Abstract
We report the isolation of TRIP-Br1, a transcriptional regulator that interacts with the PHD-bromodomain of co-repressors of Krüppel-associated box (KRAB)-mediated repression, KRIP-1(TIF1β) and TIF1α, as well as the co-activator/adaptor p300/CBP. TRIP-Br1 and the related protein TRIP-Br2 possess transactivation domains. Like MDM2, which has a homologous transactivation domain, TRIP-Br proteins functionally contact DP-1, stimulating E2F-1/DP-1 transcriptional activity. KRIP-1 potentiates TRIP-Br protein co-activation of E2F-1/DP-1. TRIP-Br1 is a component of a multiprotein complex containing E2F-1 and DP-1. Co-expression of the retinoblastoma gene product (RB) abolishes baseline E2F-1/DP-1 transcriptional activity as well as TRIP-Br/KRIP-1 co-activation, both of which are restored by the adenovirus E1A oncoprotein. These features suggest that TRIP-Br proteins function at E2F-responsive promoters to integrate signals provided by PHD- and/or bromodomain- containing transcription factors. TRIP-Br1 is identical to the cyclin-dependent kinase 4 (cdk4)-binding protein p34SEI-1, which renders the activity of cyclin D/cdk4 resistant to the inhibitory effect of p16INK4a during late G1. TRIP-Br1(p34SEI-1) is differentially overexpressed during the G1 and S phases of the cell cycle, consistent with a dual role for TRIP-Br1(p34SEI-1) in the regulation of cell cycle progression through sequential effects on the transcriptional activity of E2F-responsive promoters during G1 and S phases.
Keywords: bromodomain/cell cycle/E2F-1/gene transcription/PHD zinc finger
Introduction
The PHD zinc finger and/or bromodomain are present in a host of important transcriptional regulators (Aasland et al., 1995; Jeanmougin et al., 1997), some of which have been implicated in human disease. KRIP-1 (also known as TIF1β) is a PHD zinc finger- and bromodomain-containing co-repressor of Krüppel-associated box (KRAB)-mediated repression (Kim et al., 1996; Le Douarin et al., 1996; Moosmann et al., 1996). KRIP-1 and the structurally related proteins TIF1α (Le Douarin et al., 1995) and PML (Goddard et al., 1991) are members of the RBCC (RING finger–B boxes–coiled coil) subfamily of the RING finger family of zinc-binding proteins (Saurin et al., 1996). KRIP-1 and TIF1α interact with and function as co-repressors of the KRAB-A repression domain present in the N-terminal regions of approximately one-third of all vertebrate Krüppel-type zinc finger proteins (Witzgall et al., 1994). TIF1α and TIF1β (KRIP-1) have also been proposed to play a dual role in the control of transcription, being involved in both co-repression through their interactions with the KRAB-A domain, and hormone-dependent co-activation through their interactions with various nuclear hormone receptors (Le Douarin et al., 1996; Chang et al., 1998).
The C-termini of KRIP-1, TIF1α and the nuclear body-associated protein SP140 (Bloch et al., 1996) encode a PHD zinc finger followed closely by a bromodomain, which we refer to as the composite PHD-bromodomain. We now report the identification and characterization of a novel family of transcription factors, the TRIP-Br proteins, which possess the unique ability to interact with the PHD zinc fingers and/or bromodomains of a host of proteins involved in transcriptional regulation, including KRIP-1 and p300 (Eckner et al., 1994). TRIP-Br proteins make a direct functional contact with DP-1, stimulating E2F-1/DP-1 transcriptional activity. We suggest a model in which TRIP-Br proteins function at E2F-responsive promoters to integrate signals provided by PHD zinc finger- and/or bromodomain-containing transcription factors.
Results
Isolation of cDNAs whose products interact with the PHD-bromodomain of KRIP-1
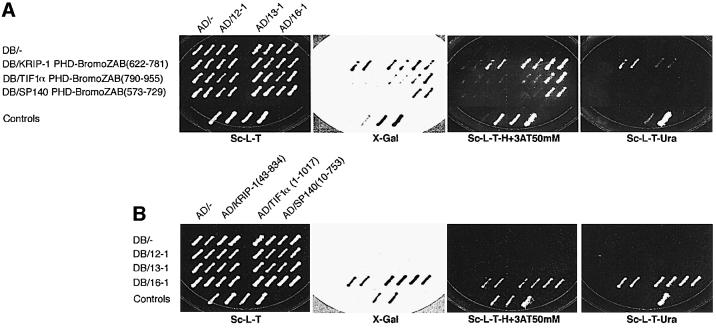
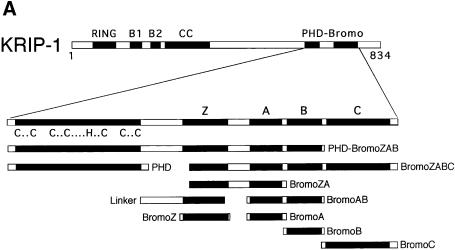
The yeast two-hybrid system (Kim et al., 1996; Vidal et al., 1996) was used to identify proteins that interact with the composite PHD-bromodomain of KRIP-1. The bromodomain has been proposed to encode four conserved subdomains that are predicted to form α-helical structures (helices Z, A, B and C) (Jeanmougin et al., 1997). A fusion between the GAL4 DNA-binding domain (DB) and the KRIP-1 PHD-bromodomain, designated DB/KRIP-1 PHD-BromoZAB (622–781), was used as a bait to screen a library of whole mouse fetus (day 12–13) cDNAs fused to the GAL4 activation domain (AD). Approximately 0.5 × 106 yeast transformants were screened. Three putative interactors were identified, which we designated 12-1, 13-1 and 16-1.
Only 16-1 also interacted with the homologous PHD-bromodomain sequences of TIF1α and SP140 (Figure 1A). Similarly, only DB/16-1 also interacted with full-length AD hybrid proteins of KRIP-1, TIF1α and SP140 (Figure 1B). Next, we tested the ability of the putative PHD-bromodomain interactors to recognize the individual PHD zinc finger or bromodomain of KRIP-1. Regions of the KRIP-1 PHD-bromodomain included in each DB hybrid construct are shown in Figure 2A. DB/16-1 interacted with AD hybrid proteins containing the PHD zinc finger domains of the structurally related proteins KRIP-1, TIF1α and SP140, as well as the PHD domains of structurally unrelated proteins NF-X1 (Aasland et al., 1995) and p300 (Eckner et al., 1994) (Figure 2B). Similarly, AD/16-1 interacted with DB hybrid proteins containing the BromoZAB domain of the structurally related proteins KRIP-1, TIF1α and SP140, as well as that of p300 (Figure 2C). In contrast, 12-1 and 13-1 failed to interact with either the individual PHD zinc finger (Figure 2B) or BromoZAB domain of KRIP-1 (Figure 2C). Indeed, 12-1 and 13-1 exhibited an unexpected specificity for interaction with the isolated TIF1α BromoZAB domain, while 12-1 also interacted with the p300 BromoZAB domain (Figure 2C). Thus, the presence of the PHD zinc finger in the TIF1α PHD-BromoZAB domain appears to inhibit 12-1 and 13-1 interactions with the TIF1α BromoZAB domain (compare Figures 1A and 2C). These results suggest that while 16-1 is a ‘general’ interactor of both the PHD zinc finger and the bromodomain, 12-1 and 13-1 are likely to interact with specific bromodomains but not PHD zinc fingers.
Fig. 1. Yeast two-hybrid cloning of interactors to the PHD-bromodomain of KRIP-1. Two independent yeast colonies co-expressing the indicated DB and AD hybrid proteins were grown on synthetic complete medium lacking leucine and tryptophan (Sc-L-T), and subsequently tested for activation of the LacZ (X-gal filter assay), HIS3 and URA3 reporters by replica-plating onto appropriate drop-out medium Sc-L-T, Sc-L-T-H+3AT50mM (supplemented with 50 mM 3-aminotriazole) and Sc-L-T-Ura, respectively. A series of four control patches co-expressing each of the following hybrid protein pairs was included on each plate (from left to right): (i) DB/– + AD/–, negative interaction; (ii) DB/RB + AD/E2F-1, weak positive interaction; (iii) DB/Fos + AD/Jun, strong positive interaction; and (iv) Gal4 (full length) + AD/–, strong positive. Expression of hybrid proteins was confirmed by western blot analysis (data not shown). (A) The ability of each putative interactor (12-1, 13-1 or 16-1) to interact with the composite PHD-bromodomain of KRIP-1, TIF1α and SP140 in the yeast two-hybrid system is shown. Amino acid residues corresponding to these protein motifs are shown in parentheses. (B) The ability of each putative interactor to interact with full-length KRIP-1, TIF1α and SP140 in the yeast two-hybrid system is shown.
