Abstract
Recently we showed that partial depletion of mitochondrial DNA (genetic stress) or treatment with mitochondrial-specific inhibitors (metabolic stress) induced a stress signaling that was associated with increased cytoplasmic-free Ca2+ [Ca2+]c. In the present study we show that the mitochondria-to-nucleus stress signaling induces invasive phenotypes in otherwise non-invasive C2C12 myoblasts and human pulmonary carcinoma A549 cells. Tumor-specific markers cathepsin L and transforming growth factor β (TGFβ) are overexpressed in cells subjected to mitochondrial genetic as well as metabolic stress. C2C12 myoblasts subjected to stress showed 4- to 6-fold higher invasion through reconstituted Matrigel membrane as well as rat tracheal xenotransplants in Scid mice. Activation of Ca2+-dependent protein kinase C (PKC) under both genetic and metabolic stress conditions was associated with increased cathepsin L gene expression, which contributes to increased invasive property of cells. Reverted cells with ∼70% of control cell mtDNA exhibited marker mRNA contents, cell morphology and invasive property closer to control cells. These results provide insights into a new pathway by which mitochondrial DNA and membrane damage can contribute to tumor progression and metastasis.
Keywords: calcium-dependent PKC/cathepsin L expression/invasive phenotypes/mitochondrial membrane potential/stress signaling
Introduction
Since Warburg’s initial hypothesis (Warburg, 1956), the role of mitochondrial dysfunction in tumorigenesis has been investigated extensively using multiple approaches (Shay and Werbin, 1987; Cavalli and Liang, 1998). Cytoplasm from tumor cells was shown to transfer tumorigenic property when fused with karyoplasts from normal cells, suggesting that unknown cytoplasmic factors may support the formation of malignant phenotypes (Jonasson et al., 1977; Howell and Sager, 1978; Giguere and Morais, 1981). Recent reports showing point mutations and deletions in mtDNA from a wide range of tumor cells (Horton et al., 1996; Polyak et al., 1998; Fliss et al., 2000; Yeh et al., 2000) have rekindled interest in Warburg’s hypothesis. Studies with mtDNA-depleted tumor cells have yielded mixed results, ranging from minimal effects to possible cooperativity between the mitochondrial and nuclear genes in tumor growth (Giguere and Morais, 1981; Morais et al., 1994; Cavalli et al., 1997; Cavalli and Liang, 1998; Hofhaus and Gattermann, 1999). Currently there is increasing evidence that mitochondrial mutations and/or structural–functional abnormality are associated with various tumors (Shay and Werbin, 1987; Cavalli and Liang, 1998), although it is not clear whether the mitochondrial lesions are contributing factors in carcinogenesis or whether they simply arise as part of the secondary effects of cancer.
The mode of mitochondria-to-nucleus communication has been intensively studied in yeast ρ° cells, in which a number of nuclear genes including those of mitochondrial rRNA and CIT2 (gene for peroxisomal citrate synthase) are overexpressed (Parikh et al., 1987; Liao et al., 1991). Overexpression of nuclear genes was also reported in ρ° avian and human cells (Marusich et al., 1997; Wang and Morais, 1997). Detailed mechanistic studies in yeast have demonstrated that upregulation of CIT2 gene expression involves the activation and nuclear translocation of heterodimeric basic helix–loop–helix (bHLH) factors, Rtg1 and Rtg3, in response to mitochondrial stress (Liao and Butow, 1993; Sekito et al., 2000). It has been suggested that mitochondrial O2 tension or oxidative stress may somehow be involved in the activation of cytoplasmic regulatory factor Rtg2 (Rothermel et al., 1995). Recently we described mitochondria-to-nucleus stress signaling in C2C12 myoblasts that were partially depleted of mtDNA or treated with mitochondrial inhibitors, conditions that disrupted mitochondrial membrane potential, Δψm. Disruption of Δψm was accompanied by elevated cytosolic free [Ca2+]i and activation of Ca2+-responsive transcription factors and calcineurin (Biswas et al., 1999). Elevated Ca2+ and increased extracellular signal-regulated kinase 1 (ERK1) and ERK2 activity were also observed in PC12 pheochromocytoma cells treated with FCCP, a mitochondria-specific ionophore (Luo et al., 1997). Results emerging from these studies, therefore, demonstrate that mitochondrial stress signaling in different cell types modulates nuclear gene expression.
In continuation of these studies, here we show that mitochondrial stress signaling induces an array of nuclear genes including cathepsin L, transforming growth factor (TGFβ), mouse melanoma antigen (MMA) and others, which are well known markers for tumorigenesis. We describe a novel mechanism whereby damage to mtDNA and mitochondrial membrane signals a change in nuclear gene expression, leading to overexpression of marker genes for tumor progression. Genetic and metabolic stress, both of which affect Δψm, convert the otherwise non-tumor-forming C2C12 myoblasts and non-invasive human lung carcinoma A549 cells to highly tumorigenic and invasive phenotypes.
Results
Effects of partial mtDNA depletion on mitochondrial membrane potential
In a recent study, we generated a series of C2C12 cell lines containing varying levels of mtDNA (20–50% of control cells) by ethidium bromide treatment for 40–50 growth cycles, followed by single cell cloning. mtDNA-depleted cells exhibited varying levels of mitochondrial transmembrane potential, Δψm, elevated cytoplasmic free [Ca2+]i and elevated nuclear genome-coded ryanodine receptor 1 (RyR1) mRNA (Biswas et al., 1999). These processes were reversed to near normal cellular levels in a ‘reverted cell line’, whose mtDNA content was brought back to >70% of the control by growing mtDNA-depleted cells (20% mtDNA content) in normal medium, lacking ethidium bromide, for ∼30 growth cycles. In the present study we used these cell lines for characterizing further the nature of mitochondrial stress signaling and its potential nuclear gene targets. The northern blot in Figure 1A shows the level of mitochondrial gene-coded tRNALeu in control, mtDNA-depleted and reverted cells. The depleted cells show ∼70% reduced tRNALeu, while the reverted cells show ∼30% reduced RNA compared with control cells. The mitochondrial RNA levels directly reflect the cellular mtDNA contents. Figure 1B shows the extent of the staining of cells with mitochondrial-specific cationic dye, Mitotracker, analyzed by flow cytometry. Results show that control cells exhibit maximum emission of 2 × 103, while mtDNA-depleted cells show a reduced emission of ∼2 × 102. The reverted cells show an intermediate intensity of 5 × 102. Cells treated with CCCP for 1 h show the least intensity of ∼5 × 101, signifying most disruption of Δψm. In confirmation of our previous results with confocal microscopy, these results show a more severe disruption of Δψm in cells with a higher level of mtDNA depletion.
Fig. 1. Changes in mitochondrial membrane potential in mtDNA-depleted and reverted cells. (A) Mitochondrial tRNALeu levels in control, mtDNA-depleted and reverted C2C12 myoblasts, determined by northern blot analysis of total RNA (20 µg in each lane) using 32P-labeled DNA probe. The same blot was stripped and reprobed with 18S rRNA probe to assess the loading level. (B) Flow cytometry analysis of cells stained with mitochondrial-specific cationic dye, Mitotracker, as described in Materials and methods.
