Abstract
Spatiotemporal regulation of protein kinase A (PKA) activity involves the manipulation of compartmentalized cAMP pools. Now we demonstrate that the muscle-selective A-kinase anchoring protein, mAKAP, maintains a cAMP signaling module, including PKA and the rolipram-inhibited cAMP-specific phosphodiesterase (PDE4D3) in heart tissues. Functional analyses indicate that tonic PDE4D3 activity reduces the activity of the anchored PKA holoenzyme, whereas kinase activation stimulates mAKAP-associated phosphodiesterase activity. Disruption of PKA– mAKAP interaction prevents this enhancement of PDE4D3 activity, suggesting that the proximity of both enzymes in the mAKAP signaling complex forms a negative feedback loop to restore basal cAMP levels.
Keywords: AKAP/cAMP/phosphodiesterase/PKA/signal transduction
Introduction
Understanding the molecular organization of intracellular signaling pathways has recently become a topic of considerable research interest (Hunter, 2000). It is now appreciated that the assembly of multiprotein signaling complexes creates focal points of enzyme activity that transduce the intracellular action of many hormones, neurotransmitters and cardiotonic agonists (Jordan et al., 2000). Accordingly, the spatiotemporal activation of protein kinases and/or phosphatases is important in controlling where and when phosphorylation events occur. Anchoring proteins and targeting subunits provide a molecular framework that orients protein kinases and phosphatases towards selected substrates (Pawson and Scott, 1997; Hunter, 2000). Prototypic examples of these ‘signal-directing molecules’ are A-kinase anchoring proteins (AKAPs), which sustain multi-component signaling complexes of protein kinase A (PKA) and other enzymes (Colledge and Scott, 1999). These protein– protein interactions not only focus PKA toward the phosphorylation of certain ion channels, cytoskeletal elements and transcription factors, but also spatially segregate parallel cAMP signaling pathways (Johnson et al., 1994; Rosenmund et al., 1994; Feliciello et al., 1997; Fraser et al., 1998; Colledge and Scott, 1999; Marx et al., 2000; Westphal et al., 2000). However, activation of each intracellular PKA pool is ultimately determined by the availability of cAMP. Accordingly, the accumulation of cAMP at sites of PKA anchoring must be tightly and dynamically regulated.
Phosphodiesterases (PDEs) are critical regulators of cyclic nucleotide homeostasis (Beavo, 1995; Conti, 2000). Together with phosphatases, they control the signal termination processes by counteracting the actions of second messenger-dependent enzymes. This is achieved by limiting the diffusion of second messengers, thereby influencing the activity state of cyclic nucleotide-gated ion channels, cAMP-GEFs, and enzymes such as PKA or PKG (Conti, 2000). Given the broad role that these enzymes play in all aspects of cellular regulation, it is not surprising that control of PDE activity is of considerable clinical importance. In fact, PDE inhibitor drugs are used to treat clinical depression, asthma and erectile dysfunction (Corbin and Francis, 1999; Soderling and Beavo, 2000). Inhibition of PDE activity is particularly relevant in the cardiovascular system where certain PDE families have been implicated in promoting vasodilatation of pulmonary arteries and aortic endothelial cells (Kessler and Lugnier, 1995; Maclean et al., 1997). Consequently, PDE inhibitors such as indolidan, enoximone and cilostazol have been developed as cardiotonic agents (reviewed by Beavo, 1995).
The PDE superfamily currently consists of 21 different genes encoding 11 families of distinct PDE subtypes (Conti, 2000). The PDE4 cAMP-specific, rolipram-inhibited PDEs are derived from four related genes that encode numerous isoforms through alternate splicing of RNA (PDE4, A–D) (Houslay et al., 1998). Each isoenzyme contains a conserved catalytic core (Xu et al., 2000), but has divergent N- and C-terminal sequences, which have been proposed to regulate enzyme activity and direct the subcellular location of individual PDE4D isoforms (Jin et al., 1998; McPhee et al., 1999; Beard et al., 2000). On the basis of these findings, we reasoned that certain PDE4 cAMP-specific PDE isoforms may be compartmentalized with the cAMP-dependent protein kinase in heart tissue. In this report, we provide evidence that the PDE4D3 isoform and the PKA holoenzyme exist in the same signaling complex through their association with the muscle-selective A-kinase anchoring protein, mAKAP. Functional studies indicate that anchored PKA enhances cAMP degradation 2-fold, indicating that mAKAP provides the molecular framework for the assembly of a PKA/PDE negative feedback loop.
Results
mAKAP associates with a type 4 PDE
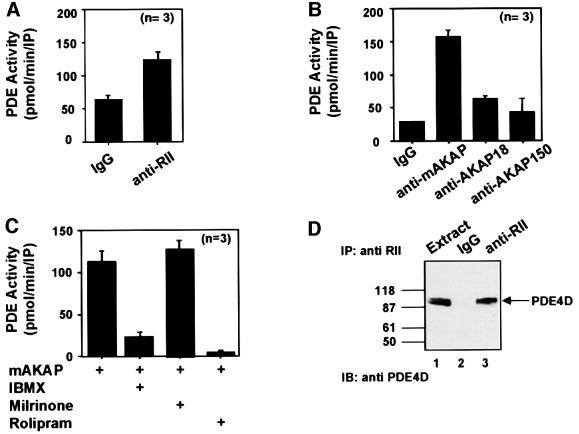
We focused initially on measuring PDE activity that co-purified with the PKA holoenzyme from rat heart extracts (Figure 1A–C). Immunoprecipitation of the type II regulatory (RII) subunit of PKA resulted in a modest increase of 1.86 ± 0.4-fold (n = 3) in PDE activity over an IgG control using [3H]cAMP as a substrate (Figure 1A). This implied that both enzymes were recruited to the same signaling complex, but did not indicate whether an anchoring protein maintained these interactions. To test this, two AKAP signaling complexes known to be present in heart were isolated from tissue extract (Fraser et al., 1998; Kapiloff et al., 1999). Immunoprecipitation of mAKAP resulted in a 5.1 ± 0.2-fold (n = 5) increase in PDE activity over an IgG control, whereas immunoprecipitation of AKAP 15/18 only elicited a 1.7 ± 0.3-fold (n = 3) increase in enzyme activity (Figure 1B). In addition, AKAP150 immune complexes isolated from brain extracts displayed little PDE activity [1.45 ± 0.7-fold (n = 3); Figure 1B]. Owing to the significant amount of PDE activity associated with mAKAP, further experiments focused upon characterizing this interaction.
Fig. 1. Type 4 PDE activity co-purifies with mAKAP. (A) Immune complexes were isolated from rat heart extracts using antibodies against the RII subunit of cAMP-dependent protein kinase or control IgG serum. Co-precipitating PDE activity [pmol/min/immunoprecipitation (IP)] was measured using [3H]cAMP as a substrate. Data are presented as the average of three independent experiments. (B) Immune complexes were isolated from rat heart extracts using antibodies against mAKAP, AKAP18 or control IgG and from brain extracts using antibodies against AKAP150. The source of the antibody is indicated below each column. Co-precipitating PDE activity (pmol/min/IP) was measured as described above. Data presented are the average of three independent experiments. (C) mAKAP immune complexes were treated with PDE inhibitors (indicated below each column) and PDE activity was measured as described above. Data presented are the average of three independent experiments. (D) Immunoprecipitations were performed from rat heart extracts using antibodies against the RII subunit of PKA or IgG control. The resulting immune complexes were separated by electrophoresis on a 7.5% SDS gradient polyacrylamide gel and electrotransferred to nitrocellulose membranes. Detection of PDE4D in the extract (lane 1), IgG control (lane 2) or the RII immunoprecipitation (lane 3) was by immunoblot analysis using a monoclonal antibody against the PDE4D family. Detection of signals was by chemiluminescence. Molecular weight markers are indicated. The migration of PDE4D is indicated.
