Abstract
Bacterial virulence proteins that are translocated into eukaryotic cells were expressed in Saccharomyces cerevisiae to model human infection. The subcellular localization patterns of these proteins in yeast paralleled those previously observed during mammalian infection, including localization to the nucleus and plasma membrane. Localization of Salmonella SspA in yeast provided the first evidence that SspA interacts with actin in living cells. In many cases, expression of the bacterial virulence proteins conferred genetically exploitable growth phenotypes. In this way, Yersinia YopE toxicity was demonstrated to be linked to its Rho GTPase activating protein activity. YopE blocked polarization of the yeast cytoskeleton and cell cycle progression, while SspA altered polarity and inhibited depolymerization of the actin cytoskeleton. These activities are consistent with previously proposed or demonstrated effects on higher eukaryotes and provide new insights into the roles of these proteins in pathogenesis: SspA in directing formation of membrane ruffles and YopE in arresting cell division. Thus, study of bacterial virulence proteins in yeast is a powerful system to determine functions of these proteins, probe eukaryotic cellular processes and model mammalian infection.
Keywords: actin cytoskeleton/cell cycle/Salmonella/type III secretion/Yersinia
Introduction
Microbial pathogens can manipulate eukaryotic host cell physiology to promote their own survival. Bacteria have evolved various protein delivery mechanisms to accomplish this task. For example, bacteria responsible for cholera, diphtheria and pertussis secrete individual proteins (toxins) into the extracellular milieu. These toxins can act at the eukaryotic host cell membrane, or bind to surface receptors that result in their internalization and subsequent binding to intracellular targets (Falnes and Sandvig, 2000). Many Gram-negative bacteria can also directly translocate sets of proteins into the cytoplasm of mammalian cells through a receptor-independent mechanism that utilizes a complex bacterial encoded ‘machine’ referred to as the type III secretion system (TTSS) (Hueck, 1998; Galán and Collmer, 1999). TTSSs are highly prevalent in both plant and animal pathogens. These virulence systems are essential for the development of typhoid fever, plague, dysentery, gastroenteritis and fire blight, and are probably required for additional diseases including trachoma, pelvic inflammatory disease and pertussis (Yuk et al., 1998; Subtil et al., 2000).
While the components of the type III secretion apparatus are relatively well conserved, each bacterium translocates a distinct set of proteins. Deciphering the mechanisms of action of these virulence proteins is vital to advancing our understanding of pathogenic bacterial strategies. In limited cases, bacterial virulence proteins encode regions of homology to mammalian enzymes, including serine-threonine kinases, tyrosine phosphatases and adenylate cyclases. In each of these cases, the purified virulence protein has been demonstrated to have the proposed biochemical activity. Functional approaches to study virulence proteins have been limited, but include phenotypic analyses conducted after exposure of mammalian cells to individual virulence proteins or to bacteria deleted for genes encoding individual virulence proteins. While the functions of many type III translocated virulence proteins remain unknown, such studies demonstrate that translocated virulence proteins can participate in many host cell processes, including cytokine production, inhibition of phagocytosis, cytoskeletal rearrangements and induction of either cytotoxicity or apoptosis (Hueck, 1998).
Analyses of bacterial virulence proteins have been limited by the lack of tractable genetic systems among higher eukaryotes. The ease of genetic analysis in the yeast Saccharomyces cerevisiae provides substantial opportunities for unraveling processes that are affected by virulence factors. Compared with higher eukaryotes, S.cerevisiae is easily transformed with DNA, undergoes homologous recombination at relatively high efficiency and grows as either a haploid or a diploid. In addition, the availability of the complete genomic sequence and numerous reagents including overexpression libraries and deletion strains contributes greatly to the ease of conducting genetic screens and selections in this system. Notably, studies in yeast demonstrate a striking degree of conservation between yeast and mammalian cellular processes, including the cell cycle, intermediary metabolism, actin cytoskeleton, pre-mRNA processing and protein sorting. In addition, many mutations implicated in complex human diseases, from deafness to cystic fibrosis, map to genes that are highly conserved between yeast and mammals (Foury, 1997). Since S.cerevisiae is a powerful system for studying human disease processes that result from inherent alterations in the basic cellular-encoded machinery, it seemed likely that yeast could also be used to study translocated bacterial virulence proteins that target well conserved basic cellular processes. By focusing on relatively well understood translocated proteins, the present study demonstrates that S.cerevisiae can be used as a model system to identify virulence protein function while advancing our understanding of basic eukaryotic host cell processes.
Results
Parallel subcellular localization patterns of Yersinia translocated virulence proteins in yeast and mammalian cells
One of the best characterized TTSSs is encoded within the virulence plasmid shared by all three pathogenic species of Yersinia (Y.pestis, Y.pseudotuberculosis and Y.enterocolitica), the causative agents of a variety of diseases including bubonic plague. All three pathogenic Yersinia species translocate at least five bacterial proteins into eukaryotic cells: YopE, YopH, YopJ (YopO), YopM and YpkA (YopO) [six in the case of Y.enterocolitica (YopT)]. These translocated proteins promote survival of Yersinia in the host, in part by inhibiting macrophage phagocytosis and cytokine production (Cornelis et al., 1998).
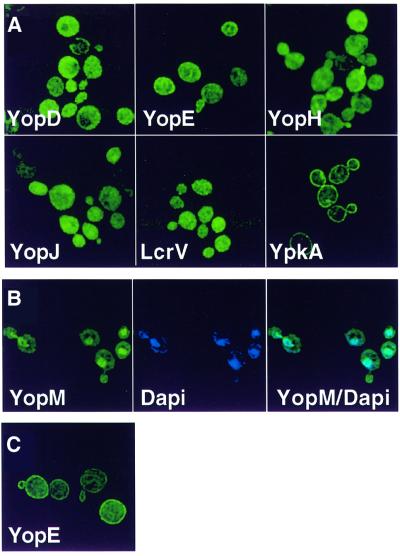
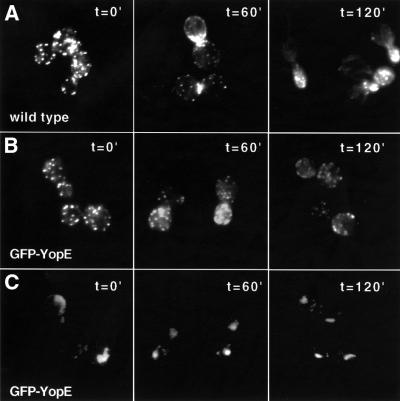
Since the eukaryotic host cell subcellular localization patterns of many of these translocated proteins were known, the first step to determine whether S.cerevisiae could serve as a model system to study the bacterial translocated proteins was to test whether these patterns were conserved. This was especially relevant as the mode of delivery of the virulence protein to eukaryotic cells was altered from direct translocation of bacterial virulence proteins across the eukaryotic plasma membrane to de novo synthesis in yeast. Therefore, green fluorescent protein (GFP) of Aqueoria victoria was fused to the N-terminus of all of the known Y.pseudotuberculosis translocated proteins (YopE, YopH, YopJ, YopM and YpkA) as well as to two proteins that are components of the translocation apparatus but are not translocated (YopD and LcrV) (Cornelis et al., 1998). The fusion genes were placed under the control of the galactose-inducible GAL10 promoter in order to regulate their expression. Yeast expressing each of the fusion proteins were observed by fluorescent microscopy 1 h after induction of fusion gene expression (from a high-copy plasmid). GFP–YopH, GFP–YopE, GFP–YopJ and GFP–LcrV each localized to the cytoplasm (Figure 1A). In contrast, GFP–YpkA localized to the plasma membrane and GFP–YopM localized to the nucleus [as confirmed by 4′,6-diamidino-2-phenylindole (DAPI) staining of DNA] (Figure 1B). Notably, the subcellular localization patterns of YopH, YopE, YopM and YpkA GFP fusion proteins paralleled those previously identified after direct delivery by bacterial translocation to mammalian cells (Rosqvist et al., 1994; Persson et al., 1995; Håkansson et al., 1996; Skrzypek et al., 1998). Interestingly, 2–3 h after induction of expression, GFP–YopE localized to the cell membrane and the cytoplasm (Figure 1C). The subcellular localization patterns of the YpkA, YopE, YopM and YopH GFP fusion proteins were unchanged when expressed from single chromosomal copies of the fusion genes (data not shown).
