Abstract
Protein targeting to the endoplasmic reticulum (ER) membrane is regulated by three GTPases, the 54 kDa subunit of the signal recognition particle (SRP) and the α- and β-subunits of the SRP receptor (SR). Using a soluble form of SR and an XTP-binding mutant of SRβ, we show that SRβ is essential for protein translocation across the ER membrane. SRβ can be cross-linked to a 21 kDa ribosomal protein in its empty and GDP-bound state, but not when GTP is bound. GTP binding to SRβ is required to induce signal sequence release from SRP. This is achieved by the presence of the translocon, which changes the interaction between the 21 kDa ribosomal protein and SRβ and thereby allows SRβ to bind GTP. We conclude that SRβ coordinates the release of the signal sequence from SRP with the presence of the translocon.
Keywords: endoplasmic reticulum/GTPase/protein translocation/ribosome/signal recognition particle receptor (SR)
Introduction
Efficient targeting of secretory and membrane proteins to the endoplasmic reticulum (ER) is dependent upon the interaction of the signal recognition particle (SRP) with its cognate receptor (reviewed by Walter and Johnson, 1994; Johnson and van Waes, 1999). Secretory proteins are synthesized with an N-terminal hydrophobic signal sequence. As the signal sequence emerges from the ribosome, it is recognized by SRP, via its 54 kDa protein (SRP54), leading to a transient retardation of translation (elongation arrest) and targeting of the ribosome–nascent chain complex (RNC) to the ER membrane (Walter and Johnson, 1994; Johnson and van Waes, 1999). At the membrane, SRP contacts the SRP receptor (SR) (Gilmore et al., 1982; Meyer et al., 1982), a heterodimer composed of a 70 kDa peripheral membrane protein (SRα) that is tightly associated with a 30 kDa integral membrane protein (SRβ) (Tajima et al., 1986). Contact between SRP54 and SRα leads to the transfer of the nascent chain from SRP54 into the translocation channel formed by the Sec61p complex (Simon and Blobel, 1991; Görlich et al., 1992; Crowley et al., 1993; Hanein et al., 1996). As the elongation arrest is released, translation resumes, with the nascent chain passing through the translocation channel into the lumen of the ER.
The targeting process is strictly dependent upon GTP (Connolly and Gilmore, 1986); three of the proteins involved in targeting are members of the GTPase superfamily (Bourne et al., 1991): SRP54, SRα and SRβ (Bernstein et al., 1989; Connolly et al., 1989; Römisch et al., 1989; Miller et al., 1995). SRP54 and SRα are similar in their GTPase domains as deduced from the molecular structures of their respective prokaryotic homologues Ffh and FtsY (Freymann et al., 1997; Montoya et al., 1997). They form a distinct subfamily of GTPases, characterized by a relatively low affinity for nucleotide, with a KD of 0.1 and 14 µM for free SRP54 and SRα, respectively, and are relatively stable in their empty states (Miller et al., 1995; Bacher et al., 1996; Rapiejko and Gilmore, 1997). In contrast, SRβ is most closely related to Sar1, a member of the Arf subfamily of GTPases that are involved in vesicle trafficking, and has a relatively high affinity for nucleotide (KD ∼20 nM for GTP) (Miller et al., 1995; Bacher et al., 1999).
The roles of SRP54 and SRα in targeting have been quite well characterized. In the presence of the ribosome, SRP54 has increased affinity for GTP (Bacher et al., 1996). Upon contact with SRα, both proteins bind GTP with high affinity, forming a rather stable complex (Rapiejko and Gilmore, 1992, 1997). The presence of the Sec61p complex is then required to promote the release of the nascent chain from SRP54 (Song et al., 2000). The details of this critical step remain largely uncharacterized. Finally, mutual GTP hydrolysis in SRα and SRP54 leads to the dissociation of SRP from SR, allowing another round of targeting (Connolly et al., 1989; Rapiejko and Gilmore, 1997).
Unlike SRP54 and SRα, which have prokaryotic homologues, SRβ appears to be present only in eukaryotic cells. Furthermore, the role of SRβ in targeting has so far remained enigmatic. Recently, it was shown in Saccharomyces cerevisiae that a null mutant of SRβ shows a slow growth phenotype and translocation defects, the same phenotype as is observed in SRP54 and SRα deletion strains (Ogg et al., 1998). This indicates an essential role for SRβ in targeting in vivo. However, the transmembrane (TM) domain and short lumenal domain of SRβ, located at the N-terminus, appear to be dispensable for function (Ogg et al., 1998). Furthermore, the binding of nucleotide to SRβ is required for complex formation with SRα (Ogg et al., 1998; Legate et al., 2000).
Previously, we have shown that the ribosome influences the GTPase domain of SRβ, leading to a reduced affinity for nucleotides and increased GTPase activity (Bacher et al., 1999). To characterize this interaction in more detail, we produced a recombinant, soluble SR, composed of full-length SRα and a truncated SRβ, lacking the N-terminal TM domain.
Here we show that nucleotide binding to SRβ is required for translocation. Furthermore, nucleotide binding to SRβ is modulated by an interaction with a component of the large ribosomal subunit, in a manner that is sensitive to the presence of the Sec61p complex. We propose that SRβ regulates the transfer of the nascent chain from SRP to the translocon, such that the signal sequence is only released from SRP when the Sec61p complex is available for insertion. SRβ therefore appears to be the missing link between the GTP-dependent interaction of SRP54 with SRα and the subsequent insertion of the nascent chain into the translocon.
Results
Expression and characterization of recombinant SR
In order to understand better the role of SR in targeting, we sought to produce a recombinant form of the SR protein complex. To overcome the difficulties in expressing and purifying integral membrane proteins, we deleted the TM domain of SRβ, which has previously been shown to be dispensable for function in S.cerevisiae (Ogg et al., 1998).
We co-expressed, in Escherichia coli, His6-tagged, full-length human SRα (hisSRα) and mouse SRβ, lacking the first 57 amino acids, which includes the TM region (SRβΔN). A complex of the two proteins could be purified to homogeneity using metal chelate, ion exchange and gel filtration chromatography. The complex migrated as a sharp single peak of ∼100 kDa in the final gel filtration step (Figure 1). The predicted mass was confirmed by analytical ultracentrifugation. This molecular weight is consistent with a heterodimer containing one hisSRα and one SRβΔN polypeptide. The recombinant form of the SR complex is referred to as SRhisα/βΔN.
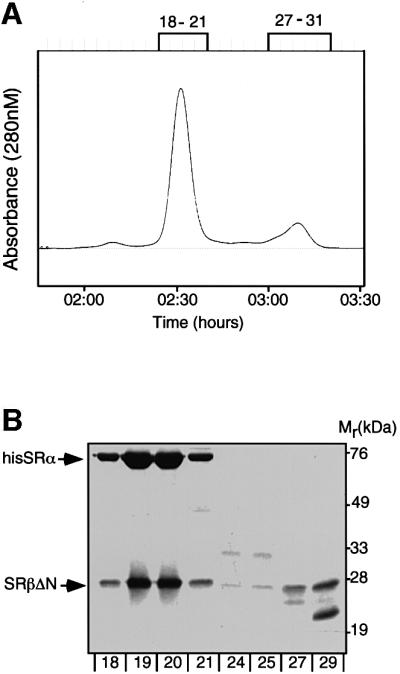
Fig. 1. Purification and characterization of recombinant SR. SRα and SRβ were co-expressed in E.coli and purified by metal chelate and ion exchange chromatography. The peak fractions were then analysed on a Superdex-200 gel filtration column (A). Samples from the two major peaks (fractions 18–21 and 29–30) were analysed by SDS–PAGE followed by staining with Coomassie Brilliant Blue (B).