PDE inhibitors were added to mAKAP immune complexes to establish which family of PDE associated with the anchoring protein (Figure 1C). The general PDE inhibitor 3-isobutyl-1-methylxanthine (IBMX; 15 µM) reduced PDE activity by 74 ± 8% (n = 3). In contrast, application of milrinone (1 µM), a selective PDE3 inhibitor, had no effect on the mAKAP-associated PDE activity (Figure 1C). However, rolipram (10 µM), a specific PDE4 inhibitor, blocked all mAKAP-associated PDE activity (Figure 1C), indicating that PDE4 activity associates with mAKAP in heart extracts. We obtained independent confirmation of this result when RII antibodies co-precipitated a 100 kDa protein recognized by monoclonal antibodies against the PDE4D gene family (Figure 1D, lane 3). On the basis of molecular weight, this protein was likely to be either PDE4D3 (97 kDa) or PDE4D5 (105 kDa). Both enzymes are expressed in cardiac tissues and PKA phosphorylation stimulates their PDE activity (Kostic et al., 1997).
PDE4D associates with mAKAP inside cells
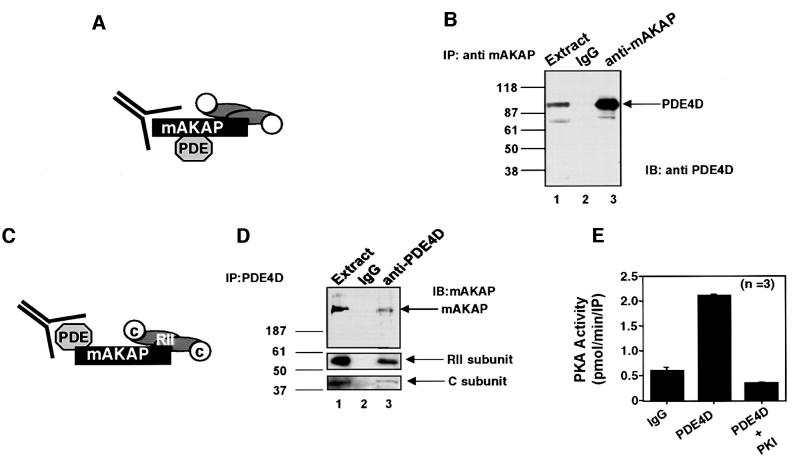
To characterize the mAKAP signaling complex biochemically, additional co-precipitation experiments and PKA activity measurements were performed (Figure 2). Immunoprecipitation of mAKAP from rat heart extracts using polyclonal antisera against the rat anchoring protein resulted in co-purification of a 100 kDa PDE4D isoform, as detected by western blotting (Figure 2B, lane 3). Identical results were obtained when experiments were repeated using antisera raised against the human mAKAP protein (data not shown). PDE immunoreactivity was not co-precipitated with a control rabbit IgG control (Figure 2B, lane 2). It was estimated that ∼5% of the total cardiac PDE4 pool was associated with mAKAP. In reciprocal experiments, immunoprecipitation of PDE4D family members resulted in the co-purification of mAKAP, as detected by western blotting (Figure 2D, top panel, lane 3). An RII binding protein corresponding in size to mAKAP was detected when the same filter was probed for AKAPs by the overlay assay (data not shown). The anchoring protein was not detected when immunoprecipitations were performed with control IgG or pre-immune serum (Figure 2D, lane 2). Further analysis confirmed that the PKA holoenzyme was co-purified with the signaling complex, as RII (Figure 2D, middle panel, lane 3) and the C subunit of PKA (Figure 2D, bottom panel, lane 3) were detected by immunoblotting.
Fig. 2. Biochemical characterization of the mAKAP signaling complex. The mAKAP signaling complex was analyzed by a series of complementary biochemical approaches. (A) A schematic diagram depicting the isolation of the mAKAP immune complexes. (B) Immunoprecipitations were performed from rat heart extracts using antibodies against rat mAKAP or control IgG. The resulting immune complexes were separated by electrophoresis on a 7.5% SDS–polyacrylamide gel and electrotransferred to nitrocellulose membranes. Immunoblot analyses using monoclonal antibodies against the PDE4D family were used to identify the PDE in heart extracts (lane 1) and immunoprecipitations with the IgG control (lane 2) or anti-mAKAP antisera (lane 3). Detection of signals was by chemiluminescence. Molecular weight markers are indicated. The migration of PDE4D is indicated. (C) A schematic diagram depicting immunoprecipitation of PDE4D and associated proteins. (D) Immune complexes were isolated from rat heart extracts (lane 1) using goat polyclonal antibodies against PDE4D isoforms (lane 3) or control goat IgG (lane 2). Immune complexes were separated by electrophoresis on a 7.5% SDS–polyacrylamide gel and electrotransferred to nitrocellulose membranes. The filter was subjected to immunoblot analysis using rabbit polyclonal antibodies against rat mAKAP (top panel), RII regulatory subunit (middle panel) and catalytic subunit of PKA (bottom panel). Detection of signals was by chemiluminescence. Molecular weight markers and the migration position of mAKAP are indicated. (E) PKA activity in PDE4D immune complexes was measured using the heptapeptide Kemptide as a substrate. PKA specific activity (pmol/min/IP) was measured from PDE4D immune complexes, the IgG control and in the presence of the PKI 5–24 peptide, a specific inhibitor of PKA. The source of the sample is indicated below each lane. The accumulated data from three experiments are presented.
Additional experiments were performed to establish whether PDE4D could immunoprecipitate the PKA holoenzyme through the association with mAKAP (Figure 2C). The presence of the PKA catalytic subunit was detected by activity measurements using the heptapeptide Kemptide as a substrate (Corbin and Reimann, 1974). The specific activity of PKA was enriched 3.3 ± 0.6-fold (n = 3) over the IgG control in PDE4D immunoprecipitates from rat heart extracts (Figure 2E). Furthermore, PKA activity was reduced to baseline levels in the presence of the PKI 5–24 peptide, a specific inhibitor of PKA (Scott et al., 1985) (Figure 2E). Collectively, these results demonstrate that mAKAP maintains a signaling complex of a cAMP-selective type 4D PDE and the PKA holoenzyme in vivo.
PDE4D3 binds directly to mAKAP
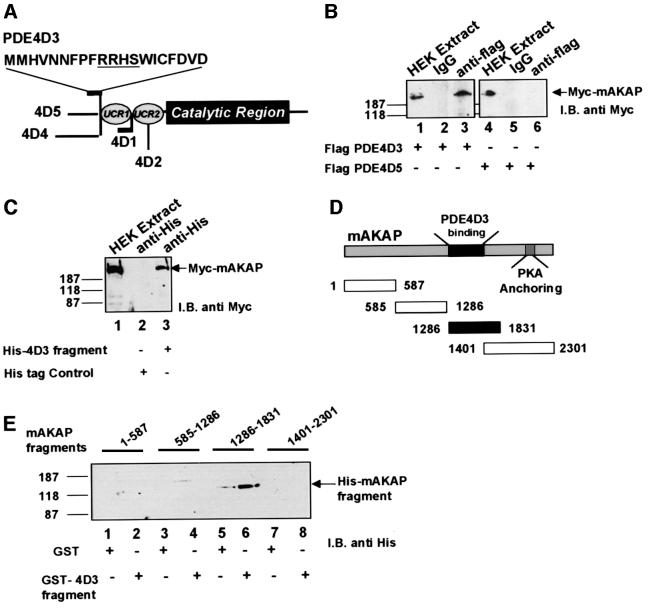
The PDE4D3 and PDE4D5 isoforms are splice variants of the same gene that differ only in their extreme N-terminal sequences (Beavo et al., 1994) (Figure 3A). In order to establish which PDE4D isoform associated with the anchoring protein, HEK 293 cells were transfected with pcDNA3 expression vectors encoding myc-tagged mAKAP and Flag-tagged versions of either PDE4D3 or PDE4D5. The epitope-tagged PDEs were immunoprecipitated from cell extracts with anti-Flag-tag antibodies, and co-purification of the anchoring protein was assessed by western blotting using anti-Myc antibodies (Figure 3B and C). Co-precipitation of mAKAP was only seen with co-expression of PDE4D3 (Figure 3B, lanes 1–3). In contrast, the anchoring protein did not co-precipitate with PDE4D5 (Figure 3B, lanes 4–6). Thus, PDE4D3 appears to associate with mAKAP selectively.