Fig. 1. Immunofluorescence microscopy of yeast expressing Y.pseudotuberculosis virulence proteins. Yeast (W303a) carrying high-copy-number plasmids encoding galactose-inducible GFP fusion genes to each of the indicated bacterial virulence proteins were visualized 1 h (A and B) or 3 h (C) after the addition of galactose. (B) Yeast were fixed in ethanol and stained with DAPI to visualize DNA. GFP fused to the virulence protein (green) is shown in the first panel, DNA (blue) is shown in the second panel and the third panel (YopM/DAPI) represents the first two panels merged together such that the turquoise features represent co-localization of GFP–YopM (green) and DNA (blue).
Inhibitory growth phenotypes conferred by expression of bacterial virulence proteins in yeast
One of the major advantages of utilizing S.cerevisiae to study bacterial virulence proteins is the genetic tractability of this model system. Thus, after observing the remarkably parallel subcellular localization patterns of the Yersinia translocated proteins in yeast and mammals, the next step was to test whether expression of the Yersinia virulence proteins in yeast would confer genetically manipulable growth phenotypes. Yeast transformed with each of the GFP–Yersinia fusion genes were assayed for potential growth phenotypes conferred by expression of the fusion proteins. Yeast expressing GFP fusion proteins to two of the translocated virulence proteins, YopH and YopJ, or the translocation apparatus proteins, YopD and LcrV, grew similarly to a strain expressing GFP alone. However, GFP–YopM expression conferred an intermediate inhibitory growth phenotype, and GFP–YopE or GFP–YpkA expression conferred a severe inhibitory growth phenotype (Figure 2). Strikingly, expression of three of the five translocated virulence proteins, but neither of the translocase apparatus proteins, conferred an inhibitory growth phenotype.
Fig. 2. Yeast growth is inhibited by expression of YpkA or YopE. Yeast (W303a) carrying high-copy-number plasmids encoding galactose-inducible GFP fusion genes to the indicated bacterial protein were grown overnight in non-inducing selective synthetic media containing raffinose as a carbon source. Cultures were then normalized to OD600 = 1 and serial 10-fold dilutions were spotted onto selective media plates containing raffinose (1%) and galactose (2%). Photographs were taken after 2 days of growth at 30°C.
YopE toxicity was dependent on RhoGAP activity
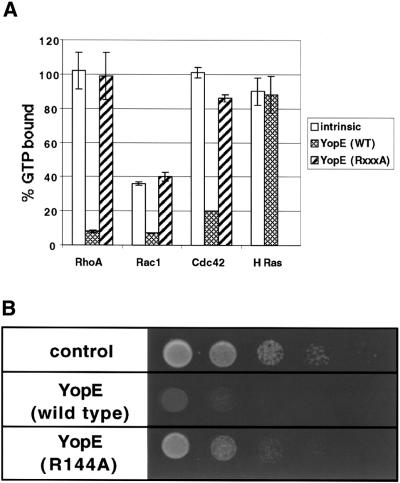
The observation that YopE, a mammalian cell cytotoxin (Rosqvist et al., 1990), severely inhibited S.cerevisiae growth suggested that this bacterial virulence protein functioned similarly in both eukaryotic systems. YopE shares a region of homology (GxxRxxGS) to ExoS of Pseudomonas and SptP of Salmonella, two effector proteins that are Rho GTPase activating proteins (RhoGAPs). This region of homology is essential for RhoGAP activity, as mutation of the invariant arginine to alanine results in inactivation of the purified bacterial RhoGAPs. YopE was recently demonstrated to have RhoGAP activity (Black and Bliska, 2000; Von Pawel-Rammingen et al., 2000). To confirm these observations, His-tagged YopE was purified and incubated with purified mammalian G-proteins [glutathione S-transferase (GST)–RhoA, GST–Rac1, GST–Cdc42 or GST–H Ras] that had been pre-loaded with [γ-32P]GTP. Reactions were stopped after 5 min and the amount of GTP hydrolysis was determined. As was previously observed (Black and Bliska, 2000; Von Pawel-Rammingen et al., 2000), YopE significantly stimulated GTP hydrolysis of all members of the Rho family of G-proteins (RhoA, Rac1 and Cdc42), but not H-Ras (Figure 3A). Mutation of the invariant arginine of the RhoGAP consensus sequence to alanine (YopE-R144A) resulted in loss of in vitro RhoGAP activity with all three Rho G-protein substrates (Figure 3A). To test whether the in vitro RhoGAP activity of YopE was linked to the in vivo yeast toxicity, a plasmid encoding the inactive allele of YopE (YopE-R144A) fused to GFP under the control of the GAL10 promoter was introduced into yeast. Indeed, as was recently demonstrated (Von Pawel-Rammingen et al., 2000), expression of this mutant allele greatly alleviated the growth phenotype conferred by expression of GFP–YopE (Figure 3B).
Fig. 3. YopE toxicity is dependent on RhoGAP activity. (A) Purified His-tagged YopE and mutant (R144A) YopE-R144A (5.5 pmol) were incubated with G-proteins (GST–Rac1, GST–RhoA, GST–Cdc42 and GST–H-Ras) (5.5 pmol) that were pre-loaded with [γ-32P]GTP. (His6-tagged YopE-R144A was not incubated with GST–H-Ras.) GTPase activity was assayed after 5 min. Values represent the mean ± SD of three separate experiments. (B) Growth assays (as described in Figure 2) were conducted on cells transformed with a high-copy-number plasmid encoding galactose-inducible mutant GFP-yopE (R144A) and compared to cells transformed with empty vector or wild-type GAL10-GFP-yopE.
