Abstract
The nuclear-encoded Sup35p protein is responsible for the prion-like [PSI+] determinant of yeast, with Sup35p existing largely as a high molecular weight aggregate in [PSI+] strains. Here we show that the five oligopeptide repeats present at the N-terminus of Sup35p are responsible for stabilizing aggregation of Sup35p in vivo. Sequential deletion of the oligopeptide repeats prevented the maintenance of [PSI+] by the truncated Sup35p, although deletants containing only two repeats could be incorporated into pre-existing aggregates of wild-type Sup35p. The mammalian prion protein PrP also contains similar oligopeptide repeats and we show here that a human PrP repeat (PHGGGWGQ) is able functionally to replace a Sup35p oligopeptide repeat to allow stable [PSI+] propagation in vivo. Our data suggest a model in which the oligopeptide repeats in Sup35p stabilize intermolecular interactions between Sup35p proteins that initiate establishment of the aggregated state. Modulating repeat number therefore alters the rate of yeast prion conversion in vivo. Furthermore, there appears to be evolutionary conservation of function of the N-terminally located oligopeptide repeats in prion propagation.
Keywords: oligopeptide repeats/prion/PrP/Sup35p/yeast
Introduction
The Sup35p protein of Saccharomyces cerevisiae is an essential component of the translation termination machinery (Stansfield et al., 1995b) and is also responsible for the prion-like [PSI+] determinant (Doel et al., 1994; Ter-Avanesyan et al., 1994; Wickner, 1994). In [PSI+] cells, Sup35p is found predominantly as part of non-functional high molecular weight complexes. This results in a low level of functional, soluble Sup35p protein in [PSI+] cells and leads to a decrease in the efficiency of translation termination that can be readily detected in vivo using a nonsense suppressor-based assay (Cox, 1965). In [psi–] cells, where Sup35p is mostly soluble and hence able to interact with eRF1 to mediate translation termination (Stansfield et al., 1995b), termination is not impaired (Patino et al., 1996; Paushkin et al., 1996). A considerable body of data exists that points to the Sup35p prion being propagated by a mechanism that is similar to that by which the infectious mammalian prion protein PrP is propagated (reviewed in Serio and Lindquist, 2000).
Sup35p consists of a 114 amino acid N-terminal prion-forming domain (PrD), a charged M (middle) domain of unknown function and a C-terminal domain that provides the essential translation termination activity (Kushnirov et al., 1988; Wilson and Culbertson, 1988; Ter-Avanesyan et al., 1993, 1994). The Sup35p-PrD is essential for the establishment and maintenance of the [PSI+] phenotype (Ter-Avanesyan et al., 1993; Doel et al., 1994). A second well-characterized yeast prion, the Ure2p protein, which gives rise, in its prion form, to the [URE3] determinant (Wickner, 1994), also has an N-terminally located PrD that is essential for the establishment and maintenance of the [URE3] determinant (Masison and Wickner, 1995). The Sup35p-PrD is modular and can be used to impose prion-like behaviour on a protein that normally does not show such properties, e.g. a Sup35p-PrD–rat glucocorticoid receptor fusion can behave as a prion when expressed in S.cerevisiae (Li and Lindquist, 2000).
Overexpression of the PrD of Sup35p and Ure2p can induce de novo the establishment of the [PSI+] and [URE3] determinants, respectively (Chernoff et al., 1993; Masison and Wickner, 1995; Derkatch et al., 1996; Patino et al., 1996). This occurs at a much higher frequency than is observed when the corresponding full-length proteins are overexpressed, indicating that regions outside the PrD may also modulate protein–protein interactions necessary for prion conversion. While the structure of the aggregated form of Sup35p or Ure2p in vivo remains to be defined, in vitro both proteins can be self-seeded to form large regular fibres analogous to those found in mammalian amyloid- and prion-related diseases. These fibres can be seeded from soluble protein by pre-formed fibres (Glover et al., 1997; King et al., 1997) using either full-length protein or the PrD polypeptides. These data are consistent with a model in which both [PSI+] and [URE3] arise as a consequence of oligomerization, driven by the respective PrDs, into amyloid-like aggregates.
The Sup35p-PrD has an unusual amino acid composition, being composed of 43% glutamine/asparagine residues as compared with 9% for the average yeast protein (Santoso et al., 2000). At the extreme N-terminus (amino acids 8–33) is a region particularly rich in glutamine/asparagine that has been implicated in both prion conversion (DePace et al., 1998) and in providing a species specificity determinant (Santoso et al., 2000). C-terminal to this region is a region composed of five complete copies (R1–R5) and one partial copy (R6) of an imperfect oligopeptide repeat with the consensus sequence PQGGYQQ-YN. Deletions removing these repeat regions abolish [PSI+] (Ter-Avanesyan et al., 1994; Derkatch et al., 1996; Kochneva-Pervukhova et al., 1998) while a glycine to aspartic acid substitution in one of the repeats (repeat 2, R2) gives rise to the PNM2 allele, which leads to a dominant [PSI+] maintenance defect (PNM) phenotype (Doel et al., 1994). The dominance of the PNM mutation indicates that the ability of a wild-type Sup35p protein to maintain [PSI+] aggregates is dramatically reduced when wild-type Sup35p is synthesized in the presence of the mutated Sup35p (Ter-Avanesyan et al., 1993; Doel et al., 1994; Kochneva-Pervukhova et al., 1998).
When two extra copies of the R2 repeat were introduced into the Sup35p-PrD, the spontaneous conversion from [psi–] to [PSI+] increased some 5000-fold over the rate of conversion for wild-type Sup35p (Liu and Lindquist, 1999). In contrast, a Sup35p-PrD with only a single intact repeat is not sufficient for prion conversion of Sup35p (Liu and Lindquist, 1999). This in vivo behaviour is paralleled by the ability of the PrD repeat variant Sup35p to form amyloid fibres from unfolded protein in vitro. For example, a Sup35p variant with two additional R2 repeats showed a seeded in vitro prion conversion rate significantly faster than that for the wild-type protein, while a Sup35p variant with only one repeat formed fibres at a much slower rate than wild type (Liu and Lindquist, 1999). While these data suggest that the number of oligopeptide repeats in the Sup35p-PrD can modify the rate of conversion of Sup35p in vivo, the role of the oligopeptide repeats in the Sup35p-PrD remains to be established, although they are most probably important in mediating the crucial protein–protein interactions that lead to conversion to the high molecular weight oligomeric form of Sup35p found in [PSI+] strains.
