Abstract
FadR is an acyl-CoA-responsive transcription factor, regulating fatty acid biosynthetic and degradation genes in Escherichia coli. The apo-protein binds DNA as a homodimer, an interaction that is disrupted by binding of acyl-CoA. The recently described structure of apo-FadR shows a DNA binding domain coupled to an acyl-CoA binding domain with a novel fold, but does not explain how binding of the acyl-CoA effector molecule >30 Å away from the DNA binding site affects transcriptional regulation. Here, we describe the structures of the FadR–operator and FadR– myristoyl-CoA binary complexes. The FadR–DNA complex reveals a novel winged helix–turn–helix protein–DNA interaction, involving sequence-specific contacts from the wing to the minor groove. Binding of acyl-CoA results in dramatic conformational changes throughout the protein, with backbone shifts up to 4.5 Å. The net effect is a rearrangement of the DNA binding domains in the dimer, resulting in a change of 7.2 Å in separation of the DNA recognition helices and the loss of DNA binding, revealing the molecular basis of acyl-CoA-responsive regulation.
Keywords: acyl-CoA/fatty acid/protein structure/regulation/transcription
Introduction
The anabolic and catabolic pathways of fatty acid metabolism in Escherichia coli consist of enzymes that are significantly different from their mammalian counterparts (DiRusso et al., 1999). The E.coli fatty acid synthase system is of type II, consisting of individual dissociated enzymes (Rock and Cronan, 1996), and has been a successful target for the development of antibiotics such as triclosan and thiolactomysin (Jackowski et al., 1989; Waller et al., 1998; Levy et al., 1999). Degradation of fatty acids occurs through a multifunctional enzyme complex (DiRusso et al., 1999) coupled to a unique fatty acid uptake pathway, including an active membrane transporter (FadL) and an acyl-CoA synthetase (FadD), allowing the bacterium to grow on long-chain fatty acids as a sole energy source (DiRusso et al., 1999). To allow adaptation to various stages in the cell cycle and concentrations of fatty acids in the media, these anabolic and catabolic routes need to be balanced through precise transcriptional regulation. This is achieved by the acyl-CoA-responsive transcription factor FadR (DiRusso et al., 1999). In the absence of long-chain acyl-CoAs, FadR directly binds to specific DNA sequences (DiRusso et al., 1999) and acts as a repressor for transcription of genes involved in fatty acid degradation (fadE, fadF, fadG and fadBA) and import (fadD and fadL). Simultaneously, transcription of biosynthetic genes (fabA and fabB) is activated. The products of these genes, β-hydroxydecanoyl-acyl carrier protein dehydrase and β-ketoacyl-acyl carrier protein synthase II, respectively, play a crucial role in the synthesis of unsaturated long-chain fatty acids (Rock and Cronan, 1996). Apart from binding DNA, FadR is able to bind long-chain acyl-CoAs directly with a Kd of 50–400 nM (Raman and DiRusso, 1995). Binding of acyl-CoA is presumed to result in a conformational change and leads to inhibition of DNA binding (DiRusso et al., 1992, 1993). This loss of DNA binding then results in derepression of the catabolic genes, and deactivation of the anabolic genes. The exact mechanism and structural details of effector binding have been the subject of speculation (DiRusso et al., 1999), but so far have not been studied experimentally.
Recently, the structure of FadR has been determined in the absence of both DNA and acyl-CoA (van Aalten et al., 2000a). This study revealed an unusual overall fold, not previously observed in currently known regulator structures. The N-terminal DNA binding domain is of the winged helix–turn–helix (wHTH) type, and the C-terminal domain contains a new fold consisting of a seven-helical bundle with cross-over topology. This bundle was subsequently shown to contain a large cavity (142 Å3), lined by several residues that were shown, by mutagenesis studies, to be involved in acyl-CoA binding (Raman and DiRusso, 1995). This cavity, located >30 Å away from the DNA binding site, was thus suggested to be the putative site of binding of the effector molecule, long-chain acyl-CoA. Although this putative site of binding was identified, it remained unclear what were (i) the structural details of the FadR–DNA interaction, (ii) the mode of binding of the effector and (iii) the nature and mechanism of effector-induced conformational changes leading to disruption of the FadR–DNA complex.
Here, we describe the crystal structures of FadR in complex with an operator DNA duplex at 3.25 Å resolution, and of FadR bound to an effector, myristoyl-CoA, at 2.1 Å resolution, which define the FadR–DNA interaction, the site and mode of effector binding, and demonstrate the presence of large effector-induced conformational changes leading to disruption of the protein– DNA complex.
Results and discussion
Overall structure of the FadR–operator complex
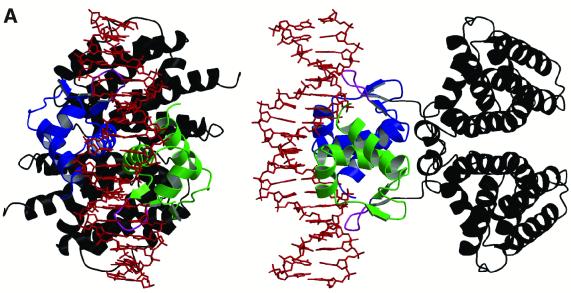
The FadR–operator complex was solved by molecular replacement (aided by the anomalous signal from an incorporated gold atom) and refined to 3.25 Å resolution (Table I). The structure (Figure 1A) reveals dimeric FadR bound to a DNA duplex, corresponding to the natural fadB operator, on which FadR acts as a transcriptional repressor. Each FadR monomer binds to a semi-symmetrical half of the operator through its wHTH motif. Apart from interacting with the DNA (total buried surface 1892 Å2), there are also interactions between the DNA binding domains themselves (buried surface 266 Å2). The orientation of the DNA binding domains with respect to the effector binding domains is similar to the apo-structure [root mean square deviation (r.m.s.d.) on all Cα atoms in the dimer = 0.8 Å], suggesting that the apo-protein is able to bind DNA, which is expected as the acyl-CoA- and DNA-bound states are mutually exclusive. The structure of the operator appears to be affected by binding to the protein. The central 3 bp are bulged out, as evidenced by a translation of ∼5 Å of the helical axis compared with the axis defined by the base pairs at the ends of the 19meric operator. This is further supported by a roll (7° and 9°, away from the minor groove) of the two base-pair steps neighbouring the central base pair, and a significant widening of the minor groove (to 9.1 Å) opposite the central base pairs. Overall, however, the DNA helix corresponds to canonical B-DNA, with proper Watson–Crick hydrogen bonding for all 19 bp, and no intercalated protein side chains.