Since S.cerevisiae encodes six genes (Rho1, Rho2, Rho3, Rho4, Rho5 and Cdc42) with strong homology (50–80% identity) to the mammalian family of Rho G-proteins, each or all of these were potential targets of YopE. If YopE targeted only one member of this family of proteins, it most likely targeted one of the essential members of this family, Rho1 or Cdc42. Since overexpression of the target(s) of YopE might suppress YopE toxicity by saturation of RhoGAP activity, Rho1 and/or Cdc42 were overexpressed in YopE-expressing yeast. In addition, as RhoGAPs inactivate G-proteins by accelerating the exchange of active GTP-bound G-proteins to their inactive GDP-bound forms, constitutively activated alleles of either Rho1 or Cdc42, which are always in the active GTP-bound state, were also overexpressed in YopE-expressing yeast. None of these alleles alleviated YopE toxicity (data not shown). These results differed from recent observations by Von Pawel-Rammingen et al. (2000), where overexpression of either Rho1, Rho4 or Ste20 (a downstream effector of Cdc42) suppressed YopE toxicity when YopE was expressed from the methionine-inducible promoter (MET1). Notably, proteins are expressed at lower levels from the MET1 promoter as compared with the GAL10 promoter. Thus, if YopE exerts saturable RhoGAP activity on all or multiple members of the Rho family of G-proteins, then when expressed from the relatively weak MET1 promoter it would be possible to titrate out the RhoGAP activity of YopE by overexpressing any one of its multiple targets, but toxicity caused by higher levels of expression from the galactose-inducible promoter could not be overcome by overexpressing any individual substrate. Thus, YopE is likely to be a RhoGAP for all or multiple members of the yeast Rho family of G-proteins.
YopE disrupted actin cytoskeletal polarity
The actin cytoskeleton is a common target of type III translocated virulence proteins. Bacterial pathogens reorganize the actin cytoskeleton to either promote uptake of the bacteria into the host cell (Salmonella, Shigella) (Bourdet-Sicard et al., 2000; Galán and Zhou, 2000) or prevent phagocytosis of the bacteria by macrophages (Yersinia) (Cornelis, 2000). Since many of the components of the yeast actin cytoskeleton are well conserved between yeast and mammals, yeast should serve as a model system to dissect the interactions between bacterial proteins and the cytoskeleton. Since injection of purified YopE into epithelial cells results in disruption of actin stress fibers (Rosqvist et al., 1991a), studies were directed to determine whether YopE would mediate similar alterations in the yeast actin cytoskeleton. Yeast do not contain actin stress fibers; rather, their actin cytoskeleton is composed of two distinct components that have been identified by fluorescence microscopy: cortical patches and actin cables. Cortical patches are punctate actin-rich bodies, while cables are bundles of actin filaments that are oriented along the mother–bud axis. Structures of both types are located at the cell cortex and their localization patterns change in a cell cycle-dependent manner (Adams et al., 1995).
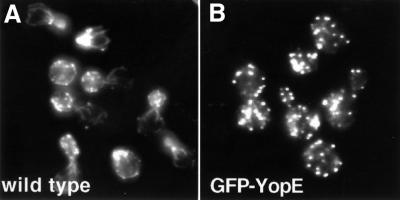
To determine whether YopE disrupts the yeast actin cytoskeleton, yeast containing GFP–yopE integrated into the chromosome (W303::GAL10-GFP-yopE) were stained with an actin-staining dye, rhodamine-labeled phalloidin, 2 h after induction of YopE expression. Notably, while the actin cytoskeleton of YopE-expressing cells was not totally disrupted (Figure 4B), it appeared distinctly different from that of wild-type cells grown under identical conditions (Figure 4A). The majority of wild-type yeast displayed a polarized actin cytoskeleton. Cortical patches were observed to concentrate in the new bud or at the site of probable impending bud formation, and actin cables were observed to be polarized along the mother–bud axis. However, almost none of the YopE-expressing yeast demonstrated a polarized actin cytoskeleton. Rather, these cells appeared to have cortical patches dispersed throughout the cells, regardless of their stage in the cell cycle. These results strongly suggested that expression of YopE resulted in loss of actin cytoskeletal polarity. The yeast actin cytoskeleton is normally polarized at several points in the cell cycle. Thus, to determine whether this was a direct effect of YopE expression, yeast were synchronized at several points in the cell cycle and subsequently analyzed for alterations in cytoskeletal polarity in the presence or absence of GFP–YopE.
Fig. 4. YopE expression disrupts polarity of the actin cytoskeleton. Wild-type (A) and W303::GAL10-GFP-yopE (B) yeast were grown in non-inducing selective liquid media containing raffinose (2%) and sorbitol (1 M) to OD600 = 0.3–0.6. Galactose was added to these asynchronous cultures to a final concentration of 2%. Yeast were then harvested after 2 h, fixed with formaldehyde and stained with rhodamine-labeled phalloidin to visualize the actin cytoskeleton.
YopE expression blocked bud formation and activated the morphogenesis checkpoint of the cell cycle
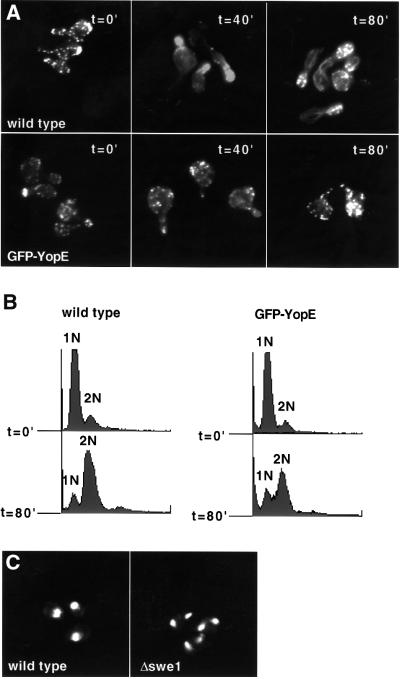
The actin cytoskeleton is first polarized early in the cell cycle when cortical patches accumulate at one pole of the cell, the presumed site of impending bud formation. The cortical patches then move into the growing bud and the actin cables extend from the mother cell into the bud. Notably, the Rho family of G-proteins, in part, mediate actin cytoskeletal rearrangements associated with bud formation (Cabib et al., 1998). Since YopE is a RhoGAP, YopE toxicity in yeast could be due to disruption of the normal budding pathway through inhibition of the Rho family of G-proteins. To test this hypothesis, wild-type and W303::GAL10-GFP-yopE yeast were incubated with α-factor for 2.5 h such that they arrested in G1 of the cell cycle and formed mating projections (schmoos). Galactose was then added to the media in the presence of addi tional α-factor for 1 h to induce YopE expression in W303::GAL10-GFP-yopE yeast. Immediately, after removal of the α-factor, the cortical patches in both wild-type and W303::GAL10-GFP-yopE schmoos were randomly distributed (Figure 5A). However, in wild-type cells, 20 min after the removal of α-factor, cortical patches accumulated at sites of new bud formation, and 40 min after the removal of α-factor, cortical patches localized to the new bud and polarized actin cables were visualized. In contrast, cortical patches in YopE-expressing yeast remained depolarized (Figure 5A) and, even 2 h after the removal of α-factor, these cells did not proceed with bud formation (data not shown). Thus, YopE expression inhibited reorganization/polarization of the actin cytoskel eton and bud formation. To confirm that these cells were still viable and progressing through the cell cycle, their DNA content was analyzed by flow cytometry analyses at 20 min intervals. Notably, the majority of both wild-type and YopE-expressing yeast replicated their DNA by 80 min after the removal of α-factor (Figure 5B). Thus, despite the block to polarization of the actin cytoskeleton and bud formation, YopE-expressing cells underwent DNA replication.