The mammalian prion protein PrP also contains five copies of an oligopeptide repeat (PHGGGWGQ) related in sequence to the Sup35p-PrD repeats (Kretzschmar et al., 1986; Tuite, 1994). Expansion of these repeats is found in certain familial prion diseases such as Gerstmann– Straussler–Scheinker syndrome (GSS) and Creutzfeldt– Jakob disease (CJD) (Prusiner and Scott, 1997; Chiesa et al., 1998). For both Sup35p and PrP, therefore, expansion of their N-terminal repeats leads to an increased frequency of conversion from [prion–] to [PRION+]. Deletion of the repeats from PrP does not prevent scrapie infection in mice; however, in such animals, prion titres and levels of aggregated PrP are lower than in wild-type mice (Flechsig et al., 2000). In addition, disease incubation times are longer and no histopathological signs of scrapie infection are found in the brains of terminally diseased animals expressing the repeat-truncated PrP. These findings led to the conclusion that repeat-truncated PrP shows a decrease in the prion conversion rate when compared with wild-type protein (Flechsig et al., 2000). Thus, there is evidence that the N-terminal PrP repeats contribute to prion conversion, but as with the Sup35p-PrD repeats, their role remains to be fully established.
In this study, we have used progressive deletion of the repeats in the S.cerevisiae Sup35p-PrD to probe their contribution in vivo to prion conversion and propagation in order to understand further how expansion and deletion of the repeats can modulate the propagation of the [PSI+] determinant. Furthermore, we show that the related PrP ‘octarepeat’ can functionally replace a Sup35p-PrD repeat, providing evidence that the N-terminal oligopeptide repeats in both proteins control the rate of prion conversion, most probably by modulating the stability of the protein–protein interactions that lead to protein polymerization.
Results
A plasmid-based assay for [PSI+] maintenance
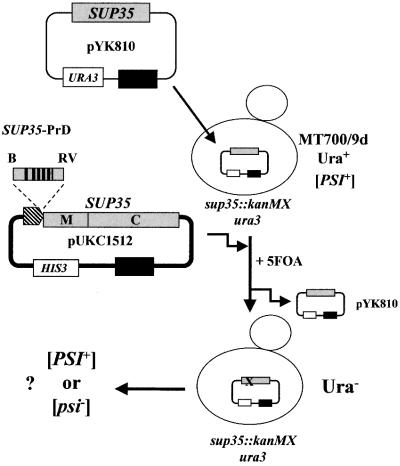
In order to assess the consequences of manipulations within the non-essential PrD region of Sup35p on [PSI+] maintenance, a plasmid-based assay was developed (Figure 1). A strain of S.cerevisiae (MT700/9d) was constructed in which the chromosomal copy of SUP35 was disrupted by the kanMX gene (sup35::kanMX; see Materials and methods) with viability being supported by the URA3-CEN plasmid pYK810 carrying the wild-type SUP35 gene. To test whether this strain could be used to monitor the ability of different SUP35 alleles to maintain the [PSI+] determinant, both [PSI+] and [psi–] derivatives of MT700/9d [pYK810] were transformed with HIS3-based plasmids carrying either the wild-type SUP35 gene (pSUP), the PNM2 allele (pPNM; Doel et al., 1994) or a SUP35 allele lacking the PrD domain critical for [PSI+] maintenance (pRΔN). 5-Fluoro-orotic acid (5-FOA)-containing medium was used to select for cells that had lost the pYK810 plasmid, and single Ura– colonies were then tested for suppression of the ade2-1 marker to assess termination efficiency. In this suppressor tRNA-mediated assay, [PSI+] colonies have a white/pink, Ade+ phenotype, while [psi–] colonies have a red, Ade– phenotype (Cox, 1965).
Fig. 1. A plasmid-based assay for [PSI+] maintenance by N-terminally truncated forms of Sup35p. Strain MT700/9d contains a sup35::kanMX disruption and harbours a copy of the wild-type SUP35 gene on plasmid pYK810. Derivatives of plasmid pUKC1512, carrying N-terminally modified alleles of SUP35, are then transformed into this strain and cells lacking the pYK810 plasmid subsequently selected on 5-FOA-containing medium. The [PSI] phenotype of the resulting haploid strain is then assessed qualitatively using the suppression of the ade2-1 allele by the SUQ5-encoded tRNA suppressor. In plasmid pUKC1512, the N-terminal domain variants are introduced via a BamHI (B)–EcoRV (RV) fragment.
Endogenous wild-type Sup35p was able to seed conversion of Sup35p expressed from pSUP prior to loss of pYK810 because all His+ Ura– transformants remained [PSI+] (Figure 2A). The [PSI+] phenotype of the pSUP transformants could be eliminated permanently by growth on a rich medium containing 4 mM guanidine hydrochloride (Tuite et al., 1981). In contrast, MT700/9d His+ Ura– transformants containing either pPNM or pRΔN became [psi–] after loss of pYK810 (Figure 2A). This was expected, since pRΔN lacks the PrD and is therefore unable to support [PSI+], while the PNM2 allele of SUP35 contains an amino acid substitution in the PrD that ablates prion propagation of the wild-type Sup35p (Doel et al., 1994; Kochneva-Pervukhova et al., 1998). MT700/9d [pYK810] [psi–] derivatives transformed with either pSUP, pPNM or pRΔN each gave rise to red, Ade– [psi–] colonies after plasmid shuffling (data not shown). These data demonstrate that the plasmid-based assay can be used as an in vivo test of the ability of mutants of Sup35p to propagate [PSI+] when ‘seeded’ by wild-type Sup35p aggregates, and then, subsequently, in the absence of ongoing synthesis of the wild-type Sup35p.
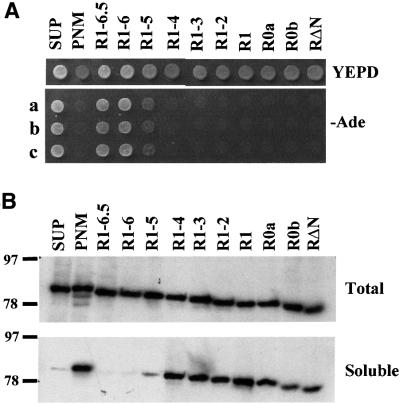
Fig. 2. The ability of N-terminally truncated Sup35p to maintain the [PS1+] determinant in the absence of endogenous wild-type Sup35p synthesis. (A) Cell suspensions of individual transformants post-5-FOA selection were plated onto either YEPD (to determine colony colour) or defined medium lacking adenine (–Ade) to test for suppression of the ade2-1 marker. Three independent transformants (a–c) are shown on the –Ade plates. (B) Western blot analysis of either total cell-free extracts (upper panel) or a soluble protein fraction (lower panel; prepared as described in Materials and methods) using an anti-Sup35p polyclonal antibody. Details of the various Sup35p derivatives tested are given in Figure 3.
Deletion of one or more of the oligopeptide repeats from the Sup35p-PrD inhibits the establishment of the [PSI+] determinant
To define the regions in the Sup35p N-terminal PrD essential for prion propagation, a series of deletions was constructed that sequentially removed the oligopeptide repeats from the C-terminal end of the N-terminal domain (Figure 3). Following transformation of the SUP35 gene constructs into the MT700/9d [PSI+] sup35::kanMX strain, all transformants gave rise to viable Ura– colonies on medium containing 5-FOA, confirming that the PrD-truncated Sup35p proteins were able to support viability in the absence of wild-type Sup35p (Figure 2). Ura– transformants obtained from parallel experiments with MT700/9d [psi–] remained [psi–], thereby demonstrating that none of the deleted proteins was functionally impaired in the Sup35p-dependent translation termination function (data not shown). Ura– transformants obtained from MT700/9d [PSI+] were analysed similarly (Figure 2A) and western blot analysis showed that the steady-state levels of the various truncated Sup35p proteins were approximately equivalent to the levels of the wild-type Sup35p (Figure 2B, upper panel).