Fig. 3. PDE4D3 directly binds a central region of mAKAP. Mapping studies were performed to determine which PDE4D isoform interacts with mAKAP. (A) A schematic diagram of the PDE4D gene family highlights the conserved catalytic core and upstream conserved regions (UCR). The locations of divergent sequences in each isoform are indicated. The unique first 15 residues of PDE4D3 are presented using the one letter amino acid code. (B) Recombinant Flag-tagged PDE4D3 or Flag-tagged PDE4D5 was expressed in HEK293 cells. Anti-Flag antibodies or control IgG were used to isolate immune complexes. The precipitates were separated by gel electrophoresis on a 7.5% SDS–polyacrylamide gel and electrotransferred to nitrocellulose membranes. Co-precipitation of epitope-tagged mAKAP from cell extracts expressing PDE4D3 (lanes 1–3) or PDE4D5 (lanes 4–6) was assessed by immunoblotting (I.B.) using monoclonal antibodies against the myc tag. Detection of the signal was by chemiluminescence. Molecular weight markers and the migration position of Myc-mAKAP are indicated. (C) A His-tagged fusion protein encompassing the unique region of PDE4D3 or a His-tagged control fragment was co-expressed with mAKAP in HEK293 cells. Anti-His-tag antisera were used to isolate immune complexes. The presence of mAKAP was assessed by immunoblotting using anti-Myc monoclonal antibodies in HEK 293 cell extracts (lane 1) and in immune complexes from cells expressing His-PDE4D3 fragment (lane 3) or control fragment (lane 2). Detection of the signal was by chemiluminescence. Molecular weight markers and the migration position of Myc-mAKAP are indicated. A family of His-tagged-mAKAP fragments encompassing distinct regions of the anchoring protein were expressed in Escherichia coli. (D) A schematic diagram shows the size of each mAKAP fragment and its location within the linear sequence of the anchoring protein. The first and last amino acids of each fragment are indicated. The region of the anchoring protein that interacts with the PDE (closed box) and the binding site for PKA are highlighted. (E) A GST fusion protein including the unique N-terminal sequence of PDE4D3 was used to screen the purified mAKAP fragments by in vitro pull-down assay (lanes 2, 4, 6 and 8). Control experiments were performed with GST alone (lanes 1, 3, 5 and 7). The isolated materials were separated by gel electrophoresis on a 7.5% SDS–polyacrylamide gel and electrotransferred to nitrocellulose membranes. Isolation of mAKAP fragments was assessed by immunoblotting (I.B.) using monoclonal antibodies against the His tag. Detection of the signal was by chemiluminescence. Molecular weight markers are indicated.
Given that PDE4D3 and PDE4D5 differ only in their extreme N-termini, we hypothesized that association between mAKAP and PDE4D3 was due to the unique sequence at the extreme N-terminus of PDE4D3 (Figure 3A). A His-tagged fusion protein including this region was expressed in HEK 293 cells and used as a ligand to isolate recombinant mAKAP (Figure 3C). Immunoprecipitation of the His-tagged 4D3 fragment resulted in co-purification of recombinant mAKAP (Figure 3C, lane 3). The anchoring protein was not detected upon immunoprecipitation of a control His-tagged fragment (Figure 3C, lane 2). To confirm these results, a glutathione S-transferase (GST) fusion protein encompassing the 15-amino-acid unique region of PDE4D3 was used to screen a family of mAKAP fragments in an in vitro pull-down assay (Figure 3D). The GST–PDE4D3 fragment bound to a central portion of mAKAP encompassing residues 1286–1831 (Figure 3E, lane 6), but did not bind adjacent regions of the anchoring protein including a fragment encompassing residues 1401–2301 (Figure 3E, lane 8). GST alone did not bind to any of the mAKAP fragments (Figure 3E). Control experiments confirmed that approximately equal amounts of each bacterially purified mAKAP-His-tag fragment were used in the pull-downs (data not shown). Taken together, these mapping studies indicate that there is a direct interaction between the N-terminus of PDE4D3 and a site located between residues 1286 and 1401 of mAKAP.
Redistribution of PDE4 upon induction of mAKAP expression in cultured rat neonatal ventriculocytes
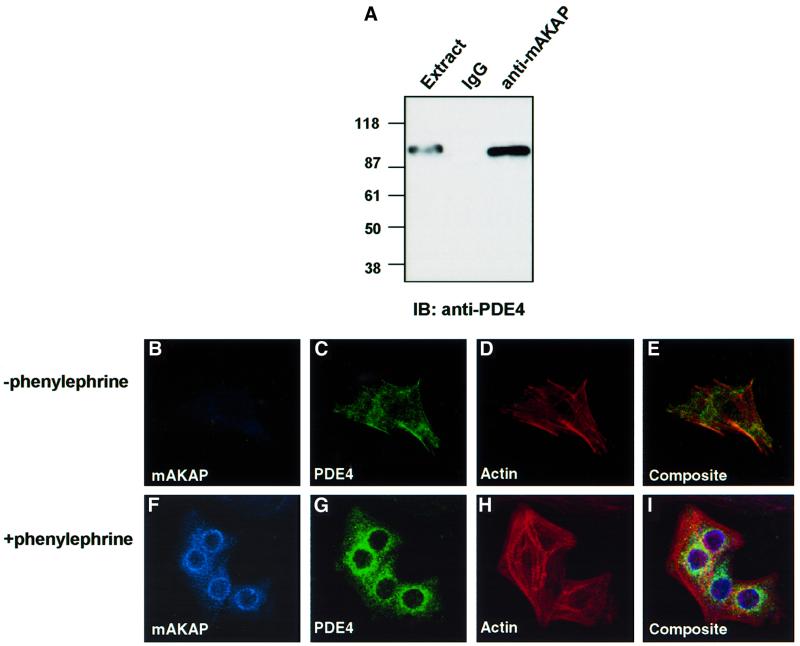
We have demonstrated previously that mAKAP expression is induced in rat neonatal ventriculocytes (RNVs) by treatment with hypertrophic stimuli. This promotes accumulation of the anchoring protein at perinuclear sites and movement of the PKA holoenzyme to this region (Kapiloff et al., 1999). It seemed likely that PDE4D3 localization would be altered if mAKAP expression levels were manipulated. Immunofluorescence techniques were used to visualize PDE4 location in primary cultures of RNV before and after induction of mAKAP expression following treatment with phenylephrine (100 µM) for 24 h (Figure 4). Since the antibodies used in our biochemical analyses did not work for immunocytochemical detection of the enzyme, it was necessary to use a monoclonal antibody that recognized all PDE4 subtypes for these experiments. Control experiments confirmed that this antibody detected a 100 kDa PDE4 species upon immunoprecipitation of mAKAP from RNV (Figure 4A).