Nuclear gene targets of mitochondrial stress signaling
To gain insight into the nature of nuclear gene targets of the mitochondrial stress signaling, we carried out differential display of RNA from control, mtDNA-depleted and reverted cells. We observed that 96 display products were either overexpressed or suppressed compared with control cell products. Of these, 35 display products were analyzed by DNA sequencing and northern hybridization; 17 products, listed in Table I, were positive. RNA from three different depleted cell lines reproducibly showed 2.5- to 18-fold increased mRNA for RyR1 Ca2+ channel, calreticulin, calsequestrin, MMA, cathepsin L, TGFβ and phosphoenol pyruvate (PEP) carboxykinase, in addition to two novel genes with no homology to sequences in the database (Table I). We also observed uniformly reduced mtDNA-encoded, as well as nuclear-encoded cytochrome oxidase VIIa and insulin receptor substrate 1 mRNAs. The altered mRNA levels for all the listed genes was due to mitochondrial genetic stress, since they were restored to near normal levels (1- to 2.5-fold of control cells, Table I) in reverted cells with 70% restored mtDNA and a steeper membrane potential (Figure 1).
Table I. Differential expression of nuclear genes in mtDNA-depleted C2C12 cells.
| Genes identified | mRNA levels (fold of control cells) |
|
|---|---|---|
| Depleted | Reverted | |
| RyR1 | 18.0 | 3.0 |
| Calsequestrin | 4.1 | 1.1 |
| Calreticulin | 2.5 | 1.2 |
| Cathepsin L | 5.0 | 2.0 |
| Mouse melanoma antigen | 16.0 | 2.5 |
| PEP carboxykinase | 3.0 | 1.5 |
| Two novel genes with no homology to published sequence | 4.0–5.3 | 2.0–3.0 |
| TGFβ1 | 5.0 | 1.1 |
| Insulin receptor substrate 1 | 0.5 | 0.7 |
| mtDNA transcripts | 0.2 | 0.7–0.9 |
Primer extension products showing increased or decreased RNA levels in mtDNA-depleted cells were PCR amplified and sequenced to determine the identity of genes. The DNA fragments were also used as probes in northern blot hybridization with RNA from control, depleted and reverted cells. The RNA levels are expressed as fold of control cell level, considered to be 1.
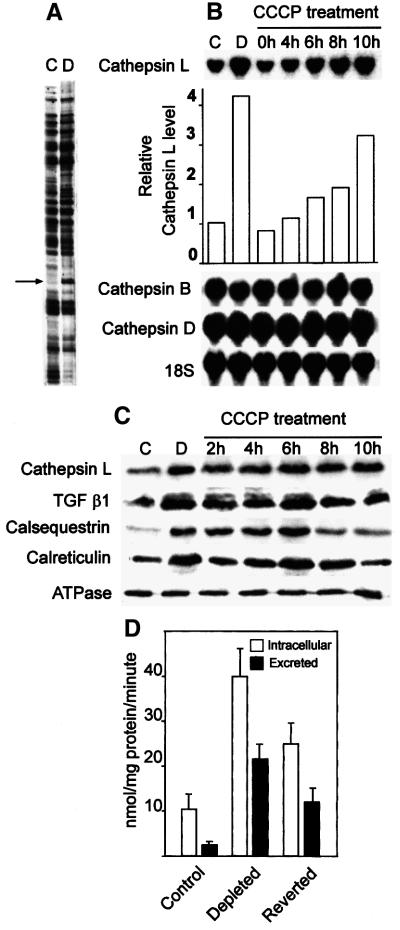
The effects of mitochondrial stress on the expression of well known tumor marker genes cathepsin L and TGFβ were investigated using CCCP, a mitochondrial-specific ionophore, as a model metabolic inhibitor. The gel pattern in Figure 2A shows a 6- to 8-fold increased differential display product, which was found to be cathepsin L specific. The northern blot in Figure 2B (top panel, first two lanes) shows a >4-fold higher level of cathepsin L mRNA in mtDNA-depleted cells compared with control C2C12 cells. Additionally, cathepsin L mRNA is increased 2- to 3-fold in cells treated with CCCP for 6–10 h (top panel of Figure 2B, last five lanes). However, cathepsin B and D mRNA levels remained the same in cells subjected to both types of stress, suggesting specificity of nuclear gene targets affected by the stress signaling. Consistent with the mRNA level, the western blot in Figure 2C shows a 3- to 5-fold increased cathepsin L protein in both mtDNA-depleted cells and cells treated with CCCP. The level of TGFβ protein increased by ∼4.5-fold in cells subjected to both genetic and metabolic stress. In further support of the differential display data in Table I, the levels of calsequestrin and calreticulin were increased 4-fold in mtDNA-depleted cells, as well as in CCCP-treated cells. For unknown reasons, the level of calsequestrin was reduced to near normal levels after 8 h of CCCP treatment. The levels of plasma membrane ATPase used as a loading control, on the other hand, did not change due to mitochondrial stress. Unpublished results (G.Biswas, H.Anandatheerthavarada and N.Avadhani) also show that mtDNA-depleted C2C12 cells contain increased BCl2 levels and show resistance to apoptosis. These results suggest that mitochondrial genetic stress signaling regulates the expression of nuclear genes involved in a wide range of cellular locations and functions, in addition to those targeted to mitochondria.
Fig. 2. Induction of cathepsin L during mitochondrial genetic and metabolic stress. (A) Differential display pattern showing induced level of cathepsin L mRNA in mtDNA-depleted cells. (B) Northern blots using cathepsin L cDNA probe (upper panel) and cathepsin B and D cDNA probes (lower panels) from control C2C12 cells (C) and mtDNA-depleted cells (D), and also cells treated with CCCP (25 µM) for the indicated time. In all panels 20 µg RNA was hybridized with 32P-labeled DNA probes under standard conditions. The blots were reprobed with 18S rRNA probe to ensure the RNA loading. (C) Immunoblot analysis of total cell extracts (50 µg protein in each case) from control and treated cells as indicated at the top. The blots were probed with indicated antibodies (1:1000 dilution) and developed using the Superglo kit from Pierce (Rockford, IL). The blots were reprobed with antibody against Na+/K+ ATPase to determine the protein loading level. The RNA and protein blots were quantified using a BioRad radiometric imager. (D) The level of intracellular and extracellular cathepsin L activity was assayed in total cell extract or concentrated culture fluid as described in Materials and methods. The values represent mean ± SEM of four experiments.
Elevated secretion of matrix protease cathepsin L is an important marker for invasive tumors (Cuvier et al., 1997; Kim et al., 1998). We therefore determined the levels of cathepsin L secreted in the culture media of normal and mtDNA-depleted cells. Figure 2D shows that control C2C12 cells secreted a relatively small amount of cathepsin L, while mtDNA-depleted cells secreted ∼10-fold more than control cells. The reverted cells secreted significantly lower levels of cathepsin L, which amounted to ∼5-fold higher than control cells. These results show that both intracellular as well as the excreted cathepsin L level is increased in mtDNA-depleted cells.