Fig. 3. Construction of a series of N-terminally truncated alleles of the SUP35 gene. (A) Sequences of the oligopeptide repeats within the N-terminal domain of Sup35p (R1–R6) and comparison with the consensus sequence of the mammalian PrP ‘octarepeat’. The conserved residues between the Sup35p and PrP repeats are underlined. (B) Deletion series encompassing the oligopeptide repeats of Sup35p (R1–R6). The construct numbers are shown on the left while the amino acids deleted are indicated on the right.
A deletion spanning amino acids 98–112 of the Sup35p-PrD had no effect on [PSI+] maintenance, while removal of the half-repeat R6 (construct pR1-5) led to an increase in termination efficiency, as defined by a weak Ade+ phenotype (Figure 2A). When streaked for single colonies on rich YPD medium, the pR1-5 transformants produced a number of pink/red sectored colonies, with ∼10% of the colonies being red, Ade– (data not shown). Therefore, removal of the half-repeat R6 causes weakening of the [PSI+]-mediated termination defect and a reduction in the ability to maintain the [PSI+] determinant. A larger deletion that also removed the R5 repeat (pR1-4) resulted in termination efficiency equal to that seen in the control [psi–] strain, with further deletions giving the same phenotype as pR1-4 transformants (Figure 2A). The [PSI+] determinant was not maintained in any strain expressing Sup35p lacking one or more copies of the oligopeptide repeats in the PrD. These data demonstrate that deletions C-terminal to the R6 repeat of the Sup35p-PrD do not affect [PSI+] stability, thereby defining the minimal Sup35p-PrD as amino acids 1–97.
To determine whether the various mutant Sup35p proteins expressed in MT700/9d [PSI+] were aggregated or soluble, cell-free lysates were subjected to differential centrifugation and the resulting supernatant fraction analysed by immunoblotting (Figure 2B). In the control [PSI+] strain pSUP, only a very low level of soluble Sup35p could be detected, while in the pPNM and pRΔN transformants, Sup35p remained largely soluble, as would be expected of a [psi–] strain. The Sup35p pR1-6.5 and pR1-6 derivatives both pelleted, confirming that they behaved as wild-type Sup35p with respect to aggregation, consistent with their [PSI+] Ade+ phenotype. Deleting the half-repeat R6 (pR1-5) caused partial solubilization of Sup35p, accounting for the increase in termination efficiency (i.e. phenotype) observed in this strain (Figure 2A). Further deletions resulted in high levels of soluble Sup35p, consistent with these strains showing restored termination efficiency due to a failure to maintain the [PSI+] determinant. The absence of Sup35p in the supernatant fractions of the pR1-6.5, pR1-6 and pR1-5 strains, which was observed consistently, was unlikely to be due to proteolysis, since the absence of soluble Sup35p in vitro correlated exactly with the in vivo termination phenotype (compare Figure 2B, soluble, with A). Further more, lower molecular weight fragments of Sup35p were not detected in these samples.
It remained possible that the strains showing efficient termination contained ‘cryptic’ [PSI+] seeds that were not pelleted by centrifugation. To test this possibility, the various Ura– MT700/9d transformants were mated to BSC783/4c [psi–]. Only the pSUP, pR1-6.5, pR1-6 and pR1-5 strains gave rise to [PSI+] diploids, indicating that the red, Ade– MT700/9d transformants carrying deletion of one or more of the complete oligopeptide repeats in the Sup35p-PrD were bona fide [psi–].
These data confirm that Sup35p has an absolute requirement for all five complete oligopeptide repeats for stable [PSI+] maintenance in the absence of wild-type Sup35p and that the minimal Sup35p-PrD is encompassed within the first 97 amino acids of Sup35p.
Sup35p PrD repeats stabilize protein–protein interactions
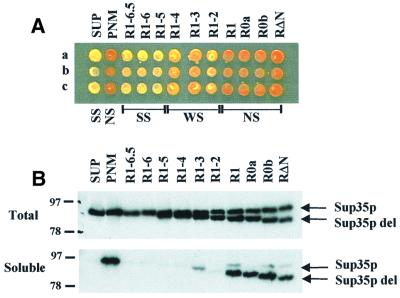
Expression of Sup35p lacking the PrD in a [PSI+] strain (Ter-Avanesyan et al., 1994) leads to an efficient termination phenotype probably due to the truncated Sup35p protein being unable to interact with the wild-type [PSI+] aggregates, resulting in a pool of N-terminally truncated, soluble Sup35p that is functional in translation termination. To test whether the repeat-truncated Sup35p mutants could interact stably with endogenous wild-type Sup35p [PSI+] aggregates, the Sup35p deletion plasmid series was introduced into strain BSC783/4a [PSI+] and a [psi–] derivative thereof. While stable interactions would be expected to result in a termination defect (i.e. white, Ade+), moderately stable interactions would be expected to give rise only to an intermediate level of nonsense suppression. All transformants obtained with the BSC783/4a [psi–] strain were red, Ade–, while the resulting BSC783/4a [PSI+] transformants showed a range of termination phenotypes (Figure 4A). In the control experiment using the BSC783/4a [PSI+] strain, the wild-type Sup35p (pSUP) gave no increase in termination efficiency while the pRΔN and pPNM control constructs resulted in restoration of an efficient termination phenotype, as expected.
Fig. 4. The incorporation of N-terminally truncated Sup35p derivatives into aggregates containing wild-type Sup35p. (A) Cell suspensions of three independent transformants of BSC783/4a were plated on YEPD to determine qualitatively the degree of SUQ5-mediated ade2-1 suppression. The three different phenotypes of the deletants are indicated as follows: SS, strong suppression; WS, weak suppression; NS, no suppression. (B) Western blot analysis of either total cell-free extracts (upper panel) or a soluble protein fraction (lower panel; prepared as described in Materials and methods) using an anti-Sup35p polyclonal antibody. The positions of the full-length wild-type Sup35p (Sup35p) and the co-expressed, N-terminally truncated form (Sup35pdel) are indicated. Details of the various derivatives tested are given in Figure 3.
The N-terminal mutants of Sup35p gave rise to a gradual increase in the efficiency of termination as the repeats were removed progressively from the C-terminus. BSC783/4a [PSI+] transformed with pR1-6.5, pR1-6 and pR1-5 retained the white, Ade+ phenotype, while removal of repeats R3–R5 caused a detectable increase in termination efficiency (Figure 4A). Removal of repeat R2 (pR1) resulted in termination efficiencies comparable to that obtained with the pRΔN and pPNM controls. Thus, Sup35p is able to interact efficiently with wild-type Sup35p to generate [PSI+] aggregates provided two or more repeats are present.