Fig. 4. Recruitment of mAKAP and PDE4D3 to the perinuclear regions of RNVs upon induction of hypertrophy. Expression of mAKAP is upregulated upon the induction of hypertrophy in RNV, resulting in accumulation of the anchoring protein at perinuclear regions (Kapiloff et al., 1999). (A) As a prelude to immunocytochemical analysis, control experiments confirmed that a monoclonal antibody against PDE4 family members recognized the PDE that co-precipitated with mAKAP from cardiomyocyte extracts. Immunoprecipitations were performed using antibodies against rat mAKAP or control IgG. The resulting immune complexes were separated by electrophoresis on a 7.5% SDS–polyacrylamide gel and electrotransferred to nitrocellulose membranes. Immunoblot analysis using a monoclonal antibody that recognizes the PDE4 family was used to identify the PDE in immunoprecipitations with anti-mAKAP antisera or the IgG control (indicated above each lane). This antibody was subsequently used in immunofluorescence experiments. Dissociated RNVs were plated on coverslips and grown in the absence (upper panels) or presence (lower panels) of phenylephrine (100 µM) for 24 h at 37°C. Ventriculocytes were fixed by incubation in 3.7% formaldehyde for 10 min at room temperature, permeablized with 0.3% Triton X-100, and subjected to labeling with primary antibodies against mAKAP (1:500 dilution) and PDE4 family(1:500 dilution) for 16 h at 4°C. The intracellular location of mAKAP (blue; B and F) was detected with Cy5-labeled secondary antibodies (1:100 dilution), PDE4 (green; C and G) was detected with fluorescein-labeled secondary antibodies (1:100 dilution) and the actin cytoskeleton (red; D and H) was detected with Texas red–phalloidin. Immunofluorescent detection of each signal was performed by confocal microscopy on a BioRad 1024 UV/Vis confocal microscope. Composite images are presented in (E) and (I).
mAKAP was barely detected in undifferentiated RNV (Figure 4B), and PDE4 exhibited a general cytoplasmic staining pattern (Figure 4C). Actin staining defined cell morphology and indicated a moderate level of stress fiber formation (Figure 4D). Prominent mAKAP staining was detected at the perinuclear regions upon the induction of cardiomyocyte hypertrophy with phenylephrine (Figure 4F). Significantly, a proportion of the PDE4 signal was now detected at these sites (Figure 4G). In addition, stress fiber formation was more pronounced (Figure 4H), which is a recognized hallmark of RNV hypertrophy (Thorburn et al., 1995). These results suggest that induction of mAKAP promotes the recruitment of PDE4 to the perinuclear membranes of RNV. This is best illustrated in a composite image showing that a proportion of the mAKAP and PDE4 signals overlap (Figure 4I). This finding is consistent with subcellular fractionation data indicating that PDE4D subtypes are present in the outer perinuclear membranes of isolated cardiac nuclei (Lugnier et al., 1999).
Regulation of the mAKAP signaling complex
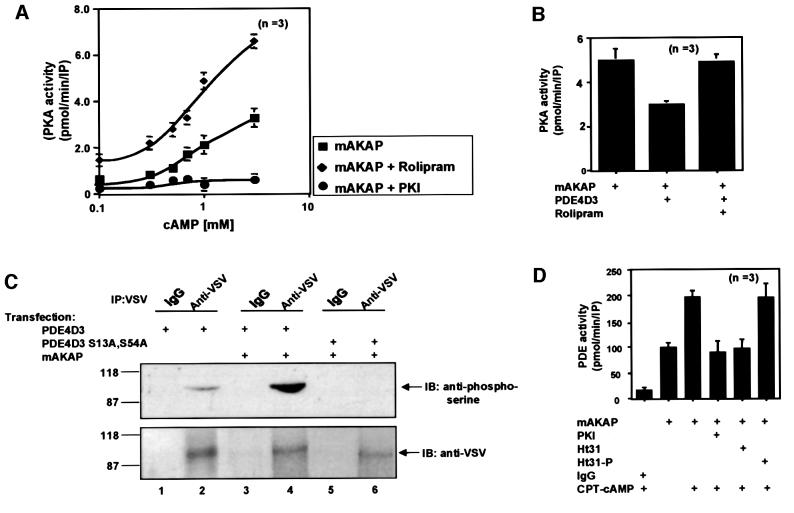
Our combined biochemical, enzymological and immunofluorescence data suggest that mAKAP maintains a signaling complex of active PDE4D3 and the PKA holoenzyme. A functional consequence of these interactions could be to control the activity status of the anchored kinase through regulation of local cAMP concentrations. To test this hypothesis, the dose dependency of PKA activation within the mAKAP signaling complex was measured over a range of cAMP concentrations. Untreated mAKAP immunoprecipitates from heart extracts displayed a dose-dependent increase in PKA activity (Figure 5A, squares) that resulted in a 7.8 ± 0.9-fold (n = 3) maximal stimulation of the kinase (Figure 5A). Addition of the PDE4 inhibitor rolipram enhanced sensitivity to cAMP (Figure 5A, diamonds), resulting in a 15.4 ± 2-fold (n = 3) stimulation of kinase activity at the highest concentration of cAMP (Figure 5A). Control experiments using the specific PKA inhibitor peptide PKI 5–24 confirmed that all of the anchored kinase activity was PKA (Figure 5A, circles). These data indicate that the pool of PDE4D3 associated with mAKAP influences the activity status of the anchored PKA. This result was confirmed by assembly of the signaling complex with recombinant components in HEK 293 cells (Figure 5B). Following immunoprecipitation of the mAKAP signaling complex, PKA activity was decreased 40 ± 10% (n = 3) in cells co-transfected with PDE4D3 (Figure 5B). Upon application of rolipram, the mAKAP-associated PKA activity was restored to a level equivalent to that measured from cells expressing mAKAP alone (Figure 5B). Thus, recruitment of the PDE into the mAKAP signaling complex attenuates PKA activation. Furthermore, these experiments indicate that mAKAP organizes two functionally coupled enzymes in a cAMP-responsive signaling scaffold to locally regulate PKA activity.
Fig. 5. mAKAP assembles a PDE/PKA negative feedback loop. The functional consequences of PDE4D3 localization through association with mAKAP were assessed on PKA activity. (A) The mAKAP signaling complex was immunoprecipitated from rat heart extracts using antibodies against the mouse anchoring protein. PKA activity (squares) was measured over a range of cAMP concentrations (0.1–10 mM) as described in Materials and methods, upon inhibition of PDE4D3 by rolipram (diamonds) and in the presence of the PKI 5–24 peptide (circles). The specific activity of the PKA catalytic subunit is presented as pmol/min/IP. The averaged data from three experiments are presented. (B) Recombinant myc-tagged mAKAP was expressed in HEK293 cells in the absence or presence of PDE-4D3 (indicated below each lane). Anti-Myc antibodies were used to isolate immune complexes and PKA activity was measured in the presence of 1 mM cAMP. The specific activity of the PKA catalytic subunit is presented as pmol/min/IP. The averaged data from three experiments are presented. Rolipram (10 µM) was used to selectively inhibit PDE activity. (C) Expression vectors encoding VSV-tagged PDE4D3 (lanes 1–4) or the phosphorylation site mutant VSV-tagged PDE4D3 S13A,S54A (lanes 5 and 6) were co-expressed with myc-tagged mAKAP in HEK293 (lanes 2–6). The transfected plasmids are indicated above each lane. The cells were incubated with 1 mM CPT-cAMP for 15 min before immunoprecipitation with anti-VSV antibodies or control IgG. The immune complexes were separated by gel electrophoresis on a 7.5% SDS–polyacrylamide gel and electrotransferred to nitrocellulose membranes. Phosphorylation of PDE4D3 was assessed by immunoblotting (IB) using a polyclonal phosphoserine-specific antibody (top panel). Equal loading of PDE4D3 was confirmed by reprobing the blot with VSV-specific monoclonal antibody (bottom panel). Detection of the signals was by chemiluminescence. Molecular weight markers and the migration position of PDE are indicated. This figure is representative of three independent experiments. (D) In reciprocal experiments, the functional consequences of PKA anchoring were assessed on PDE4D3 activity. The mAKAP signaling complex was immunoprecipitated from rat heart extracts using antibodies against the rat anchoring protein. After a 16 h incubation, the immunoprecipitates were incubated in the presence or absence of CPT-cAMP for 30 min (indicated below). PDE4 activity was then measured under conditions where PKA activity was inhibited (PKI), in the presence of a PKA anchoring inhibitor peptide (Ht31) or a control peptide (Ht31p). PDE activity was presented as pmol/min/IP. The data averaged from three experiments are presented.