Effects of mitochondrial stress on tumor progression
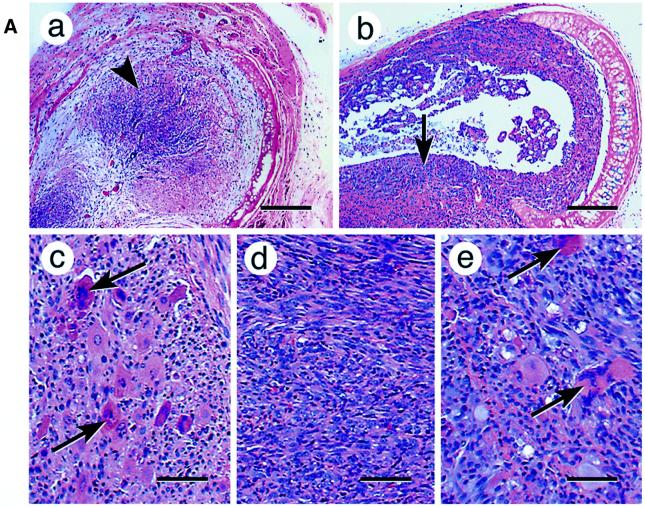
Overexpression of procathepsin L converted human melanoma cells from a non-metastetic to highly metastetic state (Denhardt et al., 1987; Yagel et al., 1989; Welch et al., 1990; Frade et al., 1998; Kirschke et al., 2000), while its inhibition by antisense RNA reversed the invasive property of tumor cells (Kirschke et al., 2000). We therefore tested the role of mitochondrial stress on tumor progression using a xenotransplantation assay (Momiki et al., 1991), which involves growth of tumor cells inside de-epithelialized rat tracheas, transplanted subcutaneously into Scid mice (Figure 3A). Histological examination of transplants showed that control C2C12 cells (Figure 3A) grew mostly in the middle of the tracheal transplant with no detectable invasion of the tracheal wall. mtDNA-depleted cells, on the other hand, exhibited vastly increased invasiveness (Figure 3B). In the latter case, cells destroyed the tracheal wall and its cartilage, spreading into the host tissue.
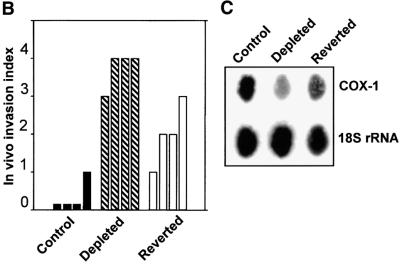
Fig. 3. Induced tumorigenicity of mtDNA-depleted C2C12 cells in vivo in Scid mice. (A) Hematoxylin–eosin stained sections of xenotransplants are shown. Control cells (a) that are growing inside the former tracheal lumen are shown by an arrowhead (invasion level 0). Panel b shows a representative in vivo growth pattern of mtDNA-depleted cells in a tracheal transplant. Note that the cells are growing inside the trachea as well as invading the external tracheal wall, as indicated by the arrow (invasion level 3–4). Panels c, d and e show the cellular differentiation pattern of control, mtDNA-depleted and reverted cells, respectively. The giant multinucleated rhabdomyoblasts (shown by arrows) are clearly seen in the control (c) and reverted cells (e). Such multinucleated cells are not seen in mtDNA-depleted cells (d). Bars in a and b represent 800 µm and those in c, d and e represent 100 µm. (B) The invasion levels of control, depleted and reverted cells from four different double blind assays are presented as bar diagrams. (C) Mitochondrial DNA contents of cells recovered from the representative sections of tracheal transplants as in (A). Total cell DNA (5 µg each) were subjected to Southern blot hybridization with mitochondrial-specific cytochrome oxidase I DNA probe. The blot was stripped and rehybridized with labeled probe for 18S rRNA probe to determine the loading level.
We used an arbitrary invasion scale based on histopathological examination of xenotransplants as described (Momiki et al., 1991). According to the scheme, level 0 is no invasion, where cells remain confined to the luminal ring; level 1 is minimal invasion of the subepithelial tissue; level 2 is moderate invasion, where cells reach the pars membranes; level 3 is marked invasion, where cells are outside the tracheal wall, covering and infiltrating the adventitia; and level 4 is maximal invasion, where cells are in the adventitia growing in large nodules or masses larger than the intratracheal tumor. Results of four independent double blind assays in Figure 3B show that the control cells showed an invasion level of 0–1, the mtDNA-depleted cells showed the highest level of 4, in three out of four cases, while the reverted cells showed a medium range of 1–3. Although not shown, three different reverted cell lines showed uniformly reduced invasion.
At higher magnification, the xenotransplanted control cells (Figure 3A, c) were seen to contain giant multinucleated rhabdomyoblasts, which are characteristic of differentiated skeletal myocytes. The mtDNA-depleted cells were poorly differentiated with no multinucleated structures (d). The reverted cells (e), on the other hand, showed a number of multinucleated structures similar to that seen in normal cells. It is therefore remarkable that reverted cells not only show an altered morphology closer to control untreated cells, but also exhibit a marked reduction in their invasive behavior.
Since the reverted cells showed a significantly lower invasive behavior compared with mtDNA-depleted cells, the mitochondrial DNA content of the various cell types during the 5 weeks of growth in the xenotransplants was an important issue. This question was addressed by assessing the mtDNA levels in the total DNA isolates from companion sections of xenotransplants shown in Figure 3A. A large majority of cells from these tracheal sections (>80%) were myoblasts, and 15–20% were fibroblasts from the host tissue. Results of Southern hybridization in Figure 3C reveal that explants of depleted cells show ∼40% mtDNA content, while those of reverted cells show ∼70% of control, suggesting the mtDNA levels change only marginally during the time frame of xenotransplantation assay.
Role of cathepsin L in mitochondrial stress-induced invasion
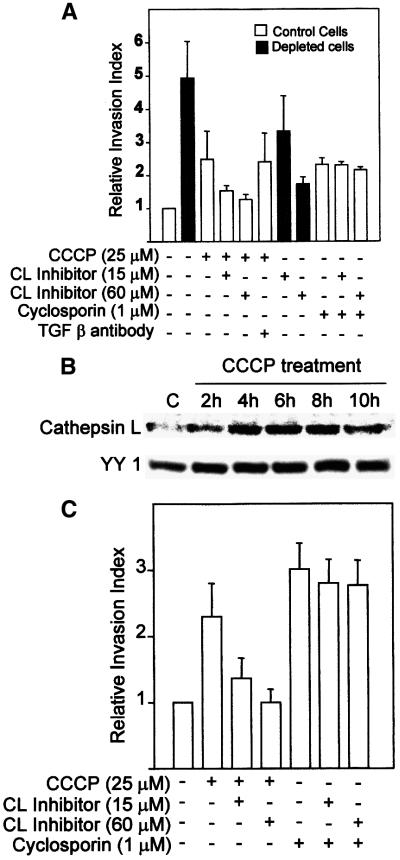
The role of cathepsin L and TGFβ in mitochondrial stress-induced invasiveness was investigated further using an in vitro Matrigel assay system (Yagel et al., 1989). This system measures the extent of cell migration through reconstituted biologically active basement membrane matrix, coated on microporous membrane. From Figure 4A it can be seen that mtDNA-depleted cells show >5-fold higher invasion, while cells treated with CCCP for 5 h show a 2.5-fold higher invasion as compared with untreated cells. Interestingly, a peptide inhibitor of cathepsin L reduced the invasive behavior of cells under both types of stress conditions in a concentration-dependent manner, while antibody to TGFβ had no effect. Cyclosporin, an immunosuppressive drug, was recently shown to induce the invasiveness of A549 human pulmonary carcinoma cells, through the TGFβ pathway (Welch et al., 1990; Hojo et al., 1999). In C2C12 cells, cyclosporin induced 2-fold higher invasion, which was not reversed even by 60 µM cathepsin L inhibitor peptide (see Figure 4A). These results show that induced cathepsin L gene expression is associated with tumor invasion during both mitochondrial genetic and metabolic stress conditions.
Fig. 4. Mitochondrial stress-induced cell invasion in C2C12 myoblasts and A549 lung carcinoma cells. In vitro Matrigel assays were carried out using 3H-labeled cells as described in Materials and methods. Treatment with indicated levels of CCCP and cyclosporin was carried out for 5 h, and other treatments were as described in Materials and methods. (A) In vitro invasion of C2C12 cells in Matrigel chambers. (B) Effects of CCCP on the induction cathepsin L protein in A549 cells. Immunoblot analysis of whole-cell extracts (50 µg protein each) was carried out with antibody to human cathepsin L (Santacruz Biotech., 1:1000 dilution). The blot was also reprobed with antibody to ubiquitous transcription factor YY1 as a loading control. (C) In vitro Matrigel invasion with treated and untreated A549 cells. The values represent mean ± SEM of four independent experiments.