Table I. Details of data collection and refinement.
| FadR–myristoyl-CoA | FadR–operator | |
|---|---|---|
| Spacegroup | P6122 | P6122 |
| Unit cell (Å) | a = 59.48 | a = 93.00 |
| b = 59.48 | b = 93.00 | |
| c = 290.48 | c = 334.83 | |
| Resolution, last bin (Å) | 30–2.1 (2.18–2.1) | 25–3.25 (3.37–3.25) |
| Unique reflections | 18 556 | 14 138 |
| Redundancy | 4.7 (4.9) | 6.2 (4.5) |
| Completeness (%) | 97.8 (95.5) | 98.0 (98.6) |
| Rmerge | 0.038 (0.292) | 0.143 (0.458) |
| I/σI | 17.9 (5.1) | 6.6 (2.5) |
| Rcryst | 0.226 | 0.271 |
| Rfree | 0.256 | 0.309 |
| No. of atoms | 1781 protein | 3631 protein + 2 Au + 3 Cl |
| 134 water | 28 water | |
| 63 ligand | 773 DNA | |
| Wilson B-factor (Å2) | 39.4 | n.d. |
| <B> protein (Å2) | 42.5 | 67.8 |
| <B> DNA (Å2) | – | 61.8 |
| <B> (adenosine + pyrophosphate) | 65.9 | – |
| <B> (pantetheine + myristate) | 48.8 | – |
| <B> (water) | 50.5 | 35.2 |
| R.m.s.ds from ideal geometry | ||
| bond lengths (Å) | 0.009 | 0.013 |
| bond angles (°) | 1.46 | 1.7 |
| main chain B-factors (Å2) | 1.7 | 1.1 |
Data were collected at 100 K (λ = 0.92 Å for FadR–myristoyl-CoA and λ = 1.025 Å for the FadR–operator complex). All measured data were included in the refinement.
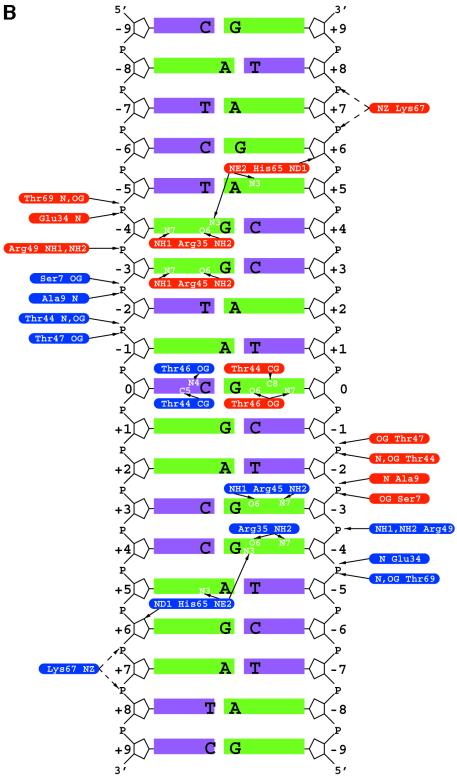
Fig. 1. (A) Structure of the FadR–DNA complex. The FadR–DNA complex is shown in two orientations (along and perpendicular to the pseudo 2-fold axis). The DNA binding domains in the FadR homodimer are coloured green and blue, the effector binding domains are shown in black. The tips of the wing in the wHTH motif are coloured magenta and the DNA duplex is shown as a red sticks model. (B) Schematic overview of FadR–DNA contacts. The 19 bp DNA duplex is shown with purines represented by long green bars and pyrimidines by short purple bars. The rounded boxes represent side chains on the protein interacting with the DNA (blue, monomer A; red, monomer B), with interactions shown by black arrows. (C) Stereo image of the DNA binding domains bound to DNA. Cα traces of the DNA binding domains in the dimer are shown in grey and blue. For the blue domain, the wing and the loop in the wHTH motif are coloured green, and side chains contacting the DNA (defined as contact distance less than the sum of van der Waals radii + 0.5 Å) are shown in sticks representation, with carbons coloured black. Ten base pairs of the DNA duplex are shown as sticks, with the final 2Fo – Fc electron density map contoured at 1.25 σ (magenta). Hydrogen bonds between FadR and DNA are shown as dotted black lines, and can also be identified in Figure 1B. Hydrogen bonds originating from FadR backbone nitrogens are drawn as originating from the corresponding Cα atom.