Fig. 2. Yeast two-hybrid analyses of the role of the isolated PHD zinc finger and isolated bromodomain in protein–protein interactions. (A) Schematic of a series of DB hybrid constructs encoding subregions of the KRIP-1 PHD-bromodomain. The ability of each putative interactor to interact with the PHD zinc fingers (B), bromodomains (C) or individual bromodomain α-helices (D) of both related and unrelated proteins is shown. Methods and controls are as for Figure 1. DB/RB (378–793), encoding the retinoblastoma gene product ‘pocket domain’, was also included as a control for non-specific interactions.
16-1 interacted with the ‘full’ bromodomain DB/KRIP-1 BromoZABC hybrid protein, containing all four putative α-helices of the bromodomain (Figure 2D). The presence of either α-helix A or C alone appeared to be sufficient for interaction. Helix Z has been hypothesized to define distinct subfamilies of bromodomain-containing proteins, perhaps by specifying different protein–protein interactions (Jeanmougin et al., 1997). Although AD/16-1 failed to demonstrate bona fide interaction with DB/KRIP-1 BromoZ, it interacted strongly with the DB/TIF1α BromoZ, DB/SP140 BromoZ and DB/p300 BromoZ proteins, resulting in activation of all three reporter genes (Figure 2D, lower panel). Like helices A and C of KRIP-1, helix Z of TIF1α, SP140 and p300 appears to be sufficient for interaction with 16-1. The failure of the KRIP-1 helix Z to interact with 16-1 despite significant primary sequence homology to the TIF1α and SP140 helix Z, and the ability of the unrelated p300 helix Z to interact with 16-1, suggests that the ability of certain bromodomain helix Z sequences to form specific α-helical secondary structures determines their ability to interact with 16-1.
The fact that 16-1 interacts with PHD zinc fingers and bromodomains of related and unrelated primary sequence suggests that 16-1 binds to protein motifs defined by secondary structural predictions, rather than by primary sequence homology. In this report we focus on the further characterization of 16-1.
Domain structure of the TRIP-Br protein family
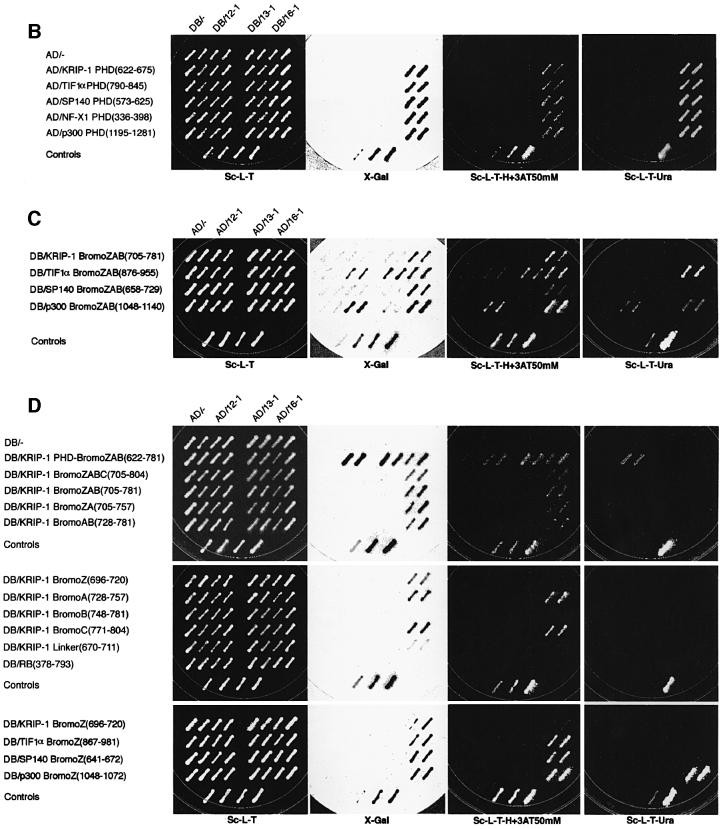
The open reading frame (ORF) of the 701-nucleotide 16-1 cDNA is fused in-frame with the N-terminal GAL4-AD sequence. The first methionine at nucleotide 34 is encoded within the context of an imperfect Kozak sequence (TGTAGGATGC). The 16-1 ORF ends at an endogenous SpeI restriction site, consistent with truncation of the 3′ end during cloning of the cDNA library into the EcoRI and SpeI sites of the AD hybrid vector (Kim et al., 1996). A BLAST search of the EST database using the entire 16-1 sequence identified a likely candidate for a full-length mouse cDNA (DDBJ/EMBL/GenBank accession No. W82777), which we have designated TRIP-Br1 (transcriptional regulator interacting with the PHD-bromodomain). During review of this manuscript, the isolation and functional characterization of a human cyclin-dependent kinase 4 (cdk4)-binding protein designated SEI-1 (identical to TRIP-Br1) was reported (Sugimoto et al., 1999).
The murine TRIP-Br1 cDNA (mTRIP-Br1) encodes an ORF of 236 amino acids, extending the 16-1 sequence by 14 additional C-terminal residues. This ORF is predicted to encode a protein of 25.1 kDa. A single non-conserved amino acid difference from 16-1 [Glu (GAG)→Lys (AAG)] occurs at amino acid 12. The latter is most likely to represent an allelic polymorphism, since sequence analysis of four independent cDNA clones confirmed this difference (data not shown). mTRIP-Br1 represents a full-length cDNA since the putative initiator methionine at nucleotide 173 is the first methionine occurring after an in-frame stop codon (TAG) at nucleotide 92. A full-length human orthologous cDNA obtained from the EST database (DDBJ/EMBL/GenBank accession No. W52056), which we designate hTRIP-Br1, also encodes an ORF of 236 amino acids whose sequence is 86% identical to that of mTRIP-Br1.
A BLAST search also identified KIAA0127 (Nagase et al., 1995; DDBJ/EMBL/GenBank accession No. D50917), which encodes a 314 amino acid ORF and shares three large regions of homology with TRIP-Br1 (see Figure 3A). The ORF of KIAA0127, which we have redesignated hTRIP-Br2, is predicted to encode a protein of 33.9 kDa. Comparison of the sequence of KIAA0127 (TRIP-Br2) and a partial mouse cDNA obtained from the EST database (DDBJ/EMBL/GenBank accession No. AA183772) revealed 81% identity.