The generality of the mitochondrial stress-induced tumor invasion was tested using human pulmonary A549 cell line, which has been shown to be non-invasive under in vivo and in vitro conditions (Frade et al., 1998). This cell line was recently used as a model system to study cyclosporin-mediated tumorigenesis (Hojo et al., 1999). The state of mitochondrial membrane potential in A549 cells treated with CCCP was assessed by the extent of staining with Mitotracker. The western blot of whole-cell extracts in Figure 4B shows that the level of cathepsin L is induced 3- to 4-fold in cells treated with CCCP for 4–10 h in comparison with untreated normal cells. Figure 4C shows that 5 h of CCCP treatment induced the invasiveness to ∼2.5-fold of control, which was reduced to 1.5-fold level by treatment with cathepsin L inhibitor peptide. In support of a recent study (Hojo et al., 1999), cyclosporin treatment also induced the invasive behavior of A549 cells by ∼3-fold, which was refractory to treatment with cathepsin L inhibitor peptide. Although not shown, the cyclosporin-induced invasion, but not that induced by mitochondrial stress, was partially reversed by monoclonal antibody to TGFβ.
Mechanism of stress-induced upregulation of cathepsin L gene expression
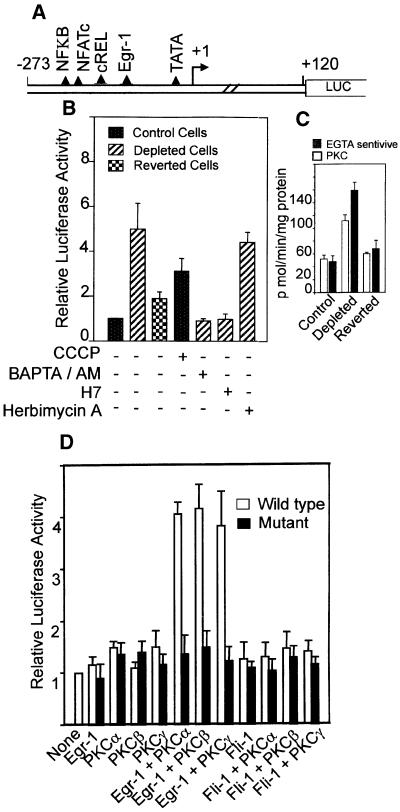
The murine cathepsin L promoter (Troen et al., 1991) cloned in a luciferase reporter vector, pGL3, was used to investigate the mechanism of mitochondrial stress-mediated transcription activation. Figure 5A shows the protein binding motifs of the promoter based on sequence analysis (Troen et al., 1991). In transient transfection assays, the promoter DNA construct (Figure 5B) showed a 5.5-fold higher transcription activity in mtDNA- depleted cells compared with control C2C12 cells. The reverted cells showed ∼1.7-fold higher activity than the control cells. Treatment of control C2C12 cells for 5 h with CCCP also resulted in a 3- to 4-fold induced transcription of the promoter construct. The increased transcription in mtDNA-depleted cells was due to increased intracellular Ca2+, since the activity was sensitive to the calcium chelator BAPTA/AM. Transcription of the promoter construct in mtDNA-depleted cells was selectively sensitive to added PKC inhibitor H7, but not to tyrosine kinase inhibitor, herbimycin A. These results are in line with the reported effects of kinase inhibitors on cathepsin L mRNA levels in intact cells (Atkins and Troen, 1995).
Fig. 5. Mechanism of activation of cathepsin L promoter by mitochondrial stress. (A) Structure of murine cathepsin L promoter and potential protein binding motifs based on nucleotide sequence. (B) Transcriptional activity of promoter construct in pGL3 reporter vector (Promega, Madison, WI). Details of transfection with reporter and renila luciferase as internal control and analysis of luciferase activity were as described in Materials and methods. Various agents as indicated were added to transfected cells also as described in Materials and methods. (C) PKC activity was assayed based on the level of inhibition with a PKC α, β and γ-specific pseudosubstrate inhibitor peptide. Ca2+-sensitive activity represents the level of inhibition with 5 mM EGTA. (D) Effects of coexpression with Egr-1 and PKC isoform-specific cDNAs on the transcriptional activity of the cathepsin L promoter in control C2C12 cells. Values represent the average ± SEM of 4–6 transfection assays in all cases.
Figure 5C shows PKC activity assayed based on the extent of inhibition by a pseudosubstrate inhibitor peptide that is specific for Ca2+-dependent PKC α, β and γ isoforms. Results show that PKC activity is induced 2-fold in mtDNA-depleted cells, which returns to near control cell levels (1.1- to 1.2-fold of control) in reverted cells. A comparable change in the total Ca2+-sensitive kinase activity is also observed in these cells (Figure 5C). Thus, there is close correlation between the PKC activity, transcriptional activity of the promoter and the cathepsin L mRNA levels in the three cell types. A possible link between these two factors was investigated by co-transfection with Egr-1 cDNA and isoform-specific PKC cDNAs. Additionally, the role of Egr-1 in transcription activation of cathepsin L promoter was tested by using promoter constructs with point mutations targeted to the Egr-1 binding site. As shown in Figure 5D, co-transfection with Egr-1 cDNA alone or PKC cDNAs alone had no effect on the activity of the wild-type as well as Egr-1 mutant promoter constructs in control C2C12 cells. A combination of Egr-1 and PKC α, β or γ cDNAs, however, yielded ∼4-fold higher transcription activities with the wild-type promoter construct but not with the mutant construct. Similarly, co-transfection with Fli cDNA, a close relative of Egr-1 factor with 90% sequence conservation had no stimulatory effect even in the presence of PKC α, β or γ cDNAs.
The levels of Egr-1-specific DNA binding proteins in the nuclear extracts from various cell types were examined by gel mobility shift assays. As shown in Figure 6A, nuclear extract from control cells formed two protein complexes marked as complex I and II. The nuclear extract from mtDNA-depleted cells formed these same complexes at 4- to 5-fold higher intensity, in addition to a slower migrating complex. The latter complex, but not complexes I and II, was supershifted by Egr-1 antibody. All of the complexes with control and depleted nuclear extract were competed by unlabeled excess Egr-1 DNA. Furthermore, nuclear extract from reverted cells showed reduced levels of antibody-reactive complex. These results suggest that Egr-1 is indeed induced in mtDNA-depleted cells. The nature of similarly migrating complexes I and II obtained with all three nuclear extracts remain unclear, although they may represent related protein(s) with similar DNA-binding specificity. Although not shown, the levels of ubiquitous factors Sp1 and YY-1 were similar in nuclear extracts from all three cell types.
Fig. 6. Role of Ca2+-activated PKC on the activation of Egr-1 and endogenous cathepsin L gene expression. (A) Levels of Egr-1 DNA binding factors in the nuclear extracts from control, depleted and reverted (R) cells by gel mobility shift analysis (P.S, pre-immune serum; SS, supershifted). (B) Gel shift patterns with nuclear extracts from cells transfected with Egr-1 alone, and with PKCα, β or γ cDNAs. In each case, 7.5 µg of protein was used for DNA binding with Egr-1 DNA (left panel) and Sp1 DNA (right panel). (C) The western blot analysis of whole-cell extracts (50 µg each) from control C2C12 cells transfected with Egr-1 cDNA alone and with either PKC α, β or γ cDNAs. The blot was co-developed with antibody to cathepsin L and antibody to Na+/K+ ATPase as a loading control (C, control extracts; D, depleted extract).