Details of the FadR–DNA interaction
The contacts between FadR and the DNA helix are summarized in Figure 1B and shown in Figure 1C. All contacts are symmetrical, with the exception of the Arg35 interaction with a guanine (Figure 1B). FadR side chains interacting with the DNA, and the DNA helix itself, were well defined in the electron density maps (Figure 1C). No water-mediated protein–DNA contacts were observed, although this may be a function of the relatively low resolution of the diffraction data. Five different areas of each FadR monomer interact with the DNA. Residues 7–9, just prior to the start of helix α1 in the wHTH motif, make several non-specific contacts with the phosphate backbone, possibly providing support to the more specific contacts downstream. Mutation of Ala9 to valine disrupts the ability of FadR to interact with DNA (Raman et al., 1997). The start of helix α2 (the first helix in the classical HTH motif) provides a contact between the backbone of Glu34 and the phosphate backbone, and contains Arg35, which donates two specific hydrogen bonds to acceptor atoms on the major groove side of guanine at the –4 position (Figure 1B and C). Mutation of Arg35 abrogated the FadR–DNA interaction, as shown by alanine-scanning mutagenesis and random hydroxylamine mutagenesis (which generated the Arg35Cys mutant) (Raman et al., 1997). The third area of interaction constitutes the turn in the HTH motif. The side chain of Thr44 hydrogen bonds to the phosphate backbone and provides a hydrophobic contact to the cytosine/guanine in the central base pair. Arg45 donates hydrogen bonds to the guanine at the –3 position, comparable to the Arg35–guanine interaction (Figure 1C). The fourth and fifth areas of FadR–DNA contacts are somewhat different from previously observed wHTH–DNA interactions (Gajiwala and Burley, 2000). Helix α3, normally termed the recognition helix, contacts the major groove using residues on the first helical turn only. Thr46 provides specific contacts with the central G-C base pair, and Thr47 donates a hydrogen bond to a nearby backbone phosphate group. The last residue on helix α3 to interact with the DNA helix is Arg49, which donates two hydrogen bonds to the phosphate backbone. The Arg49Ala mutation was shown to inhibit DNA binding completely (Raman et al., 1997). The last FadR–DNA contact area constitutes residues 63–69, which correspond to wing W1 in the standard wHTH nomenclature (Gajiwala and Burley, 2000) as they form the loop between β-strands β2 and β3, which, together with β1, form a small β-sheet. This loop deeply invades the minor groove, with the Cα atom of Gly66 lying approximately on an axis connecting the ring oxygens of the ribose units forming the minor groove (Figure 1A and C). Thr69 hydrogen bonds to the phosphate backbones, whereas Lys67 points between two phosphates on the backbone, but does not approach them closely enough (4.3–6.2 Å) to form proper hydrogen bonds. It is possible that there is still a small electrostatic contribution to the binding from this residue, as evidenced from mutagenesis studies which showed a small loss of binding for the Lys67Ala mutant, as qualitatively assessed by gel shift assays (Raman et al., 1997). An unusual protein–DNA contact is formed by His65, located at the tip of the wing, which extends even further into the minor groove. The imidazole ring is located in the minor groove bordering base pairs 4/5. His65 hydrogen bonds with guanine at –4 and adenine at +5, through its Nε2 atom. In addition (or alternatively, depending on the protonation state of this histidine), Nδ2 atom forms a hydrogen bond with the oxygen in the ribose ring at guanine +6 (Figure 1B and C). Substitution of His65 and Gly66 for other amino acid residues inhibits DNA binding completely (Raman et al., 1997).
The FadR–DNA contacts that may determine sequence specificity are found on the central C-G base pair (0), the two guanines at –3 and –4, and the adenine at +5. It has been shown that FadR protects its fadB operator from treatment with dimethylsulfonate at guanines 0, –3 and –4 (the latter two on both strands) (C.C.DiRusso and N.Raman, unpublished results), which is in agreement with our structural data. The DNase I footprint of FadR was determined to be 17 bp (DiRusso et al., 1992). In our structure, the contact farthest away from the central binding site is the Lys67 electrostatic interaction with the backbone phosphate connecting base pairs –7 and –8, which would thus agree approximately with the measured footprint. Although fadB is the strongest known binding site for FadR [Kd = 0.2 nM (DiRusso et al., 1992)], seven other fad binding sites are known, with the consensus 5′-NRCTGGT(A/C)YGAY(G/C)(T/A)N(T/A)N-3′ (N, any; R, purine; Y, pyrimidine). Although it is worth noting that this consensus is not symmetrical (see also Figure 1B), the two guanines that interact tightly with Arg35 and Arg45 on FadR are conserved.
FadR is a member of the wHTH family, the topology of which normally consists of α1–β1–α2–turn–α3–β2–W1– β3–W2, where W denotes a ‘wing’ (Lai et al., 1993). Recent structures have emphasized the diverse ways in which these domains can interact with DNA. Helix α3, termed the recognition helix, normally occupies the major groove defining the larger part of the specificity of the protein–DNA interaction (Gajiwala and Burley, 2000). In the recently published hRFX1 structure, however, a new mode of DNA binding was observed, with the recognition helix contacting the minor groove, whereas W1 occupied the major groove, making the most of specific contacts (Gajiwala et al., 2000). The structure of a heat shock transcription factor DNA binding domain complexed with DNA revealed another orientation, with weak recognition helix–DNA contacts and a wing primarily involved in protein–protein interactions (Littlefield and Nelson, 1999). While describing the structure of apo-FadR (van Aalten et al., 2000a), we proposed a model of the FadR–DNA interaction that was based on the structure of the E2F-DP–DNA complex (Zheng et al., 1999). The approximate areas of interaction agree between the proposed model and our present structure. However, compared with the FadR–DNA complex described here, the recognition helix (and the DNA binding domain) in the model was rotated too far towards the DNA helix. E2F4 binds DNA with its recognition helix pushed into the major groove, and with its single wing barely touching the outside of the phosphate backbone. Here, we have shown that the FadR–operator complex represents an unusual wHTH protein–DNA interaction, in which only the tip of the recognition helix α3 is involved in specificity contacts and wing W1 penetrates the minor groove of the helix, where it can make specific contacts with two bases (Figure 1B and C).
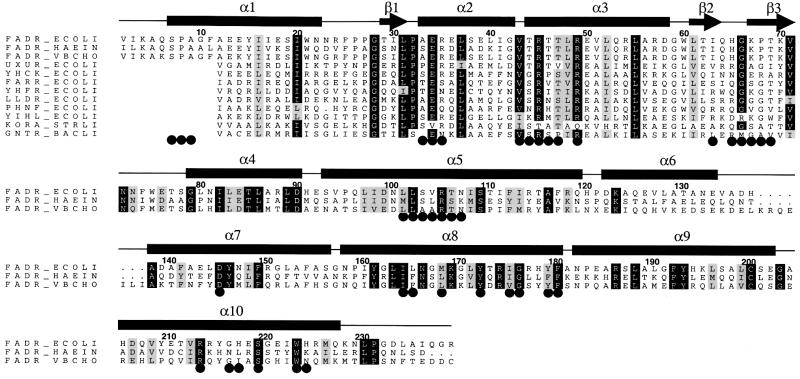
FadR is a member of the GntR family of prokaryotic transcription factors, for which no structures were available before the determination of the apo-FadR crystal structure (van Aalten et al., 2000a). We have aligned a representative set of sequences from this family to FadR and evaluated sequence conservation in the context of the present FadR–operator complex structure (Figure 2). The first 10 residues of FadR are not part of the GntR signature, which basically consists of the secondary structure elements from helix α1 to the last strand β3. Glu34 appears to be conserved, and although its side chain does not interact with DNA, it seems to serve to keep Arg45 and Arg49 together (Figure 1C). Arg35, involved in specific contacts, occurs in other GntR sequences, but is not well conserved. Two other important arginines, Arg45 (specific contacts) and Arg49 (phosphate backbone contacts), are both conserved. The threonines interacting with the central base pair (Thr44, Thr46 and Thr47) are highly variable in the GntR family, but completely conserved in the FadRs. Although His65, which interacts with the minor groove (Figure 1B and C), is not found in other GntR sequences, Gly66 is highly conserved. Inspection of the FadR–DNA complex (Figure 1C) reveals the reason for this: the Gly66 backbone is located deep within the minor groove, and any side chain atoms incorporated at this position would introduce steric clashes with the DNA. Taken together, it seems likely that the overall GntR–DNA interactions will be similar to the FadR–operator complex.
