Shinagawa,H. (1997) Roles of the recG gene product of Escherichia coli in recombination repair: effects of the ΔrecG mutation on cell division and chromosome partition. Genes Genet. Syst., 72, 91–99. [DOI] [PubMed] [Google Scholar]
- Justice S.S., Garcia-Lara,J. and Rothfield,L.I. (2000) Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol. Microbiol., 37, 410–423. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. (1993) A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol., 229, 1165–1174. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lenzen C.U., Steinmann,D., Whiteheart,S.W. and Weis,W.I. (1998) Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell, 94, 525–536. [DOI] [PubMed] [Google Scholar]
- Leslie A. (1991) Macromolecular data processing. In Moras,D., Podjarny,A.D. and Thierry,J.C. (eds), Crystal Computing V. Oxford University Press, Oxford, UK, pp. 27–38.
- Lutkenhaus J. (1993) FtsZ ring in bacterial cytokinesis. Mol. Microbiol., 9, 403–409. [DOI] [PubMed] [Google Scholar]
- Margolin W. (1998) A green light for the bacterial cytoskeleton. Trends Microbiol., 6, 233–238. [DOI] [PubMed] [Google Scholar]
- Merrit E.A. and Bacon,D.J. (1997) Raster3D—photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Mulder E., Woldringh,C.L., Tetart,F. and Bouche,J.P. (1992) New minC mutations suggest different interactions of the same region of division inhibitor MinC with proteins specific for minD and dicB coinhibition pathways. J. Bacteriol., 174, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–325. [DOI] [PubMed] [Google Scholar]
- Pai E.F., Krengel,U., Petsko,G.A., Goody,R.S., Kabsch,W.K. and Wittinghofer,A. (1990) Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J., 9, 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R., Pagan,J. and Senior,A.E. (1988) Directed mutagenesis of the strongly conserved lysine 175 in the proposed nucleotide-binding domain of α-subunit from Escherichia coli F1-ATPase. J. Biol. Chem., 263, 15957–15963. [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A. (1997) The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E.coli. Cell, 91, 685–694. [DOI] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A. (1999a) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA, 96, 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A. (1999b) MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol., 181, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L.I. and Justice,S.S. (1997) Bacterial cell division: the cycle of the ring. Cell, 88, 581–584. [DOI] [PubMed] [Google Scholar]
- Saraste M., Sibbald,P.R. and Wittinghofer,A. (1990) The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci., 15, 430–434. [DOI] [PubMed] [Google Scholar]
- Schindelin H., Kisker,C., Schlessman,J.L., Howard,J.B. and Rees,D.C. (1997) Structure of ADP × AIF4(–)-stabilized nitrogenase complex and its implications for signal transduction. Nature, 387, 370–376. [DOI] [PubMed] [Google Scholar]
- Schlessman J.L., Woo,D., Joshua-Tor,L., Howard,J.B. and Rees,D.C. (1998) Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum. J. Mol. Biol., 280, 669–685. [DOI] [PubMed] [Google Scholar]
- Shahrestanifar M., Saha,D.P., Scala,L.A., Basu,A. and Howells,R.D. (1994) Cloning of a human cDNA encoding a putative nucleotide-binding protein related to Escherichia coli MinD. Gene, 147, 281–285. [DOI] [PubMed] [Google Scholar]
- Sondek J., Lambright,D.G., Noel,J.P., Hamm,H.E. and Sigler,P.B. (1994) GTPase mechanism of Gproteins from the 1.7-Å crystal structure of transducin α-GDP-AIF-4. Nature, 372, 276–279. [DOI] [PubMed] [Google Scholar]
- Story R.M. and Steitz,T.A. (1992) Structure of the recA protein-ADP complex. Nature, 355, 374–376. [DOI] [PubMed] [Google Scholar]
- Teather R.M., Collins,J.F. and Donachie,W.D. (1974) Quantal behavior of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli.J. Bacteriol., 118, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale G., Fabre,E. and Hurt,E.C. (1996) NBP35 encodes an essential and evolutionarily conserved protein in Saccharomyces cerevisiae with homology to a superfamily of bacterial ATPases. Gene, 178, 97–106. [DOI] [PubMed] [Google Scholar]
- Walker J.M., Saraste,M., Runswick,M. and Gay,N. (1982) Distantly related sequence in α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer A. and Nassar,N. (1996) How Ras-related proteins talk to their effectors. Trends Biochem. Sci., 21, 488–491. [DOI] [PubMed] [Google Scholar]
- Yamauchi M. and Baker,T.A. (1998) An ATP–ADP switch in MuB controls progression of the Mu transposition pathway. EMBO J., 17, 5509–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.R., de Boer,P.A. and Rothfield,L.I. (1995) Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl Acad. Sci. USA, 92, 4313–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]