To understand further the role of PKC isoforms on the activity of Egr-1, nuclear extracts from control cells transfected with Egr-1 cDNA alone and in combination with PKCα, β or γ cDNAs were analyzed by gel shift with Egr-1 and Sp1 DNA probes (Figure 6B). Results show that nuclear extract from Egr-1 cDNA-transfected cells formed a complex that was marginally supershifted with Egr-1 antibody, while nuclear extracts from cells transfected with Egr-1 and any of the three PKC cDNAs yielded prominent supershifting complexes. Although not shown, transfection with PKC cDNAs alone did not yield antibody supershifting complex. The level of Sp1 DNA binding complex remained the same in these cells. Additionally, complexes with Sp1 DNA were not supershifted with Egr-1 antibody, suggesting specificity. These results demonstrate that all three Ca2+-activated PKC isoforms increase the DNA binding ability of Egr-1. The immunoblot in Figure 6C shows that transfection conditions that increased the formation of antibody supershifting complexes also increased the cellular cathepsin L protein levels by 4- to 5-fold over control cells. Although not shown, C2C12 cells co-transfected with Egr-1 and any of the three Ca2+-specific PKC cDNAs showed 5- to 6-fold higher invasiveness through the Matrigel membrane system. These results therefore provide direct evidence for the role of Ca2+-dependent PKC in the transcription regulation of cathepsin L gene expression. Results also indicate that the Ca2+-specific PKC effects are mediated through Egr-1 factor binding to the cathepsin L promoter.
Discussion
Mitochondrial DNA in cultured cells as well as in intact animals has been shown to be a direct and preferential target of attack by varied carcinogens (Allen and Coombs, 1980; Backer and Weinstein, 1980; Niranjan et al., 1982). Mitochondria are important cellular sites for the production of reactive O2 species (ROS) under different physiological and pathological conditions (Kehrer, 1993; Lenaz, 1998). These observations, coupled with the occurrence of DNA excision repair in mitochondria (Pinz and Bogenhagen, 1998), suggest that mtDNA is a likely target of mutational damage under both oxidative and chemical stress conditions. In support of this view, a variety of homoplasmic, as well as heteroplasmic mtDNA mutations have been reported in a number of human tumors (Horton et al., 1996; Polyak et al., 1998; Fliss et al., 2000; Yeh et al., 2000). It is, however, unclear whether these mutations are simply the consequence of increased oxidative stress, known to occur during tumor progression, or whether they have any direct role in the progression of cancer. Results presented in this paper provide a mechanistic insight into a pathway of cancer progression and tumor invasion resulting from mitochondrial DNA and/or membrane damage.
While investigating the potential nuclear gene targets of the mitochondria-to-nucleus stress signaling (Table I), we found that some of the genes implicated in tumor progression are also overexpressed in C2C12 myoblasts subjected to mitochondrial genetic stress. Growth factor receptor, TGFβ and extracellular matrix protease cathepsin L are among the well known tumor markers induced in many invasive cancers (Denhardt et al., 1987; Yagel et al., 1989; Welch et al., 1990; Frade et al., 1998; Hojo et al., 1999; Kirschke et al., 2000). Our results show that inhibition of cathepsin L, but not TGFβ, causes a partial reversal of mitochondrial stress-induced invasive behavior, both in C2C12 myoblasts and lung carcinoma A549 cells. These results show that cathepsin L gene expression is important in mitochondrial stress-induced in vitro invasion and is likely to be important in in vivo invasion as well.
Results of this study also show that, in addition to the Ca2+-dependent calcineurin pathway described before, the Ca2+-dependent PKC pathway is also activated as part of mitochondrial genetic and metabolic stress signaling. A recent study showed that mitochondrial oxidative stress through overproduction of H2O2 induced the Jun kinase pathway (Nemoto et al., 2000). These results are consistent with our previous study, which showed that mitochondrial genetic and metabolic stress induced JNK (c-Jun NH2-terminal kinase)-dependent activation and associated nuclear localization of activated transcription factor 2 (ATF2) (Biswas et al., 1999). In the present study we demonstrate that the ability of mtDNA-depleted cells and CCCP-treated cells to support higher transcription activity of the cathepsin L promoter is directly related to PKC activation. Mutational analysis together with DNA–protein binding also suggest the importance of Egr-1 factor for promoter activity. Egr-1, a member of immediate early Jun/Fos family genes, was suggested to be activated by phosphorylation (Cao et al., 1990; Huang and Adamson, 1994). Rat cathepsin L gene is reported to be activated by Egr family proteins (Ishidoh et al., 1997). Results of this study suggest for the first time a direct link between Ca2+-dependent PKC and the DNA-binding activity of Egr-1 in modulating cathepsin L gene expression. Similar PKC-mediated activation of other members of immediate early family transcription factors has been reported (Grover-Bardwick et al., 1994). Consistent with this possibility, murine Egr-1 contains four consensus motifs for PKC, although the precise phosphorylation site critical for activation and the precise effects of phosphorylation on DNA binding remain unclear. Transcription activation of cathepsin L gene expression is highly dependent on Egr-1 binding, since other distant members of this family, Jun/Fos, and the very close relative Fli are unable to induce the transcription of the promoter construct or the endogenous Egr-1 gene.
A remarkable observation of this study is that mitochondrial stress-induced phenotypic changes of invasive behavior, patterns of gene expression and ability to differentiate into multinucleated rhabdomyocytes are reversed, albeit partly, in reverted cells containing 70% of control cell mtDNA content. A partial recovery of invasiveness in reverted cells under both in vivo and in vitro conditions is consistent with their higher level of marker gene expression and Ca2+-dependent PKC activity compared with control C2C12 cells. Nevertheless, reduced invasive behavior in cells with reduced stress signaling provides a direct link between these two processes. Although not shown, Ca2+-dependent PKC activity and Egr-1-specific DNA binding activity are returned to near normal levels (1.2- to 1.3-fold of control cells) in three different reverted cell lines containing ∼70% of mtDNA content.
Acquired resistance to apoptosis in response to various apoptotic stimuli or cytotoxic drugs is the hallmark of many invasive tumors. In keeping with these, mtDNA-depleted C2C12 myocytes and also CCCP-treated A549 cells that show high invasive behavior are resistant to apoptosis in response to varied agents including staurosporine, etoposide and CCCP (G.Biswas, G.Amuthan and N.Avadhani, unpublished results). A close association between disrupted Δψm and release of cytochrome c and procaspases has been noted in many studies (Kluck et al., 1997; Yang et al., 1997). However, some recent studies show that the release of cytochrome c and procaspases is not directly dependent on mitochondrial membrane depolarization (Bossy-Wetzel et al., 1998). Similarly, membrane depolarization alone appears insufficient for induction of apoptosis, since mtDNA-depleted osteosarcoma cells with altered Δψm and reduced ATP synthesis are more resistant to apoptotic stimuli than normal cells (Dey and Moraes, 2000). In support of these observations, recent results show 3- to 4-fold higher Bcl2 in mtDNA-depleted C2C12 cells and also A549 lung carcinoma cells, which might be the basis for the observed resistance of these cells to apoptosis by CCCP treatment (G.Biswas, H.Anandatheerthavarada and N.Avadhani, unpublished results).