Fig. 2. Sequence alignment of E.coli FadR (FADR_ECOLI) and putative FadRs from Haemophilus influenzae (FADR_HAEIN) and Vibrio cholerae (FADR_VBCHO), together with a representative group of signature sequences from the GntR family, indicated by their standard SwissProt accession codes. FadR sequence numbering is used, and the secondary structure elements are indicated by bars and arrows. Conserved residues are indicated by black boxes. Residues touching the acyl-CoA ligand or the DNA helix are indicated by black circles.
Binding of myristoyl-CoA to FadR
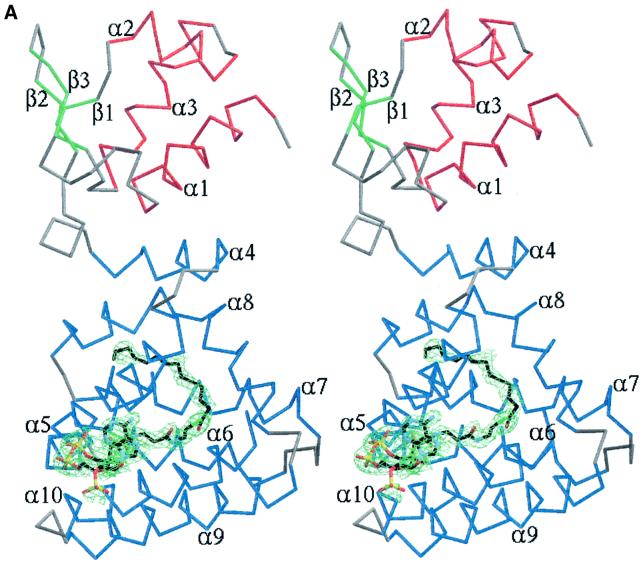
The structure of FadR complexed with myristoyl-CoA was solved by molecular replacement and refined to 2.1 Å resolution. The electron density for the effector was located in the C-terminal acyl-CoA binding domain (Figure 3A), as predicted from the native structure (van Aalten et al., 2000a). The adenosine 3′-phosphate moiety is partially solvent exposed and covers some of the more hydrophobic atoms on the pantothenic acid moiety, as seen in many other acyl-CoA binding proteins (Engel and Wierenga, 1996; Lerche et al., 1997; van Aalten et al., 2001). The pantothenic acid moiety penetrates the seven-helix bundle through a channel between helices α5 and α10, such that the acyl chain is buried deep inside the acyl-CoA binding domain (Figure 3A). Three main areas of the ligand show interactions with the protein (Figure 3B). The adenosine 3′-phosphate and pyrophosphate units form several hydrogen bonds (some water mediated) with solvent-exposed protein residues, most notably Arg213 (conserved in other FadRs; Figure 2), whose positively charged side chain is well positioned to interact with the negative charges on the pyrophosphate. Polar atoms on the pantothenic acid moiety form hydrogen bonds to residues on the protein (Thr106 and Ser219, conserved in other FadRs; Figure 2). The thioester oxygen accepts a hydrogen bond from Arg105 and the C14 acyl chain is buried in a pocket surrounded by hydrophobic residues (Figure 3B). The acyl chain adopts a bent conformation and terminates in a pocket mainly formed by residues from helices α5 and α8 (Ile108, Ile164, Leu165). The size of the pocket appears to leave room for additional methylene groups. Long-chain acyl-CoAs have been shown to bind FadR, with Kds for binding of C14:0-, C16:0- and C18:1-CoA of 59, 369 and 63 nM, respectively, measured by isothermal titration calorimetry (DiRusso et al., 1998). FadR is apparently able to bind acyl-CoAs containing an acyl chain longer than C14:0, with high affinity, in agreement with the extra space observed in the present structure.
Fig. 3. (A) Stereo image of FadR–myristoyl-CoA structure and ligand electron density. A Cα trace is shown, coloured according to secondary structure. Helices are coloured red (DNA binding domain) or blue (acyl-CoA binding domain), strands green and turns grey. Secondary structure elements are labelled. The ligand, myristoyl-CoA, is shown as a stick model with carbons coloured black. A simulated annealing Fo – Fc map is also shown, contoured around the ligand at 2.25 σ (green). (B) Stereo image of protein–ligand interactions. The protein backbone is shown as a blue trace. Side chains touching the ligand (contact distance less than the sum of van der Waals radii + 0.5 Å) are shown as sticks coloured by atom type. Myristoyl-CoA is shown as a stick model with carbons coloured black. Water molecules interacting with the ligand are shown as green spheres. Hydrogen bonding with the ligand is indicated by black dashed lines.