HPLC analysis after denaturation revealed that <20% of the complex had bound GTP at the end of the purification procedure; however, ∼80% of the complexes were competent to rebind nucleotide (see Supplementary figure 1, available at The EMBO Journal Online).
SR lacking a TM domain is functional in translocation
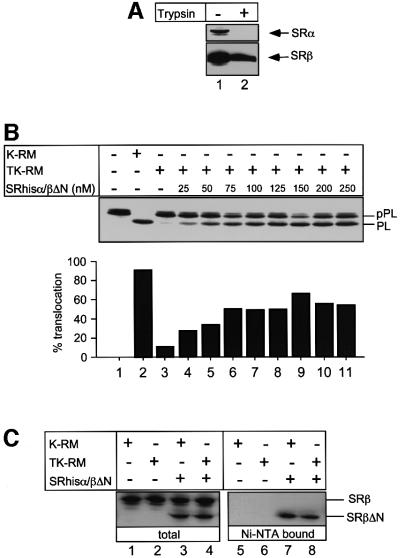
To test whether the SRhisα/βΔN is functional, we assayed its ability to complement membranes from which SRα had been removed by treatment with low concentrations of trypsin in the presence of high salt (Meyer and Dobberstein, 1980) (Figure 2A). Pre-prolactin (pPL) was translated in a reticulocyte lysate in the presence of either high-salt-washed membranes (K-RM) or trypsinized K-RM (TK-RM). As expected, K-RM supported efficient translocation as indicated by processing of pPL to prolactin (PL). In contrast, TK-RM were unable to promote translocation (Figure 2B). However, if the TK-RM were supplemented with increasing concentrations of the SRhisα/βΔN, translocation activity could be restored to between 50 and 60% of the activity of the endogenous receptor in the K-RM. This reactivation occurred in a dose-dependent manner, with the activity saturating at a SRhisα/βΔN concentration between 50 and 75 nM, which is slightly higher than that of the endogenous membrane-bound receptor originally present in the TK-RM (30 nM) (Tajima et al., 1986).
Fig. 2. Functional analysis of recombinant SR (SRhisα/βΔN). (A) High-salt-washed membranes (K-RM; lane 1) were treated with 5 µg/ml trypsin (TK-RM; lane 2) to inactivate the endogenous SR. Equal aliquots of each membrane (10 eq.) were analysed by SDS–PAGE followed by immunoblotting with antibodies against the SRα and SRβ subunits. (B) Co-translational translocation of pre-prolactin (pPL) was assayed in a reticulocyte lysate supplemented with either K-RM (lane 2), TK-RM (lanes 3) or TK-RM with increasing concentrations of SRhisα/βΔN receptor (25–250 nM; lanes 4–8). The reactions were analysed by SDS–PAGE and phosphorimaging. The positions of the unprocessed pPL and the signal sequence-cleaved prolactin (PL) are indicated. The efficiency of translocation was deduced from quantification of the amounts of cleaved and non-cleaved pPL. (C) Translocation reactions were performed as in (B) with K-RM (lanes 1, 3, 5 and 7) or TK-RM (lanes 2, 4, 6 and 8) in the presence (lanes 3, 4, 7 and 8) or absence (lanes 1, 2, 5 and 6) of 50 nM SRhisα/βΔN. After translocation, the samples were analysed by immunoblotting with SRβ antibodies either directly (lanes 1–4) or after purification on Ni-NTA agarose (lanes 5–8). The positions of the endogenous SRβ and SRβΔN are indicated.
One caveat of this assay is that the endogenous SRβ is still present. To test whether hisSRα could exchange between endogenous SRβ and SRβΔN, hisSRα was re-isolated at the end of the translocation assay by purification on Ni-NTA resin, following membrane solubilization with Triton X-100. The eluted material was analysed by immunoblotting with anti-SRβ antibodies. Although equimolar amounts of SRhisα/βΔN and endogenous SRβ are present in the assay, only the SRβΔN form co-purified with the His6-tagged SRα (Figure 2C). This indicates that hisSRα does not exchange between endogenous SRβ and SRβΔN. Thus, it is unlikely that the endogenous SRβ present in the TK-RM is functional in the assay.
We can therefore show that the recombinant SR, lacking a TM domain, is functional in translocation, in good agreement with the in vivo results shown in the yeast system (Ogg et al., 1998). However, the soluble receptor is clearly less efficient than the endogenous membrane-integrated receptor, most probably due to the lack of the TM domain.
SRβ interacts with a ribosomal protein in a GTP-dependent manner
It was shown previously that an interaction between RNC–SRP complexes and SRβ reduces the affinity of SRβ for both GTP and GDP (Bacher et al., 1999). This effect was also observed with ribosomes that lack SRP and nascent chains, suggesting that it is mediated by a ribosomal component (G.Bacher and B.Dobberstein, unpublished observations). To characterize this interaction in more detail, we looked for ribosomal proteins that contact SRβ using a cross-linking assay.
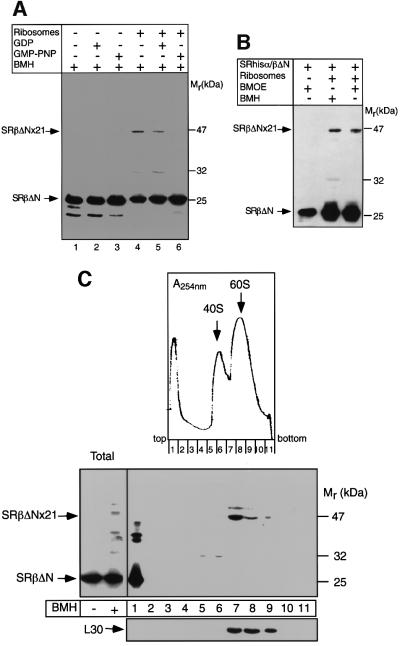
SRhisα/βΔN was incubated with purified canine pancreas ribosomes and then cross-linking was induced with the cysteine-reactive reagent bis-maleimidohexane (BMH). Following cross-linking, reactions were analysed by SDS–PAGE and immunoblotting using anti-SRβ antibodies (Figure 3A). In the absence of ribosomes, no higher molecular weight cross-link products were formed (SRα–SRβ cross-links are formed only at relatively high BMH concentrations; data not shown). However, in the presence of ribosomes, a cross-link product of ∼47 kDa was formed corresponding to a cross-link adduct of 21 kDa. A similarly sized cross-link product could also be formed with the shorter cysteine-reactive cross-linker bis-maleimidoethane (BMOE), which has a spacer length of only 7 Å (Figure 3B), suggesting a relatively close positioning of the two proteins. To test for the effect of nucleotide on this SRβ–ribosome interaction, cross-linking was performed either without added nucleotide or in the presence of either GDP or the non-hydrolysable GTP analogue, GMP-PNP (Figure 3A). The strongest cross-link was observed without added nucleotide, GDP slightly reduced the cross-link efficiency, whereas with GMP-PNP no cross-link product was observed.