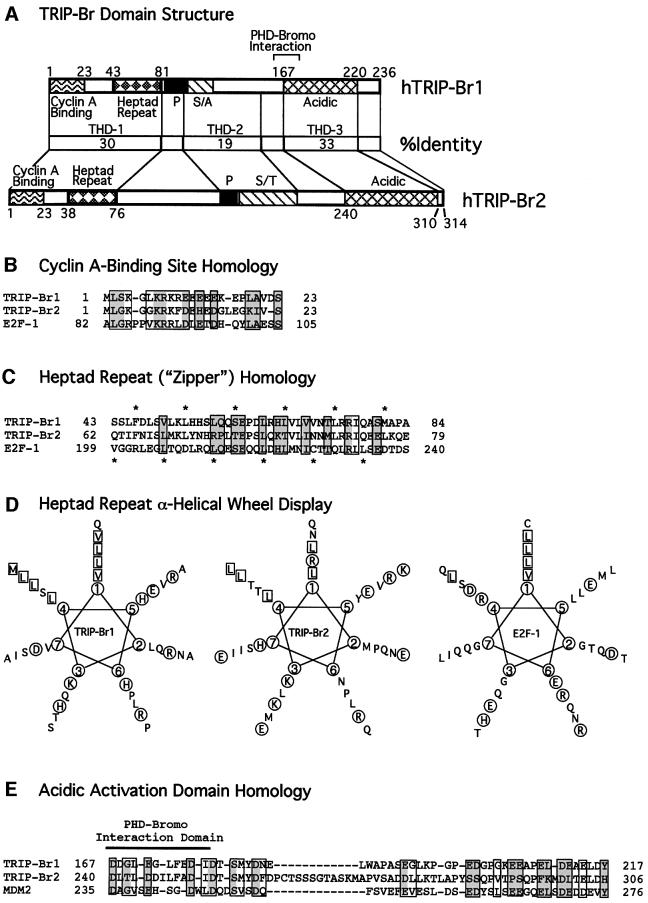
Fig. 3. Human TRIP-Br domains with homology to E2F-1 and MDM2. (A) Comparison of the domain structure and amino acid identity of the TRIP-Br proteins. The minimal region of TRIP-Br1 (aa 161–178) required for interaction with the PHD-bromodomain is indicated by brackets. P, proline-rich; S/A, serine/alanine-rich; S/T, serine/threonine-rich. (B) N-terminal sequences of the human TRIP-Br proteins are shown aligned with the cyclin A-binding domain of E2F-1. Boxed residues indicate amino acids that are similar between E2F-1 and at least one of the TRIP-Br proteins, and shaded boxes indicate residues that are identical between E2F-1 and at least one of the TRIP-Br proteins. (C) N-terminal sequences of the human TRIP-Br proteins are shown aligned with the heptad repeat region of E2F-1. Amino acids are boxed and shaded as in (B). Asterisks shown below indicate the heptad periodicity characteristic of E2F-1 and DP-1, which is also shared by the TRIP-Br proteins. Asterisks shown above indicate a novel heptad periodicity, which occurs only in the TRIP-Br proteins. (D) Amino acids in the putative heptad repeat regions of the human TRIP-Br proteins and E2F-1 are displayed in an α-helical wheel. Hydrophobic residues that occur in positions 1 and 4, corresponding to the two heptad periodicities shown in (C), are boxed. Charged residues are circled. (E) C-terminal sequences of the human TRIP-Br proteins are shown aligned with the MDM2 transactivation domain. Amino acids are boxed and shaded as in (B).
We identified three potential functional protein domains shared by TRIP-Br1 and TRIP-Br2 (Figure 3A). THD-1 (TRIP homology domain-1) is 30% identical between hTRIP-Br1 and hTRIP-Br2, and contains two domains that are similar to those found in members of the E2F family of transcriptional activators: a cyclin A-binding domain and a hydrophobic heptad repeat. Figure 3B shows an alignment of residues 1–23 of hTRIP-Br1 and hTRIP-Br2 with the cyclin A-binding domain of E2F-1. Residues 67–108 of E2F-1 have been shown to be necessary and sufficient for functional interaction with cyclin A/cyclin-dependent kinase 2 (cdk2) (Krek et al., 1994). Although the putative cyclin A-binding domains of the TRIP-Br proteins do not have the conserved HXL motif, which has been reported to be required to bind a hydrophobic patch in cyclin A (Schulman et al., 1998), glutathione S-transferase (GST) binding studies and co-immunoprecipitation studies in mammalian cells demonstrate that the TRIP-Br proteins interact with cyclin A both in vitro and in vivo (our unpublished data).
The heptad hydrophobic repeat (zipper) of THD-1 resembles a similar sequence found in all members of the E2F and DP families (Lam and La Thangue, 1994) (Figure 3C), although the heptad periodicity of TRIP-Br proteins is novel (Figure 3C). When displayed as α-helical wheels, the putative heptad repeat domains of hTRIP-Br1 and hTRIP-Br2, like those of E2F-1 (Kaelin et al., 1992) and DP-1 (Girling et al., 1993), have the potential to form amphipathic α-helices (Figure 3D). The TRIP-Br2 heptad periodicity at position 1 (Figure 3D) is disrupted by an arginine residue at amino acid 76. Nevertheless, the proposed heptad repeats mediate both homodimeric and heterodimeric interactions between TRIP-Br1 and TRIP- Br2 (our unpublished data).
THD-2 is 19% identical between hTRIP-Br1 and hTRIP-Br2 and is moderately rich in proline, acidic, serine and threonine residues—hallmarks of the PEST sequence frequently found in proteins with short half-lives. THD-3, a sequence rich in acidic amino acids, is the region with the highest degree of homology between hTRIP-Br1 and hTRIP-Br2 (Figure 3A and E). This region of hTRIP-Br1 also exhibits a substantial degree of conservation (30% identity and 45% similarity) with the MDM2 transactivation domain (Figure 3E). MDM2 is a p53-associated oncoprotein, which has been shown to both inhibit p53-mediated transactivation (Momand et al., 1992) and stimulate E2F-1/DP-1-mediated transactivation (Martin et al., 1995). The hTRIP-Br2 acidic region is also similar to the MDM2 transactivation domain but contains an additional central spacer of 12 amino acids.
Deletion analysis revealed that amino acid residues 161–178 at the beginning of the acidic THD-3 domain (Figure 3A and E) are critical for interaction with the KRIP-1 PHD-bromodomain. This region is the most highly conserved among all TRIP-Br proteins and is predicted to encode an α-helical structure. These residues are also required for interaction with the PHD-bromodomain of TIF1α and SP140 (data not shown). Despite the significant homology between the MDM2 and TRIP-Br transactivation domains in the region involved in interaction with the PHD-bromodomain (Figure 3E), the MDM2 transactivation domain did not interact with the KRIP-1 PHD-bromodomain (data not shown).
TRIP-Br gene expression and evolutionary conservation
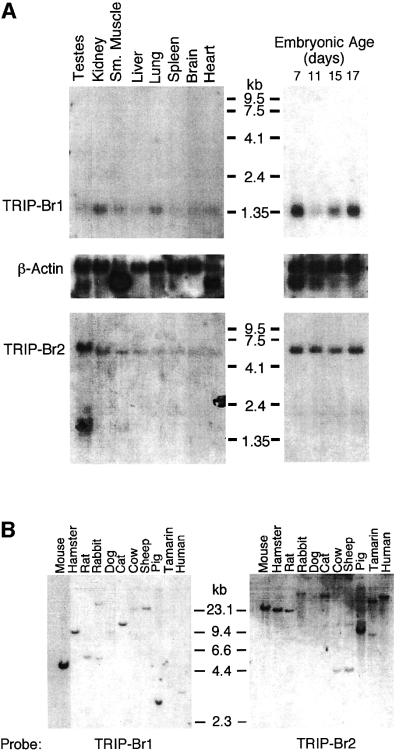
mTRIP-Br1 and hTRIP-Br2 cDNA probes detected transcript sizes of ∼1.35 and 6 kb, respectively (Figure 4A). The mTRIP-Br1 transcript was expressed at low levels in all tissues examined, with slightly higher expression in kidney and lung. A BLAST search of the EST database revealed that the hTRIP-Br1 gene is also expressed in human colon, foreskin, heart, kidney, lung, ovary, pancreas, prostate and uterus. Hybridization to poly(A)+ RNA isolated from whole mouse embryonic fetus showed high mTRIP-Br1 gene expression on day 7, subsequent down-regulation of expression by day 11, followed by progressive increases on days 15 and 17. In contrast, mTRIP-Br2 gene expression levels were similar in all tissues and developmental stages examined. KIAA0127 (hTRIP-Br2) mRNA has also been reported to be ubiquitously expressed at low levels in human tissues, with slightly higher levels in thymus and peripheral blood leukocytes (Nagase et al., 1995). In contrast, SEI-1 (hTRIP-Br1) mRNA has been reported to be absent in thymus (Sugimoto et al., 1999).