Several amino acid substitutions have been reported that cause a FadR ‘super-repressor’ phenotype, i.e. rendering FadR unable to dissociate from DNA in the presence of long-chain acyl-CoAs. These substitutions, namely Tyr179→Ala, Gly216→Ala, Ser219→Asn and Trp223→ Ala, were suggested to lie at positions lining the acyl-CoA binding site (Raman and DiRusso, 1995), although they could also affect the conformational changes necessary to transduce the signal to the DNA binding domains. Analysis of the location of these residues with respect to myristoyl-CoA bound to FadR shows that they all line the acyl-CoA binding pocket (Figure 3B) and that most of them are conserved in the sequence of other FadRs (Figure 2). Tyr179 changes conformation upon acyl-CoA binding, lying tightly packed against the acyl chain, and substitution to alanine would disrupt this interaction. Trp223 has only weak direct contacts with the ligand, but may act as a ‘seal’ at the bottom of the hydrophobic binding pocket, protecting it from the solvent. Mutation to alanine would disrupt the contacts with the ligand and would expose the binding pocket to the solvent (Figure 3B). Fluorescence studies have confirmed a role of Ser219 in ligand binding (DiRusso et al., 1998), and it is worth noting that this is one of the few residues conserved in the effector binding domain (Figure 2). Introduction of any side chain atom beyond Cα on Gly216 or a larger side chain instead of Ser219 would change the tight interactions these residues have with the pantothenic acid moiety into severe steric clashes. Thus, our structural data are in agreement with available experimental data and point to several other key residues whose mutation could have significant effects on acyl-CoA binding (Figure 3B).
Effector-induced conformational changes
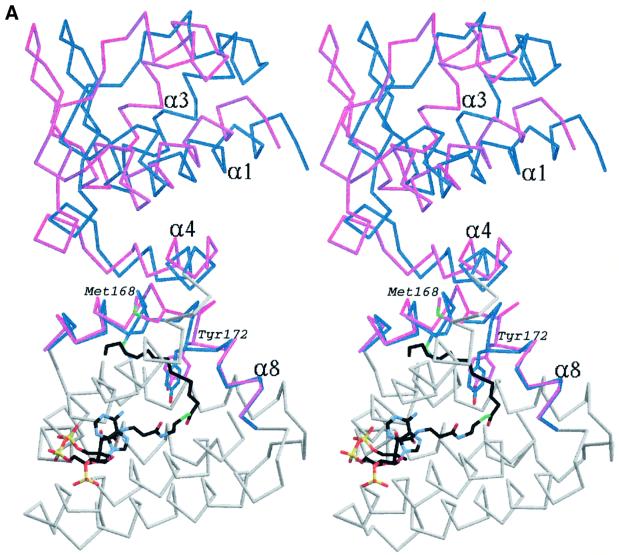
As pointed out earlier, the protein in the apo-FadR and FadR–DNA complexes is in a similar conformation. The effector binding domains are slightly more similar, and superimpose with an r.m.s.d. of 0.57 Å on the Cα atoms. Given the higher resolution data for the apo-FadR structure (2.0 Å, compared with 3.25 Å for the FadR–operator structure), here we study the finer details of the acyl-CoA-induced conformational changes by comparing the apo-FadR and myristoyl-CoA–FadR structures, and subsequently discuss the large conformational changes by comparison of the FadR–operator and FadR–myristoyl-CoA complexes. Although myristoyl- CoA appears to fit into a pocket within the acyl-CoA binding domain in the apo-FadR and FadR–operator stuctures, this is not possible without significant conformational changes. Prior to effector binding, Met168 and Tyr172 partially occupy the pocket, as shown in Figure 4A. Upon binding of myristoyl-CoA, these residues undergo dramatic conformational changes. Met168 is forced to rotate 92° around χ2 and ends up pointing towards the outside of the helical bundle. Tyr172 rotates 44° around χ2, but even in that position has many close contacts with the methylene groups of the acyl chain (Figure 4A). While Tyr172 is conserved in other FadRs (Figure 2), Met168 is less conserved and a leucine also appears possible at that position. This may reflect a tuning of specificity towards length or saturation of the fatty acid. To further relieve their steric clashes with the bound effector, these two ‘sensor’ residues induce conformational changes in the backbone (Cα positional shifts of up to 3.5 Å; Figure 4A). Helix α8 is already bent in the apo-FadR and FadR–operator structures, and almost forms two separate helices. In the structure of the complex with myristoyl- CoA, the effector-induced conformational changes in the sensor residues kink helix α8, in effect forming two separate helices (Figure 4A). As a result of the kink in helix α8, some of the side chains on this helix are pushed towards the neighbouring helix α4, which responds by shifting away from helix α8 towards the N-terminal DNA binding domain (Figure 4A). This shift of helix α4 introduces additional contacts with helix α1 in the N-terminal domain, which tilts away, taking the entire DNA binding domain with it in a rigid body motion (Figure 4A). Thus, it appears that binding of myristoyl-CoA in the effector binding domain, >30 Å away from the site of DNA binding, causes a hinge-bending motion of the entire DNA binding domain over an angle of 13°, resulting in backbone shifts of up to 4.5 Å.
Fig. 4. (A) Stereo image of the superposition of a monomer from the apo-FadR structure onto the FadR–myristoyl-CoA complex. For areas showing relatively little conformational change, only the backbone trace of the monomer from the apo-FadR structure is shown in grey. Helices α4 and α8 in the acyl-CoA binding domain and the entire DNA binding domain are show in blue for apo-FadR, and in magenta for the FadR–myristoyl-CoA complex. Side chains of residues Met168 and Tyr172 are shown with blue carbons for apo-FadR and magenta carbons for the FadR–myristoyl-CoA complex. Myristoyl-CoA is shown as a stick model, with carbons coloured black. (B) Stereo image of the superposition of the FadR–operator complex onto the dimer of the FadR–myristoyl-CoA complex. The FadR–operator structure is coloured grey and FadR–myristoyl-CoA is coloured blue, with red DNA-recognition helices. Residues Met168 and Tyr172 are shown with carbons coloured orange (FadR–operator) or magenta (FadR–myristoyl-CoA). Myristoyl-CoA is shown as a stick model, with carbons coloured black. The DNA helix is shown as a thin black sticks model.
Apart from the large rigid body movements described above, the effector binding domain does not show other large shifts. When the effector binding domains of the FadR–myristoyl-CoA and apo-FadR structures, excluding helices α4 and α8, are superposed, this results in an r.m.s.d. of 0.3 Å on all atoms. Perhaps this is not surprising, as it was already possible to detect nearly the entire ligand binding pocket in the apo structure (van Aalten et al., 2000a). The largest side chain conformational change in the pocket, apart from Met168 and Tyr172, is Arg105, which rotates around χ2–χ4 to form a hydrogen bond with the thioester oxygen (Figure 3B). This residue is conserved in other FadR sequences (Figure 2).