Fig. 3. Cross-linking of SRβΔN to a ribosomal protein. (A) SRhisα/βΔN was incubated in the presence or absence of high-salt-washed canine ribosomes either with no nucleotide, or with 200 µM GMP-PNP or GDP, and cross-linking was then induced with the reagent BMH (20 µM). The samples were analysed by SDS–PAGE followed by immunoblotting with anti-SRβ antibodies. The positions of the uncross-linked recombinant SRβ (SRβΔN) and the major ribosome-dependent cross-link of 47 kDa (SRβΔNx21) are indicated. (B) Cross-linking was performed as in (A) but with 20 µM of either BMH or BMOE. (C) Cross-linking reactions as in (A) with no nucleotide present (total fractions) were treated with puromycin and high salt and then separated by sucrose density gradient centrifugation. The gradients were then fractionated with continuous monitoring of the absorbance at 254 nm (upper panel) to determine the positions of the 40S and 60S subunits (arrows). The fractions were then analysed by SDS–PAGE followed by immunoblotting with antibodies against ribosomal protein L30 (lower panel) and against SRβ (middle panel).
The nucleotide-dependent cross-linking of SRβ to a 21 kDa protein was also observed with RNC–SRP complexes bearing pPL86mer, indicating that this effect is relevant to functional targeting of substrates (see Supplementary figure 2). This agrees with the previously observed effect of RNC complexes on the affinity of SRβ for GTP (Bacher et al., 1999).
The cross-linking of SRβΔN to the 21 kDa protein was not restricted to the soluble form of the receptor. A 21 kDa ribosome-dependent cross-link adduct was also observed when endogenous SR, purified from dog pancreas microsomes and incorporated into proteoliposomes, was used in the cross-link assay (see Supplementary figure 3).
In order to determine whether the cross-link adduct is a bona fide ribosomal protein, we treated the cross-link reactions with puromycin and high salt, to dissociate the ribosomal subunits and to remove any residual peripheral proteins, and then analysed them by sucrose density gradient centrifugation (Figure 3C). The uncross-linked SRβΔN migrated at the top of the gradient. In contrast, the 47 kDa cross-link product co-migrated exclusively with the 60S fractions, as deduced from the distribution of the large subunit protein L30 and alignment with the A254 nm profile. The 21 kDa protein is, therefore, a component of the 60S subunit.
Binding of Sec61p to ribosomes attenuates SRβ–ribosome cross-linking
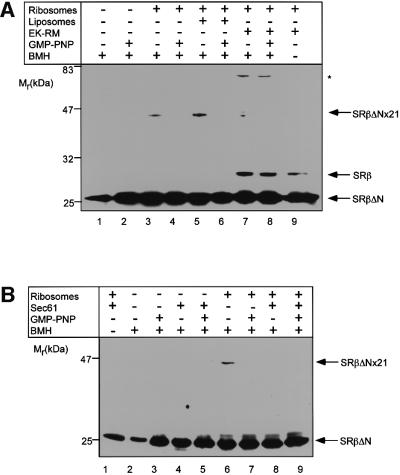
During the targeting reaction, ribosomes become bound to the Sec61p complex (Görlich et al., 1992; Kalies et al., 1994). We therefore asked whether binding of ribosomes to ER membranes affects cross-linking of SRβΔN to the 21 kDa ribosomal protein. Addition of membranes stripped of ribosomes by EDTA and high-salt treatment (EK-RM) led to a strong reduction in cross-linking of SRβ to the 21 kDa protein (Figure 4A). This was not due to inhibition of the cross-linker per se, as a cross-link between SRβ and an unidentified membrane species of 45 kDa was observed.
Fig. 4. Binding of ribosomes to Sec61p blocks cross-linking of SRβ to the ribosome. (A) SRhisα/βΔN was incubated with pancreatic ribosomes and/or EDTA high-salt-washed membranes (EK-RM) or liposomes. Where indicated, 200 µM GMP-PNP was also present in the reaction. Cross-linking was induced with BMH and the samples were analysed by SDS–PAGE and immunoblotting for SRβ. The positions of SRβΔN, the endogenous SRβ, the major ribosomal cross-link product (SRβΔNx21) and a cross-link to an unidentified protein of EK-RM (*) are indicated. (B) Cross-linking reactions were performed and analysed as in (A), except that purified Sec61p reconstituted into liposomes was used in place of EK-RM.
To test whether the effect of EK-RM was due to binding of the ribosomes to Sec61p, cross-linking was performed in the presence of purified Sec61p complex reconstituted into proteoliposomes. In contrast to the control liposomes (Figure 4A), Sec61p proteoliposomes prevented cross-linking of SRβΔN to the 21 kDa protein in the presence and absence of GMP-PNP (Figure 4B). Thus, it appears that the presence of Sec61p alone is sufficient to attenuate the cross-linking of SRβ to the ribosome.
As SRhisα/βΔN is present in molar excess over Sec61p and ribosomes in the assay, it is most likely that the effect is due to the interaction of Sec61p with the ribosomes rather than an interaction between SRhisα/βΔN and Sec61p.
The 47 kDa cross-link adduct observed with EK-RM was not formed in the presence of Sec61p proteoliposomes, suggesting that it is unlikely to be a cross-link to a component of the Sec61p complex.
GTP binding to SRβ is required for translocation
Since the binding of SR to the ribosome regulates the GTP-bound state of SRβ and binding to Sec61p interferes with this interaction, we reasoned that the nucleotide-bound state of SRβ might be influencing the targeting reaction. One would then predict that GTP binding to SRβ is required for translocation.
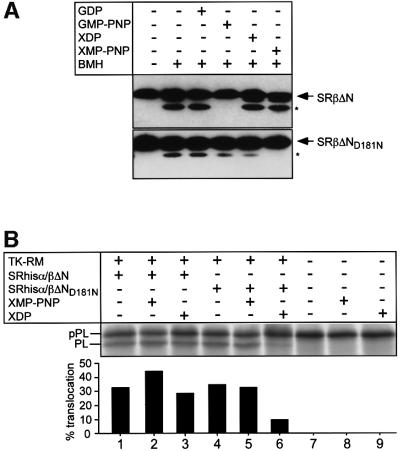
To test this directly, a point mutant of SRβ was constructed in the G4 consensus element, in which the aspartic acid (D) residue at position 181 was exchanged for an asparagine (N), to give rise to SRβΔND181N. Such a mutation has been shown for many other GTPases to lead to an increase in the affinity for xanthosine nucleotides as compared with guanosine nucleotides (Weijland and Parmeggiani, 1993; Powers and Walter, 1995; Moser et al., 1997). In fact, it has been reported recently that this mutation in SRβ also leads to an increased affinity for XNP over GNP (Legate et al., 2000), albeit only by a single order of magnitude.