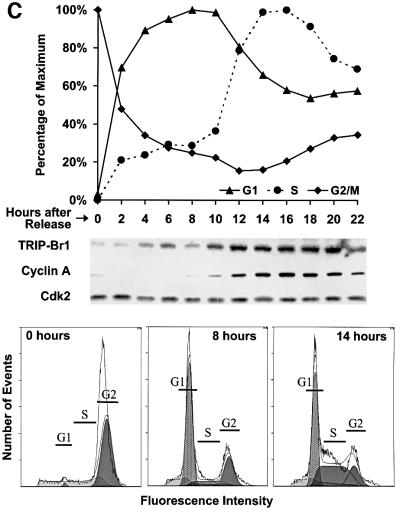
Fig. 4. TRIP-Br gene expression and evolutionary conservation. (A) Mouse multiple tissue and developmental northern blots were sequentially probed with an mTRIP-Br1 probe (entire 701-bp 16-1 insert containing 33 bp of 5′-UTR and encoding amino acids 1–222), an hTRIP-Br2 probe (419-bp insert encoding amino acids 1–136) and a β-actin probe. Each lane contains 2 µg of poly(A)+ RNA isolated from adult tissues or from whole fetuses on the indicated days of embryonic development. Size markers are indicated in kilobases. (B) A mammalian zoo blot of EcoRI-digested genomic DNA was sequentially probed with the same TRIP-Br1- and TRIP-Br2-specific probes as in (A). Each lane contains 25 µg of DNA from the indicated species. (C) hTRIP-Br1 protein expression and cell cycle distribution were monitored in U2OS cells (see Materials and methods) following release from nocodazole block. The fraction of cells in G1, S and G2/M phase are expressed as a percentage of the maximum. Histograms of FACS analysis at 0, 8 and 14 h are shown below.
Southern blot analysis of EcoRI-digested genomic DNA using mTRIP-Br1 and hTRIP-Br2 cDNA probes detected hybridization signals in all species examined (Figure 4B). A human TRIP-Br1 STS marker (sts-N31928) has been mapped to chromosome 19q13, between the markers D19S421 and D19S223 (Deloukas et al., 1998), within several centimorgans centromeric to the AKT2 oncogene, which is frequently amplified in ovarian carcinomas (Cheng et al., 1992). A human TRIP-Br2 STS marker (sts-G44420) has been mapped to chromosome 2p13, between the markers D2S337 and D2S147 (Deloukas et al., 1998).
Synchronized U2OS2 cells, released from nocodazole-induced M phase block, exhibited a biphasic expression pattern of TRIP-Br1, with increased expression in mid-G1 (4–6 h), transient decreased expression during late G1 (8 h), and a high expression level upon S-phase entry (10 h, coincident with the first appearance of cyclin A protein), which persists throughout S phase (Figure 4C).
TRIP-Br proteins interact with the PHD-bromodomain of KRIP-1
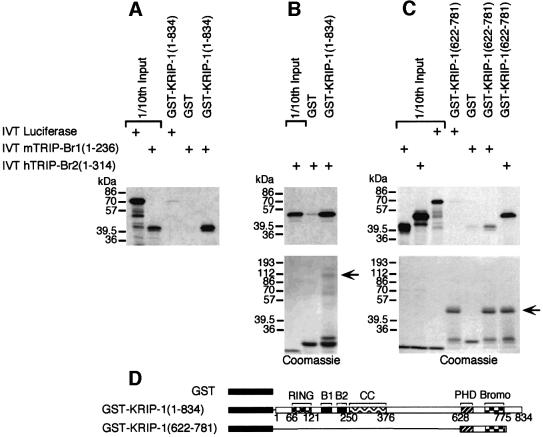
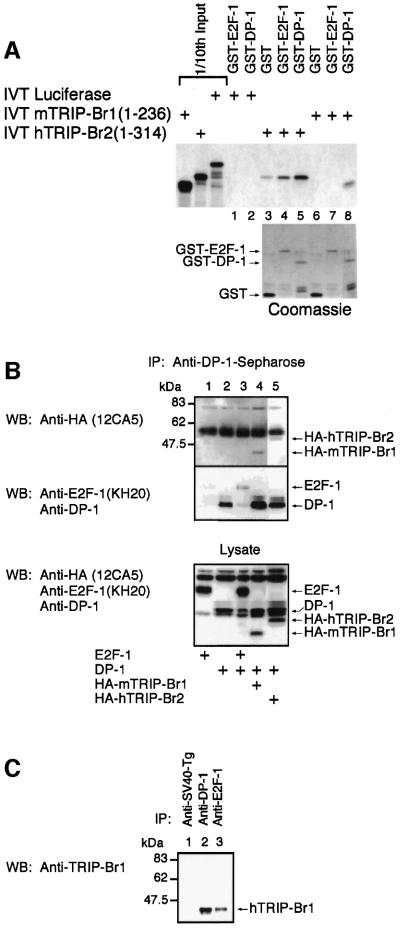
The mTRIP-Br1 in vitro translation (IVT) product migrates on SDS–PAGE as a 39 and 40 kDa doublet (Figure 5A), while the hTRIP-Br2 IVT product migrates as a single 50 kDa band (Figure 5B). Approximately 20% of input mTRIP-Br1 and hTRIP-Br2 proteins were bound by GST–KRIP-1 (1–834), but not GST alone (Figure 5A and B). GST–KRIP-1 (622–781), encoding the KRIP-1 PHD-bromodomain (Figure 5D), interacts with mTRIP-Br1 or hTRIP-Br2 IVT products, but not the control luciferase IVT product (Figure 5C). These results indicate that a direct interaction between KRIP-1 and the TRIP-Br proteins is mediated at least in part by the PHD-bromodomain of KRIP-1.
Fig. 5. Specific in vitro interaction of TRIP-Br1 and TRIP-Br2 proteins with full-length KRIP-1 and the PHD-bromodomain. (A–C) In vitro-translated (IVT), [35S]methionine-labeled full-length mTRIP-Br1 and hTRIP-Br2 (2.5 µl) were incubated with 2 µg of various GST fusion proteins immobilized on glutathione–agarose beads. Bound proteins were eluted and resolved by SDS–PAGE. Gels were stained with Coomassie. Arrows indicate the position of GST fusion proteins. One-tenth of input IVT products were included in separate lanes. (D) Schematic of the GST fusion proteins used in the above in vitro binding studies.
TRIP-Br proteins possess potent acidic transactivation domains
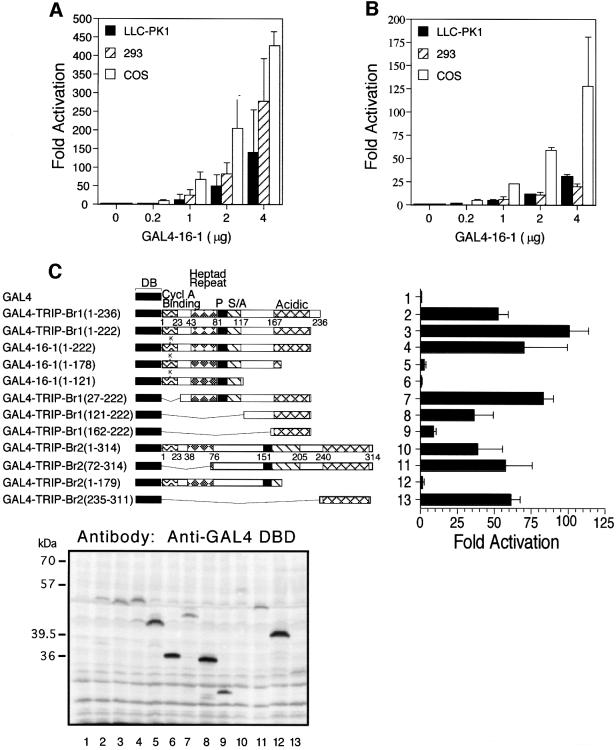
A GAL4–16-1 fusion protein stimulates both basal and enhancer-activated transcription, when recruited to a heterologous promoter bearing GAL4 DNA-binding sites, in both oncoprotein-transformed cells (293 and COS cells) and spontaneously immortalized LLC-PK1 porcine epithelial cells (Figure 6A and B). The transactivation domains of mTRIP-Br1 and hTRIP-Br2 were mapped by deletion analysis (Figure 6C). Both full-length GAL4–TRIP-Br1 (1–236) and GAL4–TRIP-Br2 (1–314) stimulated transcription by ∼25% of that observed for the acidic transactivation domain of the herpes simplex virus VP16 protein (data not shown). Deletion of the terminal 14 amino acids of TRIP-Br1 [construct GAL4–TRIP-Br1 (1–222)], which contain a putative class II (PPXPXR) SH3 (Src-homology-3) binding site (Cohen et al., 1995), resulted in a significant increase in transcriptional activity, even when adjusted for differences in protein expression (Figure 6C, lower panel). A single non-conserved amino acid substitution (E12K) in the putative cyclin A-binding domain of GAL4–16-1 (1–222) reduced transcriptional activation (Figure 6C, compare lanes 3 and 4).