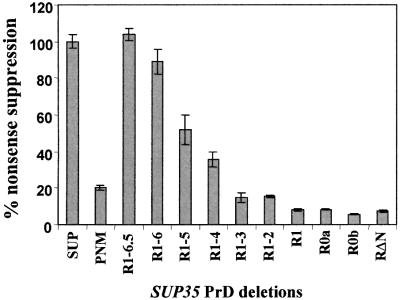
To provide an accurate quantitative measure of the termination defect in the BSC783/4a [PSI+] transformants, the plasmids pUKC815 and pUKC817 (Stansfield et al., 1995b) were used. Plasmid pUKC815 contains the PGK– β-galactosidase fusion gene, while pUKC817 encodes the same fusion but interrupted at the junction of the two coding sequences with a UAA stop codon. β-galactosidase production thus requires the host strain to read through the UAA stop codon, and the level at which this occurs can be used to quantify the efficiency of termination (Firoozan et al., 1991). The results of these assays (Figure 5) were consistent with those of the qualitative suppression assay (Figure 4A).
Fig. 5. Quantification of the degree of nonsense suppression in the BSC783/4a transformants co-expressing N-terminally truncated and wild-type Sup35p. The various transformants were transformed with the PGK–lacZ fusion plasmids pUKC815 and pUKC817, and the levels of expressed β-galactosidase were used to determine the percentage nonsense suppression, as described in Materials and methods. The level of nonsense suppression in the pSUP transformant was taken as 100% and all other transformants were compared with that strain. For each transformant, four samples were assayed independently and the error bars represent standard deviation.
To assess whether the PrD-defective Sup35p mutant proteins were able to interact with wild-type Sup35p in vivo, protein samples were prepared from the BSC783/4a transformants and fractionated as described in Materials and methods. The wild-type Sup35p was present at similar levels in all the strains tested, with the truncated proteins being present at equivalent levels (Figure 4B). In the pSUP-transformed strain, no soluble Sup35p could be detected, while in the pPNM-transformed strain high levels of soluble Sup35p were detected. In the pRΔN-transformed strain, the wild-type Sup35p pelleted as [PSI+] aggregates, but the truncated RΔN protein remained soluble. The truncated Sup35p-PrD proteins lacking repeats R3–R6 were largely absent from the soluble fraction, although the pR1-3 construct did show reduced levels of interaction with wild-type [PSI+] aggregates, since it was partially soluble (Figure 4B, lower panel). Further deletions resulted in the truncated Sup35p being found almost exclusively in the soluble fraction, with the majority of the wild-type protein remaining aggregated. Thus, when only one or no Sup35p-PrD repeats were present, Sup35p was unable to interact with wild-type Sup35p in [PSI+] aggregates, while Sup35p with between two and six repeats was able to interact.
To determine whether the Sup35p repeat deletants acted as dominant [PSI+] maintenance (PNM) mutants, the various BSC783/4a transformants were grown under non-selective conditions, and His– colonies that had lost the pUKC1512 plasmid carrying the mutant SUP35 gene were identified. All such His– transformants were found to revert to the white, Ade+, guanidine HCl-curable [PSI+] phenotype, with the exception of pPNM, which remained red, Ade–. Therefore, co-expression of the wild-type and mutant Sup35p did not result in elimination of the [PSI+] determinant. This behaviour was as described for Sup35p lacking the complete N-terminal domain (Ter-Avanesyan et al., 1994) and as confirmed here with our pRΔN construct.
The PrP octarepeat can functionally replace a Sup35p-PrD oligopeptide repeat in [PSI+] propagation
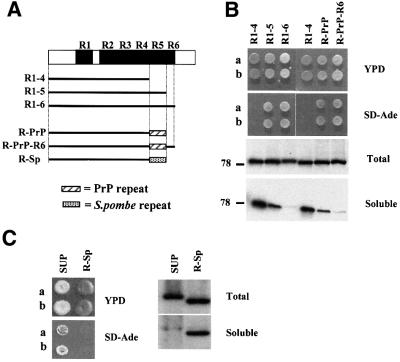
The five octarepeats present in mammalian PrP proteins show amino acid sequence similarity to the repeats present in the Sup35p-PrD (Figure 3A). We therefore tested whether the consensus PrP octarepeat PHGGGWGQ was able to function in the propagation and maintenance of [PSI+] in place of the essential Sup35p oligopeptide repeat R5. Starting with pR1-4, we re-inserted the PrP octarepeat C-terminal to repeat R4, to generate pR-PrP. The partial repeat R6 was then added back to generate pR-PrP-R6 (Figure 6A). These various plasmids were then shuffled into both the MT700/9d [PSI+] and [psi–] strains in place of the plasmid-borne wild-type SUP35 gene, and their [PSI+] phenotype determined.
Fig. 6. Replacement of the R5 repeat in the Sup35p-PrD with a copy of the mammalian PrP octarepeat allows for propagation of the [PSI+] determinant. (A) The various constructs shown were generated as described in Materials and methods. (B) Analysis of two independent transformants (a and b) in which repeat R5 is replaced by the PrP octarepeat (R-PrP/R-PrP-R6) (see the legend to Figure 2). (C) Analysis of two independent transformants (a and b) in which repeat R5 is replaced by an octapeptide from the S.pombe Sup35p N-terminus (R-Sp).
All MT700/9d [psi–] transformants gave rise to red, Ade– colonies, indicating that the PrP repeat-containing Sup35p was able to supply full termination function in a [psi–] background (data not shown). As described previously, the pR1-5 transformants showed a weak unstable [PSI+] phenotype in the MT700/9d [PSI+] strain, while the pR1-6 construct generated stable [PSI+] strains. With pR-PrP, the resulting colonies showed a weak Ade+ phenotype (Figure 6B), indicating that the PrP repeat can function in place of R5 to support [PSI+] propagation. The addition of R6 increased the stability of [PSI+] with the pR-PrP-R6 constructs similarly to the pRI-6 construct (Figure 6B). The presence of the [PSI+] determinant in these strains was confirmed by showing that growth in 4 mM guanidine HCl converted the colonies to a stable red, Ade– [psi–] phenotype. Neither the pR-PrP [psi–] nor the pR-PrP-R6 [psi–] strains showed an abnormally high rate of de novo induction of the [PSI+] determinant.
The subcellular localization of the PrP repeat-containing Sup35p protein was compared with that of the pR1-4, pR1-5 and pR1-6 variants. All mutant proteins showed similar steady-state levels (Figure 6B). When the PrP octarepeat was added to pR1-4, soluble Sup35p was found at an intermediate level, whereas the majority of the R-PrP-R6 Sup35p pelleted with little detectable in the soluble fraction. Thus, Sup35p containing the PrP repeat can be seeded by wild-type Sup35p to propagate [PSI+]. However, the PrP repeat did not function as efficiently as a Sup35p-R5 repeat in [PSI+] maintenance, since the pR-PrP-R6 [PSI+] transformants showed a lower level of nonsense suppression than the wild-type Sup35p, as defined by the qualitative colony phenotype (Figure 6B). Furthermore, a small proportion of the Sup35p R-PrP-R6 protein remained soluble, whereas no soluble Sup35p was found in the pR1-6 transformants.