Our anchoring hypothesis proposes that AKAPs function to position PKA in close proximity to substrates (Colledge and Scott, 1999). Interestingly, PDE4D3 is phosphorylated on Ser13 and Ser54 by PKA (Sette and Conti, 1996; Hoffmann et al., 1998; Oki et al., 2000). Phosphorylation of Ser54 enhances PDE4D3 activity 2- to 3-fold in vitro and inside cells (Sette and Conti, 1996; Lim et al., 1999), explaining earlier reports that elevation of intracellular cAMP stimulates PDE4 activity (Sette and Conti, 1996). Transfection experiments in HEK293 cells were performed to confirm that the mAKAP-associated pool of PDE4D3 is preferentially phosphorylated by PKA. All cells were treated with CPT-cAMP for 15 min prior to immunoprecipitation of the PDE, and phosphate incorporation was monitored by immunoblotting using anti-phosphoserine antibodies (Figure 5C). Low level PDE4D3 phosphorylation was detected in cells transfected with the PDE alone (Figure 5C, lane 2). However, PDE4D3 phosphorylation was markedly increased in cells co-transfected with the anchoring protein (Figure 5C, lane 4). Importantly, phosphorylation was not detected in cells co-transfected with the anchoring protein and PDE4D3 S13A, S54A, a mutant form of the enzyme lacking both PKA phosphorylation sites (lane 6). This shows that the mAKAP-associated pool of PDE4D3 is a substrate for the kinase.
On the basis of the experiments described above, it is possible that mAKAP maintains a pool of PKA that could upregulate PDE4D3 activity in heart cells. Immuno precipitation of endogenous mAKAP from heart extracts resulted in a 3.5 ± 0.5-fold (n = 3) increase in basal PDE activity over the IgG control (Figure 5D). However, in the presence of CPT-cAMP, the mAKAP-associated PDE activity was increased further to 7.0 ± 0.3 (n = 3). This 2-fold increase in activity is consistent with previous reports demonstrating that elevated cAMP increases PDE4D3 activity (Sette and Conti, 1996). Control experiments confirmed that PDE activity was decreased 53 ± 4% (n = 3) in the presence of PKI 5–24 peptide, a specific inhibitor of the PKA catalytic subunit (Figure 5D). This strongly suggests that PKA regulates PDE4D3 activity within the mAKAP signaling complex.
The anchoring inhibitor peptide Ht31 is a competitive inhibitor of PKA–AKAP interaction and is routinely used to release PKA from signaling complexes inside cells (Carr et al., 1992; Rosenmund et al., 1994). Treatment with Ht31 prior to immunoprecipitation of the mAKAP signaling complex decreased cAMP-stimulated PDE activity by 47 ± 0.3% (n = 3), a similar decrease to that seen with PKI (Figure 5B). In contrast, treatment with a control peptide, Ht31-proline, had no effect on cAMP-stimulated PDE activity (Figure 5D). Thus, we conclude that displacement of PKA from the mAKAP signaling complex prevents cAMP-dependent modulation of PDE activity. These findings provide a molecular explanation for the negative feedback effects on cAMP levels ascribed to the PKA phosphorylation of PDE4D3 (Oki et al., 2000).
Discussion
In this report, we present evidence that the muscle-selective anchoring protein mAKAP assembles a signaling complex including the PKA holoenzyme and a cAMP PDE at perinuclear regions in hypertrophic cardiomyocytes. Our mapping studies indicate that the first 15 residues of PDE4D3 contain determinants for association with the anchoring protein. This finding is consistent with evidence that the divergent N-terminal regions of the ‘long form’ PDE4D isozymes associate with signaling molecules (Conti, 2000). (The domain organization of the PDE4D gene family is presented in Figure 3A.) A 152-residue unique region of PDE4D4 interacts with SH3 domains of the tyrosine kinases src, lyn and fyn, whereas the first 88 amino acids of PDE4D5 bind to the scaffold protein RACK1 with high affinity (Beard et al., 1999; Yarwood et al., 1999). RACK1 also binds to activated PKC, although isolation of a signaling complex containing both enzymes has not been possible (Ron et al., 1994; Yarwood et al., 1999). These and other PDE4D isoforms are also retained at specific subcellular sites in macrophages and FRTL-5 cells, possibly via the interaction with specific binding proteins (Jin et al., 1998; Pryzwansky et al., 1998). Myomegalin, a Golgi/centrosomal protein with homology to Drosophila centrosomin, has recently been isolated by two-hybrid analysis using the PDE4D upstream conserved region 2 (UCR2), a regulatory domain common to all long isozymes, as bait (Verde et al., 2001). Although direct interaction with the PDE has yet to be formally proven inside cells, it seems likely that association with myomegalin provides a means to target certain PDE4D isoforms to the Golgi and centrosomes (Verde et al., 2001). Interestingly, two centrosomal AKAPs, AKAP350/450/CG-NAP and pericentrin, target PKA to this region (Schmidt et al., 1999; Takahashi et al., 1999; Witczak et al., 1999; Diviani et al., 2000). Thus, alternative targeting mechanisms may permit the clustering of PKA and PDE4D isoforms at other intracellular locations.
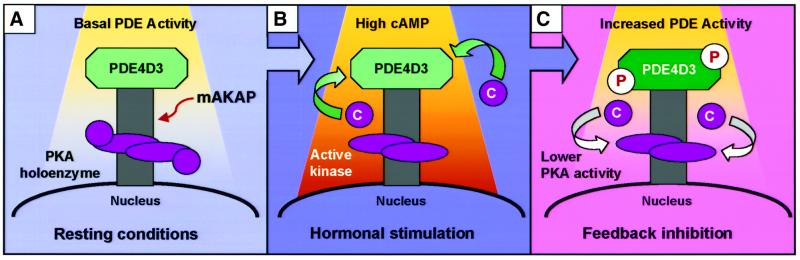
Our studies with the PDE4 inhibitor rolipram suggest that the close proximity of the mAKAP-associated PDE controls the activation status of anchored PKA. To our knowledge, this provides the first evidence for a cAMP-dependent kinase and a cAMP-metabolizing enzyme in the same macromolecular unit. The functional ramifications of such a tightly coupled cAMP signaling module may be significant for feedback inhibition of PKA phosphorylation events. A model highlighting this process is presented in Figure 6. Under basal conditions, the tethered PDE4D3 metabolizes cAMP diffusing into the local environment at a rate that is sufficient to prevent activation of PKA (Figure 6A). However, upon hormonal stimulation, the increased flow of cAMP is sufficient to override the local PDE activity (Figure 6B). This releases active C subunit from the mAKAP signaling complex and permits PKA phosphorylation (Figure 6B). Two important regulatory factors built into the mAKAP signaling complex favor the signal termination process. First, the tethered PDE is constitutively active and will rapidly restore basal cAMP levels when the flow of second messenger is turned off. Secondly, elegant experiments by Conti and others (Sette and Conti, 1996; Hoffmann et al., 1998; Lim et al., 1999; Conti, 2000; Oki et al., 2000) have demonstrated that PKA phosphorylation of PDE4D3 on Ser54 increases the Vmax of the enzyme 2- to 3-fold over basal conditions. Phosphorylation of PDE4D3 increases cAMP degradation to favor re-formation of the PKA holoenzyme (Figure 6C). PKA anchoring is a unique and critical element in this PKA/PDE4D3 feedback loop, as displacement of the kinase with the anchoring inhibitor peptide Ht31 prevents cAMP-dependent stimulation of the mAKAP-associated PDE4D activity. This not only emphasizes the importance of PDE compartmentalization in the preservation of cAMP homeostasis, but also highlights that multiple regulatory processes control where and when PKA activation occurs inside cells.