To show the effect of XNP on SRβΔND181N, we made use of an intramolecular cross-link product that was formed in the presence of BMH and was sensitive to the presence of different guanosine nucleotides. With wild-type SRhisα/βΔN, the cross-link is observed with GDP and in the empty state, but it is dramatically reduced in the presence of GMP-PNP (Figure 5A). This would be consistent with cross-linking between two cysteine residues whose relative positioning is altered upon binding of GTP. Neither XDP nor XMP-PNP affects the cross-link formation in the wild-type receptor. In contrast, with SRhisα/βΔN containing the D181N mutation within SRβ (SRhisα/βΔND181N), XMP-PNP leads to a strong reduction of the internal cross-link, unlike GMP-PNP, which led only to a small reduction in cross-linking. These observations are consistent with the previous report that the D181N mutant of SRβ has increased affinity for XTP and a residual affinity for GTP (Legate et al., 2000).
Fig. 5. Characterization of a mutant SR with altered nucleotide specificity in SRβΔN. (A) SRhisα/βΔN (upper panel) or SRhisα/βΔND181N (lower panel) was treated with BMH (20 µM) in the presence of different guanosine and xanthosine nucleotides (200 µM) as indicated. The reactions were then analysed by SDS–PAGE and immunoblotting with SRβ antibodies. Note the difference in internal cross-link (*) formation in the presence of the indicated nucleotides. (B) Co-translational translocation of pre-prolactin was assayed in a reticulocyte lysate supplemented with TK-RM and either 100 nM SRhisα/βΔN (lanes 1–3) or 100 nM SRhisα/βΔND181N (lanes 4–6). Where indicated, 500 µM XMP-PNP (lanes 2, 5 and 8) or 500 µM XDP (lanes 3, 6 and 9) was added in addition to the 100 µM GTP already present in the lysate.
We next tested the ability of SRhisα/βΔN and SRhisα/βΔND181N to complement translocation with TK-RM in the presence of different xanthosine nucleotides (Figure 5B), added in excess over GTP. The presence of XDP strongly inhibited the mutant but had no effect on the wild-type receptor (compare lanes 3 and 6). This indicates that the nucleotide-bound state of SRβ regulates the targeting reaction. Furthermore, it confirms that the endogenous SRβ still present in the membrane, which binds GTP and should therefore be insensitive to XDP, is not functional in the assay.
In the absence of added XNP, both wild-type and mutant SRhisα/βΔN could promote translocation of pPL with TK-RM. This is because the affinity of the mutant for GTP (Legate et al., 2000) is still high enough to promote GTP binding at the concentrations present in the translocation reaction (0.1 mM). The presence of XMP-PNP led to a slight stimulation of translocation with wild-type SR but not with the mutant.
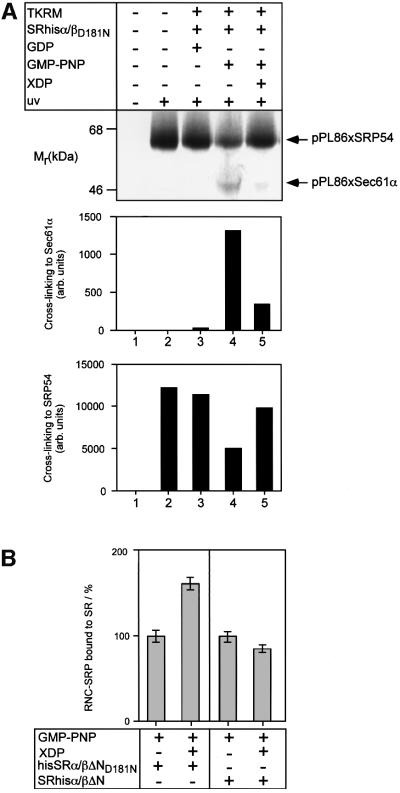
We next looked at which step in the targeting process GTP binding to SRβ is required, again using SRhisα/βΔND181N in the presence and absence of XDP. We first monitored the transfer of the nascent chain from SRP54 to the translocon by site-specific photocross-linking. RNC complexes were prepared by translating, in the presence of SRP, an mRNA that encodes the first 86 amino acids of pPL (pPL86mer) and does not contain a stop codon. A photoactivatable amino acid {l-4′-(3-[trifluoromethyl]-3H-diazirin-3-yl) phenylalanine; (Tmd)Phe} was incorporated into the signal peptide of the nascent chain (High et al., 1993). The resulting RNC–SRP complexes were then used in targeting assays with TK-RM and SRhisα/βΔND181N, in the presence of different nucleotides, and cross-linking was then induced by irradiation with UV light. Transfer of the nascent chain from SRP to the translocon was revealed by a reduction in cross-linking between pPL86 and SRP54 and the concomitant appearance of cross-links to Sec61α (High et al., 1993; Martoglio et al., 1995) (Figure 6A). In the absence of TK-RM, or in the presence of GDP, the nascent chains remained associated with SRP. When GMP-PNP was added, a significant proportion of the nascent chains was found to be cross-linked to Sec61α. As judged from the reduction in cross-linking to SRP54, this corresponded to the insertion of ∼60% of the nascent chains into the translocon. However, if GMP-PNP was supplemented with an excess of XDP, a marked reduction in cross-linking to Sec61α was observed together with an increase in SRP54 cross-linking. This indicates that XDP leads to a block in translocation at a step before the transfer of the signal peptide from SRP to the translocon.
Fig. 6. SRβ regulates a specific step in targeting. (A) Targeting reactions were performed with RNC–SRP complexes containing pPL86mer with a photoactivatable cross-linker ([Tmd]Phe) incorporated at position 18, TK-RM supplemented with SRhisα/βΔND181N (100 nM), 25 µM GMP-PNP or GDP, and, where indicated, 250 µM XDP. After targeting, the reactions were chilled on ice and irradiated with UV light (364 nm) prior to analysis by SDS–PAGE and phosphorimaging (upper panel). The positions of the pPL86xSRP54 and pPL86xSec61α cross-link products are indicated. The identity of the cross-links was confirmed by immunoprecipitation (data not shown). The amount of cross-linking to Sec61α and SRP54 was quantified directly from the phosphorimager and is displayed graphically (lower panel). (B) Binding of RNC–SRP complexes to SRhisα/βΔN. Targeting reactions were performed with purified pPL86mer RNC–SRP complexes, TK-RM supplemented with either SRhisα/βΔN or SRhisα/βΔND181N (100 nM), 100 µM GMP-PNP and, where indicated, 1 mM XDP. After targeting, the reactions were solubilized with 1% Nikkol and then the SRhisα/βΔN was re-isolated on an Ni-NTA matrix in the presence of high salt (300 mM KOAc). The amount of bound pPL86 was then quantified by SDS–PAGE and phosphorimaging; binding in the absence of XDP was set to 100%. Error bars indicate the standard deviation from experiments carriedout in triplicate.
To look directly at whether the block in translocation occurred after stable binding of SRP54 to SRα (Song et al., 2000), we asked whether RNC–SRP complexes accumulated in a high-salt-resistant manner with SRhisα/ βΔND181N in the presence of XDP, as compared with SRhisα/βΔN. The recombinant SR was re-isolated from targeting assays, using Ni-NTA chromatography under high-salt conditions, and the amount of bound RNC–SRP was quantified (Figure 6B). In the presence of wild-type receptor, addition of XDP had little effect on the amount of nascent chains re-isolated. In contrast, with the mutant receptor, addition of XDP led to an almost 2-fold increase in nascent chain binding. This strongly suggests that GTP binding to SRβ is required to release the nascent chain from the SRP–SR complex.