One possible explanation for our findings with the PrP repeat is that it is a sequence non-specific effect. The R1–R4 deletion may simply have brought the key N-terminal region of the Sup35p-PrD too close to the M/C region for efficient prion propagation, and the addition of any eight amino acid sequence would restore prion propagation. We therefore introduced an eight amino acid sequence taken from the region of the Schizo saccharomyces pombe Sup35p (Ito et al., 1998) N-terminal domain at a position (amino acids 74–81) equivalent to that of the R5 repeat in the S.cerevisiae Sup35p. There are no oligopeptide repeats in this region of the S.pombe Sup35p, nor is there any evidence that it is able to form a prion. The resulting R-Sp Sup35p protein could not be seeded by wild-type Sup35p and the resulting strain showed all the characteristics of a [psi–] strain (Figure 6C). This negative control thus validates our conclusion that the PrP octarepeat allowed the Sup35p- R1-4 mutant to be seeded to the prion form, indicating that the mammalian repeat is able to mediate protein–protein interactions almost as effectively as the S.cerevisiae Sup35p oligopeptide repeats in vivo.
Discussion
Role of Sup35p-PrD oligopeptide repeats in prion conversion
Both the mammalian and yeast prion proteins PrP and Sup35p contain oligopeptide repeats of similar sequence at their N-termini (Kretzschmar et al., 1986; Tuite, 1994). Expansion of repeat copy number in both these proteins is associated with an increased rate of spontaneous prion induction, demonstrating the importance of these sequences in the molecular mechanism of prion generation and implying a conserved function for the repeat regions in these two prions (Prusiner and Scott, 1997; Chiesa et al., 1998; Liu and Lindquist, 1999). A number of studies have shown that the repeat region of Sup35p is required for prion conversion since gross deletions in this region prevent [PSI+] maintenance (Ter-Avanesyan et al., 1993; Liu and Lindquist, 1999). The Sup35p repeats may modulate the rate of prion conversion from partially unfolded protein to a β-sheet-rich structure (Liu and Lindquist, 1999), although how this is accomplished is unclear. One of the major enigmas in prion biology is the process whereby oligopeptide repeat expansions increase de novo prion formation. We have carried out a detailed in vivo analysis of the repeat region of Sup35p, dissecting the role of these repeats in the establishment and maintenance of the [PSI+] determinant, leading us to suggest a molecular mechanism that explains how such repeats mediate prion formation.
Using a plasmid-based assay, we defined the minimum length of the Sup35p-PrD as amino acids 1–97, up to and including repeat R6. Remarkably, deletion of only the four amino acids that constitute R6 leads to reduced maintenance of [PSI+], partial solubilization of the mutant protein and an increase in termination efficiency. Deletion of more than one repeat prevents the resulting mutant Sup35p from maintaining [PSI+] in the absence of wild-type Sup35p, nor do they form ‘cryptic’ [PSI+] determinants as they are unable to seed the conversion of wild-type Sup35p.
Our deletion studies suggest that the Sup35p-PrD has evolved to contain the optimal number of oligopeptide repeats required for the stable maintenance of either the [PSI+] or [psi–] states, with minimal switching between the two. A Sup35p protein containing more than six repeats cannot maintain [psi–] stably (Liu and Lindquist, 1999), whereas a PrD containing fewer than six repeats cannot maintain [PSI+] stably (this study). This evolution towards optimal repeat number suggests that the [PSI] phenotype has a biological role where the stable propagation of the [PSI] phenotype is important. This is consistent with the recent proposal that [PSI] acts to uncover hidden genetic variation in yeast populations (True and Lindquist, 2000). If either [PSI] state confers an evolutionary advantage to a yeast population, then it can be maintained due to the optimal PrD repeat number which minimizes [PSI] state switching.
Expression of repeat truncated Sup35p in the presence of wild-type Sup35p allowed us to determine whether the mutant proteins could become incorporated into existing [PSI+] aggregates. The strength of the mutant–wild-type Sup35p interaction was determined by analysing the amount of soluble mutant Sup35p in cells directly by western blot analysis and indirectly by measuring termination efficiency due to increased soluble mutant Sup35p. Deletions C-terminal to the repeat region (i.e. amino acids 98–114) did not significantly impair Sup35p–Sup35p interactions, demonstrating that this region is not required for [PSI+] propagation. As the repeats were deleted sequentially, there was a corresponding decrease in the ability of the mutant proteins to become incorporated into aggregates. A Sup35p-PrD containing only repeat R1 behaved essentially as a protein without a PrD (Figure 4). Thus, in vivo, the oligopeptide repeats most probably affect the strength of Sup35p–Sup35p interactions and alter the relative amount of protein competent for prion conversion.
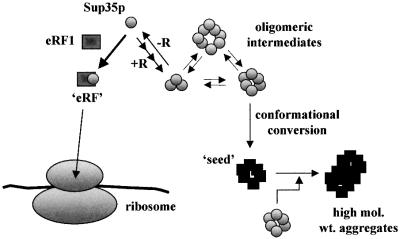
Interestingly, Sup35p mutant proteins containing two or more repeats could be incorporated into wild-type aggregates, yet were unable to maintain a [PSI+] state in the absence of wild-type Sup35p. These mutant proteins are therefore able to undergo prion conversion, but their decreased rate of conversion prevents them from maintaining the [PSI+] determinant. This behaviour can be explained in the light of the recently proposed ‘nucleated conformational conversion’ model (Serio et al., 2000) for [PSI+] replication. In this model, the first (and rate-limiting) step is the assembly of Sup35p monomers into oligomeric complexes, which can then undergo conformational conversion to form oligomeric seeds. These seeds are then able to recruit other oligomers, which in turn undergo conformational conversion, ultimately leading to the formation of amyloid-like aggregates in the cell (Figure 7). We propose that the oligopeptide repeats in the Sup35p-PrD stabilize the Sup35p–Sup35p interactions needed to produce the Sup35p oligomers. Mutant Sup35p lacking one or more of the full repeats thus form oligomers at a reduced rate compared with wild-type Sup35p, leading to a decrease in the overall rate of seed formation as oligomer concentration becomes limiting. If the rate of prion formation falls below a critical level, the [PSI+] determinant will not be maintained, presumably because the rate of prion clearance by cell division, possibly through the action of molecular chaperones (Kushnirov and Ter-Avanesyan, 1998), exceeds the rate of prion formation. Co-expression of repeat-truncated Sup35p with wild-type Sup35p results in [PSI+] aggregates containing the wild-type protein, with the level of the mutant protein incorporated into oligomeric complexes, and hence Sup35p aggregates, depending on the number of repeats present. Thus, as repeat number is reduced, more mutant Sup35p remains soluble.