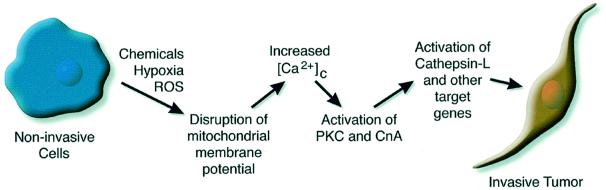
A model for the proposed mitochondrial stress-induced cancer progression is presented in Figure 7. Although a complete spectrum of nuclear gene targets affected by this stress signaling and their functional significance remain unclear, results in Table I demonstrate the overexpression of putative tumor marker genes. We propose that ROS, activated carcinogens, various drugs and anti-viral/anti-tumor agents (Shigenaga et al., 1994; Sokolove, 1994; Melov et al., 1999) that affect mitochondrial genetic and membrane systems may induce mitochondrial stress signaling. The stress signaling may therefore be a critical factor in the ability of some cells within a tumor mass to acquire invasive phenotypes and undergo morphological changes leading to tumor progression and metastasis. In summary, mitochondrial dysfunction and the resulting alterations in nuclear gene expression through mitochondria-to-nucleus stress signaling might be an important factor in cancer progression and tumor cell metastasis. The mitochondrial stress signaling described in this and our previous study (Biswas et al., 1999) may also help explain mechanistic details of various degenerative diseases associated with mitochondrial dysfunction, in addition to pathophysiological processes of myogenic differentiation (Rochard et al., 2000) and impaired insulin secretion (Tsuruzoe et al., 1998) by mtDNA-depleted cells. Finally, mitochondrial stress-induced cathepsin L expression and tumor invasion appear to be a common phenomenon in many cell types, since CCCP treatment induced the cathepsin L mRNA level and invasive property in various established tumor lines, including PC12 pheochromocytoma, C6 glioma, HepG2 cells and squamous cell carcinoma SCC9 and SCC12.
Fig. 7. A model for the mitochondrial genetic and metabolic stress-mediated tumor invasion.
Materials and methods
Cell lines and culture
Murine C2C12 skeletal myoblasts (ATCC CRL1772) and human pulmonary carcinoma A549 cells (ATCC CCL 185) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. Depletion of mtDNA by ethidium bromide treatment and isolation of C2C12 clone 2 with 20% of control cell mtDNA content was as described before (Biswas et al., 1999). Reverted cells represent clone 2 grown for 17 division cycles in the absence of ethidium bromide and contain 70% of control cell mtDNA. To ensure steady mtDNA levels, aliquots of the same cell isolate were used in all experiments and the mtDNA contents were routinely assayed in each experiment.
Flow cytometry
Mitotracker green (40 µM, Molecular Probes, Inc., Eugene, OR), diluted in serum-free DMEM (pre-warmed to 37°C), was added to CCCP-treated (20 µM, 1 h) and untreated cells in fresh medium, and incubated for 45 min with the dye. The adhered cells were washed twice with cold phosphate-buffered saline (PBS) and suspended in 1 ml PBS for analysis using a Becton Dickinson FACSCalibur Flow Cytometer. Mitotracker green was excited at 488 nm and fluorescence was detected at 525 nm.
Differential display
Differential display of RNA was carried out essentially as described (Liang and Pardee, 1992). Total RNA (400 ng each) from control and mtDNA-depleted C2C12 cells were reverse transcribed and the primer extension products were amplified using primers supplied in a kit from Display Systems Biotech (Vista, CA). The primer extension products showing different intensities on 8% sequencing gels were amplified and the PCR products were cloned in pCR2 vector (Invitrogen, Carlsbad, CA), sequenced and used as probes for northern analysis. Northern blot hybridization was carried out using 32P-labeled DNA probes and 20 µg RNA under standard conditions.
In vitro invasion assay
The in vitro invasion assays were carried out as described previously (Yagel et al., 1989). The Matrigel invasion chambers were prepared at 1:2 dilution of Matrigel (Beckton Dickinson, Belford, MA) according to the manufacturer’s protocol. Cells (5 × 104), uniformly labeled with [3H]dU (American Radiochemicals Inc.) for 24 h, were loaded on top of the Matrigel layer. After incubation for 24–72 h at 37°C, non-invading cells and the Matrigel layer were quantitatively removed and the microporous membrane containing the invaded cells was counted in a scintillation counter to determine the extent of invasion. While testing the inhibitor or antibody-treated cells, the Matrigel also contained the appropriate amounts of inhibitors or antibodies.
In vivo xenotransplantation assay
Assays were carried out essentially as described before (Momiki et al., 1991). Briefly, 5 × 105 cells in 100 µl of tissue culture medium were seeded into the lumen of de-epithelialized rat tracheal tubes and sealed at both ends. The sealed tubes were transplanted into dorsal mouse subcutaneous tissue (Momiki et al., 1991). Transplants (4–8 for each cell type) were recovered after 5–6 weeks, cross-sectioned into 3 mm thick rings, fixed in 10% neutral buffered formaldehyde (NBF) and embedded in paraffin. The paraffin blocks were sectioned and stained with hematoxylin and eosin.
Cathepsin L activity
Cathepsin L activity was determined as described (Sheahan et al., 1989) using the synthetic substrate benzyl-phenylalanine-arginine-4-methoxy-2-naphthylamide (Z-Phe-Arg-MNA) (Enzyme Systems products, Dublin, CA). Total cell lysates or concentrated culture medium (∼150 µg of protein) were assayed for cathepsin L activity in 50 µl of reaction mixture containing 0.1 M morpholinethanesulfonic acid (MES) buffer pH 3.5, 1 mM Z-Phe-Arg-MNA, 1 mM EDTA, 1 mM dithiothreitol, at 37°C for 10 min. The reaction was terminated by addition of 50 µl of 1 N HCl containing 2% Triton X-100. Fast blue (o-dianisidine tetrazotized from Sigma Chemicals Co., St Louis, MO) was added, the color developed for 15 min and OD520 determined. Culture medium was concentrated by filtration through Ambion Filtration system using 10 kDa cut-off membranes.
Assay for PKC activity
The PKC activity was measured using the PKC Assay System kit from Life Technologies (Grand Island, NY), which employs a PKC-specific peptide substrate and a pseudosubstrate inhibitor specific for Ca2+-dependent PKC isoforms. The Ca2+-sensitive kinase activity was assayed by adding 5 mM EGTA to the assay mix. PKC enzyme was partially purified by chromatography on DEAE–Sephacel microcolumns by a protocol described in the kit and assays were carried out in 50 µl volumes using 1 µCi of [γ-32P]ATP.
Transcription analysis of promoter constructs
Cathepsin L promoter DNA (sequence –221 to +47) was amplified from the mouse genomic DNA based on the published sequence (Troen et al., 1991) and cloned in pGL3 vector (Promega, Madison, WI). Mutations at the Egr-1 binding site were targeted by substituting the two GG residues from the Erg-1 binding site with TA residues (wild type: CGGGGCGGGGGCGGGC; mutant: CGGGGCTAGGGCTAGC) by primer-mediated mutagenesis (Higuchi et al., 1988), and the mutant promoter DNA was cloned in pGL3 vector. Transfections were carried out in control C2C12 and mtDNA-depleted cells using Fugene 6 reagent (Roche Molecular Biochemicals, Indianapolis, IN) by the manufacturer’s suggested protocol. Promoter (1 µg) and 0.1 µg of renila luciferase construct as an internal control were used in each transfection. The luciferase activity was assayed using Dual-Luciferase reporter assay system from Promega. Co-transfection with various cDNA constructs was carried out using 0.2 µg of expression construct.