Fig. 6. Transcriptional activation by TRIP-Br proteins recruited to a heterologous promoter. (A) Cell lines were transiently transfected with the indicated amounts of the SV40 expression vector pBXG1/16-1 encoding the entire 16-1 insert fused in-frame with GAL4-DB, 1.5 µg of the luciferase minimal reporter plasmid pG5-GL3 bearing five copies of the GAL4 DNA-binding sequence followed by the E1b TATA box, and 0.1 µg of pCMV/β-galactosidase expression vector. Fold activation refers to luciferase activity normalized to β-galactosidase activity, and is expressed relative to the activity observed with transfection of the reporter alone. Values represent the average ± standard deviation of three or four independent experiments. (B) Transient transfections were performed as in (A) except that the upstream SV40 enhancer reporter plasmid pG5 (SV)-GL3 was used in 293 cells, while the downstream SV40 enhancer reporter plasmid pG5-GL3 (SV) was used in COS and LLC-PK1. (C) Schematic of various GAL4/TRIP-Br deletion mutants (left) and their respective abilities to activate a heterologous minimal promoter (right) are shown. Transient transfections were performed as in (A) with 0.5 µg of each expression construct. Western blot analysis (below) of 50 µg of total cell lysate from each transfection was performed with anti-GAL4 DB mouse monoclonal antibody (Santa Cruz). The GAL4/16-1 (1–222) fusion protein (lane 4) migrates more slowly than the GAL4/TRIP-Br1 (1–222) fusion protein (lane 3), due to the presence of a ‘linker’ (LRSDWPRVICR) encoded by the short 5′-UTR of the original 16-1 clone.
Removal of the acidic region [construct GAL4–16-1 (1–178)] eliminated nearly all activation (Figure 6C). Further C-terminal deletion to residue 121 completely abolished activation. Progressive N-terminal deletion of mTRIP-Br1 residues 27–161 resulted in a progressive decline in transcriptional activity, although GAL4– TRIP-Br1 (162–222) retained ∼20% activity compared with the full-length protein. Amino acids 27–161 of mTRIP-Br1 may therefore either be directly involved in activation or favorably affect the structure and activation function of the C-terminal acidic region. A similar deletion analysis of hTRIP-Br2 also implicated the acidic region (residues 235–311) as a transactivation domain.
The TRIP-Br proteins co-activate E2F-1/DP-1
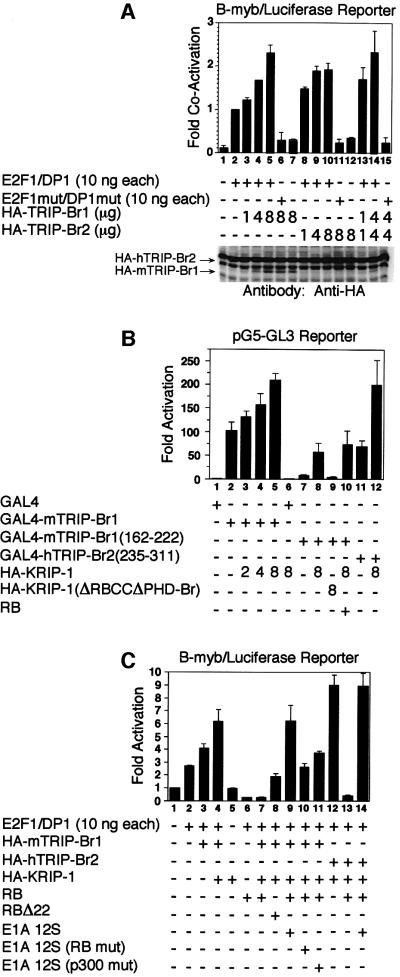
Approximately 10-fold activation of a native B-myb promoter region containing a single E2F-1-binding site with dyad symmetry (Lam and Watson, 1993) was observed when SAOS2 cells were transfected with 10 ng each of wild-type pCMV/E2F-1 and pCMV/DP-1 (Figure 7A, lanes 1 and 2). Co-expression of increasing amounts of hemagglutinin (HA)-tagged TRIP-Br proteins resulted in further stimulation of transcriptional activity (Figure 7A, lanes 3–5 and 8–10). The magnitude of the co-activation by TRIP-Br proteins (∼2.5-fold) is similar to that observed for MDM2 co-activation of E2F-1/DP-1 in SAOS2 cells (Martin et al., 1995). This effect was not observed in the absence of exogenous E2F-1 and DP-1 or when DNA binding mutants of E2F-1 and DP-1 were co-expressed (Figure 7A, lanes 6, 7, 11, 12 and 15). Co-stimulatory effects of HA-mTRIP-Br1 and HA-hTRIP-Br2 were additive (Figure 7A, compare lanes 5 and 14). Western blot analysis confirmed expression of HA-mTRIP-Br1 and HA-hTRIP-Br2, although the latter is partially obscured by a non-specific band (Figure 7A, lower panel, uppermost band), which appears as an increasingly wider band with co-expression of increasing amounts of HA-hTRIP-Br2.
Fig. 7. Cooperative stimulation of E2F-1/DP-1 transcriptional activity by KRIP-1 and the TRIP-Br proteins. (A) SAOS2 cells were transiently transfected with the indicated expression constructs, 3 µg of the B-myb luciferase reporter plasmid pGL2– (–536), and 0.5 µg of pCMV/β-galactosidase expression vector. Fold co-activation refers to luciferase activity normalized to β-galactosidase activity, and is expressed relative to the activity observed with expression of wild-type E2F-1 and DP-1 alone. Values represent the average ± standard deviation of three or four independent experiments. Western blot analysis of 50 µg of total cell lysate from each transfection was performed with the anti-HA mouse monoclonal antibody 12CA5. Arrows mark the position of HA-mTRIP-Br1 and HA-hTRIP-Br2. (B) SAOS2 cells were transfected with 10 ng of the indicated GAL4 expression contructs and various combinations of HA-KRIP-1, HA-KRIP-1 (ΔRBCCΔPHD-Br) and 0.5 µg of RB expression constructs, along with 3 µg of the luciferase minimal reporter pG5-GL3. Fold activation refers to luciferase activity normalized to β-galactosidase activity, and is expressed relative to the activity observed with transfection of the reporter alone. Values represent the average ± standard deviation of two independent experiments. (C) SAOS2 cells were transfected as in (A) with various combinations of 10 ng each of pCMV/E2F-1 and pCMV/DP-1, 4 µg of pSV/HA-mTRIP-Br1, 4 µg of pSV/HA-hTRIP-Br2, 4 µg of pSV/HA-KRIP-1, 0.5 µg of pCMV/RB, 0.5 µg of pCMV/RBΔ22, 0.5 µg of pRSV/E1A 12S, 0.5 µg of pRSV/E1A 12S (RB mut) and 0.5 µg of pRSV/E1A 12S (p300 mut), along with 3 µg of the B-myb luciferase reporter plasmid pGL2– (–536).
KRIP-1 potentiates the ability of TRIP-Br proteins to co-activate E2F-1/DP-1
Co-expression of increasing amounts of HA-tagged KRIP-1 led to dose-dependent potentiation of transcriptional activation by full-length GAL4–mTRIP-Br1 (Figure 7B, lanes 1–6), and GAL4–mTRIP-Br1 (162–222) and GAL4– hTRIP-Br2 (235–311), encoding only the acidic activation domains of mTRIP-Br1 and hTRIP-Br2, respectively (Figure 7B, lanes 7, 8, 11 and 12). Co-activation was not observed with HA-KRIP-1 (ΔRBCCΔPHD-Br), an HA-tagged KRIP-1 mutant in which both the RBCC tripartite motif and the PHD-bromodomain have been deleted (Figure 7B, lane 9). The α-helix Z of both the KRIP-1 and TIF1α (but not SP140) bromodomains encodes the LXCXE motif important for binding to the ‘pocket’ region of the RB protein (Defeo-Jones et al., 1991). However, RB had no effect on KRIP-1 co-activation of GAL4– mTRIP-Br1 (162–222) (Figure 7B, compare lanes 8 and 10). In addition, GST–RB (379–928), encoding the RB pocket region, did not bind full-length mTRIP-Br1 in vitro (data not shown).