Fig. 7. The nucleated conformational conversion model of Serio et al. (2000). By increasing the number of oligopeptide repeats (+R), Sup35p–Sup35p interactions are favoured over Sup35p–Sup45p interactions. Similarly, deletion of oligopeptide repeats (–R) leads to reduced efficiency of Sup35p–Sup35p interactions. For further discussion of the model, refer to the text.
This model can also explain why an expansion in Sup35p-PrD repeat number leads to a dramatic increase in the de novo appearance of [PSI+] cells, namely that the strength of the Sup35p–Sup35p interaction is increased further by the additional repeats such that the balance of Sup35p interactions shifts more towards Sup35p–Sup35p rather than the Sup35p–Sup45p interaction required to constitute a functional release factor (eRF). Thus, there would be a corresponding increase in the concentration of the conformationally converted intermediates, leading to an increase in the rate of prion conversion in the absence of a pre-existing or transmitted seed.
Evolutionary conservation of function of N-terminal oligopeptide repeats in prion conversion
A copy of the mammalian PrP octapeptide repeat was able to functionally replace S.cerevisiae Sup35p-PrD repeat R5 to allow stable [PSI+] propagation in vivo. However, the PrP repeat did not function as efficiently as the Sup35p R5 in prion propagation (Figure 6), suggesting that the Sup35p–Sup35p interactions are not as strong when the PrP repeat is present. The reduced level of interaction may simply reflect the size of the inserted peptide since R5 is 10 amino acids in length compared with the eight amino acid PrP repeat. Small changes in amino acid length in the Sup35-PrD can affect [PSI+] propagation; for example, the pR1-6 and pR1-5 variants differ by only four amino acids, yet the pR1-6 variant can maintain [PSI+] whereas pR1-5 shows reduced maintenance and increased Sup35p solubility (Figure 2). The observation that an unrelated octapeptide taken from the S.pombe Sup35p N-terminal domain did not allow conversion to [PSI+] when added to pR1-4 (Figure 6C) would suggest that it is the amino acid sequence or composition, rather than the addition of any eight amino acid sequence to this region, that is the crucial factor for prion conversion.
Since a PrP octarepeat is able to support [PSI+] propagation, it is possible that the octapeptide repeats present in PrP are functionally analogous to the Sup35p repeats in that they may stabilize protein–protein interactions that permit prion formation. This hypothesis is supported by the observation that expansion of the oligopeptide repeats in both Sup35p and PrP causes an increased frequency of induction of the [PRION+] state (Prusiner and Scott, 1997; Chiesa et al., 1998; Liu and Lindquist, 1999). As we, and others, have demonstrated, deletion of the repeats from Sup35p prevents stable prion formation, whereas deleting the octapeptide repeats from PrP does not totally block scrapie replication (Flechsig et al., 2000). In fact, in vitro studies by Jackson et al. (1999) have shown that the switch between a soluble form of PrP and a fibrilogenic conformation can occur with PrP variants lacking the octarepeats. However, there is no direct evidence that the amyloid form of PrP so generated represents the transmissible component of the prion disease. The apparently different requirements, for oligopeptide repeats in prion conversion, between yeast and mammals, could be explained if there were additional PrP–PrP interactions site, outside the octapeptide repeat region. Thus, removal of the PrP repeats would not eliminate prion replication, although the rate of prion conversion would be expected to decrease. Indeed, a decrease in the rate of prion conversion of octapeptide-deleted PrP was proposed to explain the slow scrapie pathogenesis in mice expressing PrP lacking the octarepeats (Flechsig et al., 2000). Also consistent with this idea is the identification of a goat PrP allele containing only three instead of the usual five octarepeats (Goldmann et al., 1998). Goats heterozygous for this truncated allele show long incubation periods for disease, consistent with a slower rate of prion conversion.
It is tempting to speculate that PrPSc formation occurs by a process analogous to the nucleated conformational conversion model proposed for Sup35p (Serio et al., 2000). Initial PrP–PrP interactions would occur to form oligomeric intermediates involving, in part, the octapeptide repeat region. Conformational conversion could then produce a species that is stabilized by PrP–PrP interactions, but that does not involve the octapeptide repeats. This would explain why the octapeptide repeat region is not found in the aggregated proteinase K-resistant PrP fragment (Prusiner et al., 1984; Basler et al., 1986). In such a model, repeat expansions would increase the concentration of an oligomeric intermediate able to undergo aggregation.
In summary, our results demonstrate that the rate of prion replication in yeast can be controlled by the number of oligopeptide repeats. We propose that repeats act to stabilize Sup35p–Sup35p interactions required for the formation of oligomeric intermediates that are competent for conformational conversion and subsequent polymerization to produce amyloid-like aggregates. This may also be the role, at least in part, of the octarepeats in mammalian PrP.
Materials and methods
Yeast strains
The S.cerevisiae strain BSC783/4a (MATα, ade2-1UAA, leu2-3,112, ura3-1, his3-11,15, SUQ5 [PSI+]) was used in prion interaction studies. [psi–] derivatives were obtained on 4 mM guanidine hydrochloride-containing media (Tuite et al., 1981). Strain MT700/9d (MATα, sup35::kanMX4, SUQ5, ade2-1UAA, his3-11,15, ura3-1, leu2-3,112) transformed with pYK810 (a centromeric plasmid containing SUP35; Kikuchi et al., 1988) was used in prion propagation assays (C.G.Resende and M.F.Tuite, unpublished data).
Media and yeast genetic methods
Standard yeast media, cultivation and genetic methods were used (Kaiser et al., 1994). Zygotes were picked on a Singer Micromanipulator model MSM System and diploids verified by sporulation. Yeast cells were grown at 30°C on complete glucose medium (YPD) unless stated otherwise. [PSI+] strains were cured of the prion by growth on YPD containing 4 mM guanidine hydrochloride (Tuite et al., 1981). Trans formants were grown on synthetic glucose medium selective for a plasmid-borne marker, e.g. SD-His for HIS3-containing plasmids. [PSI+] strains were monitored phenotypically by suppression of the ade2-1UAA mutation in strains also containing the weak suppressor tRNASer encoded by the gene SUQ5. This allows direct visualization of nonsense suppression by colour and/or adenine prototrophy (Cox, 1965). [PSI+] strains are white on 1/4 YPD [4% (w/v) glucose, 1% (w/v) bactopeptone, 0.25% (w/v) yeast extract, 2% (w/v) agar] and able to grow on SD-Ade medium, while [psi–] strains are red on 1/4 YPD and unable to grow on SD-Ade medium. Transformations were carried out as described by Schiestl et al. (1989). When assaying by colony colour, yeast were grown in 1/4 YPD. The presence of sup35::kanMX4 was monitored by resistance to geneticin (G418; 200 µg/ml).