Fig. 5. YopE blocks bud formation and arrests cells at the morphogenesis checkpoint. Yeast were grown in non-inducing YEP (W303 strains) or selective (S288C strains) liquid media containing raffinose (2%) and sorbitol (1 M) to OD600 = 0.3–0.6. The cultures were then synchronized with α-factor for 2.5 h such that they arrested at G1 and formed mating projections (schmoos). Galactose (2%) was added to the synchronized cultures. After 1 h, α-factor was removed and cells were harvested at 40 min intervals. (A) Wild-type and W303::GAL10-GFP-yopE yeast were fixed with formaldehyde and stained with rhodamine-labeled phalloidin to visualize the actin cytoskeleton. (B) Wild-type and W303::GAL10-GFP-yopE yeast were fixed with ethanol and stained with PI, and subjected to flow cytometry analysis. 1N and 2N represent the DNA content of the two peaks. (C) Wild-type or Δswe1 yeast (S288Ca background) carrying galactose-inducible GFP-yopE encoded on a low-copy plasmid (pCFL112) were fixed with ethanol and stained with PI to visualize the DNA/nuclei.
This phenotype of small-budded or unbudded cells that have undergone DNA replication has previously been observed under conditions where the actin cytoskeleton is perturbed such that bud development is delayed. Perturbations of the actin cytoskeleton activate the morphogenesis checkpoint of the cell cycle, which delays nuclear division in the absence of an adequate bud (McMillan et al., 1998). Swe1 regulates the morphogenesis checkpoint. Δswe1 yeast normally display no discernible phenotype; however, when bud formation is delayed, these cells are unable to delay mitosis appropriately and undergo nuclear division (endomitosis) to form unbudded binucleate cells. To test whether YopE expression activated the morphogenesis checkpoint, YopE was expressed from a low-copy plasmid in wild-type and Δswe1 yeast. Prior to expression of YopE, the yeast were exposed to α-factor for 2.5 h such that they synchronized at G1 and formed schmoos. Galactose was then added to the media in the presence of additional α-factor for 1 h to allow for GFP–YopE expression in the appropriate cells. After removal of the α-factor, cells were examined at 1 h intervals by both microscopic and cytometric analyses. Three hours after YopE expression, ∼60% of both the wild-type and Δswe1 cells underwent DNA replication (data not shown). Nevertheless, after 3 h, over half of the YopE-expressing Δswe1 cells were binucleate, in con trast to <1% of the YopE-expressing wild-type yeast (Figure 5C). Thus, the growth phenotype conferred by YopE expression in yeast is, at least in part, a result of activation of the morphogenesis checkpoint.
YopE blocks formation of actin rings and cytokinesis
As demonstrated in Figure 4, the actin cytoskeleton of YopE-expressing cells was disrupted in both budded and unbudded cells. This observation raised the question as to whether YopE expression affects actin cytoskeletal polarity at later steps in the cell cycle (after bud formation). After bud formation and nuclear division proceed, cortical patches normally randomly distribute throughout the mother and bud. However, prior to cytokinesis, the actin cytoskeleton is again polarized. At this point in time, the cortical patches form one or two actin rings at the bud neck, the future sight of cytokinesis (Pruyne and Bretscher, 2000). Little is known about the role of the Rho G-proteins at this point in the cell cycle. To determine whether YopE affects actin cytoskeleton polarity observed in ring formation and cytokinesis, wild-type and W303::GAL10-GFP-yopE yeast were exposed to low-dose nocodazole, a microtubule-destabilizing drug, for 2 h. The cells uniformly arrested as large budded cells with duplicated DNA at the spindle assembly checkpoint of the cell cycle (which takes place after the morphogenesis checkpoint). Galactose was added to these cultures, in the presence of nocodazole, to induce YopE expression in W303::GAL10-GFP-yopE yeast. After 1 h, the nocodazole was removed from the galactose-containing media and the yeast were analyzed at 1 h intervals for alterations in cytoskeletal polarity and cell cycle progression. As demonstrated in Figure 6A, by 1 h after removal of nocodazole from the wild-type yeast, the actin cytoskeleton was polarized. In many cells, cortical patches were observed to form a single or double actin ring at the bud neck. In contrast, even by 2 h, the YopE-expressing cells did not polarize their actin cytoskeleton or undergo cytokinesis (Figure 6B). Despite the absence of a polarized actin cytoskeleton, the YopE-expressing cells underwent nuclear division (Figure 6C). Thus, these cells were viable and able to reconstitute a microtubule spindle, allowing for nuclear division after removal of the nocodazole. Since YopE is likely to be a GAP for the entire family of Rho G-proteins, these observations implicate a previously unidentified role for Rho G-proteins in the polarization of the actin cytoskeleton observed with the formation of actin rings.
Fig. 6. YopE blocks cytokinesis and formation of actin rings. Wild-type and W303::GAL10-GFP-yopE yeast were grown in non-inducing YEP liquid media containing raffinose (2%) and sorbitol (1 M) to OD600 = 0.3–0.6. The cultures were then synchronized with nocodazole for 2 h such that they arrested as large budded cells at the spindle checkpoint. Galactose (2%) was added to the synchronized cultures. After 1 h, nocodazole was removed and cells were harvested at 60 min intervals. Cells were fixed with formaldehyde and stained with rhodamine-labeled phalloidin to visualize the actin cytoskeleton (A and B) and with DAPI (C) to visualize DNA/nuclei.