KRIP-1 potentiated the ability of TRIP-Br proteins to stimulate E2F-1/DP-1 transactivation function in the context of the B-myb promoter (Figure 7C, lanes 1–5). As before, overexpression of HA-mTRIP-Br1 resulted in co-activation of E2F-1/DP-1 (Figure 7C, lanes 1–3), although the level of co-activation was lower than previously observed under similar conditions (Figure 5A, lane 2). This is likely to be due to a decline over time in the transfection efficiency achieved with cells in continuous passage. Nonetheless, this level of transcriptional activity was further augmented by the overexpression of HA-KRIP-1 (Figure 7C, lane 4). As expected, in SAOS2 cells that lack endogenous functional RB, the overexpression of RB was associated with repression of E2F-1/DP-1 transcriptional activity (Figure 7C, compare lanes 2 and 6). Furthermore, overexpression of RB also resulted in potent repression of HA-mTRIP-Br1 and HA-KRIP-1 co-activation (Figure 7C, compare lanes 4 and 7). Overexpression of RBΔ22, an RB mutant unable to bind in vitro to E2F-1/DP-1 or to repress its transactivation function, was less able to repress the co-activation function of HA-mTRIP-Br1 and HA-KRIP-1 (Figure 7C, compare lanes 7 and 8). The repression activity of wild-type RB was completely reversed by co-expression of the E1A (12S) oncoprotein (Figure 7C, compare lanes 7 and 9). Likewise, RB completely repressed HA-hTRIP-Br2 and HA-KRIP-1 co-activation functions, which were restored by co-expression of E1A (Figure 7C, lanes 12–14). The ability of E1A to fully activate E2F-1/DP-1 is dependent on its ability to bind both RB and p300 (Trouche et al., 1996). E1A mutants defective in their ability to bind only RB or p300 are only able to partially restore HA-mTRIP-Br1 and HA-KRIP-1 co-activation function (Figure 7C, compare lanes 9–11).
The TRIP-Br proteins interact with DP-1
TRIP-Br1 specifically bound GST–DP-1 but not GST–E2F-1 (Figure 8A, lanes 6–8). Although some non-specific background binding of hTRIP-Br2 to GST was evident, binding to GST–DP-1 (and to a lesser extent GST–E2F-1) was observed to be significantly greater (Figure 8A, lanes 3–5). Neither GST–E2F-1 nor GST– DP-1 bound the IVT product of the luciferase gene (Figure 8A, lanes 1 and 2).
Fig. 8. The TRIP-Br proteins interact with DP-1 in vitro and in vivo. (A) The ability of the IVT products of the TRIP-Br proteins to bind GST–DP-1 specifically is shown. GST binding studies were performed as described in Figure 5. Arrows mark the position of various GST fusion proteins. (B) The ability of TRIP-Br proteins to associate with DP-1 in vivo is demonstrated. SAOS2 cells were transfected with various combinations of expression vectors and lysed as described in Materials and methods. An aliquot of whole-cell lysate was analyzed by western blot analysis to confirm protein expression (lower panel). Lysates were immunoprecipitated with anti-DP-1–Sepharose, and analyzed sequentially by western blot analysis with anti-HA (12CA5) and anti-DP-1/anti-E2F-1 (KH20) antibodies (upper panels). Arrows mark the position of expressed proteins. ‘IP’ and ‘WB’ indicate antibodies used for immunoprecipitation and western blot analysis, respectively. (C) Whole-cell lysates from human ML-1 cells were immunoprecipitated with anti-DP-1, anti-E2F-1 (KH20) or control anti-SV40-Tg monoclonal antibodies. The immunoprecipitates were analyzed by western blot analysis with a rabbit anti-TRIP-Br1 polyclonal antibody.
DP-1 (Figure 8B, lanes 2–5) co-immunoprecipitates with E2F-1 (Figure 8B, lane 3), HA-mTRIP-Br1 (Figure 8B, lane 4) or HA-hTRIP-Br2 (Figure 8B, lane 5). Endogenous hTRIP-Br1 was also detected in immunoprecipitates formed from whole-cell lysates of untransfected ML-1 cells using either anti-DP-1 or anti-E2F-1 (but not control anti-SV40-Tg) monoclonal antibodies (Figure 8C).
Discussion
TRIP-Br1 and TRIP-Br2 engage in a novel interaction with the PHD zinc fingers and/or bromodomains of a number of important transcriptional regulators. TRIP-Br proteins also bind directly to DP-1, resulting in stimulation of E2F-1/DP-1 transcriptional activity in a manner that is regulated by the RB and E1A oncoproteins. Endogenous hTRIP-Br1 is differentially expressed during the cell cycle and exists in a multiprotein complex with endogenous E2F-1 and DP-1. We propose that TRIP-Br proteins are transcriptional ‘integrators’ that couple regulatory signals provided by PHD zinc finger- and/or bromodomain-containing transcription factors with the well defined transcriptional and cell cycle regulatory pathway mediated by E2F-1/DP-1 and RB.
The bromodomain of the histone acetyltransferase (HAT) co-activator P/CAF (p300/CBP-associated factor) interacts specifically with acetylated lysine (Dhalluin et al., 1999), while the bromodomain of the yeast co-activator Gcn5p mediates specific protein–protein interactions with the N-terminal tails of histones H3 and H4 in vitro (Ornaghi et al., 1999). These data provide a functional link between HAT activity of co-activators and chromatin remodeling and gene activation. Our data suggest that bromodomains of KRIP-1, TIF1α, SP140 and p300 also mediate additional protein–protein interactions that do not involve acetylated lysine residues (Figure 2C and D).
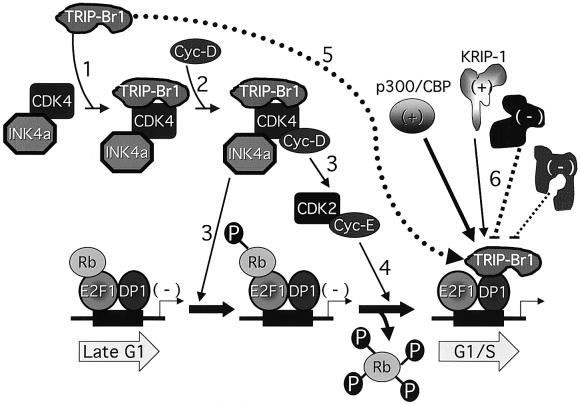
p34SEI-1 makes functional contact with cdk4 through a region distinct from the PHD-bromodomain interaction domain of TRIP-Br1(p34SEI-1), rendering the cyclin D– cdk4 complex resistant to the inhibitory effect of p16INK4a. The interaction of TRIP-Br proteins with DP-1, and their ability to stimulate E2F-1/DP-1 transcriptional activity in a manner that is regulated by RB and E1A, suggest that TRIP-Br proteins also function as co-activators of E2F-1/DP-1. Both U2OS cells released from M-phase block (Figure 4C) and TIG3 human primary fibroblasts released from serum starvation (Sugimoto et al., 1999) exhibit a biphasic expression pattern of TRIP-Br1(p34SEI-1), with increased expression in mid-G1 (at the peak of G1 in U2OS cells and 2 h after release from G0 in TIG3 fibroblasts), transient decreased expression during late G1, and high expression levels upon entry and throughout S phase. TRIP-Br1(p34SEI-1) may thus play a dual role in the regulation of cell cycle progression through sequential effects on the transcriptional activity of E2F-responsive promoters during G1 and S phases (Figure 9).
Fig. 9. Proposed model for the dual function of TRIP-Br1(p34SEI-1) in the regulation of cell cycle progression. p16INK4a inhibits CDK4 activity, rendering the DNA-bound RB/E2F-1/DP-1 transcriptional complex inactive. In response to mitogenic stimulation and the presence of TRIP-Br1(p34SEI-1) (step 1), cyclin D is able to bind CDK4 simultaneously with p16INK4a (step 2) to form a functionally active quaternary complex that is resistant to the inhibitory effects of p16INK4a (Sugimoto et al., 1999). This complex contributes both to cyclin E–CDK2 activation and the generation of hypophosphorylated Rb (step 3). The hypophosphorylated RB bound to transcriptionally inactive E2F-1/DP-1 complexes serves as a target for hyperphosphorylation by functionally activated cyclin E–CDK2 (step 4), resulting in the dissociation of RB and release of cells from the late G1 checkpoint. Upon Rb dissociation, TRIP-Br1(p34SEI-1) is recruited to the E2F-1/DP-1 complexes of E2F-responsive promoters through physical association with DP-1 (step 5), resulting in the assembly of an E2F-1/DP-1/TRIP-Br1 ternary complex. PHD zinc finger- and/or bromodomain-containing proteins such as p300/CBP and KRIP-1, present in differing amounts and possessing differing binding affinities (thick and thin straight arrows), compete for binding to TRIP-Br proteins and confer positive (+) or negative (–) regulatory signals to E2F-1/DP-1 transcription complexes assembled on E2F-responsive promoters (step 6). An ‘integrated’ transcriptional read-out is achieved by effects on the basal transcriptional machinery.