DNA–protein binding by gel mobility shift
Nuclear extracts from control, depleted and reverted C2C12 myocytes were prepared as described (Dignam et al., 1983). DNA–protein binding was assayed by using 0.1 to 0.2 ng (20 000 c.p.m.) of 32P-end-labeled double-stranded DNA probes essentially as described (Singh et al., 1986). The following synthetic double-stranded DNAs were used either as probes or competitors: SP1 (5′-ATTCGATCGGGGCGGGGCGAAGC-3′), Egr-1 (5′-GACGGGGCGGGGGCGGGCCCTGT-3′) and Egr-1MUT (5′-GACGGGGCTAGGGCTAGCCCTGT-3′). Binding reactions (20 ml final volumes) were run at room temperature for 25 min using 1–3 ml of nuclear extracts (7.5 µg of protein in each case) and 1 mg of poly (dI–dC) to minimize non-specific protein binding. The DNA bound complexes were resolved by electrophoresis through 4% polyacrylamide gels using 0.25× TBE (1× TBE = 89 mM Tris base, 89 mM boric acid, 2 mM EDTA) as the running buffer. Competition with 30 molar excess unlabeled DNA and antibody supershift with 1–2 µg of either pre-immune IgG, Sp-1 (Santa Cruz Biotechnology) or Egr-1 (Geneka Biotechnology) antibodies were carried out under standard conditions.
Acknowledgments
Acknowledgements
This research was supported in part by NIH grant CA-22762. We thank Drs Ralph Brinster, Michael Atchison, Peter Nowell and Andrie Thomas-Thikonenko for critically reading the manuscript, and Mr James Haydon for helping with illustrations. We also thank Dr Shaym E.P.Reddy for the Egr-1 and Fli expression cDNAs, and Dr Peter M.Blumberg for PKC cDNAs.
References
- Allen J.A. and Coombs,M.M. (1980) Covalent binding of polycyclic aromatic compounds to mitochondrial and nuclear DNA. Nature, 287, 244–245. [DOI] [PubMed] [Google Scholar]
- Atkins K.B. and Troen,B.R. (1995) Phorbol ester stimulated cathepsin L expression in U937 cells. Cell Growth Differ., 6, 713–718. [PubMed] [Google Scholar]
- Backer J.M. and Weinstein,I.B. (1980) Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science, 209, 297–299. [DOI] [PubMed] [Google Scholar]
- Biswas G., Adebanjo,O.A., Freedman,B.D., Anandatheerthavarada, H.K., Vijayasarathy,C., Zaidi,M., Kotlikoff,M. and Avadhani,N.G. (1999) Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J., 18, 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy-Wetzel E., Newmeyer,D.D. and Green,D.R. (1998) Mitochondrial cytochrome c release in apoptosis occurs upstream of DEVD-specific caspase activation and independently of mitochondrial transmembrane depolarization. EMBO J., 17, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X.M., Koski,R.A., Gashler,A., McKiernan,M., Morris,C.F., Gaffney,R., Hay,R.V. and Sukhatme,V.P. (1990) Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol. Cell. Biol., 10, 1931–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli L.R. and Liang,B.C. (1998) Mutagenesis, tumorigenicity and apoptosis: are the mitochondria involved? Mutat. Res., 398, 19–26. [DOI] [PubMed] [Google Scholar]
- Cavalli L.R., Varella-Garcia,M. and Liang,B.C. (1997) Diminished tumorigenic phenotype after depletion of mitochondrial DNA. Cell Growth Differ., 8, 1189–1198. [PubMed] [Google Scholar]
- Cuvier C., Jang,A. and Hill,R.P. (1997) Exposure to hypoxia, glucose starvation and acidosis: effect on invasive capacity of murine tumor cells and correlation with cathepsin (L + B) secretion. Clin. Exp. Metastasis, 15, 19–25. [DOI] [PubMed] [Google Scholar]
- Denhardt D.T., Greenberg,A.H., Egan,S.E., Hamilton,R.T. and Wright,J.A. (1987) Cysteine proteinase cathepsin L expression correlates closely with the metastatic potential of H-ras-transformed murine fibroblasts. Oncogene, 2, 55–59. [PubMed] [Google Scholar]
- Dey R. and Moraes,C.T. (2000) Lack of oxidative phosphorylation and low mitochondrial membrane potential decrease susceptibility to apoptosis and do not modulate the protective effect of Bcl-x(L) in osteosarcoma cells. J. Biol. Chem., 275, 7087–7094. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliss M.S., Usadel,H., Caballero,O.L., Wu,L., Buta,M.R., Eleff,S.M., Jen,J. and Sidransky,D. (2000) Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science, 287, 2017–2019. [DOI] [PubMed] [Google Scholar]
- Frade R., Rodrigues-Lima,F., Huang,S., Xie,K., Guillaume,N. and Bar-Eli,M. (1998) Procathepsin-L, a proteinase that cleaves human C3 (the third component of complement), confers high tumorigenic and metastatic properties to human melanoma cells. Cancer Res., 58, 2733–2736. [PubMed] [Google Scholar]
- Giguere L. and Morais,R. (1981) On suppression of tumorigenicity in hybrid and cybrid mouse cells. Somatic Cell Genet., 7, 457–471. [DOI] [PubMed] [Google Scholar]
- Grover-Bardwick A., Adamson,E. and Mercola,D. (1994) Transform ation-specific pattern of phosphorylation of c-Jun, Jun-B, Jun-D and Egr-1 in v-sis transformed cells. Carcinogenesis, 15, 1667–1674. [DOI] [PubMed] [Google Scholar]
- Higuchi R., Krummel,B. and Saiki,R.K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res., 16, 7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofhaus G. and Gattermann,N. (1999) Mitochondria harbouring mutant mtDNA—a cuckoo in the nest? Biol. Chem., 380, 871–877. [DOI] [PubMed] [Google Scholar]
- Hojo M., Morimoto,T., Maluccio,M., Asano,T., Morimoto,K., Lagman, M., Shimbo,T. and Suthanthiran,M. (1999) Cyclosporine induces cancer progression by a cell-autonomous mechanism. Nature, 397, 530–534. [DOI] [PubMed] [Google Scholar]
- Horton T.M., Petros,J.A., Heddi,A., Shoffner,J., Kaufman,A.E., Graham, S.D.J., Gramlich,T. and Wallace,D.C. (1996) Novel mitochondrial DNA deletion found in a renal cell carcinoma. Genes Chromosomes Cancer, 15, 95–101. [DOI] [PubMed] [Google Scholar]
- Howell A.N. and Sager,R. (1978) Tumorigenicity and its suppression in cybrids of mouse and Chinese hamster cell lines. Proc. Natl Acad. Sci. USA, 75, 2358–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.P. and Adamson,E.D. (1994) The phosphorylated forms of the transcription factor, Egr-1, bind to DNA more efficiently than non-phosphorylated. Biochem. Biophys. Res. Commun., 200, 1271–1276. [DOI] [PubMed] [Google Scholar]
- Ishidoh K., Taniguchi,S. and Kominami,E. (1997) Egr family member proteins are involved in the activation of the cathepsin L gene in v-src-transformed cells. Biochem. Biophys. Res. Commun., 238, 665–669. [DOI] [PubMed] [Google Scholar]
- Jonasson J., Povey,S. and Harris,H. (1977) The analysis of malignancy by cell fusion. VII. Cytogenetic analysis of hybrids between malignant and diploid cells and of tumours derived from them. J. Cell Sci., 24, 217–254. [DOI] [PubMed] [Google Scholar]
- Kehrer J.P. (1993) Free radicals as mediators of tissue injury and disease. Crit. Rev. Toxicol., 23, 21–48. [DOI] [PubMed] [Google Scholar]