SspA co-localized with the actin cytoskeleton and inhibited actin depolymerization
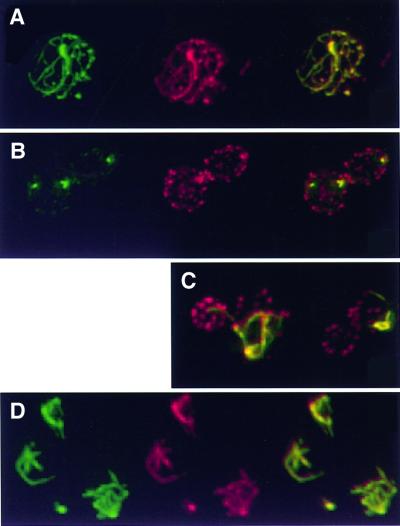
Many bacterial virulence proteins, in addition to YopE, are likely to mediate alterations in the mammalian actin cytoskeleton. Since YopE exerted similar effects on the actin cytoskeleton of both S.cerevisiae and mammals, it seemed likely that yeast would provide a fruitful system to characterize additional interactions between the cytoskeleton and bacterial virulence proteins. For example, SspA (SipA), a Salmonella translocated protein, had previously been demonstrated to bind filamentous actin, decrease the critical concentration required for actin polymerization and inhibit depolymerization of actin filaments in vitro (Zhou et al., 1999); however, little was known about the function or subcellular localization of SspA in living cells, where numerous endogenous actin-binding proteins interact dynamically with the cytoskeleton. Thus, SspA was expressed in yeast. When expressed at high levels in yeast, GFP–SspA co-localized with the majority of the actin cytoskeleton, both cortical patches and actin cables (Figure 7A). Notably, the actin cables in these cells (Figure 7A) lost their normal cellular polarity and appeared thicker than wild-type cables (Figure 4A). In contrast, when expressed at relatively low levels, the subcellular localization pattern of GFP– SspA was altered. One hour after induction, GFP–SspA localized to a small subset, 1–4, of the ∼50 actin cortical patches in the cell (Figure 7B). This subset of cortical patches appeared larger in size than those that did not associate with GFP–SspA (Figure 4A). Three hours after induction, GFP–SspA still did not co-localize with additional cortical actin patches, but rather co-localized with actin cables that appeared to emanate from the enlarged actin cortical patches associated with GFP–SspA (Figure 7C). These cables, like those observed with high levels of GFP–SspA, appeared thicker than normal and lost their normal cellular polarity.
Fig. 7. GFP–SspA co-localizes with and reorganizes the actin cytoskeleton. Each panel represents immunofluorescent images of yeast (W303a) carrying plasmids encoding galactose-inducible GFP–sspA. Yeast were fixed with formaldehyde at the designated time points and stained with rhodamine-labeled phalloidin to visualize actin. In (A, B and D), GFP–SspA (green) is shown in the first panel, actin (red) is shown in the second panel and the third panel represents the first two panels merged together such that yellow features indicate co-localization of GFP–SspA (green) and actin (red). In (C), each panel represents merged GFP–SspA (green) and actin (red) images such that the yellow features represent co-localization of GFP–SspA and actin. Six hours after induction, GFP–SspA expressed from a high-copy plasmid (pCFL170) co-localized with all of the actin in the yeast and disrupted normal cell polarity. (B) One hour after induction, GFP–SspA, expressed from a single copy of the integrated fusion gene (YCL170), co-localized with a small subset of yeast cortical patches. (C) Three hours after induction, GFP–SspA, expressed from a single copy of the integrated fusion gene (YCL170) co-localized with actin cables that appear to originate from GFP–SspA-containing cortical patches. (D) Twelve hours after induction of GFP–SspA expression from a low-copy plasmid (pCFL172), only the actin cytoskeleton that co-localized with GFP–SspA was resistant to a 60 min exposure to LatA (200 µM).
The prominent actin cables and thickened cortical patches that co-localized with GFP–SspA appeared similar to those previously observed in yeast expressing an actin allele that inhibits depolymerization. Yeast expressing this actin allele are resistant to depolymerization of filamentous actin by Latrunculin A (LatA) (Belmont and Drubin, 1998), a drug that binds to and sequesters actin monomers. Since the turnover rate of the yeast actin cytoskeleton is rapid and LatA prevents the formation of new actin polymers, 5–10 min after exposure of yeast to LatA the entire yeast actin cytoskeleton is disrupted (Ayscough et al., 1997). Since LatA does not alter the off rate of actin from filaments, it can be used to measure the rate of filament turnover in vivo (Belmont and Drubin, 1998). Yeast expressing GFP–SspA for 12 h, such that the fusion protein co-localized with a subset of the actin cytoskeleton, were exposed to LatA for 1 h. Remarkably, under these conditions, all of the actin structures in the cell were disrupted, except the actin cables and cortical patches that co-localized with GFP–SspA (Figure 7D). Thus, in vivo, as in vitro (Zhou et al., 1999), SspA inhibited actin depolymerization.
Discussion
Saccharomyces cerevisiae is a powerful model system that can be used to elucidate functions of bacterial virulence proteins
Remarkably, the subcellular localization patterns of all the Yersinia translocated virulence proteins previously identified in mammalian cells (YopE, YopH, YopM and YpkA) paralleled those observed in yeast. Notably, these evolutionarily divergent cells target bacterial proteins to the same subcellular compartments, independent of the mode of delivery: de novo synthesis in yeast or bacterial translocation of pre-formed proteins through the mammalian plasma membrane. In both mammals and yeast, YopM is targeted to the nucleus in the absence of any known nuclear localization signal and YpkA localizes to the plasma membrane despite the lack of an apparent transmembrane domain. Thus, these proteins are likely targeted to their specific subcellular localizations through components conserved among higher and lower eukaryotes. Rapid identification of these components in yeast will probably provide clues as to the roles of these proteins in pathogenesis. In addition, the strong conservation of subcellular localization patterns observed among the Yersinia proteins suggests that expression of GFP fused to translocated virulence proteins in yeast will provide an initial means to characterize these proteins and provide potential clues as to their function. For example, expression of SspA, a Salmonella translocated protein known to bind to actin in vitro, provides the first direct evidence that SspA interacts with the actin cytoskeleton in living cells.
One of the main advantages of using yeast is the ease with which genetic analyses can be conducted. Thus, it was exciting to observe that expression of the majority of the bacterial proteins (YopE, YopM and YpkA) conferred inhibitory growth phenotypes that can be exploited. Genetic screens can be conducted to identify eukaryotic proteins or cellular pathways that are targeted by these virulence proteins, or individual virulence proteins can be mutagenized and analyzed by growth and/or localization assays to identify functional domains. For example, mutation of the invariant arginine of the YopE RhoGAP consensus sequence suppressed YopE toxicity, thus implying that YopE toxicity in yeast is likely to be a direct result of its RhoGAP activity. YopM and YpkA can be mutagenized in vitro and then analyzed in yeast to identify domains required for correct subcellular localization and/or toxicity. Once characterized in yeast, mutagenized virulence proteins can be re-introduced into bacteria and tested for the roles of these domains in virulence/pathogenesis.