This proposed model is functionally analogous to that proposed for the p53-associated protein MDM2, which releases cells from the proliferative block at G1–S imposed by p53-mediated transactivation (Momand et al., 1992) while augmenting S-phase entry by stimulating E2F-1/DP-1 transactivation (Martin et al., 1995). Their ability to interact in vitro with cyclin A suggests that the TRIP-Br proteins may also serve as substrates for phosphorylation by specific cyclin–cdk complexes. The ability of RB to completely inhibit the transcriptional activity of a complex containing E2F-1, DP-1, TRIP-Br1 and KRIP-1 suggests that the TRIP-Br1 and KRIP-1 co-activation functions require that the E2F-1 transactivation domain is not bound by RB.
Notably, both p300/CBP and hbrm possess bromodomains, while p300/CBP also possesses an atypical PHD zinc finger. RB cooperates with hbrm to repress the transcriptional activity of E2F/DP-1 (Trouche et al., 1997). Both the bromodomain and the PHD zinc finger of p300 interact with TRIP-Br1 in the yeast two-hybrid system; in addition, full-length p300 interacts in vitro with sequences in the TRIP-Br1 activation domain (our unpublished data). TRIP-Br proteins may bind to these regulatory proteins in vivo through PHD zinc finger and/or bromodomain interactions and, in so doing, further stabilize the known direct contact of p300 and hbrm with E2F-1/DP-1 (Trouche et al., 1996, 1997). Indeed, we envisage that the TRIP-Br proteins function as ‘integrators’ of both positive and negative regulatory signals conferred by multiple PHD finger- and/or bromodomain-containing transcription factors, such as KRIP-1 and p300/CBP (Figure 9).
In summary, we have identified and partially characterized two proteins, TRIP-Br1(p34SEI-1) and TRIP-Br2, which interact with the PHD zinc fingers and/or bromodomains of a number of important transcription factors, including KRIP-1 and p300. TRIP-Br proteins interact directly with DP-1 and stimulate the transcriptional activation function of E2F-1/DP-1. KRIP-1 potentiates the transcriptional effect of TRIP-Br proteins on E2F-1/DP-1. This co-activation by the TRIP-Br proteins and KRIP-1 occurs within the established framework of regulation of E2F-1/DP-1 by RB and E1A. TRIP-Br1(p34SEI-1) has also been shown to render the activity of cyclin D–cdk4 resistant to the inhibitory effect of p16INK4a, consistent with a dual role for TRIP-Br1(p34SEI-1) in the regulation of cell cycle progression through sequential effects on the transcriptional activity of E2F-responsive promoters during G1 and S phases. We propose that the TRIP-Br proteins function physiologically as ‘integrators’ of the actions of PHD zinc finger- and/or bromodomain-containing proteins on E2F-responsive promoters, in a manner that contributes to the regulation of transcriptional activity.
Materials and methods
All recombinant DNA work was performed using standard techniques (Sambrook et al., 1989). All PCR-generated constructs were confirmed by sequence analysis.
Yeast two-hybrid system
Full-length mouse KRIP-1, mouse TIF1α and human SP140 cDNA inserts were ligated in-frame into the GAL4-AD plasmid pPC86 (Vidal et al., 1996). Yeast two-hybrid screening of a whole mouse fetus day 12–13 cDNA expression library (a gift from Joshua La Baer) cloned into the AD hybrid vector pPC86 was performed as previously described (Kim et al., 1996; Vidal et al., 1996).
DNA sequence analysis
The cDNA inserts of the yeast AD hybrid contruct pPC86/16-1 and various TRIP-Br EST clones (American Type Culture Collection) were sequenced in their entirety on both strands using Sequenase (US Biochemicals). These sequence data have been submitted to the DDBJ/EMBL/GenBank database under accession Nos AF366400–AF366403.
Northern and Southern blot analysis
Hybridization of mouse multiple tissue and embryonic development northern blots was performed at 55°C with 2.5 × 105 c.p.m./ml of probe as described (Clontech Multiple Tissue Northern Blot User Manual). Blots were washed at 65°C to a final stringency of 0.1× SSC/0.1% SDS for the mTRIP-Br1 probe and 2× SSC/0.05% SDS for the hTRIP-Br2 probe. The lower stringency of the latter was employed to detect cross-hybridization. Southern blot hybridization of a genomic zoo blot (Bios) was performed at 65°C with 2.5 × 105 c.p.m./ml of probe in 5× SSC/1%SDS/10× Denhardt’s solution/100 µg/ml denatured salmon sperm DNA. Blots were washed at 65°C to a final stringency of 0.5× SSC/0.5% SDS.
Cell synchronization and flow cytometric DNA content analysis
U2OS cells were plated at 5 × 105 per 60 mm plate 24 h prior to incubation with 75 ng/ml nocodazole for 22 h. Cells were released from M phase block and harvested at the corresponding time points by scraping in 100 µl of 1× SDS–PAGE loading buffer. Whole-cell lysate (10 µl) was analyzed by SDS–PAGE, followed by sequential western blot analysis with anti-mTRIP-Br1, anti-cyclin A (Santa Cruz) and anti-cdk2 (Santa Cruz) antibodies. Flow cytometry was done on a Coulter EPICS® ELITE ESP instrument at a wavelength of 488 nm.
DNA transfection and sequential β-galactosidase/luciferase assay
Cell lines were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum. COS cells and LLC-PK1 cells were transfected using the DEAE–dextran method (Witzgall et al., 1994). SAOS2 cells were transfected by calcium phosphate precipitation (Martin et al., 1995). 293 cells were transfected by enhanced calcium phosphate precipitation with 10 µg/ml CalPhos Maximizer (Clontech). Sequential measurement of β-galactosidase and luciferase activity was performed using the Dual Light System (Tropix).
Expression plasmids pCMV/E2F-1, pCMV/E2F-1 (E132), pCMV/DP-1, pCMV/DP-1 (Δ103–126), pCMV/RB, pCMV/RBΔ22, pRSV/E1A 12S, pRSV/E1A 12S (Rb mut), pRSV/E1A 12S (p300 mut) and pSV/HA-KRIP-1 have been described (Qin et al., 1992; Cress et al., 1993; Helin et al., 1993a,b; Kim et al., 1996; Trouche et al., 1996; Wu et al., 1996). The total amount of CMV, RSV and SV40 promoter DNA transfected was kept constant. The luciferase reporter plasmid pGL2– (–536), driven by E2F-responsive promoter sequences from the mouse B-myb gene, has been described (Lam and Watson, 1993).
In vitro binding studies
pGEXE2F-1 and pGST-DP-1, which encode full-length GST fusion proteins of human E2F-1 and DP-1, have been described (Helin et al., 1993a,b). pGT RB (379–928), which encodes GST fused to the RB ‘pocket’ domain, has been described (Kaelin et al., 1991). [35S]methionine-labeled IVT products were synthesized from pCMV/mTRIP-Br1, pCMV/hTRIP-Br2 and T7/luciferase plasmid templates using the TNT T7 coupled reticulocyte lysate system (Promega). Binding reactions using 50 µl of 50% (v:v) glutathione–agarose beads in 1× NTNZ-M binding buffer (100 mM NaCl, 20 mM Tris–HCl pH 8.0, 0.5% NP-40, 10 µM ZnCl2, 10 mM MgCl2) containing a protease inhibitor cocktail (Boehringer) were performed as described (Grieco et al., 1992).