Fig. 6. mAKAP maintains an anchored PKA/PDE4D3 negative feedback loop. The cartoon depicts the proposed feedback inhibition of anchored PKA by mAKAP-associated PDE4DE3 activity. (A) Under basal conditions, the PKA holoenzyme is dormant and the localized constitutively active PDE maintains cAMP concentrations below an activation threshold for the kinase. (B) Upon hormonal stimulation, the increased flow of second messenger overcomes the rate of PDE-mediated cAMP degradation. This promotes release of the PKA catalytic (C) subunit. (C) PKA phosphorylation of mAKAP-associated PDE4D3 stimulates PDE activity, thereby driving cAMP levels back to basal. This favors reformation of the PKA holoenzyme.
Assembly of the mAKAP signaling complex at perinuclear regions occurs upon catecholamine-induced hypertrophy in RNVs (Kapiloff et al., 1999). Ventricular hypertrophy involves the coordinated mobilization of various signal transduction pathways and represents a cellular adaptation to the requirement for increased contractile power in the heart (Sugden and Clerk, 1998). PKA phosphorylation induces certain cAMP-responsive genes that propagate the hypertrophic phenotype (Zimmer, 1997). Thus, a perinuclear pool of PDE4 may control the release of the C subunit into the nucleus. However, this may represent only one site of action for the mAKAP signaling complex, as the anchoring protein has also been detected in the sarcoplasmic reticulum (SR) and at intercalated discs in adult heart tissue sections (Yang et al., 1998; Marx et al., 2000). Furthermore, Marx et al. (2000) have recently reported that mAKAP is recruited into an SR-associated macromolecular complex that includes the calcium release channel ryanodine receptor. An important conclusion of their study is that defects in PKA signaling at the SR promote hyperphosphorylation of the ryanodine receptor in failing hearts. One explanation is that PKA activity at the SR is aberrantly high during cardiac failure. According to the feedback mechanism we have proposed in Figure 6, any reduction in mAKAP-associated PDE4D3 activity would favor prolonged activation of PKA. This raises the intriguing possibility that local inhibition of PDE activity or disruption of an SR-associated pool of PDE contributes to this disease state. Interestingly, ERK 2 phosphorylation of PDE4D3 inhibits enzyme activity (Hoffmann et al., 1999; MacKenzie et al., 2000), while the synergistic actions of PKA and ERK 2 favor translocation of the PDE from membranes to the cytosol in rat aortic endothelial cells (Liu and Maurice, 1999). Future studies are planned to determine whether mAKAP-associated PDE4D3 activity is reduced in failing heart tissue.
Materials and methods
Antibodies
The following primary antibodies were used for immunoblotting and immunocytochemistry: mouse monoclonal pan-PDE4D (clone 61D10E from ICOS; 2.3 mg/ml, 1:5000 dilution), mouse monoclonal pan-PDE4 family (clone 96F2G from ICOS; 4.8 mg/ml, 1:1000 dilution), rabbit polyclonal to mAKAP (clone VO54; 5 mg/ml, 1:1000 dilution), mouse monoclonal PKA catalytic and regulatory subunit (Transduction Laboratories; 1:1000 dilution), rabbit phosphoserine (Zymed; 1:1000). For immunoprecipitations, the following antibodies were used: mAKAP (VO54 and VO56; 5 mg/ml, 5 µl), mouse monoclonal pan-PDE4D (clone 61D10E from ICOS; 2.3 mg/ml, 5 µl), goat pan-PDE4D (8 µl) and PKA RII subunit (Transduction Laboratories; 4 µg).
Heart extract preparations and immunoprecipitations
For immunoprecipitations of mAKAP from heart extract, rat hearts (PelFreeze) were homogenized in 10 ml of LSE buffer (20 ml HEPES pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, protease inhibitor cocktail) and centrifuged at 15 000 g for 30 min. Soluble extracts were used for immunoprecipitations. Antibodies (mAKAP, PDE4D and PKA regulatory subunit) or control IgG were added to 500 µl of extract (1 mg) with 30 µl of protein A– or G–agarose beads and incubated at 4°C with shaking. Following an overnight incubation, pellets were washed twice with HSE buffer (10 mM Na2PHO4, 150 mM NaCl, 5 mM EGTA, 5 mM EDTA) with 1% Triton X-100, twice in the same detergent with 600 mM NaCl, and twice in HSE buffer without detergent. Bound proteins were detected by immunoblotting.
For immunoprecipitations from heterologous cells, HEK 293 cells were transfected at 50% confluency in 10 cm plates using the LipofectAMINE PLUS kit (Gibco-BRL) according to the manufacturer’s instructions, using 5 µg of each cDNA construct per plate. Cells were harvested and lysed 24 h after transfection in 1 ml of HSE buffer containing 1% Triton X-100 and protease inhibitors. Supernatants were incubated with antibodies or control non-immune IgG and 30 µl of pre-washed protein A– or G–agarose beads. Following an overnight incubation at 4°C, the immunoprecipitates were washed as described above. Bound proteins were analyzed by immunoblotting.
PDE assay
Immunoprecipitations of heart extract using 5 µl of mAKAP VO54 antibody were performed as described above. PDE activity was assayed using 1 µM cAMP as a substrate according to the method by Beavo et al. (1974). Samples were assayed in 45 µl of PDE buffer A [100 mM MOPS pH 7.5, 4 mM EGTA, 1.0 mg/ml bovine serum albumin (BSA)] and 50 µl of PDE buffer B {100 mM MOPS pH 7.5, 75 mM MgAc, 100 000 c.p.m. of [3H]cAMP (Dupont, NEN)}. To identify the specific PDE activity associated with mAKAP, PDE inhibitors were added (10 µM rolipram or 1 µM milrinone; Sigma) to the solution.
PKA assay
Immunoprecipitations of heart extract using 5 µl of mAKAP VO54 antibody were performed as described above, and PKA activity was assayed by the filter binding method of Corbin and Reimann (1974). Pellets were incubated in kinase buffer (50 mM Tris–HCl pH 7.5, 5 mM MgCl2) containing 30 mM Kemptide, 100 µM ATP, 5 µM [γ-32P]ATP and increasing concentrations of cAMP. After a 15 min incubation at 30°C, the reaction mixture was spotted onto phosphocellulose strips and washed five times in 75 mM phosphoric acid and once in 95% ethanol. Filters were air dried and counted by liquid scintillation.
For experiments involving PKA phosphorylation of PDE, immunoprecipitation of the mAKAP complex from heart extract was performed as described above. After immune complexes were washed, 200 µl of HSE buffer containing either Ht31 or Ht31-proline were added, and the complexes were incubated for 30 min at 4°C with shaking. Pellets were then washed twice with HSE buffer. To stimulate PKA bound to the complex, 20 µl of kinase buffer containing 0.75 mM CPT-cAMP and 4 mM Mg-ATP were added. After a 30 min incubation at 37°C, the pellets were washed and PDE activity assays were performed as described above.