Discussion
Three components of the ER protein targeting machinery are GTPases. SRP54 is the signal sequence-binding protein and is loaded with GTP upon contact with the ribosome (Bacher et al., 1996). After contact of SRP54 with SRα, both molecules bind GTP with high affinity (Rapiejko and Gilmore, 1997). Release of the signal sequence from SRP54 requires the presence of a membrane component, which is most probably Sec61α (Görlich and Rapoport, 1993; Song et al., 2000). Here we show that the third GTPase, SRβ, must also bind GTP in order for targeting to occur. GTP binding to SRβ is suggested to occur after the Sec61p complex has associated with the ribosome, resulting in the release of the signal peptide from SRP. SRβ therefore appears to coordinate the release of the signal sequence from SRP54 with the subsequent insertion of the nascent chain into the translocon.
A soluble form of SR promotes ER protein translocation
A tagged and soluble form of SR, SRhisα/βΔN, was obtained by expression in E.coli. The complex was soluble at rather high concentrations and contained only low amounts of bound GTP. The low level of occupancy is most likely to be due to release of nucleotide during purification. The empty receptor is still competent to bind nucleotide efficiently, as indicated in a direct binding assay. This is supported further by the observation that only the empty receptor can be cross-linked to the 21 kDa protein and that addition of GMP-PNP completely abolishes cross-linking, indicating that the receptor is functional in binding nucleotide.
It was shown in yeast that a soluble form of SRβ, lacking the TM domain, is able to complement an SRβ null mutant (Ogg et al., 1998). Here we show that mammalian SR, lacking the TM domain of SRβ, is also functional in translocation. In the context of membranes in which SR has been inactivated by mild proteolysis, the soluble receptor promotes translocation at a concentration that is slightly higher than that of the endogenous membrane-bound receptor. The most likely explanation for this difference is that the recombinant SR is not tightly anchored at the membrane and may require additional factors to interact with the membrane. This interpretation is substantiated by the finding that SRβΔN could be cross-linked to an unknown membrane protein of 45 kDa. The soluble form of the yeast protein, which could complement an SRβ deletion strain, was localized mostly in the cytosol and only a small fraction was found associated with the ER membrane (Ogg et al., 1998).
The interaction of SRβ with the ribosome is modulated by the Sec61p complex
Consistent with previous observations that the ribosome reduces the affinity of SRβ for nucleotides (Bacher et al., 1999), we have now identified a molecular interaction that correlates with this observation. SRβ can be cross-linked to a 21 kDa protein of the large ribosomal subunit in a nucleotide-dependent manner. Cross-links were observed in the absence of nucleotide and in the presence of GDP, but not of GTP or GMP-PNP. The effect is not due to nucleotide binding to SRα as a complex of SRβΔN together with an N-terminal fragment of SRα, lacking the GTPase domain, gave similar nucleotide-dependent cross-links (our unpublished observation).
This differential cross-linking is probably brought about by a conformational change in SRβ, consistent with altered intramolecular cross-linking observed with a cysteine-reactive reagent. Conformational change upon nucleotide binding is characteristic of members of the GTPase superfamily, and involves mainly the consensus elements for nucleotide binding (Schlichting et al., 1990). Interestingly, all three cysteines present in SRβ are precisely located in these regions and might be differentially engaged in the cross-linking.
During the translocation process, the RNC complex is transferred from the SRP–SR to the Sec61p complex (Görlich and Rapoport, 1993). We show here that this binding of the ribosome to the Sec61p complex leads to the loss of the SRβ–ribosome cross-link. The fact that this change is observed in the presence of excess SR suggests that this is a direct result of binding of Sec61p to the ribosome and not due primarily to an interaction between Sec61p and SR, although we cannot exclude the possibility that these two complexes also interact directly. The 21 kDa ribosomal protein may be located such that it is not accessible to SRβ once the Sec61p complex has bound to the ribosome. The 21 kDa protein is therefore likely to be located on the surface of the ribosome close to the region around the exit site that forms the contact with Sec61p (Beckman et al., 1997).
Previously, two activities of the ribosome towards SRβ have been described: an increase in GTP hydrolysis by SRβ in the presence of the ribosome and a reduction in the affinity for both nucleotides. The affinity of SRβ for nucleotide in the presence (1 µM) and absence (20 nM) of ribosomes (Bacher et al., 1999) is similar to that of other small Ras-like GTPases in the presence and absence of their exchange factors (e.g. for EF-Tu: 0.5 µM and 29 nM, respectively; Romero et al., 1985). If, in the GTPase cycle of the SRβ, the nucleotide exchange step were kinetically limiting, the ribosome would then lead indirectly to an increase in GTP hydrolysis. This would explain both effects of the ribosome and is in good agreement with the BMH cross-linking data, which showed the strongest interaction between the empty state of SRβ and the 21 kDa ribosomal protein. Therefore, taking all the data together, we suggest that the ribosome most probably stabilizes SRβ in its nucleotide-free conformation.
GTP binding to SRβ is required for release of the signal sequence from SRP
SRP receptor containing the SRβD181N mutant (SRhisα/βΔND181N) is able to bind xanthosine nucleotides, as has been reported previously (Legate et al., 2000). Furthermore, binding of XTP appears to cause an alteration in internal cross-linking formation in SRβΔND181N like GTP causes in wild-type SRβΔN. This mutant receptor is functional in co-translational translocation in the presence of GTP, consistent with the previous observation that the mutant has only lowered affinity for GTP. However, in the presence of XDP, a strong inhibition of translocation is observed, which is not seen with the wild-type receptor. This demonstrates for the first time that GTP binding to SRβ is essential for translocation.
SRhisα/βΔND181N in the presence of XDP leads to accumulation of translocation intermediates in which the nascent chain remains bound to SRP54. Furthermore, the RNC–SRP complex is associated with SR in a high-salt-resistant manner. This indicates that the GTP-bound state of SRβ is required for the release of the signal sequence from the SRP–SR complex to the translocon.
SRβ coordinates the release of the signal sequence with the presence of the translocon
Here we have shown that GTP binding to SRβ is required to release the signal peptide from SRP; in the presence of SRβ-GDP, the signal sequence remains bound to SRP54. Only when SRβ-GTP is present can the nascent chain be released from SRP54 into the Sec61p complex. In the absence of Sec61p, a ribosomal component interacts with SRβ, stabilizing it in the GDP/free state. Binding of Sec61p to the ribosome modulates or destabilizes this interaction, and could thereby allow GTP binding to SRβ. This would indicate that the transfer of the nascent chain from SRP is regulated by SRβ, such that the nascent chain is only released from SRP when the translocon is present.
The involvement of a translocon component in promoting release of the signal sequence from SRP was suggested from recent observations using membranes treated with protease concentrations high enough to compromise Sec61p function (Song et al., 2000). Translocation with such membranes leads to the accumulation of RNC–SRP complexes in a high-salt-resistant complex with SR.
The proposed function of SRβ in coordinating the transfer of the nascent chain to the translocon is supported by the observation that preformed RNC–SRP complexes have a competitive advantage over RNCs for binding to the Sec61p complex in the presence of SR (Neuhof et al., 1998; Raden and Gilmore, 1998).