We have reported previously on diffraction data of a crystal grown from solutions containing FadR and octanoyl-CoA (van Aalten et al., 2000b). The unit cell dimensions and space group of this crystal form (a = b = 59.7 Å, c = 296.2 Å, P6122) are similar to those presented here for the FadR–myristoyl-CoA structure (Table I). Interestingly, a partially refined structure from a gold derivative of this crystal form (which yielded the highest resolution data: 2.6 Å, R = 0.265, Rfree = 32.9) did not reveal ordered ligand density, and the two ‘sensor’ residues Met168 and Tyr172 were in the same conformation as in the apo-FadR structure. However, the protein adopts an overall conformation similar to the myristoyl-CoA structure, although the conformational changes are not as large (superpositions on Cαs: apo-FadR – FadR–myristoyl-CoA = 2.1 Å; apo-FadR – FadR–octanoyl-CoA = 1.8 Å), so this could be considered an ‘in-between’ structure. This agrees with the biochemical data, which indicate that octanoyl-CoA may bind weakly to FadR, but is not able to inhibit DNA binding. It may be that a highly disordered, non-interpretable ligand is present in the octanoyl-CoA crystals, causing the conformational change, although a C8 acyl chain is too short to reach both sensor residues (Figure 3B). Another possibility is that apo-FadR is able to populate the ligand-bound conformation in configurational space, which is then preferred in the P6122 space group.
Acyl-CoA-induced domain motions
Having defined structures of apoFadR (van Aalten et al., 2000a) and the FadR–operator and FadR–effector complexes, it is now possible to understand the effect of acyl-CoA binding in the context of a FadR dimer interacting with DNA. As noted above, binding of myristoyl-CoA leads to a displacement of the entire DNA binding domain with respect to the effector binding domain. The function of this motion becomes clear upon inspection of the conformational change by comparing the FadR–myristoyl-CoA and FadR–operator complexes (Figure 4B). Because of the (pseudo) 2-fold axis relating the two monomers in the homodimer, the effector-induced hinge bending of the DNA binding domains occurs in opposite directions. This leads to a variation of the distance between the two recognition helices from 8 Å (FadR–operator complex; 7 Å in the apo-FadR structure) to 14 Å (FadR–myristoyl-CoA), and shifts the tip of the wing in each DNA binding domain by up to 4 Å. Thus, binding of the effector molecule, in this case myristoyl-CoA, controls the separation between the recognition helices, and thus the ability to interact with the DNA helix. This mechanism is similar to that for the tetracycline repressor (Orth et al., 2000) and the purine repressor (Friedman et al., 1995), where the ability to bind DNA is lost by an opening of the DNA binding domains.
Concluding remarks
The structures of the FadR–myristoyl-CoA and FadR– operator complexes described here have revealed the details of the FadR–effector and FadR–DNA interaction, and the nature and extent of effector-induced conformational changes. The data show that effector binding results in significant conformational changes transmitted from the effector binding domain to the DNA binding domain, leading to a conformational state that is no longer favourable for interaction with DNA. Although many structures of effector binding domains complexed to their corresponding effectors have been described (e.g. Egea et al., 2000), not many examples are known of structures of full-length transcriptional regulators bound to their effectors. One of the most well characterized systems is the catabolite gene activator protein (CAP) family (Schultz et al., 1991), with the most recent example being the structure of CooA (Chan, 2000; Lanzilotta et al., 2000). These proteins are similar to FadR to the extent that they also bind DNA as homodimers, and that regulation of DNA binding is dependent on the binding of an effector molecule. However, whereas CAP regulators acquire the ability to bind DNA upon binding of their effectors, the FadR–DNA interaction is inhibited by effector binding. Interestingly, it has been shown that for the lactose repressor family (LacI) and the purine repressor (PurR), such opposite functions can exist within the same structural framework (Schumacher et al., 1994, 1995; Friedman et al., 1995; Bell and Lewis, 2000). Binding of their respective effector molecules increases the affinity for DNA in the case of PurR, but disrupts the LacI–DNA interaction. Although the effector-bound structures are similar, the PurR–corepressor complex binds DNA, whereas the LacI–inducer complex does not. In a manner analogous to FadR, the transcription factors from the LacI family appear to regulate DNA binding by separation of their N-terminal, HTH motif-containing DNA binding domains.
The transcription factor most similar to FadR from a structural and mechanistic point of view is the tetracycline repressor, TetR. Native TetR binds tightly to DNA through the helix–turn–helix motifs in the N-terminal domains of the homodimer (Orth et al., 2000). Binding of tetracycline at the interface of the all-helical C-terminal domains induces conformational changes of side chains in the binding pocket, which are then translated into shifts of α-helices, ultimately resulting in the two DNA binding domains moving farther apart, leading to dissociation from the DNA. The chain of events upon effector binding in FadR is similar, also leading to an opening of the DNA binding domains.
The GntR family of prokaryotic transcription factors regulate pathways as diverse as gluconate synthesis (Miwa and Fujita, 1988), trehalose metabolism (Schoeck and Dahl, 1996) and glycolate oxidation (Pellicer et al., 1996). Many of these transcription factors show sequence homology to FadR’s DNA binding domain (Figure 2), and secondary structure predictions suggest that some members of the GntR family also possess seven-helical effector binding domains (van Aalten et al., 2000a). This implies that FadR might serve as a structural scaffold for this new family of regulators. Since the FadR–effector and FadR–effector complexes have defined the details of interaction with effector and DNA, it may be feasible in the future to predict these interactions for other GntR regulators.
The definition of the effector binding site in FadR, and determination of the conformational state of the protein bound to DNA, enable studies towards inhibitory molecules that would block the ligand binding site. This could open up possibilities for interfering with the normal transcription of fatty acid biosynthetic genes, which have been shown to be an excellent target for antibiotics (Jackowski et al., 1989; Waller et al., 1998; Levy et al., 1999).