Our initial studies in yeast focused on relatively well studied proteins, YopE and SspA, which have both been proposed or demonstrated to interact with the actin cytoskeleton. Notably, while SspA expression resulted in reorganization of the yeast actin cytoskeleton, YopE expression resulted in loss of polarization or disruption of the actin cytoskeleton. This effect of YopE could be the yeast functional equivalent of disruption of the mammalian actin stress fibers by injection of YopE (Rosqvist et al., 1991b). Thus, these phenotypes observed in yeast paralleled the activities of these virulence proteins previously predicted or demonstrated in higher eukaryotic cells, and provide new insights into potential mechanisms of bacterial pathogenesis. In addition, these studies predict that the yeast system should prove to be especially fruitful at dissecting interactions between bacterial virulence proteins and the highly conserved actin cytoskeleton, a common target of many virulence proteins.
Insights into mechanisms of Salmonella invasion gleaned from observations in S.cerevisiae
Salmonella are intracellular pathogens that can invade normally non-phagocytic epithelial cells by inducing the formation of membrane ruffles at sites of bacterial attachment (Francis et al., 1992). This process is dependent on the TTSS encoded within Salmonella pathogenicity island 1 (SPI1). Several of the virulence proteins translocated by the SPI1 TTSS, including SspA, have been demonstrated or proposed to mediate alterations in the actin cytoskeleton associated with membrane ruffling (Galán and Zhou, 2000). When epithelial cells are exposed to Salmonella that do not express SspA (ΔsspA), membrane ruffles form diffusely rather than at sites of bacterial attachment. This observation led to the hypothesis that SspA plays a role in directing ruffling to sites of bacterial attachment, perhaps by directing actin cytoskeleton rearrangements to these sites (Zhou et al., 1999). However, while SspA had previously been demonstrated to interact directly with actin in vitro, there was no direct evidence that SspA was able to interact with actin in living cells in the presence of the numerous endogenous actin-binding proteins. Expression of SspA in S.cerevisiae demonstrated not only that SspA interacts with the actin cytoskeleton in vivo, but also that this interaction causes gross rearrangements in the actin cytoskeleton. Notably, when expressed at relatively high levels in yeast, GFP–SspA co-localized with the entire actin cytoskeleton, including both cortical patches and actin cables. These interactions both disrupted polarity and inhibited depolymerization of the actin cytoskeleton. Interestingly, when expressed at relatively low levels in yeast, GFP–SspA preferentially localized to a subset of yeast cortical patches rather than actin cables. Over time, SspA co-localized with actin cables that appeared to emanate from the SspA-associated cortical patches rather than to additional cortical patches. This observation suggests that SspA differentially recognizes distinct components of the actin cytoskeleton such that it is able to direct actin cable growth to specific locations in the cell. Perhaps in an analogous fashion in higher eukaryotic cells, SspA directs growth of actin filament to sites of bacterial attachment to facilitate uptake of Salmonella by a membrane ruffling mechanism.
Expression of bacterial virulence proteins in yeast provides new tools to study basic eukaryotic cellular processes
A by-product of expression of bacterial virulence proteins in eukaryotic cells is that they provide new tools to study basic eukaryotic cellular processes. For example, expression of the RhoGAP YopE is likely to result in accumulation of all or multiple members of the Rho family of G-proteins in their inactive GDP-bound state. Since appreciable levels of YopE are produced 1 h after galactose induction of GAL10-regulated GFP–YopE, it is possible to closely regulate Rho G-protein inactivation. The Rho family of G-proteins has long been known to play a role in establishing and maintaining cell polarity during bud formation, but little was known about the role of these proteins in determining actin cell polarity at later points in the cell cycle. Thus, by expressing YopE after synchronizing yeast at defined points in the cell cycle, it was possible to assess the role of the Rho G-proteins at distinct steps. YopE expression in yeast synchronized prior to bud formation at G1 blocked polarization of the actin cytoskeleton and activated the morphogenesis checkpoint. These observations are consistent with the hypothesis that YopE is a RhoGAP in yeast as conditional alleles of two members of the Rho family of G-proteins, Cdc42 (Lew and Reed, 1995) and Rho1 (Yamochi et al., 1994), confer a similar arrest at the morphogenesis checkpoint. The more surprising result was observed when YopE was expressed in cells after they were arrested at the spindle checkpoint (which occurs after the morphogenesis checkpoint and bud formation). These cells were unable to polarize the actin cytoskeleton to form actin rings, thus providing the first evidence that a member or members of the Rho G-protein family mediate, at least in part, the formation of actin ring(s) at the bud neck prior to cytokinesis.
Cell cycle arrest as a bacterial virulence mechanism
YopE promotes Yersinia virulence by disrupting the actin cytoskeleton and preventing phagocytosis by macrophages (Rosqvist et al., 1990, 1991a). YopE has long been called a cytotoxin based on the observation that injection of the proteins results in rounding up of epithelial cells. However, rounding up of epithelial cells is likely to reflect disruption of the cytoskeleton by inactivation of the Rho G-proteins rather than cytotoxicity, as other conditions that down-regulate Rho activity also result in rounding up of cells (Ridley and Hall, 1992). Thus, disruption of the actin cytoskeleton by YopE might have downstream effects on mammalian cells similar to those in yeast. Specifically, disruption of the actin cytoskeleton by YopE could both block phagocytosis and arrest cell division. Since Yersinia has a propensity for colonizing lymph tissues containing rapidly dividing lymphocytes, such cells could be quite vulnerable to YopE-mediated cell cycle arrest with resultant immune system suppression. Together, YopE and YopH, which suppresses lymphocyte activation by interfering with antigen receptor signaling (Yao et al., 1999), could mediate the TTSS virulence-plasmid-dependent suppression of cellular immunity observed after infection of BALB/c mice with Yersinia (Ruiz-Bravo et al., 1996). A growing family of bacteria, including Escherichia coli, Shigella dysenteriae, Campylobacter spp. and Haemophilus ducreyi, are now known to secrete cytolethal distending toxin, a protein that blocks eukaryotic cells in the G2 phase of the cell cycle (Pickett and Whitehouse, 1999). These results, together with our data with YopE, suggest that cell cycle arrest may be a general mechanism of microbial pathogenesis.
Future implications of this work
In this post-genomic era, where new potential coding sequences of microbes are being generated daily, new approaches will be needed to define proteins that modify host processes. These approaches will be essential for understanding mechanisms of pathogenesis for organisms that, unlike Yersinia and Salmonella, are not amenable to genetic analysis. The work presented here clearly indicates that S.cerevisiae can serve as a fruitful model system to define microbial protein functions. Study of such proteins in yeast should advance our understanding of both pathogenesis and basic eukaryotic cellular physiology.
Materials and methods
Plasmids and strains
Plasmids are summarized in Table I. The open reading frame encoding each of the translocated proteins (YopD, YopE, YopH, YopJ, YopM, YpkA, LcrV and SspA) was PCR amplified from either Y.enterocolitica or Salmonella typhimurium chromosomal DNA preparations and subcloned in-frame with the C-terminus of GFP in pFUS (Johnson, 1991), a high-copy (2µ) plasmid, to create GFP fusion proteins under the control of the GAL10 promoter. All PCR-amplified fragments used for cloning were analyzed by DNA sequencing. Low-copy-number (cen) plasmid versions of GAL10-GFP-yopE and GAL10-GFP-sspA were constructed by inserting a restriction fragment that contained the Gal10 promoter, GFP fusion gene and the ADH terminator into pRS316 and pRS315 (Sikorski and Hieter, 1989). Integrating versions of the fusion genes were constructed by deleting the 2µ replication origin from pFUS backbone. YopE-R144A was generated by PCR-mediated site-directed mutagenesis. Plasmids pGAL-Rho1, pGAL-Rho1-Q68H, pGAL-Cdc42 and pGAL-Cdc42 were kind gifts of Marie Evangelista and Charles Boone (Queens University, Toronto, Canada).