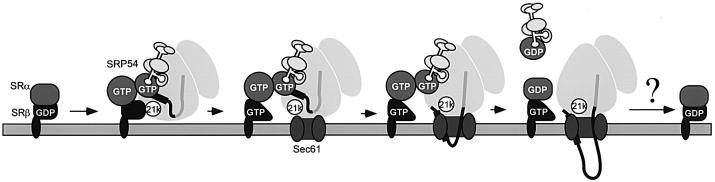
We suggest that the following stages of targeting are regulated by SRβ (Figure 7). Upon initial contact of the RNC–SRP complex with SR, via SRP54 and SRα, the ribosome interacts with SRβ and stimulates the release of the bound nucleotide; the nascent chain still remains bound to SRP. Based on our data and those of Song et al. (2000), SRα and SRP54 are, at this stage, most likely to be in their GTP-bound states. Subsequent binding of the RNC to the translocon then alters the interaction of SRβ with the ribosome and allows GTP to bind to SRβ. Whether the Sec61p complex interferes directly or indirectly with the interaction of SRβ with the 21 kDa ribosomal protein remains an open question. GTP binding to SRβ then promotes release of the signal sequence from SRP.
Fig. 7. Proposed function of SRβ in coordinating release of the signal sequence with the presence of the Sec61p complex. The RNC–SRP complex binds to the SRP receptor via an interaction between SRP54 and SRα. Concomitantly, a 21 kDa ribosomal protein (21k) interacts with SRβ, and thereby promotes release of GDP, and stabilizes the empty form. Subsequent binding of the ribosome to the Sec61p complex (Sec61p) alters the interaction of SRβ with the 21 kDa protein, allowing SRβ to bind GTP. GTP binding to SR leads to the release of the signal peptide from the SRP–SR complex and its transfer to the Sec61p complex. Finally, GTP hydrolysis in SRβ should occur. However, the precise timing and regulation of this event remain to be characterized.
This scheme proposes that SRβ coordinates SRP-dependent targeting and the faithful delivery of the nascent chain to the translocon. The interaction of the ribosome with SRβ highlights the active role of the ribosome at multiple steps in translocation. The ribosome has been shown to activate SRP54 by increasing its affinity for GTP (Bacher et al., 1996) and also to modulate the gating of the Sec61p channel (Liao et al., 1997).
In prokaryotes, only homologues of SRP (Ffh/4.5S RNA) and the α-subunit of the SR (FtsY) are found. So far, no homologues of SRβ have been detected. Bacterial SRP recognizes the signal sequence, and the subsequent interaction with FtsY is required to promote signal- sequence release (Miller et al., 1994). Release of the signal sequence is coupled to membrane binding via the interaction of FtsY with inner membrane lipids (Scotti et al., 1999; de Leeuw et al., 2000). In eukaryotes, SRP54 and SRα have roles similar to those of their prokaryotic counterparts. We suggest that the presence of SRβ ensures a higher degree of regulation for the delivery of nascent secretory and membrane proteins to the ER membrane. First, SRβ anchors SRα at the ER membrane such that substrates are targeted specifically to the ER rather than to other membrane-enclosed compartments present in the eukaryotic cell. Secondly, as we show here, the SRβ GTPase coordinates signal-sequence release with the presence of the Sec61p complex, such that substrates are transferred specifically to the translocon.
Materials and methods
Plasmid construction, expression and purification of SRhisα/βΔN
A full-length mouse SRβ clone was obtained by PCR from mouse cDNA. A fragment lacking the first 57 amino acids was then re-amplified and cloned, along with human SRα (from pGEM2hDP; Hortsch and Meyer, 1988), into pET16b (Novagen) to yield a dicistronic unit in the plasmid pSRhisαβΔN, in which the SRα is N-terminally His6 tagged. Mutagenesis of SRβ was performed using the Stratagene Quickchange kit according to the manufacturer’s instructions using the primers: 5′-GCCTGCAATAAGCAAAATATCGCAATGGC-3′ and 5′-GCCATTGCGATATTTTGCTTATTGCAGGC-3′ to create the plasmid pSRhisαβΔND181N. All constructs were verified by sequencing.
The proteins were expressed in E.coli BL21(DE3) and then purified from the corresponding lysates by metal chelate (Ni-chelating Sepharose, Pharmacia), ion exchange (Mono S, Pharmacia) and gel filtration chromatography (Superdex 200, Pharmacia).
In vitro transcription
pPPL (High et al., 1993) containing the pre-prolactin cDNA was linearized with PvuII or PstI to generate pPL86mer and full-length pPL, respectively. pPPLTAG18 (High et al., 1993), which contains an amber suppressor mutation at codon 18, was linearized similarly with PvuII. The linearized cDNAs were then used to programme in vitro transcription reactions using SP6 polymerase in the presence of Cap analogue (NEB).
In vitro translation and translocation assays
Salt-washed canine pancreatic rough microsomes (K-RM), EDTA- high-salt-stripped membranes (EK-RM), trypsin-digested membranes (TK-RM) and SRP were prepared as described previously (Gilmore et al., 1982; Walter and Blobel, 1983; Andrews et al., 1989; Martoglio et al., 1997). Sec61p was purified from dog pancreas microsomes via its association with ribosomes, followed by ion-exchange chromatography, and reconstituted into liposomes as described (Görlich and Rapoport, 1993). Mock liposomes were prepared in an identical manner but without purified protein.
Translocation assays (reaction volume of 10 µl) with nuclease-treated rabbit reticulocyte lysate (7 µl, Promega) contained full-length pPL mRNA, 3 eq. of membranes, l-[35S]methionine (7.5 µCi), unlabelled amino acids (20 µM) and supplemented, where indicated, with SRhisα/βΔN (25–250 nM). Where indicated, XDP (Sigma) or XMP-PNP (kindly provided by Roger Goody) was added to a final concentration of 0.5 mM. After incubation for 20 min at 30°C, the reactions were stopped by treatment with RNase A (0.2 mg/ml) for 5 min at 30°C and then analysed directly on 12.5% Laemmli SDS–polyacrylamide gels followed by phosphorimaging (Fuji MacBAS 1000). The percentage translocation was calculated from the relative amounts of pPL and PL, with compensation for the reduction in the number of methionines.
RNC–SRP complexes were prepared essentially as described (Hauser et al., 1995). Briefly, pPL86mer transcript was used to programme wheat germ lysate in the presence of SRP. Following synthesis, the RNC–SRP complexes were re-isolated by centrifugation (40 min at 430 000 g at 4°C) through a high-salt sucrose cushion [25 mM HEPES–KOH pH 7.6, 500 mM KOAc, 2 mM Mg(OAc)2, 1 mM dithiothreitol (DTT), 0.25 mM cycloheximide, 500 mM sucrose] and resuspended in an equal volume of RNC buffer [25 mM HEPES–KOH pH 7.6, 120 mM KOAc, 2 mM Mg(OAc)2, 1 mM cycloheximide, 1 mM DTT]. For photocross-linking, the wild-type pPL86mer transcript was replaced with that of pPL86-TAG18 and the translation reaction was supplemented with (Tmd)Phe-tRNAsup (High et al., 1993).
Targeting reactions (10 µl) were performed for 3 min at 26°C in the presence of 2 µl of RNC–SRP complexes, cytosol (4 µl of wheat germ lysate), SRhisα/βΔN (100 nM), TK-RM (3 eq.) and nucleotides [250 µM XDP and/or 25 µM GMP-PNP (Figure 6A); 1 mM XDP and/or 50 µM GMP-PNP (Figure 6B)], as indicated. For photocross-linking, the samples were then transferred to ice and UV irradiated (364 nm) for 10 min prior to analysis by SDS–PAGE and phosphorimaging.