Expression constructs
For construction of His-tagged PDE4D3 (either full length or amino acids 1–60), the domains were PCR amplified using a full-length cDNA as a template and subcloned into pcDNA3.1 V5/his-TOPO. Flag-tagged PDE4D3 was constructed by subcloning an EcoRI fragment into a pcDNA-Flag vector.
GST pull-downs
For GST pull-down experiments, GST alone or beads charged with GST fused to a fragment spanning the PDE4D3 unique region were incubated with 1 µg of purified recombinant mAKAP fragments. The incubations were performed in 400 µl of HSE buffer and incubated overnight at 4°C with shaking. The presence of mAKAP bound to the beads was detected by immunoblotting.
Cell culture of RNVs
RNVs were prepared according to the protocol of Thorburn et al. (1995). Briefly, 1- to 3-day-old rat pups were killed by decapitation, and the hearts were removed through a sternotomy. The ventricles were trimmed free of atria, fat and connective tissue, while immersed in a neutral buffer. Myocytes were dissociated by several 16 min cycles of trypsin treatment and serum neutralization. After dissociation, RNV cells were collected by centrifugation, passed through a 70-µm-mesh cell strainer to remove clumps, and pre-plated in culture dishes to remove fibroblasts. After 1 h, the medium containing the unattached RNV cells was removed, cells were collected by centrifugation and were plated again on dual-well chamberslides previously coated with 1% gelatin and 60 µg/ml laminin solution at a density of 500 000 myocytes/plate. Plating medium was Dulbecco’s modified Eagle’s medium (DMEM) with 17% Media 1999, 1% penicillin/streptomycin (Gibco-BRL), 10% horse serum (HS) and 5% fetal bovine serum (FBS). The following day, the plates were washed and then incubated with plating media containing neither HS nor FBS in the presence of 100 µM phenylephrine to induce the hypertrophic response.
Immunocytochemistry
Twenty-four hours after treatment with phenylephrine, cells were rinsed twice with phosphate-buffered saline (PBS), fixed with 3.7% formaldehyde in PBS for 10 min, and washed again with PBS. Cells were then permeablized with 0.3% Triton X-100 in PBS, washed with PBS and blocked with 0.2% BSA in PBS for 30 min. Primary antibody (mAKAP 1:500, monoclonal pan-PDE4 family; 1:500) in BSA/PBS was then added for 16 h at 4°C, and the cells were washed three times with PBS for 5 min each. Cells were then incubated with the appropriate fluorescent secondary antibodies in the same manner. After three PBS washes, the cells were rinsed with water and mounted with glass coverslips using Slowfade Light Antifade mounting medium (Molecular Probes). Texas red–phalloidin was used to visualize actin fibers.
Acknowledgments
Acknowledgements
The authors wish to acknowledge their colleagues in the Vollum Institute for critical evaluation of this manuscript, ICOS corporation for providing antibodies against PDE4 subtypes, Nicole Jackson for affinity purification of mAKAP fusion protein fragments and Scott H.Soderling for assistance with PDE assays. This work was supported by NIH grants DK54441 (to J.D.S.) and HL04229 (to M.S.K.).
References
- Beard M.B., O’Connell,J.C., Bolger,G.B. and Houslay,M.D. (1999) The unique N-terminal domain of the cAMP phosphodiesterase PDE4D4 allows for interaction with specific SH3 domains. FEBS Lett., 460, 173–177. [DOI] [PubMed] [Google Scholar]
- Beard M.B., Olsen,A.E., Jones,R.E., Erdogan,S., Houslay,M.D. and Bolger,G.B. (2000) UCR1 and UCR2 domains unique to the cAMP-specific phosphodiesterase family form a discrete module via electrostatic interactions. J. Biol. Chem., 275, 10349–10358. [DOI] [PubMed] [Google Scholar]
- Beavo J.A. (1995) Cyclic nucleotide phosphodiesterases: functional implications of multiple isoforms. Physiol. Rev., 75, 725–748. [DOI] [PubMed] [Google Scholar]
- Beavo J.A., Bechtel,P.J. and Krebs,E.G. (1974) Preparation of homogenous cyclic AMP-dependent protein kinase(s) and its subunits from rabbit skeletal muscle. Methods Enzymol., 38, 299–308. [DOI] [PubMed] [Google Scholar]
- Beavo J.A., Conti,M. and Heaslip,R.J. (1994) Multiple cyclic nucleotide phosphodiesterases. Mol. Pharmacol., 46, 399–405. [PubMed] [Google Scholar]
- Carr D.W., Hausken,Z.E., Fraser,I.D.C., Stofko-Hahn,R.E. and Scott,J.D. (1992) Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein, cloning and characterization of the RII-binding domain. J. Biol. Chem., 267, 13376–13382. [PubMed] [Google Scholar]
- Colledge M. and Scott,J.D. (1999) AKAPs: from structure to function. Trends Cell Biol., 9, 216–221. [DOI] [PubMed] [Google Scholar]
- Conti M. (2000) Phosphodiesterases and cyclic nucleotide signaling in endocrine cells. Mol. Endocrinol., 14, 1317–1327. [DOI] [PubMed] [Google Scholar]
- Corbin J.D. and Francis,S.H. (1999) Cyclic GMP phosphodiesterase-5: target of sildenafil. J. Biol. Chem., 274, 13729–13732. [DOI] [PubMed] [Google Scholar]
- Corbin J.D. and Reimann,E.M. (1974) A filter assay for determining protein kinase activity. Methods Enzymol., 38, 287–294. [DOI] [PubMed] [Google Scholar]
- Diviani D., Langeberg,L.K., Doxsey,S.J. and Scott,J.D. (2000) Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr. Biol., 10, 417–420. [DOI] [PubMed] [Google Scholar]
- Feliciello A., Li,Y., Avvedimento,E.V., Gottesman,M.E. and Rubin,C.S. (1997) A-kinase anchor protein 75 increases the rate and magnitude of cAMP signaling to the nucleus. Curr. Biol., 7, 1011–1014. [DOI] [PubMed] [Google Scholar]
- Fraser I.D., Tavalin,S.J., Lester,L.B., Langeberg,L.K., Westphal,A.M., Dean,R.A., Marrion,N.V. and Scott,J.D. (1998) A novel lipid-anchored A-kinase anchoring protein facilitates cAMP-responsive membrane events. EMBO J., 17, 2261–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R., Wilkinson,I.R., McCallum,J.F., Engels,P. and Houslay,M.D. (1998) cAMP-specific phosphodiesterase HSPDE4D3 mutants which mimic activation and changes in rolipram inhibition triggered by protein kinase A phosphorylation of Ser-54: generation of a molecular model. Biochem. J., 333, 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R., Baillie,G.S., MacKenzie,S.J., Yarwood,S.J. and Houslay,M.D. (1999) The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. EMBO J., 18, 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houslay M.D., Sullivan,M. and Bolger,G.B. (1998) The multienzyme PDE4 cyclic adenosine monophosphate-specific phosphodiesterase family: intracellular targeting, regulation and selective inhibition by compounds exerting anti-inflammatory and antidepressant actions. Adv. Pharmacol., 44, 225–342. [DOI] [PubMed] [Google Scholar]
- Hunter T. (2000) Signaling—2000 and beyond. Cell, 100, 113–127. [DOI] [PubMed] [Google Scholar]
- Jin S.L., Bushnik,T., Lan,L. and Conti,M. (1998) Subcellular localization of rolipram-sensitive, cAMP-specific phosphodiester ases. Differential targeting and activation of the splicing variants derived from the PDE4D gene. J. Biol. Chem., 273, 19672–19678. [DOI] [PubMed] [Google Scholar]