Materials and methods
Purification, crystallization and data collection
FadR was overexpressed in E.coli and purified as described previously (DiRusso et al., 1998). The resulting protein solution contained 4.6 mg/ml FadR in 50 mM KH2PO4, 10% glycerol pH 8.0. A 2-fold molar excess of myristoyl-CoA was added prior to crystallization. Hanging drop vapour diffusion crystallization experiments were set up using 1 µl of solution containing the complex and an equal volume of well solution containing 20% PEG 8000, 200 mM magnesium acetate and 100 mM sodium cacodylate pH 6.5. Crystals were grown at 20°C, and reached a size of ∼0.2 × 0.2 × 0.2 mm within 2–3 weeks. They were transferred into mother liquor containing 10% ethylene glycol, and after 60 s frozen in a nitrogen gas stream (100 K) for data collection.
For the FadR–operator complex, a 1.1-fold molar excess was added of a DNA duplex consisting of 19meric oligos 5′-CATCTGGTACGACCAGATC-3′ and 5′-GATCTGGTCGTACCAGATG-3′, which were HPLC purified prior to annealing. Hanging drop vapour diffusion crystallization experiments were set up using 1 µl of solution containing the complex and an equal volume of well solution containing 1.6 M sodium citrate pH 6.5. Small bipyramidal needles grew at 20°C, and reached a size of ∼0.1 × 0.05 × 0.05 mm within 2 weeks. The presence of DNA in the crystals was confirmed by ethidium bromide staining and mass spectrometry. A crystal was transferred into mother liquor containing 10 mM AuCN for 30 min, and then transferred to a cryoprotecting solution of mother liquor containing 5% glycerol. After 60 s, the crystal was frozen in a nitrogen gas stream (100 K) for data collection.
Data were collected on a MAR CCD detector at beamline X11, at the EMBL outstation at the DESY synchrotron, Hamburg (FadR–myristoyl- CoA), and on an ADSC-Q4 detector at beamline F2 at CHESS, Cornell University, using a wavelength (λ = 1.025 Å) optimized for a gold anomalous signal (FadR–operator). Images were processed using DENZO and reflections merged using SCALEPACK of the HKL-suite (Otwinowski and Minor, 1997). Data collection and processing statistics are summarized in Table I.
Structure determination and refinement
The FadR–DNA structure was solved by molecular replacement with AMoRe (Navaza, 1994) using the native structure of the FadR dimer [Protein Data Bank (PDB) entry 1E2X] as a search model. The correctness of the solution was verified by calculating a map from the measured anomalous signal combined with the model phases, which showed 6σ peaks for gold atoms near the single solvent-exposed cysteine in both monomers, which had previously been exploited in phasing the native structure (van Aalten et al., 2000a). These cysteines are far away from the DNA binding site, and it has been shown that reaction with the heavy atom does not result in significant conformational changes (van Aalten et al., 2000a). After rigid body refinement in CNS (Brünger et al., 1998), in which the DNA binding and effector binding domains were treated as separate rigid bodies, the R-factor was 0.421 (Rfree = 0.494, 25–3.25 Å data). The resulting maps showed density for the DNA duplex, which was built in several steps using O (Jones et al., 1991), starting with the central 5 bp. Further refinement with CNS and REFMAC included non-crystallographic symmetry restraints across the pseudo 2-fold axis relating the monomers in the homodimer. Progress was monitored by inspection of maps calculated from the anomalous signal, which verified the strength and location of the gold atoms. After completion of the DNA model, the R-factor had fallen to 0.315 (Rfree = 0.361) with 25–3.25 Å data. After further macrocycles that included adjustment of the protein model, placement of a few ordered water molecules and partially occupied gold sites, the R-factor was 0.264 (Rfree = 0.320). Statistics of the model are shown in Table I.
For the myristoyl-CoA–FadR complex, initial molecular replacement calculations using AMoRe (Navaza, 1994), with the native structure (PDB entry 1E2X) as a search model, were unsuccessful, suggesting that significant structural rearrangements might have occurred upon binding of myristoyl-CoA. Subsequent calculations in which only the C-terminal acyl-CoA binding domain (Figure 1A) was used gave a single clear solution after rigid body refinement (R-factor 0.451, correlation coefficient 0.432, 8.0–4.0 Å data). An initial round of refinement in CNS (Brünger et al., 1998) resulted in maps in which the N-terminal DNA binding domain could be positioned manually. Further rigid body refinement in CNS brought the R-factor down to 0.405 (Rfree = 44.5) with 25–3.75 Å data. Inspection of the maps revealed several regions of disorder, which could not be interpreted unambiguously. The phases of this partial solution were used as input for iterative refinement and autobuilding with warpNtrace (Perrakis et al., 1999). This resulted in a model with 189 out of 240 possible residues, giving R = 0.322, Rfree = 0.343 after simulated annealing in CNS. The maps had improved and most of the missing regions and parts of the ligand could be seen in the Fo – Fc difference maps. Iterative model building with O (Jones et al., 1991) and refinement with CNS (Brünger et al., 1998) resulted in a final model (R = 0.226, Rfree = 0.256) that included all atoms in the ligand (Figure 1A) and residues 5–227 of the protein (Table I). The coordinates and structure factors of both structures have been submitted to the PDB (entries 1H9G and 1H9T).
Acknowledgments
Acknowledgements
We thank the EMBL-Hamburg outstation at the DESY synchrotron for use of beamline X11 and the CHESS synchrotron at Cornell University for their generous allocation of time on beamlines F1/F2. D.v.A. is supported by a Wellcome Trust Career Development Research Fellowship.