Chemical cross-linking
Canine pancreas ribosomes were purified by overnight centrifugation of a post-mitochondrial supernatant fraction from a dog pancreas extract, prepared in 50 mM Tris–OAc pH 7.6, 50 mM KOAc, 6 mM Mg(OAc)2, 1 mM EDTA, 1 mM DTT, 10 µg/ml phenylmethylsulfonyl fluoride (PMSF), through a 2 M sucrose cushion (in the same buffer) at 200 000 g. The pellet was resuspended in 25 mM HEPES–KOH pH 7.6, 2 mM Mg(OAc)2, 50 mM KOAc, 250 mM sucrose and 2 mM DTT. The potassium acetate concentration was raised to 750 mM and the ribosomes were then re-centrifuged at 500 000 g for 1 h through a high-salt sucrose cushion [25 mM HEPES–KOH pH 7.6, 2 mM Mg(OAc)2, 750 mM KOAc, 1 M sucrose]. Finally, the ribosomes were re-suspended in 25 mM HEPES–KOH pH 7.6, 2 mM Mg(OAc)2, 50 mM KOAc and 250 mM sucrose at a concentration of 150 A260 U/ml.
Ribosomes (1.5 A260 U) were incubated with SRhisα/βΔN (30 pmol) in a reaction volume of 25 µl with final salt conditions: 25 mM HEPES–KOH pH 7.6, 2 mM MgCl2, 2 mM Mg(OAc)2, 50 mM NaCl, 75 mM KOAc and 200 µM nucleotide. Where indicated, microsomes or liposomes (resuspended in buffers lacking DTT) were also added at the indicated concentration (4–20 eq./25 µl). Reactions were incubated at 25°C for 15 min prior to addition of BMH or BMOE (Pierce), from a freshly prepared stock in dimethylsulfoxide at a final concentration of 20 µM. Following a further 20 min incubation at 25°C, the reactions were quenched with 2 mM DTT and then analysed by SDS–PAGE and immunoblotting.
For sucrose density gradient analysis, cross-link reactions (100 µl scale) were treated with 1 mM puromycin and 1 mM GTP and the potassium acetate concentration increased to 500 mM, prior to incubation for 30 min at 37°C. The samples were desalted in a G-25 microspin column (Pharmacia) pre-equilibrated with gradient buffer [25 mM HEPES–KOH pH 7.6, 5 mM Mg(OAc)2, 2 mM DTT, 500 mM KOAc] and then loaded onto a 4 ml 10–30% (w/v) linear sucrose gradient, made up in gradient buffer, and centrifuged for 2 h at 408 000 g. The gradients were then fractionated into 0.4 ml fractions using an ISCO 640 fractionator with continuous monitoring of absorbance at 254 nm. Proteins were precipitated with ethanol and then analysed by SDS–PAGE and immunoblotting.
Re-isolation of SRhisα/βΔN from translocation assays
Translocation assays in reticulocyte lysate (40 µl) were performed as described above but with cold methionine (20 µM). After incubation for 20 min at 25°C, the samples were diluted into ice-cold binding buffer [25 mM Tris–HCl pH 8.0, 1% (v/v) Triton X-100, 750 mM KOAc, 2 mM Mg(OAc)2 and 20 mM imidazole] and held for 10 min on ice. The samples were centrifuged for 5 min at 20 000 g in the cold. The supernatant was then incubated with 20 µl of Ni-NTA agarose (Qiagen) for 30 min at 4°C with gentle agitation. The matrix was washed four times with 1 ml of binding buffer and once with 10 mM Tris–HCl pH 7.6, 2 mM MgCl2 prior to elution with 25 µl of 2% (w/v) SDS.
For re-isolation from targeting assays, a 10 µl targeting reaction (as described above) was solubilized by addition of 10 µl of solubilization buffer [25 mM HEPES–KOH pH 7.6, 600 mM KOAc, 2 mM Mg(OAc)2 and 2% (w/v) Nikkol] and the samples incubated for 10 min on ice. Binding to Ni-NTA agarose was then performed as described above but with 25 mM HEPES–KOH pH 7.6, 300 mM KOAc, 2 mM Mg(OAc)2, 0.1% Nikkol, 20 mM imidazole and, where indicated, 200 µM XDP.
Antibodies
Antibodies against SRα and SRβ have been described previously (Görlich and Rapoport, 1993; Bacher et al., 1999). Anti-L30 antibodies were raised in rabbits using standard techniques against the peptide CPGDSDIIKTTPGEQCOOH, conjugated to keyhole limpet haemocyanin.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Roger Goody (MPI, Dortmund) for the kind gift of XMP-PNP, Josef Brunner (ETH, Zürich) for kindly providing (Tmd)Phe-pCpA, Jeannie Barrett for preparation of (Tmd)Phe-tRNAsup, Matthew Groves for the analytical ultracentrifugation data, and Ute Bach, Audrey Kuhn and Klaus Meese for excellent technical assistance. We thank Kellie Dean and members of both laboratories for critical reading of the manuscript and helpful suggestions. This work was supported by an EU TMR-network grant (ERBFMRXCT-960035; I.S. and B.D.), DFG Schwerpunkt Programm 312 (DO199/10-3; B.D.) and a Louis-Jeantet Fondation Fellowship (T.A.F.).