- Johnson B.D., Scheuer,T. and Catterall,W.A. (1994) Voltage-dependent potentiation of L-type Ca2+ channels in skeletal muscle cells requires anchored cAMP-dependent protein kinase. Proc. Natl Acad. Sci. USA, 91, 11492–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan J.D., Landau,E.M. and Iyengar,R. (2000) Signaling networks: the origins of cellular multitasking. Cell, 103, 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapiloff M.S., Schillace,R.V., Westphal,A.M. and Scott,J.D. (1999) mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J. Cell Sci., 112, 2725–2736. [DOI] [PubMed] [Google Scholar]
- Kessler T. and Lugnier,C. (1995) Rolipram increases cyclic GMP content in l-arginine-treated cultured bovine aortic endothelial cells. Eur. J. Pharmacol., 290, 163–167. [DOI] [PubMed] [Google Scholar]
- Kostic M.M. et al. (1997) Altered expression of PDE1 and PDE4 cyclic nucleotide phosphodiesterase isoforms in 7-oxo-prostacyclin-preconditioned rat heart. J. Mol. Cell. Cardiol., 29, 3135–3146. [DOI] [PubMed] [Google Scholar]
- Lim J., Pahlke,G. and Conti,M. (1999) Activation of the cAMP-specific phosphodiesterase PDE4D3 by phosphorylation. Identification and function of an inhibitory domain. J. Biol. Chem., 274, 19677–19685. [DOI] [PubMed] [Google Scholar]
- Liu H. and Maurice,D.H. (1999) Phosphorylation-mediated activation and translocation of the cyclic AMP-specific phosphodiesterase PDE4D3 by cyclic AMP-dependent protein kinase and mitogen-activated protein kinases. A potential mechanism allowing for the coordinated regulation of PDE4D activity and targeting. J. Biol. Chem., 274, 10557–10565. [DOI] [PubMed] [Google Scholar]
- Lugnier C., Keravis,T., Le Bec,A., Pauvert,O., Proteau,S. and Rousseau,E. (1999) Characterization of cyclic nucleotide phosphodiesterase isoforms associated to isolated cardiac nuclei. Biochim. Biophys. Acta, 1472, 431–446. [DOI] [PubMed] [Google Scholar]
- MacKenzie S.J., Baillie,G.S., McPhee,I., Bolger,G.B. and Houslay,M.D. (2000) ERK2 mitogen-activated protein kinase binding, phosphorylation and regulation of the PDE4D cAMP-specific phosphodiesterases. The involvement of COOH-terminal docking sites and NH2-terminal UCR regions. J. Biol. Chem., 275, 16609–16617. [DOI] [PubMed] [Google Scholar]
- Maclean M.R., Johnston,E.D., McCulloch,K.M., Pooley,L., Houslay,M.D. and Sweeney,G. (1997) Phosphodiesterase isoforms in the pulmonary arterial circulation of the rat: changes in pulmonary hypertension. J. Pharmacol. Exp. Ther., 283, 619–624. [PubMed] [Google Scholar]
- Marx S.O., Reiken,S., Hisamatsu,Y., Jayaraman,T., Burkhoff,D., Rosemblit,N. and Marks,A.R. (2000) PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell, 101, 365–376. [DOI] [PubMed] [Google Scholar]
- McPhee I., Yarwood,S.J., Scotland,G., Huston,E., Beard,M.B., Ross,A.H., Houslay,E.S. and Houslay,M.D. (1999) Association with the SRC family tyrosyl kinase LYN triggers a conformational change in the catalytic region of human cAMP-specific phosphodiesterase HSPDE4A4B. Consequences for rolipram inhibition. J. Biol. Chem., 274, 11796–11810. [DOI] [PubMed] [Google Scholar]
- Oki N., Takahashi,S.I., Hidaka,H. and Conti,M. (2000) Short term feedback regulation of cAMP in FRTL-5 thyroid cells. Role of PDE4D3 phosphodiesterase activation. J. Biol. Chem., 275, 10831–10837. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Scott,J.D. (1997) Signaling through scaffold, anchoring and adaptor proteins. Science, 278, 2075–2080. [DOI] [PubMed] [Google Scholar]
- Pryzwansky K.B., Kidao,S. and Merricks,E.P. (1998) Compartmental ization of PDE-4 and cAMP-dependent protein kinase in neutrophils and macrophages during phagocytosis. Cell Biochem. Biophys., 28, 251–275. [DOI] [PubMed] [Google Scholar]
- Ron D., Chen,C.-H., Caldwell,J., Jamieson,L., Orr,E. and Mochly-Rosen,D. (1994) Cloning of an intracellular receptor for protein kinase C: a homolog of the β subunit of G proteins. Proc. Natl Acad. Sci. USA, 91, 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C., Carr,D.W., Bergeson,S.E., Nilaver,G., Scott,J.D. and Westbrook,G.L. (1994) Anchoring of protein kinase A is required for modulation of AMPA/kainate receptors on hippocampal neurons. Nature, 368, 853–856. [DOI] [PubMed] [Google Scholar]
- Schmidt P.H., Dransfield,D.T., Claudio,J.O., Hawley,R.G., Trotter,K.W., Milgram,S.L. and Goldenring,J.R. (1999) AKAP350: a multiply spliced A-kinase anchoring protein associated with centrosomes. J. Biol. Chem., 274, 3055–3066. [DOI] [PubMed] [Google Scholar]
- Scott J.D., Fischer,E.H., DeMaille,J.G. and Krebs,E.G. (1985) Identification of an inhibitory region of the heat-stable protein inhibitor of the cAMP-dependent protein kinase. Proc. Natl Acad. Sci. USA, 82, 4379–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette C. and Conti,M. (1996) Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. J. Biol. Chem., 271, 16526–16534. [DOI] [PubMed] [Google Scholar]
- Soderling S.H. and Beavo,J.A. (2000) Regulation of cAMP and cGMP signaling: new phosphodiesterases and new functions. Curr. Opin. Cell Biol., 12, 174–179. [DOI] [PubMed] [Google Scholar]
- Sugden P.H. and Clerk,A. (1998) Cellular mechanisms of cardiac hypertrophy. J. Mol. Med., 76, 725–746. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Shibata,H., Shimakawa,M., Miyamoto,M., Mukai,H. and Ono,Y. (1999) Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the Golgi apparatus. J. Biol. Chem., 274, 17267–17274. [DOI] [PubMed] [Google Scholar]
- Thorburn J., Carlson,M., Mansour,S.J., Chien,K.R., Ahn,N.G. and Thorburn,A. (1995) Inhibition of a signaling pathway in cardiac muscle cells by active mitogen-activated protein kinase kinase. Mol. Biol. Cell, 6, 1479–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I., Phalke,G., Salanove,M., Zhang,G., Wang,S., Coletti,D., Onuffer,J., Jin,C. and Conti,M. (2001) Myomegalin: a novel protein of the Golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Westphal R.S., Soderling,S.H., Alto,N.M., Langeberg,L.K. and Scott,J.D. (2000) Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J., 19, 4589–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witczak O., Skalhegg,B.S., Keryer,G., Bornens,M., Tasken,K., Jahnsen,T. and Orstavik,S. (1999) Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J., 18, 1858–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.X. et al. (2000) Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science, 288, 1822–1825. [DOI] [PubMed] [Google Scholar]
- Yang J., Drazba,J.A., Ferguson,D.G. and Bond,M. (1998) A-kinase anchoring protein 100 (AKAP100) is localized in multiple subcellular compartments in the adult rat heart. J. Cell Biol., 142, 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood S.J., Steele,M.R., Scotland,G., Houslay,M.D. and Bolger,G.B. (1999) The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J. Biol. Chem., 274, 14909–14917. [DOI] [PubMed] [Google Scholar]
- Zimmer H.G. (1997) Catecholamine-induced cardiac hypertrophy: significance of proto-oncogene expression. J. Mol. Med., 75, 849–859. [DOI] [PubMed] [Google Scholar]