Co-immunoprecipitation and western blot analysis
SAOS2 cells were transfected with 5 µg each of various eukaryotic expression constructs. Carrier DNA (pBSK+) was added to a total of 30 µg. Cell extracts for co-immunoprecipitation were prepared by lysing cells for 30 min on ice with 1 ml of 1× lysis buffer (50 mM HEPES pH 7.4, 100 mM NaCl, 0.1% NP-40, 10% glycerol, 10 µM ZnCl2, 1 mM dithiothreitol, 1 mM EGTA, 1 mM sodium vanadate, 50 mM disodium β-glycerophosphate, protease inhibitor cocktail) per 10 cm plate. Total cell extracts were incubated with 25 µl of anti-DP-1–Sepharose for 1 h at 4°C with rotation. Anti-DP-1–Sepharose was prepared by conjugating mouse monoclonal antibodies against human DP-1 (Santa Cruz), to cyanogen bromide-activated Sepharose (Pharmacia) according to manufacturer’s instructions. Western blot analysis was performed with various antibodies using the ECL system (New England Nuclear). Co-immunoprecipitation of endogenous E2F-1, DP-1 and hTRIP-Br1 was performed on ML-1 (human myeloid leukemia) cell extracts as above. Anti-E2F-1 (KH20) was purchased from Santa Cruz. Anti-SV40-Tg (PAB 419) was a gift from Ed Harlow. Anti-TRIP-Br1 rabbit polyclonal antibody was generated against a GST fusion protein encoding N-terminal residues 1–178 of the 16-1 clone.
Acknowledgments
Acknowledgements
We are grateful to Marc Vidal, Ed Harlow, Marie Classon, Don Bloch, Takahiro Nagase, Jackie Lees, Tony Kouzarides, Pierre Chambon, David Livingston, Jack Dixon and John Kyriakis for the generous gifts of reagents, and for guidance and suggestions during the course of this work. This work was supported by NIH MERIT grant DK39773 (J.V.B.), and grants NIH T32 DK07540 and NMRC R-172-000-072-213 (S.I-H.H.). D.M.H. was supported by Boehringer Ingelheim Fonds and Deutsche Studienstiftung.
References
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Bloch D.B., de la Monte,S., Guigaouri,P., Filippov,A. and Bloch,K.D. (1996) Identification and characterization of a leukocyte-specific component of the nuclear body. J. Biol. Chem., 271, 29198–29204. [DOI] [PubMed] [Google Scholar]
- Chang C.-J., Chen,Y.-L. and Lee,S.-C. (1998) Coactivator TIF1β interacts with transcription factor C/EBPβ and glucocorticoid receptor to induce α1-acid glycoprotein gene expression. Mol. Cell. Biol., 18, 5880–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.Q., Godwin,A.K., Bellacosa,A., Taguchi,T., Franke,T.F., Hamilton,T.C., Tsichlis,P.N. and Testa,J.R. (1992) AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc. Natl Acad. Sci. USA, 89, 9267–9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G.B., Ren,R. and Baltimore,D. (1995) Modular binding domains in signal transduction proteins. Cell, 80, 237–248. [DOI] [PubMed] [Google Scholar]
- Cress W.D., Johnson,D.G. and Nevins,J.R. (1993) A genetic analysis of the E2F1 gene distinguishes regulation by Rb, p107 and adenovirus E4. Mol. Cell. Biol., 13, 6314–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeo-Jones D., Huang,P.S., Jones,R.E., Haskell,K.M., Vuocolo,G.A., Hanobik,M.G., Huber,H.E. and Oliff,A. (1991) Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature, 352, 251–254. [DOI] [PubMed] [Google Scholar]
- Deloukas P. et al. (1998) A physical map of 30 000 human genes. Science, 282, 744–746. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.-M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Eckner R., Ewen,M.E., Newsome,D., Gerdes,M., DeCaprio,J.A., Lawrence,J.B. and Livingston,D.M. (1994) Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adapter. Genes Dev., 8, 869–884. [DOI] [PubMed] [Google Scholar]
- Girling R., Partridge,J.F., Bandara,L.R., Burden,N., Totty,N.F., Hsuan,J.J. and La Thangue,N.B. (1993) A new component of the transcription factor DRTF1/E2F. Nature, 362, 83–87. [DOI] [PubMed] [Google Scholar]
- Goddard A.D., Borrow,J., Freemont,P.S. and Solomon,E. (1991). Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science, 254, 1371–1374. [DOI] [PubMed] [Google Scholar]
- Grieco F., Hay,J.M. and Hull,R. (1992) An improved procedure for the purification of protein fused with glutathione S-transferase. Biotechniques, 13, 856–857. [PubMed] [Google Scholar]
- Helin K., Harlow,E. and Fattaey,A. (1993a) Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell. Biol., 13, 6501–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K., Wu,C.-L., Fattaey,A.R., Lees,J.A., Dynlacht,B.D., Ngwu,C. and Harlow,E. (1993b) Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev., 7, 1850–1861. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F., Wurtz,J.-M., Le Douarin,B., Chambon,P. and Losson,R. (1997) The bromodomain revisited. Trends Biochem. Sci., 22, 151–153. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G. Jr, Pallas,D.C., DeCaprio,J.A., Kaye,F.J. and Livingston,D.M. (1991) Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell, 64, 521–532. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G. Jr et al. (1992) Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell, 70, 351–364. [DOI] [PubMed] [Google Scholar]
- Kim S.-S., Chen,Y.-M., O’Leary,E., Witzgall,R., Vidal,M. and Bonventre,J.V. (1996) A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc. Natl Acad. Sci. USA, 93, 15299–15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek W., Ewen,M.E., Shirodhar,S., Arany,Z., Kaelin,W.G.,Jr and Livingston,D.M. (1994) Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell, 78, 161–172. [DOI] [PubMed] [Google Scholar]
- Lam E.W.-F. and La Thangue,N.B. (1994) DP and E2F proteins: coordinating transcription with cell cycle progression. Curr. Opin. Cell Biol., 6, 859–866. [DOI] [PubMed] [Google Scholar]
- Lam E.W.-F. and Watson,R.J. (1993) An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J., 12, 2705–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B. et al. (1995) The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J., 14, 2020–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B., Nielsen,A.L., Garnier,J.-M., Ichinose,H., Jeanmougin,F., Losson,R. and Chambon,P. (1996) A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J., 15, 6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Martin K., Trouche,D., Hagemeier,C., Sorenson,T.S., La Thangue,N.B. and Kouzarides,T. (1995) Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature, 375, 691–694. [DOI] [PubMed] [Google Scholar]
- Momand J., Zambetti,G.P., Olson,D.C., George,D. and Levine,A.J. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell, 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- Moosmann P., Georgiev,O., Le Douarin,B., Bourquin,J.-P. and Schaffner,W. (1996) Transcriptional repression by RING finger protein TIF1β that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res., 24, 4859–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T., Seki,N., Tanaka,A., Ishikawa,K. and Nomura,N. (1995) Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121–KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res., 2, 167–174. [DOI] [PubMed] [Google Scholar]
- Ornaghi P., Ballario,P., Lena,A.M., Gonzalez,A. and Filetici,P. (1999) The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J. Mol. Biol., 287, 1–7. [DOI] [PubMed] [Google Scholar]
- Qin X.-Q., Chittenden,T., Livingston,D.M. and Kaelin,W.G.,Jr (1992) Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev., 6, 953–964. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Saurin A.J., Borden,K.L.B., Boddy,M.N. and Freemont,P.S. (1996) Does this have a familiar RING? Trends Biochem. Sci., 21, 208–214. [PubMed] [Google Scholar]
- Schulman B.A., Lindstrom,D.L. and Harlow,E. (1998) Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl Acad. Sci. USA, 95, 10453–10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M. et al. (1999) Regulation of CDK4 activity by a novel CDK4-binding protein, p34(SEI-1). Genes Dev., 13, 3027–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D., Cook,A. and Kouzarides,T. (1996) The CBP co-activator stimulates E2F1/DP-1 activity. Nucleic Acids Res., 24, 4139–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D., Le Chalony,C., Muchardt,C., Yaniv,M. and Kouzarides,T. (1997) RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl Acad. Sci. USA, 94, 11268–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M., Braun,P., Chen,E., Boeke,J.D. and Harlow,E. (1996) Genetic characterization of a mammalian protein–protein interaction domain by using a yeast reverse two-hybrid system. Proc. Natl Acad. Sci. USA, 93, 10321–10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzgall R., O’Leary,E., Leaf,A., Önaldi,D. and Bonventre,J.V. (1994) The Krüppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc. Natl Acad. Sci. USA, 91, 4514–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-L., Classon,M., Dyson,N. and Harlow,E. (1996) Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol. Cell. Biol., 16, 3698–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]