References
- Andrews D.W., Lauffer,L., Walter,P. and Lingappa,V.R. (1989) Evidence for a two-step mechanism involved in assembly of functional signal recognition particle receptor. J. Cell Biol., 108, 797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher G., Lütcke,H., Jungnickel,B., Rapoport,T.A. and Dobberstein,B. (1996) Regulation by the ribosome of the GTPase of the signal recognition particle during protein targeting. Nature, 381, 248–251. [DOI] [PubMed] [Google Scholar]
- Bacher G., Pool,M. and Dobberstein,B. (1999) The ribosome regulates the GTPase of the β-subunit of the signal recognition particle receptor. J. Cell Biol., 146, 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman R.D., Bubeck,R., Grassucci,P., Penczek,A., Verschoor,J., Blobel,G. and Frank,J. (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science, 278, 2123–2126. [DOI] [PubMed] [Google Scholar]
- Bernstein H.D., Poritz,M.A., Strub,K., Hoben,P.J., Brenner,S. and Walter,P. (1989) Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature, 340, 482–486. [DOI] [PubMed] [Google Scholar]
- Bourne H.R., Sanders,D.A. and McCormick,F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature, 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Connolly T. and Gilmore,R. (1986) Formation of a functional ribosome–membrane junction during translocation requires the participation of a GTP-binding protein. J. Cell Biol., 103, 2253–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T., Collins,P. and Gilmore,R. (1989) Access of proteinase K to partially translocated nascent polypeptides in intact and detergent-solubilized membranes. J. Cell Biol., 108, 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley K.S., Reinhart,G.D. and Johnson,A.E. (1993) The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell, 73, 1101–1115. [DOI] [PubMed] [Google Scholar]
- de Leeuw E., te Kaat,K., Moser,C., Menestrina,G., Demel,R., de Kruijff, B., Oudega,B., Luirink,J. and Sinning,I. (2000) Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J., 19, 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymann D.M., Keenan,R.J., Stroud,R.N. and Walter,P. (1997) Structure of the conserved GTPase domain of the signal recognition particle. Nature, 385, 361–364. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Walter,P. and Blobel,G. (1982) Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol., 95, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. and Rapoport,T.A. (1993) Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell, 75, 615–630. [DOI] [PubMed] [Google Scholar]
- Görlich D., Prehn,S., Hartmann,E., Kalies,K.-U. and Rapoport,T.A. (1992) A mammalian homolog of Sec61p and SecYp is associated with ribosomes and nascent polypeptides during translocation. Cell, 71, 489–503. [DOI] [PubMed] [Google Scholar]
- Hanein D., Matlack,K.E.S., Jungnickel,B., Plath,K., Kalies,K.-U., Miller,K.R., Rapoport,T.A. and Akey,C.W. (1996) Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell, 87, 721–732. [DOI] [PubMed] [Google Scholar]
- Hauser S., Bacher,G., Dobberstein,B. and Lütcke,H. (1995) A complex comprising the signal sequence binding protein and the SRP RNA promotes translocation of nascent proteins. EMBO J., 14, 5485–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High S. et al. (1993) Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane inserted signal sequence. J. Biol. Chem., 268, 26745–26751. [PubMed] [Google Scholar]
- Hortsch M. and Meyer,D.I. (1988) The human docking protein does not associate with the membrane of the rough endoplasmic reticulum via a signal or insertion sequence-mediated mechanism. Biochem. Biophys. Res. Commun., 150, 111–117. [DOI] [PubMed] [Google Scholar]
- Johnson A.E. and van Waes,M.A. (1999) The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol., 15, 799–842. [DOI] [PubMed] [Google Scholar]
- Kalies K.-U., Görlich,D. and Rapoport,T.A. (1994) Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J. Cell Biol., 126, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K., Falcone,D. and Andrews,D. (2000) Nucleotide-dependent binding of the GTPase domain of the signal recognition particle receptor β-subunit to the α-subunit. J. Biol. Chem., 275, 27439–27446. [DOI] [PubMed] [Google Scholar]
- Liao S., Lin,J., Do,H. and Johnson,A.E. (1997) Both lumenal and cytosolic gating of the aqueous ER translocon pore is regulated from within the ribosome during membrane protein integration. Cell, 90, 31–41. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Hofmann,M., Brunner,J. and Dobberstein,B. (1995) The protein conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell, 81, 207–214. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Hauser,S. and Dobberstein,B. (1997) In Celis,J.C. (ed.), Cell Biology: A Laboratory Handbook. Academic Press, San Diego, CA, pp. 265–273.
- Meyer D. and Dobberstein,B. (1980) A membrane component essential for vectorial translocation of nascent proteins across the endoplasmic reticulum: requirements for its extraction and reassociation with the membrane. J. Cell Biol., 87, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D.I., Louvard,D. and Dobberstein,B. (1982) Characterization of molecules involved in protein translocation using a specific antibody. J. Cell Biol., 92, 579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., Bernstein,H.D. and Walter,P. (1994) Interaction of E.coli Ffh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature, 367, 657–659. [DOI] [PubMed] [Google Scholar]
- Miller J.D., Tajima,S., Lauffer,L. and Walter,P. (1995) The β subunit of the signal recognition particle receptor is a transmembrane GTPase that anchors the α subunit, a peripheral membrane GTPase, to the endoplasmic reticulum membrane. J. Cell Biol., 128, 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya G., Svensson,C., Luirink,J. and Sinning,I. (1997) Crystal structure of the NG domain from the signal recognition particle receptor FtsY. Nature, 385, 365–368. [DOI] [PubMed] [Google Scholar]
- Moser C., Mol,O., Goody,R.S. and Sinning,I. (1997) The signal recognition particle receptor of Escherichia coli (FtsY) has a nucleotide exchange factor built into the GTPase domain. Proc. Natl Acad. Sci. USA, 94, 11339–11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhof A., Rolls,M.M., Jungnickel,B., Kalies,K.-U. and Rapoport,T.A. (1998) Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol. Biol. Cell, 9, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S.C., Barz,W.P. and Walter,P. (1998) A functional GTPase domain, but not its transmembrane domain, is required for function of the SRP receptor β-subunit. J. Cell Biol., 142, 341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. and Walter,P. (1995) Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science, 269, 1422–1424. [DOI] [PubMed] [Google Scholar]
- Raden D. and Gilmore,R. (1998) Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide-associated complex. Mol. Biol. Cell, 9, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapiejko P.J. and Gilmore,R. (1992) Protein translocation across the ER requires a functional GTP binding site in the α subunit of the signal recognition particle receptor. J. Cell Biol., 117, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapiejko P.J. and Gilmore,R. (1997) Empty site forms of SRP54 and SRα GTPases mediate targeting of ribosome–nascent chain complexes to the endoplasmic reticulum. Cell, 89, 703–713. [DOI] [PubMed] [Google Scholar]
- Romero G., Chau,V. and Biltonen,R.L. (1985) Kinetics and thermodynamics of the interaction of elongation factor Tu with factor Ts, guanine nucleotides and aminoacyl-tRNA. J. Biol. Chem., 260, 6167–6174. [PubMed] [Google Scholar]
- Römisch K., Webb,J., Herz,J., Prehn,S., Frank,R., Vingron,M. and Dobberstein,B. (1989) Homology of 54K protein of signal-recognition particle, docking protein and two E.coli proteins with putative GTP-binding domains. Nature, 340, 478–482. [DOI] [PubMed] [Google Scholar]
- Schlichting I. et al. (1990) Time-resolved X-ray crystallographic study of the conformational change in Ha-Ras p21 protein on GTP hydrolysis. Nature, 345, 309–315. [DOI] [PubMed] [Google Scholar]
- Scotti P.A., Valent,Q.A., Manting,E.H., Urbanus,M.L., Driessen,A.J., Oudega,B. and Luirink,J. (1999) SecA is not required for signal recognition particle-mediated targeting and initial membrane insertion of a nascent inner membrane protein. J. Biol. Chem. 274, 29883–29888 [DOI] [PubMed] [Google Scholar]
- Simon S.M. and Blobel,G. (1991) A protein-conducting channel in the endoplasmic reticulum. Cell, 65, 371–380. [DOI] [PubMed] [Google Scholar]
- Song W., Raden,D., Mandon,E.C. and Gilmore,R. (2000) Role of Sec61α in the regulated transfer of the ribosome–nascent chain complex from the signal recognition particle to the translocation channel. Cell, 100, 333–343. [DOI] [PubMed] [Google Scholar]
- Tajima S., Lauffer,L., Rath,V.L. and Walter,P. (1986) The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J. Cell Biol., 103, 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P. and Blobel,G. (1983) Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol., 96, 84–93. [DOI] [PubMed] [Google Scholar]
- Walter P. and Johnson,A.E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol., 10, 87–119. [DOI] [PubMed] [Google Scholar]
- Weijland A. and Parmeggiani,A. (1993) Toward a model for the interaction between elongation factor Tu and the ribosome. Science, 259, 1311–1314. [DOI] [PubMed] [Google Scholar]