References
- Bell C.E. and Lewis,M. (2000) A closer view of the conformation of the Lac repressor bound to operator. Nature Struct. Biol., 7, 209–214. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chan M.K. (2000) CooA, CAP and allostery. Nature Struct. Biol., 7, 822–824. [DOI] [PubMed] [Google Scholar]
- DiRusso C.C., Heimert,T.L. and Metzger,A.K. (1992) Characterization of FadR, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. J. Biol. Chem., 267, 8685–8691. [PubMed] [Google Scholar]
- DiRusso C.C., Metzger,A.K. and Heimert,T.L. (1993) Regulation of transcription of genes required for fatty acid transport and unsaturated fatty acid biosynthesis in E.coli by FadR. Mol. Microbiol., 7, 311–322. [DOI] [PubMed] [Google Scholar]
- DiRusso C.C., Tsvetnitsky,V., Hojrup,P. and Knudsen,J. (1998) Fatty acyl-CoA binding domain of the trancription factor FadR. J. Biol. Chem., 273, 33652–33659. [DOI] [PubMed] [Google Scholar]
- DiRusso C.C., Black,P.N. and Weimar,J.D. (1999) Molecular inroads into the regulation and metabolism of fatty acids, lessons from bacteria. Prog. Lipid Res., 38, 129–197. [DOI] [PubMed] [Google Scholar]
- Egea P.F., Mitschler,A., Rochel,N., Ruff,M., Chambon,P. and Moras,D. (2000) Crystal structure of the human RXRα ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J., 19, 2592–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel C. and Wierenga,R. (1996) The diverse world of coenzyme A binding proteins. Curr. Opin. Struct. Biol., 6, 790–797. [DOI] [PubMed] [Google Scholar]
- Friedman A.M., Fischmann,T.O. and Steitz,T.A. (1995) Crystal structure of the lac repressor core tetramer and its implications for DNA looping. Science, 268, 1721–1727. [DOI] [PubMed] [Google Scholar]
- Gajiwala K.S. and Burley,S.K. (2000) Winged helix proteins. Curr. Opin. Struct. Biol., 10, 110–116. [DOI] [PubMed] [Google Scholar]
- Gajiwala K.S., Chen,H., Cornille,F., Roques,B.P., Reith,W., Mach,B. and Burley,S.K. (2000) Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature, 403, 916–921. [DOI] [PubMed] [Google Scholar]
- Jackowski S., Murphy,C.M., Cronan,J.E. and Rock,C.O. (1989) Acetoacetyl-acyl carrier protein synthase—a target for the antibiotic thiolactomycin. J. Biol. Chem., 264, 7224–7629. [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Lai E., Clark,K.L., Burley,S. and Darnell,J.E. (1993) A family of transcription factors of diverse biological function. Proc. Natl Acad. Sci. USA, 90, 10421–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzilotta W.N., Schuller,D., Thorsteinsson,M.V., Kerby,R.L., Roberts,G.P. and Poulos,T.L. (2000) Structure of the CO sensing transcription factor CooA. Nature Struct. Biol., 7, 876–880. [DOI] [PubMed] [Google Scholar]
- Lerche M.H., Kragelund,B.B., Bech,L.M. and Poulsen,F.M. (1997) Barley lipid transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure, 5, 291–306. [DOI] [PubMed] [Google Scholar]
- Levy C.W., Roujeinikova,A., Sedelnikova,S., Baker,P.J., Stuitje,A.R., Slabas,A.R., Rice,D.W. and Rafferty,J.B. (1999) Molecular basis of triclosan activity. Nature, 398, 383–384. [DOI] [PubMed] [Google Scholar]
- Littlefield O. and Nelson,H.C.M. (1999) A new use for the ‘wing’ of the ‘winged’ helix–turn–helix motif in the HSF–DNA cocrystal. Nature Struct. Biol., 6, 464–470. [DOI] [PubMed] [Google Scholar]
- Miwa Y. and Fujita,Y. (1988) Purification and characterization of a repressor for the Bacillus subtilis gnt operon. J. Biol. Chem., 263, 13252–13257. [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Orth P., Schnappinger,D., Hillen,W., Saenger,W. and Hinrichs,W. (2000) Structural basis of gene regulation by the tetracycline inducible Tet repressor–operator system. Nature Struct. Biol., 7, 215–219. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pellicer M.T., Badia,J., Aguilar,J.T. and Baldoma,L. (1996) glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J. Bacteriol., 178, 2051–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Raman N. and DiRusso,C.C. (1995) Analysis of acyl coenzyme A binding to the transcription factor FadR and identification of amino acid residues in the carboxyl terminus required for ligand binding. J. Biol. Chem., 270, 1092–1097. [DOI] [PubMed] [Google Scholar]
- Raman N., Black,P.N. and DiRusso,C.C. (1997) Characterization of the fatty acid-responsive transcription factor FadR. J. Biol. Chem., 272, 30645–30650. [DOI] [PubMed] [Google Scholar]
- Rock C.O. and Cronan,J.E. (1996) Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta, 1302, 1–16. [DOI] [PubMed] [Google Scholar]
- Schoeck F. and Dahl,M.K. (1996). Expression of the tre operon of Bacillus subtilis 168 is regulated by the repressor TreR. J. Bacteriol., 178, 4576–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S.C., Shields,G.C. and Steitz,T.A. (1991) Crystal-structure of a CAP-DNA complex—the DNA is bent by 90°. Science, 253, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Choi,K.Y., Zalkin,H. and Brennan,R.G. (1994) Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by α-helices. Science, 266, 763–770. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Choi,K.Y., Lu,F., Zalkin,H. and Brennan,R.G. (1995) Mechanism of corepressor-mediated specific DNA binding by the purine repressor. Cell, 83, 147–155. [DOI] [PubMed] [Google Scholar]
- van Aalten D.M.F., DiRusso,C.C., Knudsen,J. and Wierenga,R.K. (2000a) Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J., 19, 5167–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aalten D.M.F., Knudsen,J., DiRusso,C.C., Kokko,T. and Wierenga,R.K. (2000b) Crystallization and X-ray diffraction studies of the fatty acid responsive transcription factor FadR from Escherichia coli. Acta Crystallogr. D, 56, 469–471. [DOI] [PubMed] [Google Scholar]
- van Aalten D.M.F., Milne,K.G., Zou,J.Y., Kleywegt,G.J., Bergfors,T., Ferguson,M.A.J., Knudsen,J. and Jones,T.A. (2001) Binding site differences revealed by crystal structures of Plasmodium falciparum and bovine acyl-CoA binding protein. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Waller R.F. et al. (1998) Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl Acad. Sci. USA, 95, 12352–12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Fraenkel,E., Pabo,C.O. and Pavletich,N.P. (1999) Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev., 13, 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]