Table I. Plasmids and yeast strains.
| Source | ||
|---|---|---|
| Plasmids | ||
| pFUS | GAL10-GFP (2µ LEU2) | 2 |
| pCFL100 | GAL10-GFP-yopD (2µ LEU2) | 1 |
| pCFL110 | GAL10-GFP-yopE (2µ LEU2) | 1 |
| pCFL120 | GAL10-GFP-yopH (2µ LEU2) | 1 |
| pCFL130 | GAL10-GFP-yopJ (2µ LEU2) | 1 |
| pCFL140 | GAL10-GFP-yopM (2µ LEU2) | 1 |
| pCFL150 | GAL10-GFP-ypkA (2µ LEU2) | 1 |
| pCFL160 | GAL10-GFP-lcrV (2µ LEU2) | 1 |
| pCFL111 | GAL10-GFP-yopE (integrating LEU2) | 1 |
| pCFL121 | GAL10-GFP-yopH (integrating LEU2) | 1 |
| pCFL141 | GAL10-GFP-yopM (integrating LEU2) | 1 |
| pCFL151 | GAL10-GFP-ypkA (integrating LEU2) | 1 |
| pCFL170 | GAL10-GFP-sspA (2µ LEU2) | 1 |
| pCFL171 | GAL10-GFP-sspA (integrating LEU2) | 1 |
| pCFL112 | GAL10-GFP-yopE (pRS316 cen URA3) | 1 |
| pCFL172 | GAL10-GFP-sspA (pRS316 cen URA3) | 1 |
| pCFL112 | GAL10-GFP-yopE-R144A (2µ LEU2) | 1 |
| Yeast strains | ||
| W303 | Mata can1-100, leu2-3,112, his3-11,-15, ura3-1, trp1-1, ade2-1 | 1 |
| YCL110 | W303a GAL-GFP-yopE::LEU2 | 1 |
| YCL120 | W303a GAL-GFP-yopH::LEU2 | 1 |
| YCL140 | W303a GAL-GFP-yopM::LEU2 | 1 |
| YCL150 | W303a GAL-GFP-ypkA::LEU2 | 1 |
| YCL170 | W303a GAL-GFP-sspA::LEU2 | 1 |
| BY4741 | S288Ca | 3 |
| ATCC1238 | S288CaΔswe1 | 3 |
Sources: 1, this study; 2, Byers laboratory; 3, American Type Culture Collection (ATCC).
Yeast strains are summarized in Table I. Integrated translocated protein strains (GFP–YpkA, GFP–YopE, GFP–YopM, GFP–YopH and GFP–SspA) were made by digestion of the integrating vector with AflII to target integration at the W303a LEU2 locus.
Microscopy techniques
Yeast carrying the plasmids of interest were grown overnight in non-inducing selective synthetic media supplemented with 2% raffinose. Yeast were diluted to OD600 = 0.5–0.6 and grown for an additional 1 h. Expression of the fusion protein was induced by addition of 2% galactose to the medium. Yeast were observed at the designated time points on a deconvolving fluorescent microscope with a 60× or 100× 1.4 numerical aperture Zeiss lens and analyzed by API Delta Vision software. Actin was visualized by staining with rhodamine-labeled phalloidin (Molecular Probes, Eugene, OR) as previously described (Pringle et al., 1989). DNA was visualized by staining with DAPI (Sigma) or propidium iodide (PI; Sigma). For DAPI staining, yeast were pelleted and resuspended in cold ethanol (70–95%) for >30 min. Fixed cells were then pelleted and incubated with DAPI (1 µg/ml) in phosphate-buffered saline. For PI staining, yeast were fixed and stained as previously described (Hutter and Eipel, 1979).
Growth assays
To compare the growth rates of different strains, saturated overnight cultures of the strains of interest were grown in non-inducing selective synthetic media supplemented with 2% raffinose. Each culture was normalized to OD600 = 1 and then serially diluted 10-fold three additional times. Aliquots (5 µl) of each of the four dilutions were spotted onto a selective medium plate supplemented with 1–2% raffinose and 2% galactose. The plates were incubated at 30°C and photographs of the plates were obtained 2 days after plating.
Overexpression and purification of YopE and YopE-R144A
YopE and YopE-R144A were PCR amplified and then subcloned into pET15b (Novagen) to generate His6-YopE and His6-YopE–R144A fusion proteins. Escherichia coli BL21 (DE3) pLysS Gold (Strategene) transformed with pET15-YopE and pET15-YopE-R144A were grown at 37°C to OD600 = 0.1 in LB media supplemented with ampicillin (100 µg/ml). Cultures were cooled on ice to lower the temperature to 20°C prior to the addition of 50 µM isopropyl-β-d-thiogalactopyranoside and incubated overnight at 25°C. Cells were pelleted and lysed by the addition of lysozyme followed by sonication. The His-tagged fusion proteins were purified by affinity chromatography on nickel resin according to the standard protocol (Qiagen). To solubilize the protein, NP-40 was added to the eluate to a final concentration of 1% prior to binding to nickel resin. The final eluate of protein was 90% pure, as estimated by Coomassie Blue staining.
GAP assays
Purified YopE and YopE-R144A were incubated with GST–RhoA, GST–Rac1, GST–Cdc42 and GST–H-Ras (Cytoskeleton, Denver, CO). Assays were conducted according to the protocol provided by the manufacturer.
Cell synchrony experiments and flow cytometry analyses
Cells were grown overnight in YEP raffinose or selective media to OD600 = 0.3–0.6 at 30°C. Sorbitol (1 M) was added to the media to suppress any cell lysis due to osmotic instability of the cells due to inactivation of Rho1. Cells were arrested at G1 by incubation with 20 µg/ml α-factor (Diagnostic Chemicals Ltd) for 2.5 h or at the spindle checkpoint by incubation with 15 µg/ml nocodazole (Sigma) for 2 h. One hour after the subsequent addition of galactose, cells were washed twice with pre-warmed medium and then resuspended in fresh medium containing galactose to release them from either the α-factor- or nocodazole-induced cell cycle arrest. Yeast were harvested at 20–30 min intervals and fixed in 70% ethanol. Yeast were stained using the DNA stain PI (Sigma), as previously described (Hutter and Eipel, 1979). Samples were analyzed on a FACscan flow cytometer using CELL QUEST software to obtain and analyze data (BDIS, San Jose, CA).