- Kim K., Cai,J., Shuja,S., Kuo,T. and Murnane,M.J. (1998) Presence of activated ras correlates with increased cysteine proteinase activities in human colorectal carcinomas. Int. J. Cancer, 79, 324–333. [DOI] [PubMed] [Google Scholar]
- Kirschke H., Eerola,R., Hopsu-Havu,V.K., Bromme,D. and Vuorio,E. (2000) Antisense RNA inhibition of cathepsin L expression reduces tumorigenicity of malignant cells. Eur. J. Cancer, 36, 787–795. [DOI] [PubMed] [Google Scholar]
- Kluck R.M., Bossy-Wetzel,E., Green,D.R. and Newmeyer,D.D. (1997) The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science, 275, 1132–1136. [DOI] [PubMed] [Google Scholar]
- Lenaz G. (1998) Role of mitochondria in oxidative stress and ageing. Biochim. Biophys. Acta, 1366, 53–67. [DOI] [PubMed] [Google Scholar]
- Liang P. and Pardee,A.B. (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science, 257, 967–971. [DOI] [PubMed] [Google Scholar]
- Liao X. and Butow,R.A. (1993) RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell, 72, 61–71. [DOI] [PubMed] [Google Scholar]
- Liao X.S., Small,W.C., Srere,P.A. and Butow,R.A. (1991) Intra mitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Bond,J.D. and Ingram,V.M. (1997) Compromised mitochondrial function leads to increased cytosolic calcium and to activation of MAP kinases. Proc. Natl Acad. Sci. USA, 94, 9705–9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusich M.F., Robinson,B.H., Taanman,J.W., Kim,S.J., Schillace, Smith,J.L. and Capaldi,R.A. (1997) Expression of mtDNA and nDNA encoded respiratory chain proteins in chemically and genetically-derived Rho0 human fibroblasts: a comparison of subunit proteins in normal fibroblasts treated with ethidium bromide and fibroblasts from a patient with mtDNA depletion syndrome. Biochim. Biophys. Acta, 1362, 145–159. [DOI] [PubMed] [Google Scholar]
- Melov S., Coskun,P.E. and Wallace,D.C. (1999) Mouse models of mitochondrial disease, oxidative stress and senescence. Mutat. Res., 434, 233–242. [DOI] [PubMed] [Google Scholar]
- Momiki S., Baba,M., Caamano,J., Iizasa,T., Nakajima,M., Yamaguchi,Y. and Klein-Szanto,A. (1991) In vivo and in vitro invasiveness of human lung carcinoma cell lines. Invasion Metastasis, 11, 66–75. [PubMed] [Google Scholar]
- Morais R., Zinkewich-Peotti,K., Parent,M., Wang,H., Babai,F. and Zollinger,M. (1994) Tumor-forming ability in athymic nude mice of human cell lines devoid of mitochondrial DNA. Cancer Res., 54, 3889–3896. [PubMed] [Google Scholar]
- Nemoto S., Takeda,K., Yu,Z.X., Ferrans,V.J. and Finkel,T. (2000) Role for mitochondrial oxidants as regulators of cellular metabolism. Mol. Cell. Biol., 20, 7311–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjan B.G., Bhat,N.K. and Avadhani,N.G. (1982) Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science, 215, 73–75. [DOI] [PubMed] [Google Scholar]
- Parikh V.S., Morgan,M.M., Scott,R., Clements,L.S. and Butow,R.A. (1987) The mitochondrial genotype can influence nuclear gene expression in yeast. Science, 235, 576–580. [DOI] [PubMed] [Google Scholar]
- Pinz K.G. and Bogenhagen,D.F. (1998) Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol. Cell. Biol., 18, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Li,Y., Zhu,H., Lengauer,C., Willson,J.K., Markowitz,S.D., Trush,M.A., Kinzler,K.W. and Vogelstein,B. (1998) Somatic mutations of the mitochondrial genome in human colorectal tumours. Nature Genet., 20, 291–293. [DOI] [PubMed] [Google Scholar]
- Rochard P., Rodier,A., Casas,F., Cassar-Malek,I., Marchal-Victorion,S., Daury,L., Wrutniak,C. and Cabello,G. (2000) Mitochondrial activity is involved in the regulation of myoblast differentiation through myogenin expression and activity of myogenic factors. J. Biol. Chem., 275, 2733–2744. [DOI] [PubMed] [Google Scholar]
- Rothermel B.A., Shyjan,A.W., Etheredge,J.L. and Butow,R.A. (1995) Transactivation by Rtg1p, a basic helix–loop–helix protein that functions in communication between mitochondria and the nucleus in yeast. J. Biol. Chem., 270, 29476–29482. [DOI] [PubMed] [Google Scholar]
- Sekito T., Thornton,J. and Butow,R.A. (2000) Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell, 11, 2103–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J.W. and Werbin,H. (1987) Are mitochondrial DNA mutations involved in the carcinogenic process? Mutat. Res., 186, 149–160. [DOI] [PubMed] [Google Scholar]
- Sheahan K., Shuja,S. and Murnane,M.J. (1989) Cysteine protease activities and tumor development in human colorectal carcinoma. Cancer Res., 49, 3809–3814. [PubMed] [Google Scholar]
- Shigenaga M.K., Hagen,T.M. and Ames,B.N. (1994) Oxidative damage and mitochondrial decay in aging. Proc. Natl Acad. Sci. USA, 91, 10771–10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Sen,R., Baltimore,D. and Sharp,P.A. (1986) A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature, 319, 154–158. [DOI] [PubMed] [Google Scholar]
- Sokolove P.M. (1994) Interactions of adriamycin aglycones with mitochondria may mediate adriamycin cardiotoxicity. Int. J. Biochem., 26, 1341–1350. [DOI] [PubMed] [Google Scholar]
- Troen B.R., Chauhan,S.S., Ray,D. and Gottesman,M.M. (1991) Downstream sequences mediate induction of the mouse cathepsin L promoter by phorbol esters. Cell Growth Differ., 2, 23–31. [PubMed] [Google Scholar]
- Tsuruzoe K. et al. (1998) Creation and characterization of a mitochondrial DNA-depleted pancreatic β-cell line: impaired insulin secretion induced by glucose, leucine and sulfonylureas. Diabetes, 47, 621–631. [DOI] [PubMed] [Google Scholar]
- Wang H. and Morais,R. (1997) Up-regulation of nuclear genes in response to inhibition of mitochondrial DNA expression in chicken cells. Biochim. Biophys. Acta, 1352, 325–334. [DOI] [PubMed] [Google Scholar]
- Warburg O. (1956) On the origin of cancer cells. Science, 123, 309–314. [DOI] [PubMed] [Google Scholar]
- Welch D.R., Fabra,A. and Nakajima,M. (1990) Transforming growth factor β stimulates mammary adenocarcinoma cell invasion and metastatic potential. Proc. Natl Acad. Sci. USA, 87, 7678–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagel S., Warner,A.H., Nellans,H.N., Lala,P.K., Waghorne,C. and Denhardt,D.T. (1989) Suppression by cathepsin L inhibitors of the invasion of amnion membranes by murine cancer cells. Cancer Res., 49, 3553–3557. [PubMed] [Google Scholar]
- Yang J., Liu,X., Bhalla,K., Kim,C.N., Ibrado,A.M., Cai,J., Peng,T.I., Jones,D.P. and Wang,X. (1997) Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science, 275, 1129–1132. [DOI] [PubMed] [Google Scholar]
- Yeh J.J., Lunetta,K.L., van Orsouw,N.J., Moore,F.D.J., Mutter,G.L., Vijg,J., Dahia,P.L. and Eng,C. (2000) Somatic mitochondrial DNA (mtDNA) mutations in papillary thyroid carcinomas and differential mtDNA sequence variants in cases with thyroid tumours. Oncogene, 19, 2060–2066. [DOI] [PubMed] [Google Scholar]