Plasmid constructs
The SUP35 PrD deletion series was constructed by cloning various PrD deletions into the plasmid pUKC1512 (Figure 1). This plasmid contains the SUP35 promoter together with the M and C domains of SUP35 in a HIS3-marked centromeric plasmid backbone (C.G.Resende and M.F.Tuite, unpublished data). The 3′ end of the promoter was engineered to contain a BamHI restriction site, and just prior to the beginning of the SUP35 M domain is a naturally occurring EcoRV site. The PrD deletions were amplified using PCR and cloned as BamHI–EcoRV fragments into pUKC1512 to reconstitute an in-frame SUP35 sequence with various sized deletions at the C-terminus of the PrD. The different deletions were made using the same forward primer and different reverse primers designed against the SUP35 sequence. All PCRs used wild-type SUP35 as a template, except the pR-PrP-R6 construction, which used pR-PrP.
The oligonucleotide primers used were as follows (restriction sites are underlined). Forward primer: SUP35 FOR1: GTAACAAAAAGGATCCTCTTCATCGACTTGCTCG. Reverse primers: REV7 (pSUP, pPNM): ACCAGCTTGATATCCTTGCA; REV12 (pR1-6.5): GGGGGGGATATCCGTAATTTCCACGGCCACCTTG; REV11 (pR1-6): GGGGGGGATATCCGCCACCTTGTGGATTGAATTG; REV10 (pR1-5): GGG GGGGATATCCATTGAATTGCTGCTGATAACC; REV2 (pR1-4): AACCGCGATATCCATTGTACTGTTGATAGCCTCCTT; REV8 (pR1-3): GGGGGGGATATCCATTATACTGTTGCTGGTAAC; REV9 (pR1-2): GGGGGGGATATCCATTGTACTGTTGATAGCCAC; REV4 (pR1): GTACCCAGGATATCCTTGGTAATTTTGGTAGTACCC; REV5 (pR0a): TGCAGGTTGATATCCAGCATTGTAAGCTTGATA; REV6 (pR0b): TGTTGGTGATATCCCTGGCTGTATTGCTGG; REV2-PrP (pR-PrP): GGGGGGGATATCCTTGACCCCAACCACCACCATGTGGATTTACTGTTGATAGCCTCC; REV-PrP-EX(pR-PrP-R6): GGGGGGGATATCCGCCACCTTGTGGTTGACCCGAACCACCACCATG; REV2-Sp (pR-Sp): GGGGGGATATCCATACTGCTGATAACTCTTCGTCGGATTGTACTGTTGATAGCCTCCTT.
Plasmid pRN was made by inserting a synthetic oligonucleotide made by annealing oligo 2A (GATCCTCTTCATCGACTTGCTCGGAATAACATCTATATCTGCCCACTAGCAACAATGTCGCGAGGAT) and oligo 2B (ATCCTCGCGACATTGTTGCTAGTGGGCAGATATAGATGTTATTCCGAGCAAGTCGATGAAGAG) into pUKC1512.
[PSI+] maintenance assay
A plasmid shuffling procedure was used to exchange SUP35 alleles in the SUP35::kanMX4 haploid strain MT700/9d and monitor the continued propagation of [PSI+]. Recombinant SUP35 genes were thus transformed into MT700/9d [pYK810]. Transformants were selected on SD-His medium and then plated to 5-FOA-containing medium to select for cells that had lost the URA3-based plasmid pYK810 (Kaiser et al., 1994). Single colonies were then picked from the FOA-containing medium and resuspended in 96-well microtitre plates. Transformants were plated onto 1/4 YPD, SD-His, SD-Ura, SD-Ade, YPD + G418 and YPD + guanidine hydrochloride, respectively, to verify phenotypes and genotypes.
Sedimentation analysis of Sup35p
Yeast protein extracts were prepared and fractionated into soluble and insoluble fractions by centrifugation as follows. Transformants were grown overnight in SD-His until the culture reached an OD600 = 0.5. Cells were then pelleted, washed with 20 ml of water, resuspended in 500 µl of lysis buffer (25 mM Tris–HCl pH 6.8, 250 mM NaCl, 5 mM EDTA, 10% glycerol) and lysed by vortexing with glass beads (40 mesh) for 4 × 30 s. Samples were then centrifuged for 3 min at 2500 r.p.m. to pellet unbroken cells, and the resulting supernatant collected. Protein concentrations were determined using the Coomassie Plus Protein assay (Pierce) and all samples adjusted to 1 mg/ml with lysis buffer. A 200 µl aliquot of this total fraction was then centrifuged at 50 000 r.p.m. for 15 min at 4°C. The resulting supernatant fraction was collected and the pellet resuspended in 200 µl of fresh lysis buffer. SDS sample buffer (2.5 ml of glycerol, 1.25 ml of 20% SDS, 250 µl of β-mercaptoethanol, 1.25 ml of 0.5 M Tris–HCl pH 6.8) was then added and all samples were boiled for 15 min. All protein samples so prepared were centrifuged for a further 10 min before being separated by 10% SDS–PAGE. To follow the fractionation of Sup35p, the proteins were transferred to nitrocellulose membranes (Sartorius) and immunoblotted with rabbit anti-Sup35p polyclonal antiserum and anti-rabbit antiserum (Dako), and visualized by chemiluminescence (ECL) according to the manufacturer’s (Pharmacia-Amersham) instructions.
Quantification of nonsense suppression
BSC783/4a transformed with the plasmid deletion series were co-transformed with the plasmids pUKC815 and pUKC817 (Stansfield et al., 1995a). Plasmid pUKC815 is a centromeric vector containing a PGK–lacZ gene fusion under the control of the constitutive PGK1 promoter. pUKC817 is identical to pUKC815 except that an in-frame TAA stop codon is located at the junction between the PGK and lacZ coding sequence. To determine suppression levels, strains were grown overnight in SD-His-Ura and the OD600 recorded. Cells were then diluted in 800 µl of Z buffer (60 mM Na2PO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 pH 7.0) supplemented with 50 mM β-mercaptoethanol. To release the β-galactosidase, 50 µl of chloroform and 20 µl of 0.1% (w/v) SDS were added, and samples were equilibrated by incubation at 37°C for 15 min. The assay was initiated by the addition of 200 µl of o-nitrophenylgalactoside (ONPG; 4 mg/ml) and incubated at 37°C for 30 min. The reaction was stopped by the addition of 500 µl of 1 M Na2CO3 and the samples clarified by centrifugation at 13 000 r.p.m. for 1 min. The absorbance of the supernatant was recorded at 420 nm and the β-galactosidase activity calculated (Miller, 1972). The average result from four different assays for each plasmid was calculated and results are expressed as a percentage of the control plasmid pUKC815.
Acknowledgments
Acknowledgements
This work was supported by project grants from the BBSRC and the Wellcome Trust (M.F.T.) and by PhD studentships from Subprograma Ciencia e Tecnologia do 2o Quadro Comunitario de apoio (C.G.R.) and the BBSRC (S.N.P.).