Acknowledgments
Acknowledgements
We thank members of the Miller laboratory, R.L.Roberts, R.E.Hughes and J.Cope for valuable discussions. We thank R.L.Roberts, B.Byers, C.A.Scherer, M.Ohl and S.L.Pennington for critically reading the manuscript. We thank L.Goetsch, L.Dirick, B.Byers, M.Evangelista and C.Boone for sharing published reagents, and S.Fields for the generous use of his microscope. C.F.L. is currently a Howard Hughes Physician Postdoctoral Fellow and was also supported by a Pfizer Postdoctoral Fellowship during this work. This work was supported by a grant to S.I.M. from the National Institutes of Health (RO1 A130479).
References
- Adams A.E., Shen,W., Lin,C.S., Leavitt,J. and Matsudaira,P. (1995) Isoform-specific complementation of the yeast sac6 null mutation by human fimbrin. Mol. Cell. Biol., 15, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough K.R., Stryker,J., Pokala,N., Sanders,M., Crews,P. and Drubin,D.G. (1997) High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol., 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L.D. and Drubin,D.G. (1998) The yeast V159N actin mutant reveals roles for actin dynamics in vivo. J. Cell Biol., 142, 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.S. and Bliska,J.B. (2000) The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol. Microbiol., 37, 515–527. [DOI] [PubMed] [Google Scholar]
- Bourdet-Sicard R., Egile,C., Sansonetti,P.J. and Tran Van Nhieu,G. (2000) Diversion of cytoskeletal processes by Shigella during invasion of epithelial cells. Microbes Infect., 2, 813–819. [DOI] [PubMed] [Google Scholar]
- Cabib E., Drgonova,J. and Drgon,T. (1998) Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu. Rev. Biochem., 67, 307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G.R. (2000) Molecular and cell biology aspects of plague. Proc. Natl Acad. Sci. USA, 97, 8778–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G.R., Boland,A., Boyd,A.P., Geuijen,C., Iriarte,M., Neyt,C., Sory,M.P. and Stainier,I. (1998) The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev., 62, 1315–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes P.O. and Sandvig,K. (2000) Penetration of protein toxins into cells. Curr. Opin. Cell Biol., 12, 407–413. [DOI] [PubMed] [Google Scholar]
- Foury F. (1997) Human genetic diseases: a cross-talk between man and yeast. Gene, 195, 1–10. [DOI] [PubMed] [Google Scholar]
- Francis C.L., Starnbach,M.N. and Falkow,S. (1992) Morphological and cytoskeletal changes in epithelial cells occur immediately upon interaction with Salmonella typhimurium grown under low-oxygen conditions. Mol. Microbiol., 6, 3077–3087. [DOI] [PubMed] [Google Scholar]
- Galán J.E. and Collmer,A. (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science, 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Galán J.E. and Zhou,D. (2000) Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc. Natl Acad. Sci. USA, 97, 8754–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkansson S., Galyov,E.E., Rosqvist,R. and Wolf-Watz,H. (1996) The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol. Microbiol., 20, 593–603. [DOI] [PubMed] [Google Scholar]
- Hueck C.J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev., 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter K.J. and Eipel,H.E. (1979) Microbial determinations by flow cytometry. J. Gen. Microbiol., 113, 369–375. [DOI] [PubMed] [Google Scholar]
- Johnson S. (1991) Structure and function analysis of the CDC4 gene product. Thesis, University of Washington, Seattle, WA.
- Lew D.J. and Reed,S.I. (1995) A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol., 129, 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J.N., Sia,R.A.L. and Lew,D.J. (1998) A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol., 142, 1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Nordfelth,R., Holmstrom,A., Håkansson,S., Rosqvist,R. and Wolf-Watz,H. (1995) Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol. Microbiol., 18, 135–150. [DOI] [PubMed] [Google Scholar]
- Pickett C.L. and Whitehouse,C.A. (1999) The cytolethal distending toxin family. Trends Microbiol., 7, 292–297. [DOI] [PubMed] [Google Scholar]
- Pringle J.R., Preston,R.A., Adams,A.E., Stearns,T., Drubin,D.G., Haarer,B.K. and Jones,E.W. (1989) Fluorescence microscopy methods for yeast. Methods Cell Biol., 31, 357–435. [DOI] [PubMed] [Google Scholar]
- Pruyne D. and Bretscher,A. (2000) Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci., 113, 365–375. [DOI] [PubMed] [Google Scholar]
- Ridley A.J. and Hall,A. (1992) The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell, 70, 389–399. [DOI] [PubMed] [Google Scholar]
- Rosqvist R., Forsberg,Å., Rimpilainen,M., Bergman,T. and Wolf-Watz,H. (1990) The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol. Microbiol., 4, 657–667. [DOI] [PubMed] [Google Scholar]
- Rosqvist R., Forsberg,A. and Wolf-Watz,H. (1991a) Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect. Immun., 59, 4562–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosqvist R., Forsberg,A. and Wolf-Watz,H. (1991b) Microinjection of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Biochem. Soc. Trans., 19, 1131–1132. [DOI] [PubMed] [Google Scholar]
- Rosqvist R., Magnusson,K.E. and Wolf-Watz,H. (1994) Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J., 13, 964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Bravo A., Jimenez-Valera,M. and Roman,S.M. (1996) Nonspecific modification of cellular immunity by Yersinia enterocolitica. Immunol. Lett., 49, 57–61. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek E., Cowan,C. and Straley,S.C. (1998) Targeting of the Yersinia pestis YopM protein into HeLa cells and intracellular trafficking to the nucleus. Mol. Microbiol., 30, 1051–1065. [DOI] [PubMed] [Google Scholar]
- Subtil A., Blocker,A. and Dautry-Varsat,A. (2000) Type III secretion system in Chlamydia species: identified members and candidates. Microbes Infect., 2, 367–369. [DOI] [PubMed] [Google Scholar]
- Von Pawel-Rammingen U., Telepnev,M.V., Schmidt,G., Aktories,K., Wolf-Watz,H. and Rosqvist,R. (2000) GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol., 36, 737–748. [DOI] [PubMed] [Google Scholar]
- Yamochi W., Tanaka,K., Nonaka,H., Maeda,A., Musha,T. and Takai,Y. (1994) Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J. Cell Biol., 125, 1077–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T., Mecsas,J., Healy,J.I., Falkow,S. and Chien,Y. (1999) Suppression of T and B lymphocyte activation by a Yersinia pseudotuberculosis virulence factor, yopH. J. Exp. Med., 190, 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuk M.H., Harvill,E.T. and Miller,J.F. (1998) The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol., 28, 945–959. [DOI] [PubMed] [Google Scholar]
- Zhou D., Mooseker,M.S. and Galán,J.E. (1999) Role of the S. typhimurium actin-binding protein SipA in bacterial internaliz ation. Science, 283, 2092–2095. [DOI] [PubMed] [Google Scholar]