References
- Basler K., Oesch,B., Scott,M., Westaway,D., Walchli,M., Groth,D.F., McKinley,M.P., Prusiner,S.B. and Weissmann,C. (1986) Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell, 46, 417–428. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Derkach,I.L. and Inge-Vechtomov,S.G. (1993) Multicopy SUP35 gene induces de-novo appearance of ψ-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet., 24, 268–270. [DOI] [PubMed] [Google Scholar]
- Chiesa R., Piccardo,P., Ghetti,B. and Harris,D.A. (1998) Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron, 21, 1339–1351. [DOI] [PubMed] [Google Scholar]
- Cox B.S. (1965) Ψ, a cytoplasmic suppressor of super-suppression in yeast. Heredity, 20, 505–521. [Google Scholar]
- DePace A.H., Santoso,A., Hillner,P. and Weissman,J.S. (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell, 93, 1241–1252. [DOI] [PubMed] [Google Scholar]
- Derkatch I.L., Chernoff,Y.O., Kushnirov,V.V., Inge-Vechtomov,S.G. and Liebman,S.W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics, 144, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doel S.M., McCready,S.J., Nierras,C.R. and Cox,B.S. (1994) The dominant PNM2- mutation which eliminates the ψ factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics, 137, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone S.S., Ruddock,L.W., Cox,B.S. and Tuite,M.F. (2000) Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoozan M., Grant,C.M., Duarte,J. and Tuite,M.F. (1991) Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast, 7, 173–183. [DOI] [PubMed] [Google Scholar]
- Flechsig E., Shmerling,D., Hegyi,I., Raeber,A.J., Fischer,M., Cozzio,A., von Mering,C., Aguzzi,A. and Weissmann,C. (2000) Prion protein devoid of the octapeptide repeat region restores susceptibility to scrapie in PrP knockout mice. Neuron, 27, 399–408. [DOI] [PubMed] [Google Scholar]
- Glover J.R., Kowal,A.S., Schirmer,E.C., Patino,M.M., Liu,J.J. and Lindquist,S. (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S.cerevisiae. Cell, 89, 811–819. [DOI] [PubMed] [Google Scholar]
- Goldmann W., Chong,A., Foster,J., Hope,J. and Hunter,N. (1998) The shortest known prion protein gene allele occurs in goats, has only three octapeptide repeats and is non-pathogenic. J. Gen. Virol., 79, 3173–3176. [DOI] [PubMed] [Google Scholar]
- Ito K., Ebihara,K. and Nakamura,Y. (1998) The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA, 4, 958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson G.S. et al. (1999) Reversible conversion of monomeric human prion protein between native and fibrilogenic conformations. Science, 283, 1935–1937. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis,S. and Mitchell,A. (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kikuchi C., Shimatake,H. and Kikuchi,A. (1988) A yeast gene required for the G1-to-S transition encodes a protein containing an A-kinase target site and GTPase domain. EMBO J., 7, 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C.Y., Tittmann,P., Gross,H., Gebert,R., Aebi,M. and Wuthrich,K. (1997) Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl Acad. Sci. USA, 94, 6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N.V., Paushkin,S.V., Kushnirov,V.V., Cox,B.S., Tuite,M.F. and Ter-Avanesyan,M.D. (1998) Mechanism of inhibition of Psi+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J., 17, 5805–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar H.A., Stowring,L.E., Westaway,D., Stubblebine,W.H., Prusiner,S.B. and Dearmond,S.J. (1986) Molecular cloning of a human prion protein cDNA. DNA, 5, 315–324. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. and Ter-Avanesyan,M.D. (1998) Structure and replication of yeast prions. Cell, 94, 13–16. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V., Ter-Avanesyan,M.D., Telckov,M.V., Surguchov,A.P., Smirnov,V.N. and Inge-Vechtomov,S.G. (1988) Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene, 66, 45–54. [DOI] [PubMed] [Google Scholar]
- Li L. and Lindquist,S. (2000) Creating a protein-based element of inheritance. Science, 287, 661–664. [DOI] [PubMed] [Google Scholar]
- Liu J.J. and Lindquist,S. (1999) Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature, 400, 573–576. [DOI] [PubMed] [Google Scholar]
- Masison D.C. and Wickner,R.B. (1995) Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science, 270, 93–95. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Patino M.M., Liu,J.J., Glover,J.R. and Lindquist,S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science, 273, 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Prusiner S.B. and Scott,M.R. (1997) Genetics of prions. Annu. Rev. Genet., 31, 139–175. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B., Groth,D.F., Bolton,D.C., Kent,S.B. and Hood,L.E. (1984) Purification and structural studies of a major scrapie prion protein. Cell, 38, 127–134. [DOI] [PubMed] [Google Scholar]
- Santoso A., Chien,P., Osherovich,L.Z. and Weissman,J.S. (2000) Molecular basis of a yeast prion species barrier. Cell, 100, 277–288. [DOI] [PubMed] [Google Scholar]
- Schiestl R.H. and Gietz,R.D. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet., 16, 339–346. [DOI] [PubMed] [Google Scholar]
- Serio T.R. and Lindquist,S.L. (2000) Protein-only inheritance in yeast: something to get [PSI+]-ched about. Trends Cell Biol., 10, 98–105. [DOI] [PubMed] [Google Scholar]
- Serio T.R., Cashikar,A.G., Kowal,A.S., Sawicki,G.J., Moslehi,J.J., Serpell,L., Arnsdorf,M.F. and Lindquist,S.L. (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science, 289, 1317–1321. [DOI] [PubMed] [Google Scholar]
- Stansfield I., Akhmaloka and Tuite,M.F. (1995a) A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr. Genet., 27, 417–426. [DOI] [PubMed] [Google Scholar]
- Stansfield I. et al. (1995b) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J., 14, 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Kushnirov,V.V., Dagkesamanskaya,A.R., Didichenko,S.A., Chernoff,Y.O., Inge-Vechtomov,S.G. and Smirnov,V.N. (1993) Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol., 7, 683–692. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Dagkesamanskaya,A.R., Kushnirov,V.V. and Smirnov,V.N. (1994) The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics, 137, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True H.L. and Lindquist,S.L. (2000) A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature, 407, 477–483. [DOI] [PubMed] [Google Scholar]
- Tuite M.F. (1994) Psi no more for yeast prions. Nature, 370, 327–328. [DOI] [PubMed] [Google Scholar]
- Tuite M.F., Mundy,C.R. and Cox,B.S. (1981) Agents that cause a high frequency of genetic change from [PSI+] to [psi–] in Saccharomyces cerevisiae. Genetics, 98, 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R.B. (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science, 264, 566–569. [DOI] [PubMed] [Google Scholar]
- Wilson P.G. and Culbertson,M.R. (1988) SUF12 suppressor protein of yeast. A fusion protein related to the EF-1 family of elongation factors. J. Mol. Biol., 199, 559–573. [DOI] [PubMed] [Google Scholar]